Abstract
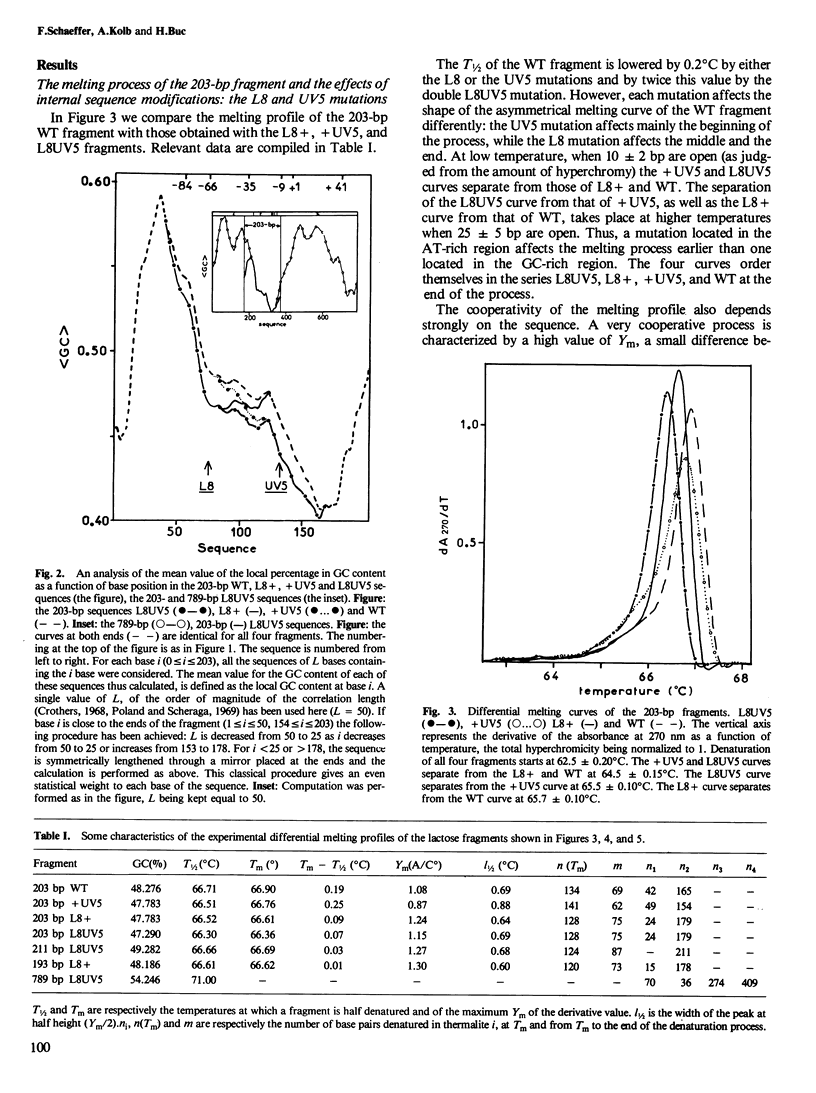
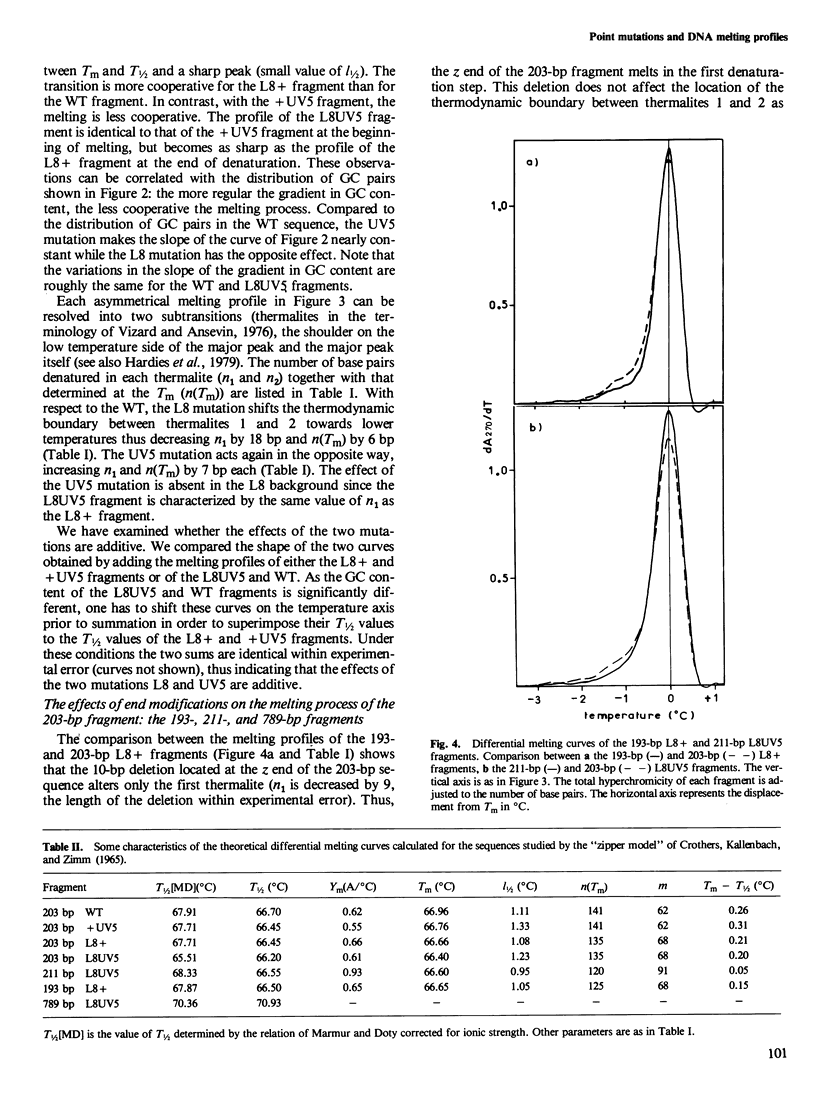
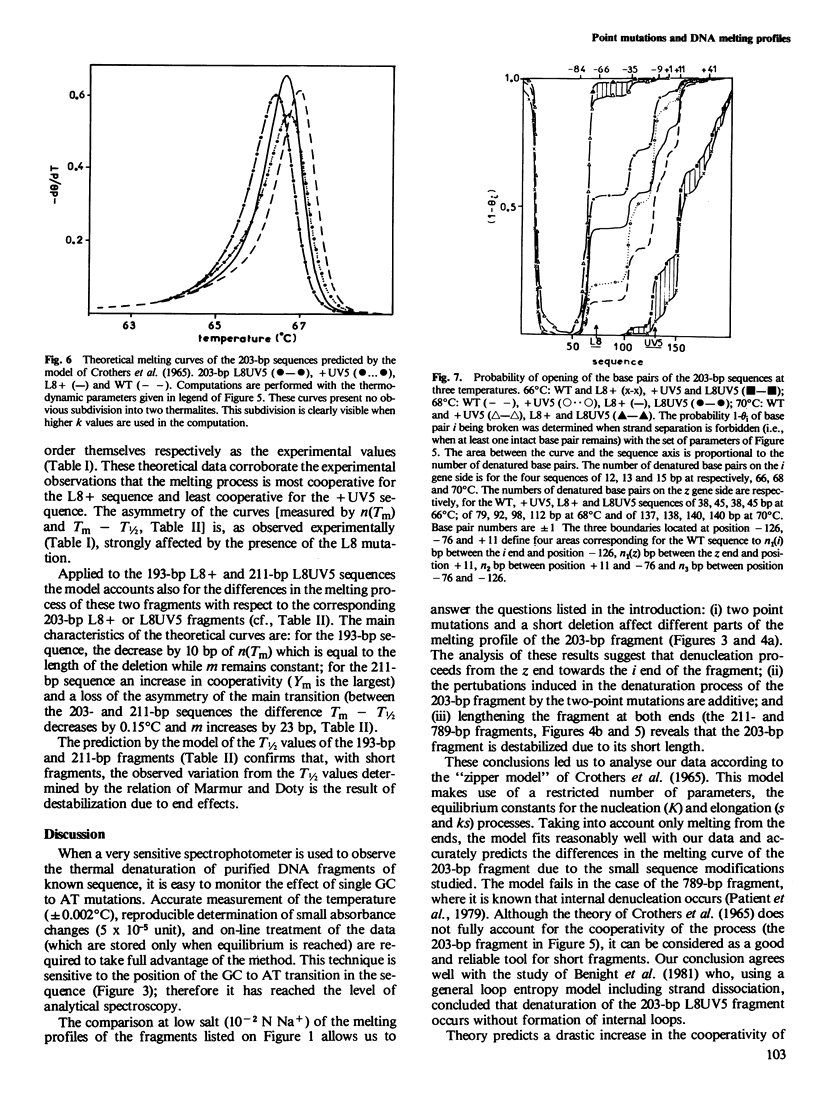
To understand the denaturation process of short DNA segments we have chosen a 203-base pair (bp) restriction fragment containing the lactose control region. A steady decrease in GC content exists between its i proximal and z proximal ends. We confirm that this fragment melts at low salt in two subtransitions. A GC to AT mutation in the AT-rich region (mutation UV5) increases the number of denatured base pairs in the first subtransition and decreases the cooperativity of the melting process. A GC to AT mutation in the GC-rich region (mutation L8) decreases the number of denatured base pairs in the first subtransition and increases the cooperativity. These mutations induce the same shift in the temperature of half denaturation. The effects of both mutations are additive. A short deletion at the z end of the fragment affects only the first subtransition. When four GC pairs are added to both end, the fragment melts in one transition. Comparison with the results obtained with a larger 789-bp lac fragment reveals strong end effects on base pair stability and suggests that denaturation of the 203-bp fragment proceeds unidirectionally from the z end. Good agreement is shown with the predictions made with the "z ipper model" of Crothers et al. (1965).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benight A. S., Wartell R. M., Howell D. K. Theory agrees with experimental thermal denaturation of short DNA restriction fragments. Nature. 1981 Jan 15;289(5794):203–205. doi: 10.1038/289203a0. [DOI] [PubMed] [Google Scholar]
- CROTHERS D. M., KALLENBACH N. R., ZIMM B. H. THE MELTING TRANSITION OF LOW-MOLECULAR-WEIGHT DNA: THEORY AND EXPERIMENT. J Mol Biol. 1965 Apr;11:802–820. doi: 10.1016/s0022-2836(65)80037-7. [DOI] [PubMed] [Google Scholar]
- CROTHERS D. M., ZIMM B. H. THEORY OF THE MELTING TRANSITION OF SYNTHETIC POLYNUCLEOTIDES: EVALUATION OF THE STACKING FREE ENERGY. J Mol Biol. 1964 Jul;9:1–9. doi: 10.1016/s0022-2836(64)80086-3. [DOI] [PubMed] [Google Scholar]
- Calos M. P. DNA sequence for a low-level promoter of the lac repressor gene and an 'up' promoter mutation. Nature. 1978 Aug 24;274(5673):762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Charnay P., Perricaudet M., Galibert F., Tiollais P. Bacteriophage lambda and plasmid vectors, allowing fusion of cloned genes in each of the three translational phases. Nucleic Acids Res. 1978 Dec;5(12):4479–4494. doi: 10.1093/nar/5.12.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of melting curves for DNA. Biopolymers. 1968 Oct;6(10):1391–1404. doi: 10.1002/bip.1968.360061003. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Hirschman S. Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J Mol Biol. 1965 Sep;13(2):407–427. doi: 10.1016/s0022-2836(65)80106-1. [DOI] [PubMed] [Google Scholar]
- Hardies S. C., Hillen W., Goodman T. C., Wells R. D. High resolution thermal denaturation analyses of small sequenced DNA restriction fragments containing Escherichia coli lactose genetic control loci. J Biol Chem. 1979 Oct 25;254(20):10128–10134. [PubMed] [Google Scholar]
- Hardies S. C., Wells R. D. Preparation and characterization of large amounts of restriction fragments containing the E. coli lac control elements. Gene. 1979 Sep;7(1):1–14. doi: 10.1016/0378-1119(79)90039-8. [DOI] [PubMed] [Google Scholar]
- Hillen W., Goodman T. C., Benight A. S., Wartell R. M., Wells R. D. High resolution experimental and theoretical thermal denaturation studies on small overlapping restriction fragments containing the Escherichia coli lactose genetic control region. J Biol Chem. 1981 Mar 25;256(6):2761–2766. [PubMed] [Google Scholar]
- Ippen K., Miller J. H., Scaife J., Beckwith J. New controlling element in the Lac operon of E. coli. Nature. 1968 Mar 2;217(5131):825–827. doi: 10.1038/217825a0. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. In vitro analysis of the Escherichia coli RNA polymerase interaction with wild-type and mutant lactose promoters. J Mol Biol. 1978 Nov 15;125(4):467–490. doi: 10.1016/0022-2836(78)90311-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient R. K., Hardies S. C., Larson J. E., Inman R. B., Maquat L. E., Wells R. D. Influence of A-T content on the fractionation of DNA restriction fragments by RPC-5 column chromatography. J Biol Chem. 1979 Jun 25;254(12):5548–5554. [PubMed] [Google Scholar]
- Reiss C., Michel F., Gabarro J. An apparatus for studying the thermal transition of nucleic acids at high resolution. Anal Biochem. 1974 Dec;62(2):499–508. doi: 10.1016/0003-2697(74)90182-1. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Silverstone A. E., Arditti R. R., Magasanik B. Catabolite-insensitive revertants of lac promoter mutants. Proc Natl Acad Sci U S A. 1970 Jul;66(3):773–779. doi: 10.1073/pnas.66.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiollais P., Rambach A., Buc H. Large scale purification of a bacterial gene. FEBS Lett. 1974 Nov 1;48(1):96–100. doi: 10.1016/0014-5793(74)81071-9. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., Ansevin A. T. High resolution thermal denaturation of DNA: thermalites of bacteriophage DNA. Biochemistry. 1976 Feb 24;15(4):741–750. doi: 10.1021/bi00649a004. [DOI] [PubMed] [Google Scholar]
- Wada A., Yabuki S., Husimi Y. Fine structure in the thermal denaturation of DNA: high temperature-resolution spectrophotometric studies. CRC Crit Rev Biochem. 1980;9(2):87–144. doi: 10.3109/10409238009105432. [DOI] [PubMed] [Google Scholar]


