Abstract
C-mannosylation is a unique type of protein glycosylation with a mannose attached to the tryptophan residue via the C-C linkage. Our previous study revealed that dpy-19 like 3 (DPY19L3) acts as a C-mannosyltransferase in human cells. The present study hypothesized that RPE-spondin (RPESP) may be a substrate protein of DPY19L3-mediated C-mannosylation. RPESP has unknown biological functions and has two putative C-mannosylation sites at the W80 and W83 residues; however, to the best of our knowledge, C-mannosylation of RPESP has not previously been investigated. The present study suggested that RPESP is C-mannosylated at W80 and W83 in human cells, whereas gain-of-function experiments using S2 cells revealed that human DPY19L3 catalyzed the C-mannosylation of RPESP at W83 but not W80, which suggested substrate specificity. In addition, the present study detected mRNA expression levels of RPESP in various types of cancer cell lines and high expression levels of RPESP were revealed in certain colorectal cancer cell lines, suggesting that RPESP may have an association with the malignancy of colorectal cancers.
Keywords: C-mannosylation, C-mannosyltransferase, Dpy-19 like 3, glycobiology, mass spectrometry, RPE-spondin
Introduction
Glycosylation, one of the post-translational modifications, commonly regulates protein stability, folding and secretion (1–3). Certain types of glycosylation exist, including N-linked and O-linked glycosylations, C-mannosylation, and the formation of glycosylphosphatidylinositol anchors. C-mannosylation is a unique type of glycosylation in which α-D-mannose is attached directly to the indole C2 carbon atom of a tryptophan residue via a C-C linkage (4,5). C-mannosylation occurs at the first tryptophan residue in the consensus sequence W-X-X-W/C (X represents any amino acid) (6). Certain C-mannosylated proteins, including ribonuclease 2 (7), interleukin-12 (8), properdin (9) and mindin (10), have been identified; however, the biological roles of C-mannosylation remain unclear. A previous study reported that Caenorhabditis elegans (C. elegans) dpy-19 is the C-mannosyltransferase for the thrombospondin type-1 repeat (TSR-1)-derived peptide (11) and that human dpy-19 like 3 (DPY19L3), one of the homologs of C. elegans dpy-19, catalyzes C-mannosylation of R-spondin (Rspo) 1 at the W156 residue (12).
RPE-spondin (RPESP) is a protein that has unknown functions and exists in the aorta extracellular matrix (13,14). RPESP has certain domains: An N-terminal signal peptide toward the secretory pathway, followed by a somatomedin B (SMB) domain and TSR-1. Specific proteins containing the SMB domain have been reported to bind plasminogen activator inhibitor-1 and ectonucleotide pyrophosphatase/phosphodiesterase 1 (15–18); however, the physiological functions of its association remain unclear. In the TSR-1 domain, RPESP has two putative C-mannosylation sites, the W80 and W83 residues. Previously, it was demonstrated that C-mannosylation of TSR-1 in Rspo1 and Rspo3 regulates these functions (12,19). The present study focused on RPESP protein and examined the existence of C-mannosylation in RPESP. The results suggested that RPESP is C-mannosylated at both prediction sites and that DPY19L3 catalyzes C-mannosylation of RPESP at W83 specifically. Therefore, it was indicated that DPY19L3 has substrate specificity and that C-mannosylation at W80 is catalyzed by an unidentified C-mannosyltransferase. Furthermore, the present study demonstrated that the expression of RPESP was observed in certain human tumor cell lines, suggesting the malignant roles of RPESP in tumorigenesis.
Materials and methods
Cell culture
Human HT1080 (JCRB Cell Bank, Osaka, Japan) fibrosarcoma, A549 (RIKEN BioResource Center, Tsukuba, Japan) non-small-cell lung cancer, HepG2 (RIKEN BioResource Center) hepatocellular cancer, HT29 [American Type Culture Collection (ATCC), Manassas, VA, USA] colon cancer and WM266-4 (ATCC) melanoma cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Nissui, Tokyo, Japan) supplemented with 10% (v/v) fetal bovine serum (FBS; BioWest S.A.S, Nuaillé, France), 100 U/ml penicillin G, 100 mg/l kanamycin, 600 mg/l L-glutamine, and 2.25 g/l NaHCO3 at 37°C in a humidified incubator with 5% CO2. Human ES2 (which was kindly donated by the Department of Obstetrics and Gynecology, Keio University School of Medicine, Tokyo, Japan) ovarian cancer and HCT116 (RIKEN BioResource Center) colon cancer cell lines were cultured in DMEM supplemented with 10% (v/v) heat-inactivated FBS, 100 U/ml penicillin G, 100 mg/l kanamycin, 600 mg/l L-glutamine, and 2.25 g/l NaHCO3 at 37°C in a humidified incubator with 5% CO2. Human COLO 205, CW-2 and LoVo colon cancer cell lines (RIKEN BioResource Center) were cultured in RPMI-1640 (Nissui) supplemented with 10% (v/v) FBS, 100 U/ml penicillin G, 100 mg/l kanamycin, 300 mg/l L-glutamine, and 2.25 g/l NaHCO3 at 37°C in a humidified incubator with 5% CO2. The human Jurkat (ATCC) acute leukemia cell line was cultured in RPMI-1640 supplemented with 10% (v/v) heat-inactivated FBS, 100 U/ml penicillin G, 100 mg/l kanamycin, 300 mg/l L-glutamine, and 2.25 g/l NaHCO3 at 37°C in a humidified incubator with 5% CO2. The S2 Drosophila melanogaster embryonic cell line (RIKEN BioResource Center) was cultured in Schneider's Drosophila medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% (v/v) heat-inactivated FBS, 100 U/ml penicillin G and 100 mg/l kanamycin at 25°C.
Plasmid construction
The synthetic DNA encoding C-terminally Myc-His6-tagged human RPESP, optimized to the human codon and introduced with N-terminal XhoI and C-terminal NotI restriction enzyme sites, respectively, was purchased from Thermo Fisher Scientific, Inc. The synthetic DNA was cloned into the XhoI/NotI restriction sites of pCI-neo vector (Promega Corporation, Madison, WI, USA). For expression in S2 cells, C-terminal Myc-His6-tagged RPESP cDNA was amplified by polymerase chain reaction (PCR) from pCI-neo-RPESP-Myc-His6 using PrimeSTAR® Max DNA Polymerase (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocol. The sequences of the primers used were as follows: Forward, 5′-TTTTAGATCTGGATGTGCCGAAGCCGGCAGAT-3′ (Eurofins Genomics, Ebersberg, Germany) and reverse, 5′-TTTTACGCGTCTAATGGTGATGGTGATGAT-3′ (Thermo Fisher Scientific, Inc.). The reaction was conducted in a C1000™ thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the cycling conditions were as follows: 30 cycles at 98°C for 10 sec, 55°C for 5 sec and 72°C for 5 sec. Subsequently, the RPESP-Myc-His6 cDNA was subcloned into the BglII/MluI restriction sites of pMT-PURO (RIKEN BioResource Center) (20), resulting in the expression of RPESP-Myc-His6 protein. Human DPY19L1, DPY19L2, DPY19L3 and DPY19L4 cDNAs, which were subcloned into pIZ/V5-his vectors (Thermo Fisher Scientific, Inc.), were constructed previously (12).
Establishment of an RPESP-overexpressing cell line
The permanent cell line expressing wild-type RPESP-Myc-His6 was established by transfecting the vectors using Lipofectamine® LTX (Thermo Fisher Scientific, Inc.) into HT1080 cells for 24 h at 37°C, followed by 400 µg/ml G418 (Wako Pure Chemical Pure Industries, Ltd., Osaka, Japan) selection. The clone that expressed high levels of Myc-His6-tagged wild-type RPESP was designated HT1080-RPESP-MH. The cells that were transfected with pCI-neo were designated HT1080-neo as control (21).
Western blotting
Western blot analysis was performed according to a previously described method with slight modification (22–26). HT1080-neo (control) and HT1080-RPESP-MH cells were cultured for 24 h at 37°C and lysed in a lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% (w/v) SDS, 1% (v/v) Triton X-100, 1% (w/v) sodium deoxycholate and 1 mM phenylmethylsulfonyl fluoride] with sonication (20 kHz, 50 W, 10 sec, twice) in an ultrasonic homogenizer (UH-50; SMT Co., Ltd., Tokyo, Japan) at 4°C. The lysates were centrifuged at 16,100 × g for 10 min at 4°C and the amount of protein in each lysate was evaluated by staining with Coomassie Brilliant Blue (CBB) G-250 (Bio-Rad Laboratories, Inc.). A loading buffer [350 mM Tris-HCl (pH 6.8), 30% (w/v) glycerol, 0.012% (w/v) bromophenol blue, 6% (w/v) SDS and 30% (v/v) 2-mercaptoethanol] was added to each lysate, which was subsequently boiled for 3 min. Proteins (15 µg per lane) were loaded and electrophoresed on 12.5% SDS-polyacrylamide gels. The proteins were transferred to polyvinylidene difluoride membranes. Membranes were blocked with TBS-Tween-20 [TBST; 20 mM Tris-HCl (pH 7.6), 137 mM NaCl and 0.1% (v/v) Tween-20] containing 5% Difco™ skim milk (cat. no. 232100; BD Biosciences, Franklin Lakes, NJ, USA) for 30 min at room temperature, and immunoblotted with anti-c-myc [dilution, 1:50 (9E10 hybridoma cultured supernatant in TBST containing 5% Difco™ skim milk; cat. no. 9E10 hybridoma; Developmental Studies Hybridoma Bank, Iowa City, IA, USA) antibody for 1 h at room temperature. Subsequently, membranes were incubated with TBST containing 5% Difco™ skim milk secondary horseradish peroxidase (HRP)-conjugated sheep polyclonal anti-mouse IgG (dilution, 1:3,000; cat. no. NA931V; GE Healthcare Life Sciences, Little Chalfont, UK) for 1 h at room temperature. Detection was performed using Immobilon Western Chemiluminescent HRP substrates (EMD Millipore, Billerica, MA, USA) on an ImageQuant LAS4000mini (GE Healthcare Life Sciences). To detect all loading control proteins, the membrane was stained by Coomassie brilliant blue solution at room temperature for 5 min (27). Western blotting was conducted in three independent experiments.
Purification of recombinant wild-type RPESP from whole-cell lysate
HT1080-RPESP-MH cells were lysed using the aforementioned lysis buffer, and the cell lysate was used for purification. Initially, solid ammonium sulfate was slowly added with agitation of the lysate preparations (125 ml) until 30% saturation was achieved. The pH of the lysate was maintained at pH 8.0 by dropwise addition of 5 N NaOH and the mixture was incubated on ice for 30 min. Subsequently, the precipitates were separated by centrifugation at 16,100 × g for 30 min at 4°C and the supernatant was collected. Solid ammonium sulfate was slowly added to the supernatant until 60% saturation was achieved and the solution was preserved on ice for another 30 min prior to centrifugation at 16,100 × g for 30 min at 4°C. The precipitates were collected and dissolved in a small volume (~2 ml) of PBS, and added to 1% (w/v) SDS. The mixture was sonicated (20 kHz, 50 W, 15 sec, 15 times) at 4°C and centrifuged at 16,100 × g for 15 min at 4°C. The supernatant was subjected to buffer exchange with PBS using PD-10 Desalting Columns (GE Healthcare Life Sciences). Following the addition of 8 M urea, the mixture was incubated with Ni-NTA agarose (Qiagen GmbH, Hilden, Germany) for 2 h at 4°C. The Ni-NTA agarose was washed three times with buffer A (900 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 and 20 mM imidazole) and Ni-NTA agarose-bound RPESP was eluted with 100 µl of 500 mM imidazole. Eluates (20 µl per lane) were loaded and electrophoresed on a 12.5% SDS-PAGE and the protein bands were visualized using CBB staining for 1 h at room temperature. The stained gel was scanned with CanoScan LiDE 200 (Canon, Tokyo, Japan) using MP Navigator EX software (Ver. 2.0.7; Canon).
Liquid chromatography-mass spectrometry (LC-MS)
In order to determine the C-mannosylation sites, the present study used the ultra-sensitive ‘Q-Exactive’ nano LC-MS/MS system (28). Purified RPESP samples were subjected to 12.5% SDS-PAGE. Following CBB staining for 1 h at room temperature, the visible band was excised and de-stained. The band was reduced using 50 mM dithiothreitol (Wako Pure Chemical Industries, Ltd.) at 37°C for 2 h, followed by carboxymethylated with 100 mM iodoacetate (Sigma-Aldrich; Merck KGaA) at 25°C for 30 min. In-gel digestion was performed using trypsin (TPCK-treated; Worthington Biochemical Corporation, Lakewood, NJ, USA) at 37°C for 12 h. The digestion mixture was separated on a nanoflow LC (Easy nLC; Thermo Fisher Scientific, Inc.) using a nano-electrospray ionization spray column (NTCC analytical C18 column, ϕ75 µm ×100 mm, 3 µm; Nikkyo Technos Co., Ltd., Tokyo, Japan) with a linear gradient of 5–60% buffer B (100% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min over 10 min and subjected on-line to a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.), which was equipped with a nanospray ion source. MS and MS/MS data were acquired using the data-dependent TOP10 method (28). Obtained MS/MS data were searched against an in-house database, including the RPESP sequence, using the MASCOT program (Matrix Science, Inc., Boston, MA) with variable modifications: Gln→pyro-Glu (N-term Q), oxidation (M), propionamide (C) and Hex (W). MS/MS chromatograms and spectra were acquired using the targeted MS/MS method (28).
Purification of RPESP from S2 cells
S2 cells were plated on 100 mm dishes and transfected with pMT-RPESP-MH and pIZ-DPY19L1, pIZ-DPY19L2, pIZ-DPY19L3, pIZ-DPY19L4 or pIZ as a control using transIT-Insect Transfection reagent (Mirus Bio, LLC, Madison, WI, USA) at 25°C. Following 24 h, the cells were washed and cultured in Schneider's Drosophila medium at 25°C with 700 µM CuSO4 to induce RPESP expression. Following 48 h of induction, the cells were lysed in 1 ml of the aforementioned lysis buffer with sonication (20 kHz, 50 W, 15 sec, four times) at 4°C and 50 µl of Ni-NTA agarose was added to the lysates. After 2 h, Ni-NTA agarose-bound RPESP was eluted with 100 µl of 500 mM imidazole. Eluates (20 µl per lane) were loaded and electrophoresed on 12.5% SDS-polyacrylamide gels. The protein bands were visualized using CBB for 1 h at room temperature and the visible band was excised and de-stained. The samples were treated with N-(iodoethyl)-trifluoroacetamide for cysteine-aminoethylation, followed by LC-MS analysis, as aforementioned.
Reverse transcription-semi-quantitative PCR (RT-sqPCR)
Reverse transcribed cDNAs of A549, ES2, HepG2, HT1080, HT29, Jurkat, LNCaP, WM266-4, COLO 205, CW-2, HCT116 and LoVo human cancer cell lines were used for PCR amplification using Quick Taq HS Dye mix (Toyobo, Co., Ltd., Osaka, Japan), according to the manufacturer's protocol. The sequences of the primers used (synthesized by Thermo Fisher Scientific, Inc.) for RT-sqPCR were as follows: RPESP forward, 5′-GACAGGGTCTACGGGACGTGTTTC-3′ and reverse, 5′-TGCAGAGGTAGTTATAAAGGCAG-3′; β-actin forward, 5′-CTTCGAGCACGAGATGGCCA-3′ and reverse, 5′-CCAGACAGCACTGTGTTGGC-3′. The cycling conditions were as follows: RPESP, 94°C for 2 min, followed by 25 cycles at 94°C for 30 sec, 60°C for 30 sec and 68°C for 30 sec; β-actin, 94°C for 2 min, followed by 20 cycles at 94°C for 30 sec, 58°C for 30 sec and 68°C for 30 sec. β-actin was used for the loading control. PCR products were electrophoresed on 2% agarose gels, stained with ethidium bromide for 10 min at room temperature and visualized using an ultraviolet illuminator. The expression levels of transfected DPY19L1-L4 genes in S2 cells were normalized to GAPDH and confirmed according to a previously described method (12). RT-sqPCR was conducted in three independent experiments.
Results
RPESP is C-mannosylated in cells
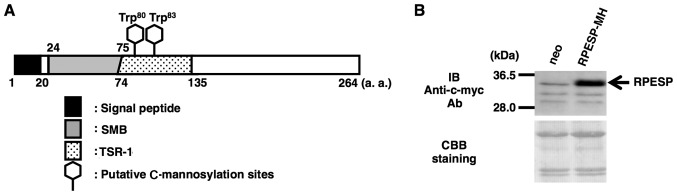
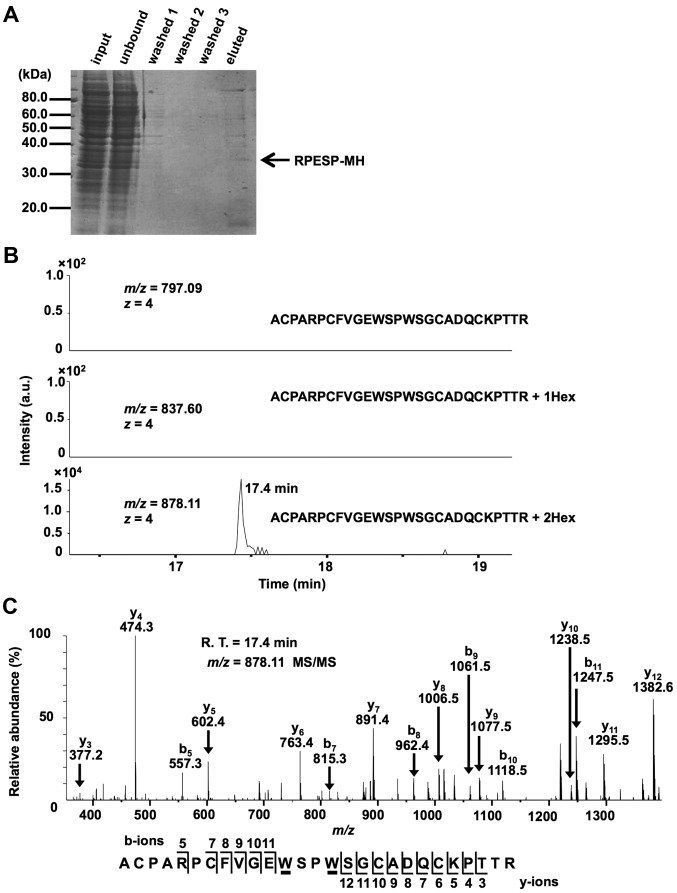
The amino acid sequence of human RPESP contains two possible C-mannosylation sites at the W80 and W83 residues in the TSR-1 domain (Fig. 1A). Although the physiological functions of RPESP remain unclear, the Rspo family and certain proteins have been reported to be C-mannosylated in the TSR-1, and C-mannosylation of these proteins regulates their functions, including secretion and Wnt/β-catenin signal-enhancing activity (12,19). Therefore, C-mannosylation of RPESP may regulate its functions. To determine whether RPESP is C-mannosylated, the present study established an RPESP-overexpressing HT1080 cell line, HT1080-RPESP-MH cells (Fig. 1B). Although RPESP has a predicted N-terminal signal peptide, the present study did not detect RPESP secretion in the conditioned medium in the HT1080-RPESP-MH cells (data not shown). Therefore, the present study purified recombinant RPESP protein from whole-cell lysates of HT1080-RPESP-MH cells for LC-MS analysis (Fig. 2A). The obtained recombinant RPESP was treated with trypsin and the resulting mixture of peptides was analyzed using the targeted MS/MS method. According to the inclusion list of three quadruple-protonated parent ions of un-mannosylated (m/z, 797.09), mono-mannosylated (m/z, 837.60) and di-mannosylated (m/z, 878.11) 69ACPARPCFVGEWSPWSGCADQCKPTTR95 peptides, MS/MS spectra were obtained. The selected ion chromatograms of y5 ion of these parent ions were determined (Fig. 2B). Fig. 2B demonstrated that un- and mono-mannosylated peptides were not detected, which suggests that all RPESP protein is C-mannosylated at W80, and W83. The MS/MS spectrum of the quadruple-charged di-mannosylated peptide ion (m/z, 878.11) is presented in Fig. 2C. LC-MS/MS analysis of the peptide, modified by two mannose residues, revealed that the differences in the theoretical m/z values for non-mannosylated tryptophan between b5-b11 and y3-y12 ions were not observed. This result indicates that modification occurs at the peptide 80WSPW83. Glycosylation at a proline residue has not been reported. Furthermore, addition of hexose at a serine residue by O-glycosylation has previously been reported (29), including O-glucosylation of epidermal growth factor-like repeats and O-mannosylation of α-dystroglycan; however, these modifications were site-specific, and RPESP did not satisfy the requirements (29). Additionally, the W80 and W83 residues met the requirements of the C-mannosylation consensus sequence W-X-X-W/C, which suggests that RPESP may be C-mannosylated at W80 and W83.
Figure 1.
Establishment of an RPESP-overexpressing cell line. (A) Schematic diagram of human RPESP. RPESP has two possible C-mannosylation sites within TSR-1. A black box, gray box, dotted box and two hexagonal shapes denote the signal peptide, SMB, TSR-1 and the putative C-mannosylation sites, respectively. (B) Establishment of an RPESP-overexpressing cell line, HT1080-RPESP-MH. HT1080-neo and HT1080-RPESP-MH cells were lysed and each cell lysate was electrophoresed, and immunoblotted with anti-c-myc antibody. The membrane was stained by CBB solution following immunoblotting. RPESP, RPE-spondin; TSR-1, thrombospondin type-1 repeat; SMB, somatomedin B; CBB, Coomassie Brilliant Blue; a.a, amino acid; neo, HT1080 cells expressing the pCI-neo vector.
Figure 2.
RPESP is C-mannosylated in cells. (A) Purification of recombinant RPESP-MH from whole-cell lysate of HT1080-RPESP-MH. HT1080-RPESP-MH cells were lysed and cell lysate was treated with ammonium sulfate. The precipitate between 30–60% saturation of ammonium sulfate was dissolved in PBS (input) and recombinant RPESP-MH was purified using Ni-NTA agarose. These samples were electrophoresed and detected by CBB staining. (B and C) Determination of C-mannosylation sites within RPESP. The samples were digested with trypsin and the resulting peptides were analyzed using the targeted MS/MS method. According to the inclusion list of three quadruple-protonated parent ions of un-mannosylated (m/z, 797.09), mono-mannosylated (m/z, 837.60) and di-mannosylated 69ACPARPCFVGEWSPWSGCADQCKPTTR95 peptides (m/z, 878.11), MS/MS spectra were obtained. Selected ion chromatograms of y5 ion (602.366±20 ppm) of these parent ions were determined (B). The MS/MS spectrum of the quadruple-charged di-mannosylated peptide ion (m/z, 878.11) is presented (C). The indicated b- and y-series ions were detected as singly charged ions and C-mannosylation at the W80 and W83 residues of RPESP were suggested. C-mannosyltryptophans are underlined. RPESP, RPE-spondin; CBB, Coomassie Brilliant Blue; MS, mass spectrometry; Hex, hexose; R.T., retention time.
Identification of DPY19L3 as the C-mannosyltransferase of RPESP at W83
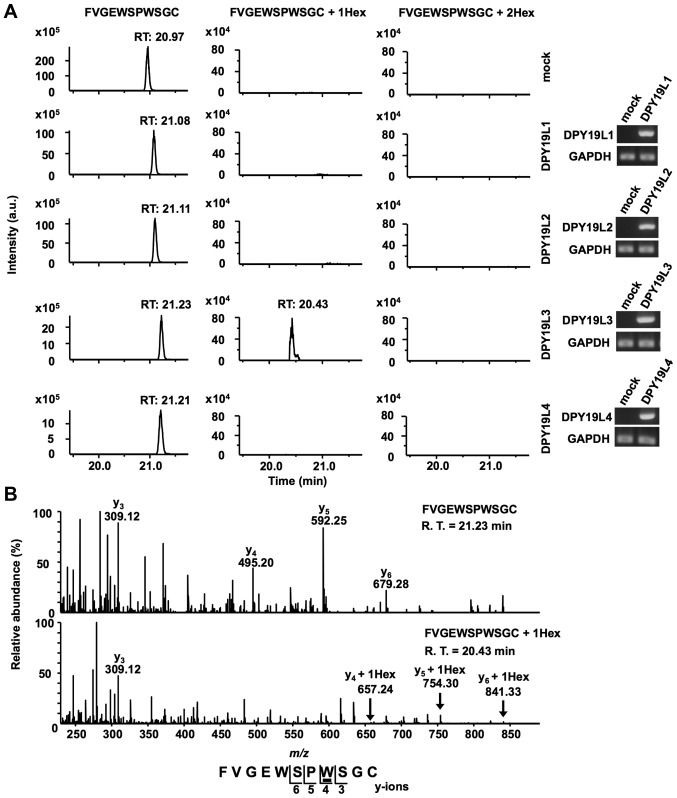
A previous report demonstrated that C. elegans dpy-19 was identified as a C-mannosyltransferase for TSR-1 (11), and that DPY19L3 catalyzes C-mannosylation of human Rspo1 at the W156 residue in human cells (12). Since C-mannosylation of RPESP occurred in TSR-1, the present study hypothesized that at least one of the human homologs (DPY19L1-L4) of C. elegans dpy-19 may be a C-mannosyltransferase of RPESP. In order to identify the C-mannosyltransferase(s) of RPESP, the present study performed gain-of-function experiments. Drosophila S2 cells have no C-mannosyltransferase activity (11,30,31) but harbor dolichol-phosphate-mannose, which is the donor substrate for C-mannosylation. The present study transiently transfected human RPESP cDNA with the probable C-mannosyltransferase DPY19L1-L4 or empty vector (mock), respectively, into S2 cells. The expression of transfected DPY19L1-L4 mRNAs in S2 cells was confirmed by RT-sqPCR (Fig. 3A). Recombinant RPESP proteins were purified from each S2 cell lysate and analyzed using LC-MS. The obtained recombinant RPESP were aminoethylated, digested with trypsin and the resulting peptides were analyzed using the targeted MS/MS method. According to the inclusion list of three doubly protonated parent ions of un-, mono- and di-mannosylated 76FVGEWSPWSGC86 peptides, MS/MS spectra were obtained. Selected ion chromatograms of y5 ions of these parent ions were determined (Fig. 3A). Mono-mannosylated peptide was observed only when RPESP was produced in DPY19L3-expressing S2 cells; conversely, di-mannosylated peptide was not observed in any of the samples (Fig. 3A). These results suggested that human DPY19L3 catalyzes C-mannosylation of RPESP at a tryptophan residue. The MS/MS spectra of the doubly charged un- and mono-mannosylated peptide ion derived from DPY19L3-expressing S2 cells were presented (Fig. 3B). The y3 ion corresponded well in un- and mono-mannosylated peptides; however, y4-y6 ions were observed at an increased position of m/z 162 in mono-mannosylated peptide compared with the unmannosylated peptide (Fig. 3B). These results suggested that DPY19L3 may be the C-mannosyltransferase of RPESP at only the W83 residue.
Figure 3.
DPY19L3 is the C-mannosyltransferase of RPESP at only W83. (A and B) Identification of C-mannosyltransferase of RPESP. Human DPY19L1-L4 or empty vector (mock) and pMT-RPESP-MH were transiently transfected into Drosophila S2 cells and protein expression was induced using treatment with 700 µM CuSO4 for 48 h. RPESP-MH protein was purified with Ni-NTA agarose. Following aminoethylation, the samples were digested with trypsin, and the resulting peptides were analyzed by targeted MS/MS method. According to the inclusion list of three doubly protonated parent ions of un-mannosylated (m/z, 649.28), mono-mannosylated (m/z, 730.31) and di-mannosylated 76FVGEWSPWSGC86 peptides (m/z, 811.34), MS/MS spectra were obtained. Selected ion chromatograms of y5 ion of these parent ions were determined (A, left panel). The expression of transfected DPY19L1-L4 mRNAs in S2 cells was confirmed by reverse transcription-semi-quantitative polymerase chain reaction (A, right panel). The MS/MS spectra of the doubly charged un- (B, upper panel) and mono- (B, lower panel) mannosylated peptide ion derived from DPY19L3-expressing S2 cells are presented. The indicated y-series ions were detected as singly charged ions and only the W83 residue of RPESP was C-mannosylated. C-mannosyltryptophan is underlined. DPY19L, dpy-19 like; MS, mass spectrometry; RPESP, RPE-spondin; Hex, hexose; R.T., retention time.
Expression level of RPESP in human tumor cell lines
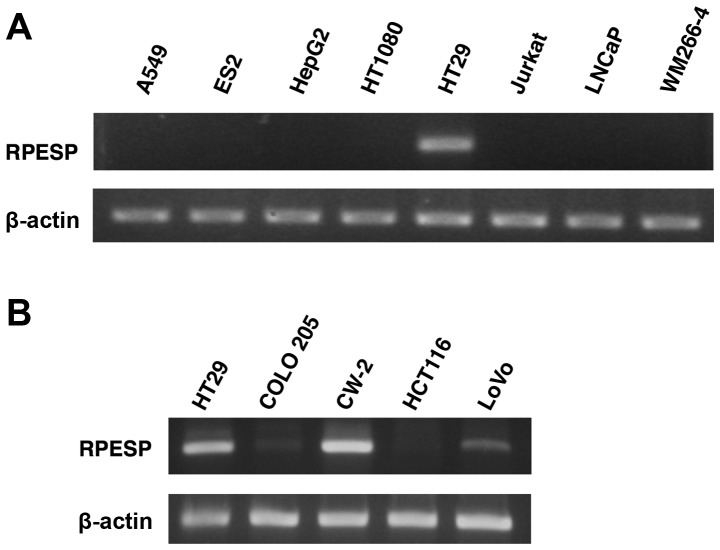
To examine the roles of RPESP in tumor malignancy, the present study analyzed RPESP mRNA expression levels using RT-sqPCR in a number of human tumor cell lines. Among 8 cell lines, the expression level of RPESP in HT29 cells was high; however, RPESP mRNA in other cell lines was undetectable under the assay conditions maintained (Fig. 4A). Since endogenous RPESP mRNA in HT29, a colon cancer cell line, was detected, the present study evaluated the expression levels of other human colon cancer cell lines. As presented in Fig. 4B, HT29, CW-2 and LoVo cells expressed RPESP mRNA, although COLO 205 and HCT116 did not. Therefore, it was suggested that RPESP may serve certain roles in tumorigenesis, particularly in colon cancer.
Figure 4.
RPESP is expressed in certain colon cancer cell lines. (A) Expression of RPESP mRNA in human tumor cell lines. Total RNAs were isolated from A549 lung adenocarcinoma, ES2 ovarian cancer, HepG2 hepatoma, HT1080 fibrosarcoma, HT29 colon cancer, Jurkat leukemia, LNCaP prostate cancer and WM266-4 melanoma cells and RT-sqPCR analysis was performed. (B) Expression of RPESP mRNA in human colon cancer cell lines. Total RNAs were isolated from human colon cancer cell lines, HT29, COLO 205, CW-2, HCT116 and LoVo, and RT-sqPCR analysis was performed. RT-sqPCR, reverse transcription-semi-quantitative polymerase chain reaction; RPESP, RPE-spondin.
Discussion
The present study suggested that human RPESP may be C-mannosylated at W80 and W83. Previous studies have suggested that C-mannosylation has potential functions, including protein folding, stability, secretion and enzymatic activity (12,19,22,32,33). Certain TSR-1 superfamily proteins are reported to be C-mannosylated, particularly in Rspo1 and Rspo3; the modifications regulate their secretion and the agonistic activity of canonical Wnt signaling pathways (12,19); however, the physiological functions of RPESP have not yet been investigated. In our preliminary experiment, the present study investigated the mRNA expression level of RPESP among various types of cancer cell lines. RPESP was expressed in certain colon adenocarcinoma cell lines (HT29, CW-2 and LoVo) but not in any other cancer cell line investigated, which suggests an importance of RPESP expression in colorectal cancer. Previously it was reported that RPESP exists in the aorta extracellular matrix (14); however, the physiological functions remain unclear. Considering RPESP is expressed in colon adenocarcinoma cell lines, RPESP may be involved in malignant alteration of cancer. As the present study was not able to elucidate the association between the physiological functions of RPESP or the role of C-mannosylation on RPESP and cancer, further studies are required.
A previous study demonstrated that C. elegans dpy-19 was a C-mannosyltransferase of TSR-1 (11). Based on this study, the present study investigated the human C-mannosyltransferase(s) of Rspo1, which has two C-mannosylation sites at W153 and W156, and identified human DPY19L3, one of the homologs of C. elegans dpy-19, as a C-mannosyltransferase of Rspo1 at W156 (12). The present study also investigated the C-mannosyltransferase(s) of RPESP, which has two C-mannosylation sites at W80 and W83, and revealed that human RPESP was C-mannosylated at W83 by DPY19L3. DPY19L3, and not DPY19L1, DPY19L2 or DPY19L4, acted as a C-mannosyltransferase of RPESP at the W83 residue, and C-mannosylation of RPESP at W80 was not catalyzed by all DPY19 family proteins, suggesting the existence of other C-mannosyltransferase member(s) in human cells. Further studies are required in order to identify the novel C-mannosyltransferase of RPESP at W80.
In conclusion, the present study demonstrated that RPESP was C-mannosylated by DPY19L3 in human cells. Although the physiological functions of RPESP and the significance of its C-mannosylation remain unclear, our previous and present studies suggest that human DPY19 family members have substrate specificity, and that DPY19L3 may have a selective amino acid sequence for C-mannosylation. These studies may aid in furthering the understanding of DPY19L3-mediated C-mannosylation and RPESP functions in cancer.
Acknowledgements
The present study was supported in part by Grants-in-Aid for Scientific Research (B) (grant no. 24310167) and Japan Society for the Promotion of Science Fellowship (grant no. 254256).
Glossary
Abbreviations
- TSR-1
thrombospondin type-1 repeat
- Rspo
R-spondin
- RPESP
RPE-spondin
- SMB
somatomedin B
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- CBB
Coomassie Brilliant Blue
- LC-MS
liquid chromatography-mass spectrometry
- ER
endoplasmic reticulum
References
- 1.Dwek RA, Butters TD, Platt FM, Zitzmann N. Targeting glycosylation as a therapeutic approach. Nat Rev Drug Discov. 2002;1:65–75. doi: 10.1038/nrd708. [DOI] [PubMed] [Google Scholar]
- 2.Simizu S, Ishida K, Wierzba MK, Osada H. Secretion of heparanase protein is regulated by glycosylation in human tumor cell lines. J Biol Chem. 2004;279:2697–2703. doi: 10.1074/jbc.M300541200. [DOI] [PubMed] [Google Scholar]
- 3.Simizu S, Takagi S, Tamura Y, Osada H. RECK-mediated suppression of tumor cell invasion is regulated by glycosylation in human tumor cell lines. Cancer Res. 2005;65:7455–7461. doi: 10.1158/0008-5472.CAN-04-4446. [DOI] [PubMed] [Google Scholar]
- 4.Furmanek A, Hofsteenge J. Protein C-mannosylation: Facts and questions. Acta Biochim Pol. 2000;47:781–789. [PubMed] [Google Scholar]
- 5.Doucey MA, Hess D, Cacan R, Hofsteenge J. Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol Biol Cell. 1998;9:291–300. doi: 10.1091/mbc.9.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krieg J, Hartmann S, Vicentini A, Gläsner W, Hess D, Hofsteenge J. Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp. Mol Biol Cell. 1998;9:301–309. doi: 10.1091/mbc.9.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Beer T, Vliegenthart JF, Löffler A, Hofsteenge J. The hexopyranosyl residue that is C-glycosidically linked to the side chain of tryptophan-7 in human RNase Us is alpha-mannopyranose. Biochemistry. 1995;34:11785–11789. doi: 10.1021/bi00037a016. [DOI] [PubMed] [Google Scholar]
- 8.Doucey MA, Hess D, Blommers MJ, Hofsteenge J. Recombinant human interleukin-12 is the second example of a C-mannosylated protein. Glycobiology. 1999;9:435–441. doi: 10.1093/glycob/9.5.435. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann S, Hofsteenge J. Properdin, the positive regulator of complement, is highly C-mannosylated. J Biol Chem. 2000;275:28569–28574. doi: 10.1074/jbc.M001732200. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Cao C, Jia W, Yu L, Mo M, Wang Q, Huang Y, Lim JM, Ishihara M, Wells L, et al. Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule. EMBO J. 2009;28:286–297. doi: 10.1038/emboj.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buettner FF, Ashikov A, Tiemann B, Lehle L, Bakker H. C. elegans DPY-19 is a C-mannosyltransferase glycosylating thrombospondin repeats. Mol Cell. 2013;50:295–302. doi: 10.1016/j.molcel.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Niwa Y, Suzuki T, Dohmae N, Simizu S. Identification of DPY19L3 as the C-mannosyltransferase of R-spondin1 in human cells. Mol Biol Cell. 2016;27:744–756. doi: 10.1091/mbc.E15-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz HL, Rahman FA, El Moula FM Fadl, Stojic J, Gehrig A, Weber BH. Identifying differentially expressed genes in the mammalian retina and the retinal pigment epithelium by suppression subtractive hybridization. Cytogenet Genome Res. 2004;106:74–81. doi: 10.1159/000078564. [DOI] [PubMed] [Google Scholar]
- 14.Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M. Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat Struct Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 16.Kamikubo Y, De Guzman R, Kroon G, Curriden S, Neels JG, Churchill MJ, Dawson P, Ołdziej S, Jagielska A, Scheraga HA, et al. Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin. Biochemistry. 2004;43:6519–6534. doi: 10.1021/bi049647c. [DOI] [PubMed] [Google Scholar]
- 17.Mayasundari A, Whittemore NA, Serpersu EH, Peterson CB. The solution structure of the N-terminal domain of human vitronectin: Proximal sites that regulate fibrinolysis and cell migration. J Biol Chem. 2004;279:29359–29366. doi: 10.1074/jbc.M401279200. [DOI] [PubMed] [Google Scholar]
- 18.Kato K, Nishimasu H, Okudaira S, Mihara E, Ishitani R, Takagi J, Aoki J, Nureki O. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling; Proc Natl Acad Sci USA; 2012; pp. 16876–16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara M, Kato S, Niwa Y, Suzuki T, Tsuchiya M, Sasazawa Y, Dohmae N, Simizu S. C-mannosylation of R-spondin3 regulates its secretion and activity of Wnt/β-catenin signaling in cells. FEBS Lett. 2016;590:2639–2649. doi: 10.1002/1873-3468.12274. [DOI] [PubMed] [Google Scholar]
- 20.Iwaki T, Castellino FJ. A single plasmid transfection that offers a significant advantage associated with puromycin selection in Drosophila Schneider S2 cells expressing heterologous proteins. Cytotechnology. 2008;57:45–49. doi: 10.1007/s10616-008-9129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa Y, Suzuki T, Dohmae N, Umezawa K, Simizu S. Determination of cathepsin V activity and intracellular trafficking by N-glycosylation. FEBS Lett. 2012;586:3601–3607. doi: 10.1016/j.febslet.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Goto Y, Niwa Y, Suzuki T, Dohmae N, Umezawa K, Simizu S. C-mannosylation of human hyaluronidase 1: Possible roles for secretion and enzymatic activity. Int J Oncol. 2014;45:344–350. doi: 10.3892/ijo.2014.2438. [DOI] [PubMed] [Google Scholar]
- 23.Goto Y, Niwa Y, Suzuki T, Uematsu S, Dohmae N, Simizu S. N-glycosylation is required for secretion and enzymatic activity of human hyaluronidase1. FEBS Open Bio. 2014;4:554–559. doi: 10.1016/j.fob.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simizu S, Suzuki T, Muroi M, Lai NS, Takagi S, Dohmae N, Osada H. Involvement of disulfide bond formation in the activation of heparanase. Cancer Res. 2007;67:7841–7849. doi: 10.1158/0008-5472.CAN-07-1053. [DOI] [PubMed] [Google Scholar]
- 25.Simizu S, Umezawa K, Takada M, Arber N, Imoto M. Induction of hydrogen peroxide production and Bax expression by caspase-3(−like) proteases in tyrosine kinase inhibitor-induced apoptosis in human small cell lung carcinoma cells. Exp Cell Res. 1998;238:197–203. doi: 10.1006/excr.1997.3823. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki S, Sasazawa Y, Mogi T, Suzuki T, Yoshida K, Dohmae N, Takao K, Simizu S. Identification of seco-clavilactone B as a novel small-molecule actin polymerization inhibitor. FEBS Lett. 2016;590:1163–1173. doi: 10.1002/1873-3468.12154. [DOI] [PubMed] [Google Scholar]
- 27.Welinder C, Ekblad L. Coomassie staining as loading control in western blot analysis. J Proteome Res. 2011;10:1416–1419. doi: 10.1021/pr1011476. [DOI] [PubMed] [Google Scholar]
- 28.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieg J, Gläsner W, Vicentini A, Doucey MA, Löffler A, Hess D, Hofsteenge J. C-mannosylation of human RNase 2 is an intracellular process performed by a variety of cultured cells. J Biol Chem. 1997;272:26687–26692. doi: 10.1074/jbc.272.42.26687. [DOI] [PubMed] [Google Scholar]
- 31.Hofsteenge J, Huwiler KG, Macek B, Hess D, Lawler J, Mosher DF, Peter-Katalinic J. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276:6485–6498. doi: 10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Vilar J, Randell SH, Boucher RC. C-mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology. 2004;14:325–337. doi: 10.1093/glycob/cwh041. [DOI] [PubMed] [Google Scholar]
- 33.Sasazawa Y, Sato N, Suzuki T, Dohmae N, Simizu S. C-mannosylation of thrombopoietin receptor (c-Mpl) regulates thrombopoietin-dependent JAK-STAT signaling. Biochem Biophys Res Commun. 2015;468:262–268. doi: 10.1016/j.bbrc.2015.10.116. [DOI] [PubMed] [Google Scholar]