Significance
Bacteria evolved molecular weapons to help them thrive in polymicrobial environments. The type VI secretion system (T6SS) is a gun loaded with a great diversity of bacterial toxins. On contact with neighboring cells, toxins are fired, and in the absence of immunity, the prey is killed, allowing the attacker to prevail. Each bacterium can be equipped with several distinct T6SSs, and it is unclear whether they are simultaneously active or whether each has a specific role in a particular environment. Here we showed that production of the three Pseudomonas aeruginosa T6SSs is orchestrated by global regulators. We suggest it may be possible for simultaneous assembly of multiple T6SSs within a single cell, priming it to fight a wide variety of organisms.
Keywords: T6SS, Pseudomonas, RsmA, AmrZ
Abstract
The type VI secretion system (T6SS) is a weapon of bacterial warfare and host cell subversion. The Gram-negative pathogen Pseudomonas aeruginosa has three T6SSs involved in colonization, competition, and full virulence. H1-T6SS is a molecular gun firing seven toxins, Tse1–Tse7, challenging survival of other bacteria and helping P. aeruginosa to prevail in specific niches. The H1-T6SS characterization was facilitated through studying a P. aeruginosa strain lacking the RetS sensor, which has a fully active H1-T6SS, in contrast to the parent. However, study of H2-T6SS and H3-T6SS has been neglected because of a poor understanding of the associated regulatory network. Here we performed a screen to identify H2-T6SS and H3-T6SS regulatory elements and found that the posttranscriptional regulator RsmA imposes a concerted repression on all three T6SS clusters. A higher level of complexity could be observed as we identified a transcriptional regulator, AmrZ, which acts as a negative regulator of H2-T6SS. Overall, although the level of T6SS transcripts is fine-tuned by AmrZ, all T6SS mRNAs are silenced by RsmA. We expanded this concept of global control by RsmA to VgrG spike and T6SS toxin transcripts whose genes are scattered on the chromosome. These observations triggered the characterization of a suite of H2-T6SS toxins and their implication in direct bacterial competition. Our study thus unveils a central mechanism that modulates the deployment of all T6SS weapons that may be simultaneously produced within a single cell.
The type VI secretion system (T6SS) is widely distributed within Gram-negative bacteria and is capable of injecting effector proteins into eukaryotic cells (1). Mounting evidence suggests the primary role of the T6SS is in bacterial warfare (2). The T6SS injects toxins (e.g., peptidoglycan hydrolases) into competing bacteria. Deployment of the T6SS provides a fitness advantage and contributes to shaping bacterial communities.
In about one third of bacterial genomes that harbor T6SS genes, multiple clusters can be found (3). Maintenance of these large operons as well as other specialized T6SS genes (e.g., vgrG or hcp islands; SI Appendix, Fig. S1) (4) suggests all these clusters are functional and give the bacteria a survival advantage. It is thought that multiple clusters are expressed and used in specific conditions, but the regulation or cross-regulation of all T6SS genes within a single bacterium is a topic of scarce knowledge.
The Gram-negative pathogen Pseudomonas aeruginosa encodes three T6SS clusters: H1-, H2-, and H3-T6SS (5). H1-T6SS transports at least seven antibacterial toxins (Tse1–Tse7) (6-8). This system has been extensively studied, as it was shown to be active in a retS mutant, whereas it is silent in the parental strain (9). This suggested a tight regulation of the T6SS, otherwise poorly expressed in in vitro growth conditions and likely triggered in a specific environment, such as during in vivo colonization. Mutation in the retS gene allows activation of the GacS/GacA two-component system, with GacA driving expression of two small noncoding RNAs, RsmY and RsmZ, that sequester RsmA and lead to de-repression of H1-T6SS (9, 10). RsmA is a translational repressor that binds on H1-T6SS messenger RNA (11) immediately upstream of the first gene in the cluster, tssA1 (12) (SI Appendix, Fig. S1). This pathway has been proposed to be required for P. aeruginosa to sense kin cell lysis and trigger the “P. aeruginosa response to antagonism,” which is an increase in H1-T6SS activity and killing of bacterial competitors (13).
Regulators and growth conditions involved in H2- or H3-T6SS expression have been proposed, including quorum sensing and iron limitation (14, 15), but none has a clear effect in vitro compared with RetS or RsmA on H1-T6SS. We addressed H2- and H3-T6SS control by using transposon (Tn) mutagenesis and reporter fusions. We demonstrate that RsmA acts on most known T6SS genes, which has been undervalued in previous studies, and show that AmrZ is another global regulator of the T6SS. We also observe assembly of different T6SSs within a single cell, suggesting they are not mutually exclusive.
Results
Global and RsmA-Dependent Control of P. aeruginosa T6SSs.
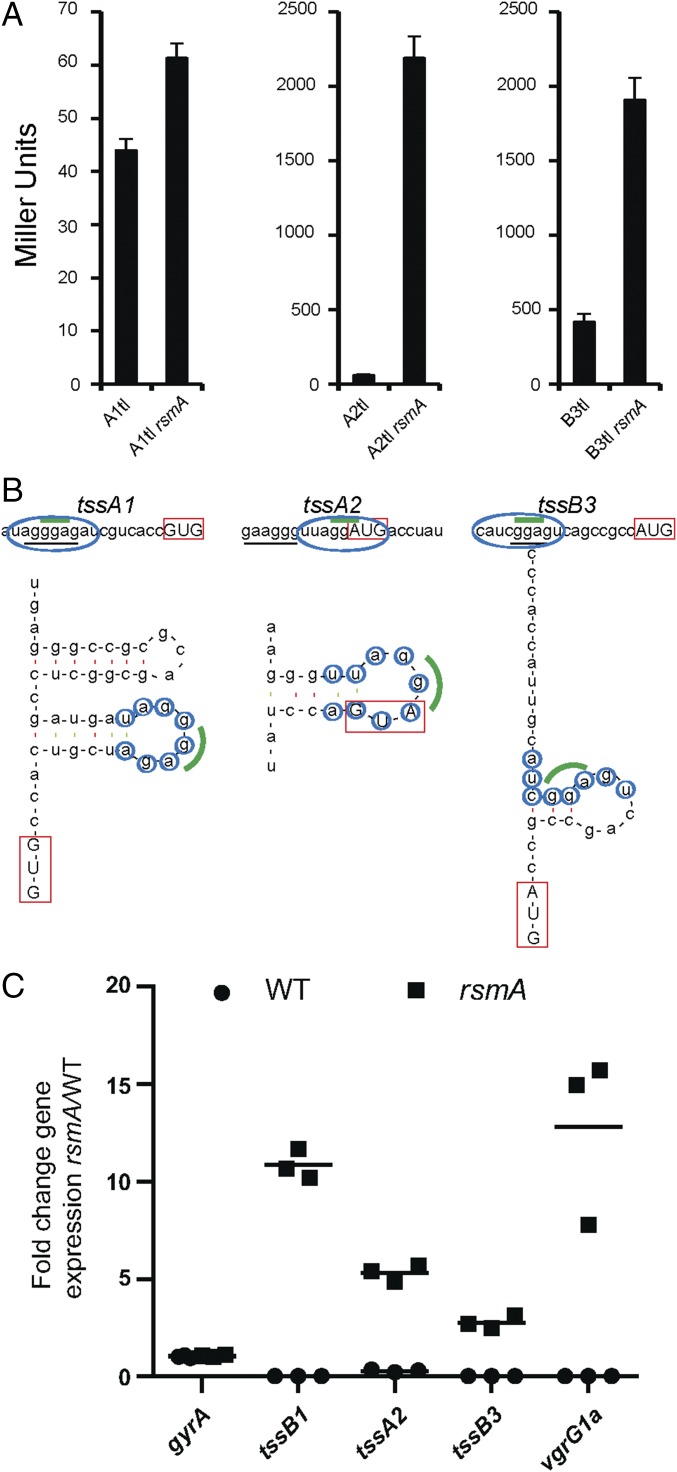
To identify regulators of H2-T6SS expression, we constructed a lacZ transcriptional fusion (A2tc) using the first gene in the H2-T6SS cluster (tssA2) (SI Appendix, Fig. S1), and inserted this reporter into the chromosome of P. aeruginosa PA14. Tn mutagenesis (16) was performed, and more than 85,000 Tn mutants were obtained. Those with altered β-galactosidase activity were isolated, and the position of the Tn insertion was mapped in 22 mutants (SI Appendix, Fig. S2 and Table S1). A Tn insertion in rsmA resulted in elevated levels of H2-T6SS expression (two- to threefold). Because RsmA is a posttranscriptional regulator, we constructed a tssA2::lacZ translational fusion (A2tl). The reporter was placed on the PA14 chromosome, and the rsmA gene was subsequently deleted. An increase of ∼36-fold in LacZ activity was observed in the rsmA background (Fig. 1A). RsmA was originally shown to repress H1-T6SS, which is confirmed here in PA14, using a tssA1::lacZ translational fusion (A1tl), although de-repression is only about 1.4-fold (Fig. 1A). We then assessed the effect of RsmA on H3-T6SS by constructing a tssB3::lacZ translational fusion (B3tl), which displays a four- to fivefold increase in activity in the rsmA background (Fig. 1A). Previous analyses have shown that RsmA binds the tssA1 mRNA in a region overlapping the ribosome binding site (RBS), which is thought to form a stem–loop structure (11) (Fig. 1B). Here, we identified putative RsmA binding sites on tssA2 and tssB3 mRNAs that both contain the core GGA motif (Fig. 1B), and Mfold analysis predicts these regions form a stem–loop structure (Fig. 1B) (17). Deletion of rsmA is likely to affect the stability of target RNA, as previously observed with H1-T6SS transcripts (11). We thus performed quantitative (q)RT-PCR analysis on tssB1, tssA2, tssB3, and vgrG1a genes (Fig. 1C), and all were up-regulated in a rsmA background from twofold (tssB3) to 12-fold (vgrG1a).
Fig. 1.
RsmA controls all three T6SSs negatively. (A) β-galactosidase assay of PA14 and rsmA mutant (where indicated) carrying the H1-, H2-, or H3-T6SS translational fusion (A1tl, A2tl, or B3tl, respectively). Graphs represent mean ± SEM of five independent replicates (paired t test, P < 0.005, P < 0.005, and P < 0.05, respectively). (B) Predicted RsmA binding sites using Mfold for tssA1, tssA2, and tssB3. Blue ovals/circles, putative RsmA binding sites; green line, core GGA motif; red box, start codon; black underline, predicted RBS. (C) qRT-PCR analysis in PA14 or rsmA mutant for tssB1 and vgrG1a (H1-T6SS), tssA2 (H2-T6SS), tssB3 (H3-T6SS), and the gyrA housekeeping gene control. Scatter plot of fold change with mean (n = 3). Statistical analysis was performed on the ΔΔCT values (ANOVA Bonferroni posttest P > 0.05, P < 0.005, P < 0.005, P < 0.005, and P < 0.005, respectively).
AmrZ Is a Global T6SS Transcriptional Regulator.
Three distinct Tn insertions in and around the amrZ gene were selected that up-regulate (blue colony, B) or down-regulate (white colony, W) tssA2 gene expression (SI Appendix, Fig. S2 and Table S1). Whereas two mutants (B1, B2) with Tn insertions close to the 5′ end of amrZ exhibited increased β-galactosidase activity, a third Tn insertion (W32), 297 bp upstream of the amrZ gene start codon, had decreased activity (SI Appendix, Figs. S2 and S3). We hypothesized that in B1/B2, the Tn insertion interrupts the gene or prevents transcription, whereas in W32, an outward reading promoter in the Tn induces amrZ expression. These results suggest that modulating AmrZ levels affects expression of H2-T6SS.
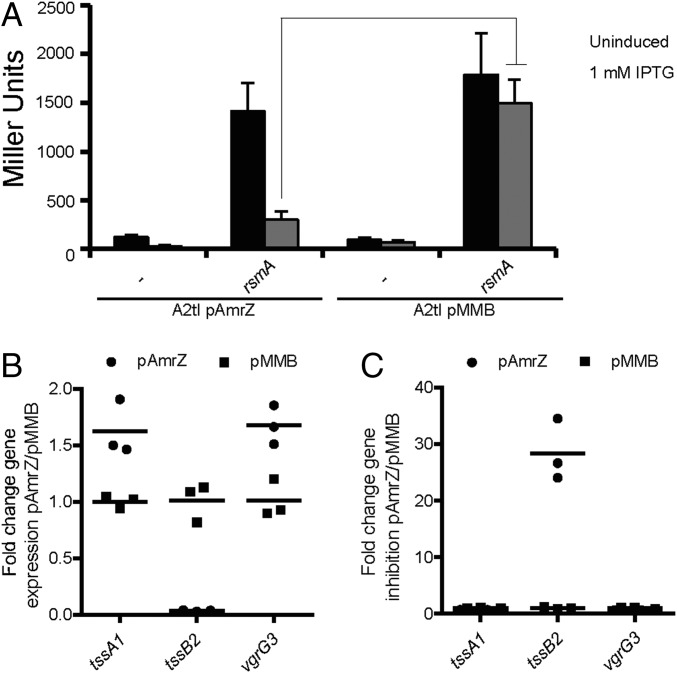
To probe this hypothesis, we engineered a deletion mutant of amrZ and a pMMB67HE-derivative overexpressing amrZ (pAmrZ). No significant difference in LacZ activity [tssA2::lacZ fusion (A2tc)] could be observed when comparing the wild-type and amrZ mutant when grown in liquid culture (SI Appendix, Fig. S3A). However, β-galactosidase assays using bacteria scrapped from plates yielded a twofold increase in LacZ activity in the amrZ mutant, comparable to activity of the original B1 mutant (SI Appendix, Fig. S3A). Conversely, AmrZ overexpression (pAmrZ) resulted in a 6- to 11-fold reduction in LacZ activity (SI Appendix, Fig. S3B). Furthermore, in a rsmA mutant, the level of LacZ activity from a tssA2::lacZ translational fusion (A2tl) is high (∼1,500 Miller units), but a fivefold reduction is seen on AmrZ overproduction (Fig. 2A), confirming AmrZ acts negatively on H2-T6SS. We conclude that two negative regulators act independently on H2-T6SS expression: AmrZ at the transcriptional and RsmA at the posttranscriptional level.
Fig. 2.
AmrZ inversely regulates H2- and H1-/H3-T6SSs. (A) AmrZ is a negative regulator for H2-T6SS. Level of LacZ activity of A2tl after overexpression of amrZ (pAmrZ) or in the presence of vector (pMMB) ± induction with IPTG. Graph represents mean ± SEM; n = 4; ANOVA Bonferroni posttest P < 0.001. (B) AmrZ represses H2-T6SS (tssB2), but activates H1-T6SS (tssA1) and H3-T6SS (vgrG3). qRT-PCR was performed on PA14rsmA overexpressing amrZ (pAmrZ) and compared with vector control (pMMB). Scatter plot of fold change gene expression with mean (n = 3). Statistical analysis was performed on the ΔΔCT values (ANOVA Bonferroni posttest P < 0.01 for all genes). (C) Fold gene repression of data shown in B.
We then analyzed the effect of AmrZ on H1- and H3-T6SS expression by performing qRT-PCR on the PA14rsmA strain overexpressing amrZ. We confirmed a significant repression of tssB2 (Fig. 2 B and C) and tssA2 (SI Appendix, Fig. S3 C and D) and observed a significant induction of tssA1 (H1-T6SS) and several genes encoding H3-T6SS components: vgrG3, tssB3, hcp3, and tssA3 (Fig. 2 B and C and SI Appendix, Fig. S3 C and D). We conclude that AmrZ acts independent of RsmA, repressing H2-T6SS and activating expression of H1-T6SS and H3-T6SS.
AmrZ Binds Directly to T6SS Promoters.
A consensus binding motif for AmrZ has been characterized in P. aeruginosa (18). We identified several degenerative versions of this motif in the upstream regions of tssA1, tssA2, and tssB3 (SI Appendix, Fig. S1 and Table S2) and performed electrophoresis mobility shift assays. A His-tagged AmrZ protein was purified, and conditions were optimized by using DNA fragments previously shown to be bound or not by AmrZ (18, 19). Binding could be observed on the tssA1 and tssA2 upstream regions (Fig. 3A), whereas a weaker band shift occurs for the tssB3 region, which is clear with 40–60 nM AmrZ (Fig. 3B). Smaller subfragments were used to demonstrate that only one of the putative binding sites for each upstream region was being bound by AmrZ (SI Appendix, Figs. S1 and S4 and Table S2) and confirmed that AmrZ binds to all three assessed T6SS promoter regions.
Fig. 3.
AmrZ binds the promoter regions of T6SS genes. Each reaction contains 5 nM 32P-labeled DNA and increasing concentrations of purified AmrZ, as indicated. Electrophoretic mobility shift assay was performed using DNA probes for (A) tssA1, tssA2, and (B) tssB3. In all cases, positive (adcA/algDl) or negative (algDs/algB) controls were used as previously published (18, 19). Asterisk indicates unspecific band.
RsmA Controls Production of Hcp Proteins Negatively.
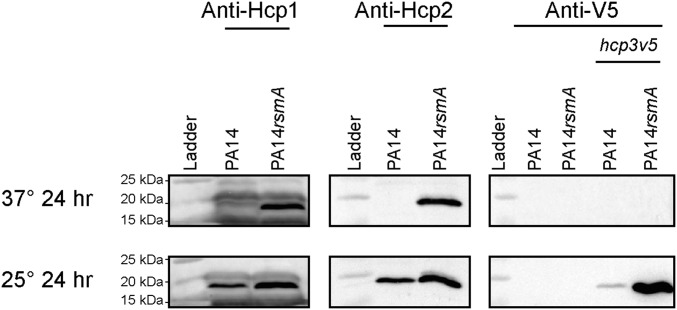
We analyzed whether control on gene expression is reflected in protein production. Western blot analysis using specific antibodies confirmed production of Hcp1 (H1-T6SS) and Hcp2 (H2-T6SS) in a rsmA mutant (Fig. 4, Upper). To probe Hcp3 production, we engineered a chimeric hcp3 gene on the PA14 chromosome, which encodes a V5-tagged version of Hcp3 (Hcp3V5). Hcp3V5 production was not detectable at 37 °C (Fig. 4, Upper), but was readily detected in a rsmA mutant when grown at 25 °C (Fig. 4, Bottom). Hcp1 and Hcp2 are also expressed in PA14 at 25 °C, with a modest increase in expression observed in a rsmA mutant (Fig. 4, Bottom).
Fig. 4.
RsmA controls all three Hcp proteins negatively. Western blot analysis comparing production of Hcp1, Hcp2, and Hcp3V5 at 37 °C (Upper) or 25 °C (Lower) in PA14 and rsmA mutant. Exposure times for Hcp blots are Hcp1, 600 s; Hcp2, 30 s; and Hcp3, 240 s.
H2- and H3-T6SS–Dependent Secretion Is Active in a rsmA Mutant.
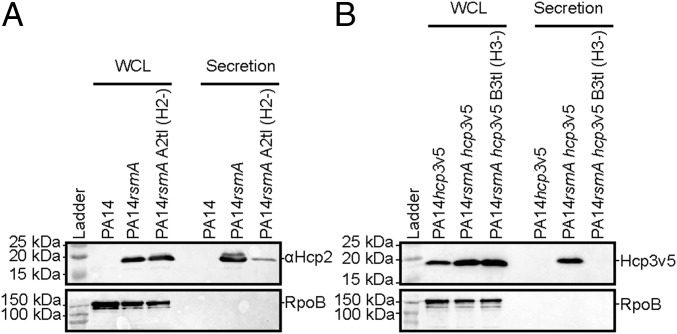
Hcp secretion is the hallmark of a functional T6SS. We assessed Hcp2 and Hcp3V5 secretion and observed that both are found in the supernatant fraction of a rsmA mutant, but were faintly detectable or absent in an H2- or H3-T6SS mutant (SI Appendix, Fig. S5 A and B). The H2-T6SS–dependent secretion of Hcp2 is very clear in both PA14 (Fig. 5A and SI Appendix, Fig. S5 A and B) and PAO1 (Fig. 6A), although low levels of Hcp2 in the supernatant of an H2-T6SS mutant were observed. Deletion of the H1- and H3-T6SS clusters (deleting both hcp1 and hcp3) does not diminish the level of protein detected with the anti-Hcp2 antibody, suggesting it is not a cross-reacting protein (SI Appendix, Fig. S5C).
Fig. 5.
H2-T6SS and H3-T6SS are functional in a rsmA mutant. (A) Hcp2 and (B) Hcp3 are used as readouts for T6SS-dependent secretion or presence in whole-cell lysate (WCL). Western blot analysis using (A) anti-Hcp2 on PA14, rsmA mutant, or rsmA H2-T6SS mutant (H2-) or (B) anti-V5 epitope to detect the tagged version of Hcp3 (Hcp3V5) in PA14, rsmA mutant, or rsmA H3-T6SS mutant (H3-). RNA polymerase (RpoB) is used as a lysis control.
Fig. 6.
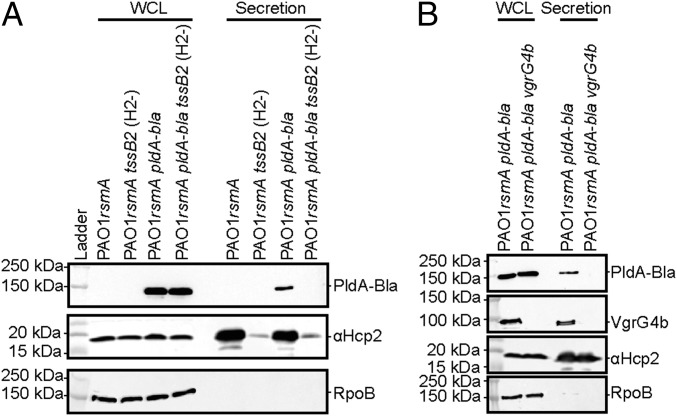
Deletion of rsmA enables PldA secretion. Western blot analysis using anti-TEM to detect PldA-Bla expression shows expression in a PAO1rsmA mutant at 25 °C and secretion in a (A) H2-T6SS–dependent manner [tssB2 mutant (H2-)] and (B) VgrG4b-depenedent manner (vgrG4b mutant). Anti-RNA polymerase (RpoB) is used as a lysis control and anti-Hcp2 as a control for H2-T6SS activity.
The identity of genuine T6SS effectors for H2- and H3-T6SS is poorly documented, but a few candidates have been described, such as PldA in the case of H2-T6SS (20, 21). PldA is encoded remotely from the H2-T6SS cluster and within the orphan vgrG4b cluster (PA3486-PA3488) (SI Appendix, Fig. S1). We engineered a chimeric gene encoding a PldA-Bla fusion and monitored its production using Western blot and a TEM β-lactamase antibody. PldA expression is increased in a rsmA background, which suggests RsmA negatively controls not only the expression of T6SS structural components but also the expression of effectors genes scattered on the chromosome (Figs. 6 and 7 and SI Appendix, Fig. S6A). We show that PldA is secreted in an H2-T6SS–dependent manner both in PAO1 or PA14 (Fig. 6A and SI Appendix, Fig. S6B) and, remarkably, in a VgrG4b-dependent manner (Fig. 6B), which suggests a direct connection with the VgrG4b spike and further validates the “à la carte delivery” concept that we previously proposed (6, 8).
Fig. 7.
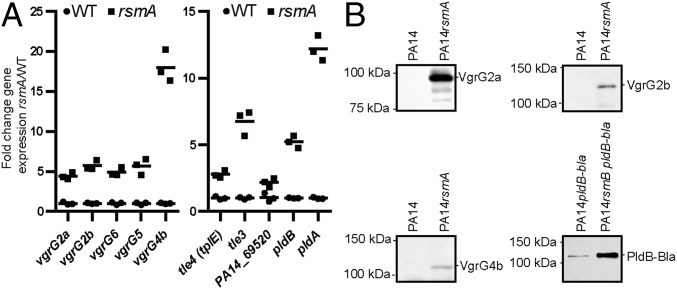
RsmA controls remote vgrG and T6SS effector genes. (A) qRT-PCR on a subset of vgrG genes (Left) and genes encoding known/putative effectors (Right) (SI Appendix, Fig. S1) in PA14 (WT) or a rsmA mutant. Scatter plot of fold change with mean (n = 3). Statistical analysis was performed on the ΔΔCT values (ANOVA Bonferroni posttest, P < 0.005 for all genes). (B) Western blot analysis using specific antibodies against selected VgrG proteins (VgrG2a, VgrG2b, and VgrG4b) and the T6SS effector PldB (PldB-Bla fusion detected with anti-TEM).
Using qRT-PCR analysis, we confirmed that genes in most of the remote vgrG islands (SI Appendix, Fig. S1), including vgrG2a, vgrG2b, vgrG4b, vgrG5, and vgrG6, are induced in a rsmA background from fivefold (vgrG2a) to about 20-fold (vgrG4b) (Fig. 7A, Left). The genes encoding the VgrG-associated effectors [tle4(tplE), tle3, pldA, pldB, and PA14_69520] were up-regulated in the rsmA mutant, ranging from twofold (PA14_69520) to 12-fold (pldA) (Fig. 7A, Right). Up-regulation coincides with protein production, as Western blot analysis using antibodies against VgrGs (VgrG2a, VgrG2b, and VgrG4b) or effectors such as PldB (PldB-Bla) showed clear de-repression in the rsmA mutant (Fig. 7B). Overall, our data demonstrate that relief of RsmA repression coordinates T6SS machinery assembly and effector delivery.
RsmA and AmrZ Repress H2-T6SS Bacterial Killing.
We assessed the phenotypic effect of H2- and H3-T6SS, using a bacterial killing assay and Escherichia coli as the prey (6). The PA14 killing induced in a rsmA background was independent of H1-T6SS (Fig. 8A and SI Appendix, Fig. S7A), which was previously shown to be an antibacterial weapon in PAK or PAO1 (7, 22). The effect of H3-T6SS is also minor, whereas interruption of the tssB2 gene in the H2-T6SS cluster abrogated killing. We have demonstrated that AmrZ negatively regulates H2-T6SS, and here show that overexpression of amrZ indeed alleviates H2-T6SS killing (Fig. 8B). We conclude that H2-T6SS is a major antibacterial weapon in PA14.
Fig. 8.
RsmA and AmrZ repress H2-T6SS–dependent bacterial killing. (A) Quantification of bacterial killing assay after coincubation of E. coli and various PA14 attackers, including H1-T6SS, H2-T6SS, and H3-T6SS mutants, as indicated by H1-, H2-, and H3-, respectively. (B) H2-T6SS–dependent bacterial killing is significantly reduced after overexpression of amrZ (pAmrZ) compared with PA14rsmA mutant carrying the empty vector (pMMB67HE). Quantification is made using colony counts. Graphs represent mean ± SEM; n = 3; statistical significance is indicated ANOVA Dunnett’s posttest P < 0.05.
Possible Assembly of Multiple T6SS Within a Single Cell.
Given that all three T6SSs are coregulated, we used fluorescence microscopy to determine whether these systems could be coassembled within one cell. Fluorescent proteins were fused to the C terminus of the sheath component TssB encoded from each of the H1-, H2-, and H3-T6SS clusters and the corresponding recombinant plasmids introduced in PA14rsmA. Each TssB fusion allowed the viewing of extended sheath assemblies (Fig. 9A). The relative number of assembled H1-, H2-, and H3-T6SS machines was determined by quantifying the fluorescent foci (TssB1-Venus, TssB2-CFP, or TssB3-CFP) per total number of cells analyzed (SI Appendix, Fig. S8). The amount of TssB2-CFP foci was more than 10 times that observed for either TssB1-Venus or TssB3-CFP (SI Appendix, Fig. S8). To ensure that the visualized foci were the result of an assembled T6SS machine, each fluorescent fusion was expressed in a strain lacking all three T6SSs. No foci were observed for TssB1-Venus or TssB3-CFP out of a total of 44,036 and 32,000 cells analyzed, respectively; for TssB2-CFP, five foci were observed out of a total of 68,208 cells, and for TssB3-sfGFP, two foci were observed out of a total of 31,666 cells. We also confirmed that using the TssB2-CFP chimera does not affect T6SS function, as H2-T6SS–dependent killing remains fully effective (SI Appendix, Fig. S7B).
Fig. 9.
Coassembly of multiple T6SS machines. (A) Assembly of extended T6SS sheaths as seen by fluorescence microscopy. Fluorescent fusions of TssB1-Venus, TssB2-CFP, TssB3-CFP, or TssB3-sfGFP were expressed in PA14rsmA. The images shown are representative of >100 fields analyzed from at least four independent experiments. (B) Fluorescent fusion combinations of TssB1-Venus with TssB2-CFP (Upper), TssB1-Venus with TssB3-CFP (Middle), or TssB2-CFP with TssB3-sfGFP (Lower) were coexpressed in PA14rsmA. (Left) Bright field channel. The arrows point to the foci of interest in cells that have two different T6SS machines assembled. The images shown are representative of ≥100 fields analyzed from at least two independent experiments. (Scale bars, 1 μm.)
We then assessed whether different T6SSs may be simultaneously assembled in a single cell by introducing, pairwise, the various plasmids into PA14rsmA: TssB1-Venus with TssB2-CFP (Fig. 9B, Upper), TssB1-Venus with TssB3-CFP (Fig. 9B, Middle), and TssB2-CFP with TssB3-sfGFP (Fig. 9B, Lower). For any combination tested, both T6SS foci types may be found within the same cell at the same time (Fig. 9B), either at very distinct positions in the cell (e.g., H1- and H2-T6SS or H2- and H3-T6SS; Fig. 9B, Upper and Lower, respectively) or in close proximity (e.g., H1- and H3-T6SS; Fig. 9B, Middle).
Discussion
The T6SS has a broad range of cellular targets and uses an armory of toxins and effectors to subvert or kill prey cells (1, 2). A bacterial species may carry several T6SSs; for example, three in P. aeruginosa (5), four in Yersinia pseudotuberculosis (23), and six in Burkholderia pseudomallei (24). In laboratory conditions, the T6SS is usually not expressed, suggesting environmental factors or host colonization, such as P. aeruginosa in the lungs of patients with cystic fibrosis (9), trigger T6SS assembly.
We demonstrate that in P. aeruginosa, the translational repressor RsmA negatively controls all T6SS clusters (H1-, H2-, and H3-T6SS) in PA14. This finding suggests H2- and H3-T6SS regulation by the RetS/Gac/Rsm cascade was undervalued and is not the privilege of H1-T6SS (25). We show that the RsmA-dependent control extends to orphan T6SS genes, such as those located in vgrG islands not associated with core T6SS clusters (6, 21). We concluded that RsmA is a central regulator imposing a tight and coordinated control that prevents T6SS-related messenger RNAs from being translated under yet-to-be-defined conditions.
We also identified AmrZ as a global transcriptional regulator of P. aeruginosa T6SSs. AmrZ acts as repressor or activator on a wide range of gene targets involved in P. aeruginosa virulence (26). Here, we showed that AmrZ positively influences both the H1- and H3-T6SS while having a negative control on the H2-T6SS. This is in agreement with available ChIP-Seq and RNA-Seq data investigating AmrZ (18). We also observed clear binding of AmrZ in the promoter region of these T6SS gene clusters. We concluded that AmrZ is a global regulator of T6SS genes, which can selectively promote or repress the transcription of a subset of T6SSs, whereas RsmA represses translation of all T6SS transcripts.
Regulatory events leading to RsmA and AmrZ expression are likely instrumental in fine-tuning expression of each individual T6SS. RsmA is downstream from a branched network in which two-component regulatory systems (25), phospho-relay (27) and c-di-GMP signaling (28), define the level of small RNAs (29), which sequester RsmA and alleviate T6SS repression. AmrZ is controlled by additional regulators such as the environmental stress sigma factor AlgU (also known as AlgT) (30). In our screen, a Tn hit in proximity to algU was identified (B100), as well as additional genes in the alg regulatory network, including algW, algC, and mucP (B26, W38b, and B39/B51; SI Appendix, Fig. S1 and Table S1). Interestingly, it was proposed that the two-component regulatory system AlgZ/AlgR manipulates the RsmAYZ pathway in P. aeruginosa (31), whereas the Gac/Rsm pathway represses the translation of AlgU (32).
The signals triggering the T6SS regulatory pathways can be many-fold. It has previously been proposed that quorum sensing and iron concentration influence H2- and H3-T6SS expression (14, 15, 33). Our study identified several genes associated with quorum sensing; for example, lasR and rhlR (B101 and W45) or pqsA (W43), which is required for the production of the 4-hydroxy-2-alkylquinoline signal (34) (SI Appendix, Fig. S1 and Table S1). We did not identify iron-related genes, but AmrZ represses many genes involved with iron procurement in Pseudomonads (18, 35). Other genes identified in our screen may have an indirect effect resulting from their role in central metabolism (pncA/B5, dut/W38b) and protein quality control (lon, W37a; SI Appendix, Fig. S1 and Table S1). We also pointed at significant differences in T6SS expression that depend on growth temperature. Whereas H1- and H2-T6SS are expressed at 37 °C, H3-T6SS is mainly produced at a lower temperature (e.g., 25 °C; Fig. 4, Lower). Furthermore, although induction of H1- and H2-T6SS is clear at 37 °C, at 25 °C, T6SS components are detectable even in a wild-type PA14 strain, indicating that expression no longer relies on deletion of rsmA (Fig. 4, Lower). An effect of temperature on T6SS expression was previously reported in P. aeruginosa (36), and one can suggest that the temperature response may be transmitted through the alternative stress sigma factor AlgU (37). In Pseudomonas fluorescens, it was also demonstrated that RetS contributes to the thermosensitivity of the Gac/Rsm-dependent gene expression (38). In Y. pseudotuberculosis, one of four T6SSs (T6SS4) displays higher level of expression at 26 °C and is under quorum sensing regulation (23).
In addition to the complexity of the regulatory network, it seems clear that expression of the different T6SSs depends on the P. aeruginosa isolate. Differences have been highlighted between the strains PAO1, PAK, and PA14, such as PA14 lacking the LadS sensor in the Gac/Rsm pathway (39). We observed differences in the expression of various T6SSs, as deletion of rsmA or retS in PA14 resulted in detectable levels of Hcp2 or TssB2 when grown at 37 °C, whereas similar deletion in PAK or PAO1 did not (SI Appendix, Fig. S9A). However, the H2-T6SS system in PAO1 is expressed and functional at 25 °C. In PA14, the role of H1-T6SS in a rsmA background is less crucial than H2-T6SS (Fig. 8A and SI Appendix, Fig. S7A), and despite Hcp1 being detectable in this background, TssB1 is not (SI Appendix, Fig. S9B). The potency of H2-T6SS in PA14 can also be suggested from the number of H2-T6SS sheaths visualized compared with H1- or H3-T6SS sheaths (SI Appendix, Fig. S8). It is also worth mentioning that in PA14, full expression of H1-T6SS involves the second RsmA-like encoding gene, rsmF (also called rsmN) (40). In the double mutant rsmA/rsmF, Hcp1 levels drastically increase, whereas TssB1 is now clearly detectable (SI Appendix, Fig. S9B). In contrast, no difference in H2-T6SS levels could be observed when comparing a rsmA or rsmA/rsmF mutant (SI Appendix, Fig. S9B). Overall, this supports the notion that different isolates of P. aeruginosa have defined networks to deploy their T6SS in different environments.
It is unclear how the different regulatory networks integrate to produce active T6SS machines (41), but in the environment or within a host, P. aeruginosa will have to face simultaneously the competition with other bacteria and resist predation from eukaryotic cells (e.g., macrophages or amoeba). Interestingly, RsmA has been suggested to be part of a bacterial danger-sensing circuit allowing P. aeruginosa to respond to kin cell lysis by enhancing the activity of H1-T6SS (13). Our data showing that RsmA represses not just H1-T6SS but all three T6SS support the idea that P. aeruginosa, on sensing kin cell lysis, relieves a RsmA posttranscriptional block of all three T6SS machines, enabling a coordinated response to attack. Our microscopy images suggesting that multiple systems may assemble within a single cell support such a defense/retaliation program in which RsmA acts to coordinate deployment of the complete arsenal of T6SS systems. In conclusion, the role of the T6SS and the complexity of the network controlling its assembly and functionality at all levels suggests it has evolved as a surveillance mechanism able to fight any organisms in any condition encountered.
Materials and Methods
Strains and plasmids are listed in SI Appendix, Table S3. Primers are listed in SI Appendix, Table S4. Gene deletions were constructed as previously described (42). Tn mutagenesis and identification of sites of integration was performed as previously outlined (16). RNA was isolated using TRIzol extraction and purified using the Qiagen RNeasy Mini kit (Qiagen). Real-time PCR was performed according to the manufacturer’s protocol (Applied Biosystems or Sigma). Electrophoretic mobility shift assays were performed as previously described, using purified AmrZ (SI Appendix, Fig. S10) (18, 19). Assays for Western blotting, T6SS secretion, and T6SS killing were performed essentially as previously explained (6). Detailed information on methods and associated references are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sara Planamente, Abderrahman Hachani, Sophie Cooke, and Kailyn Hui for constructing recombinant plasmids; Eleni Manoli for protein purification; and Martina Valentini for advice on RsmA binding sites. We thank Sivaramesh Wigneshweraraj and Daniel R. Brown for technical assistance. We thank the Facility for Imaging by Light Microscopy at Imperial College London. A.F. is supported by Medical Research Council (MRC) Grants MR/K001930/1 and MR/N023250/1 and Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/N002539/1. L.P.A. is supported by BBSRC Grant BB/N002539/1 and a Marie Curie Fellowship (PIIF-GA-2012-328261). T.E.W., S.W., and S.A.H. are recipients of PhD studentships from the Wellcome Trust, Imperial College London, and MRC, respectively. L.M.N. is supported by MRC Grant MR/N023250/1 and a Marie Curie Fellowship (PIIF-GA-2013-625318).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700286114/-/DCSupplemental.
References
- 1.Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unterweger D, Kostiuk B, Pukatzki S. Adaptor proteins of type VI secretion system effectors. Trends Microbiol. 2017;25:8–10. doi: 10.1016/j.tim.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 6.Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitney JC, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol. 2014;92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman AL, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planamente S, et al. TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 2016;35:1613–1627. doi: 10.15252/embj.201694024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeRoux M, et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife. 2015;4:4. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009;155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sana TG, et al. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem. 2012;287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulasekara HD. Transposon mutagenesis. Methods Mol Biol. 2014;1149:501–519. doi: 10.1007/978-1-4939-0473-0_39. [DOI] [PubMed] [Google Scholar]
- 17.Lapouge K, et al. RNA pentaloop structures as effective targets of regulators belonging to the RsmA/CsrA protein family. RNA Biol. 2013;10:1031–1041. doi: 10.4161/rna.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones CJ, et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan J, et al. Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J Microbiol. 2015;53:633–642. doi: 10.1007/s12275-015-0099-6. [DOI] [PubMed] [Google Scholar]
- 20.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. 2014;15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Russell AB, et al. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lossi NS, et al. The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Mol Microbiol. 2012;86:437–456. doi: 10.1111/j.1365-2958.2012.08204.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch Microbiol. 2011;193:351–363. doi: 10.1007/s00203-011-0680-2. [DOI] [PubMed] [Google Scholar]
- 24.Shalom G, Shaw JG, Thomas MS. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology. 2007;153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 25.Goodman AL, et al. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryor EE, Jr, et al. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012;8:e1002648. doi: 10.1371/journal.ppat.1002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordi C, et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 29.González N, et al. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wozniak DJ, Sprinkle AB, Baynham PJ. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol. 2003;185:7297–7300. doi: 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol. 2014;196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Granero F, et al. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One. 2012;7:e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sana TG, Soscia C, Tonglet CM, Garvis S, Bleves S. Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS One. 2013;8:e76030. doi: 10.1371/journal.pone.0076030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez-Granero F, Redondo-Nieto M, Vesga P, Martín M, Rivilla R. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics. 2014;15:237. doi: 10.1186/1471-2164-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Termine E, Michel GP. Transcriptome and secretome analyses of the adaptive response of Pseudomonas aeruginosa to suboptimal growth temperature. Int Microbiol. 2009;12:7–12. [PubMed] [Google Scholar]
- 37.Schurr MJ, Yu H, Boucher JC, Hibler NS, Deretic V. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (sigma E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J Bacteriol. 1995;177:5670–5679. doi: 10.1128/jb.177.19.5670-5679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humair B, González N, Mossialos D, Reimmann C, Haas D. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J. 2009;3:955–965. doi: 10.1038/ismej.2009.42. [DOI] [PubMed] [Google Scholar]
- 39.Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One. 2011;6:e29113. doi: 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marden JN, et al. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2013;110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–3860. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.