Abstract
Near-infrared surface plasmon resonance imaging (SPRI) microscopy is used to detect and characterize the adsorption of single polymeric and protein nanoparticles (PPNPs) onto chemically modified gold thin films in real time. The single-nanoparticle SPRI responses, Δ%RNP, from several hundred adsorbed nanoparticles are collected in a single SPRI adsorption measurement. Analysis of Δ%RNP frequency distribution histograms is used to provide information on the size, material content, and interparticle interactions of the PPNPs. Examples include the measurement of log-normal Δ%RNP distributions for mixtures of polystyrene nanoparticles, the quantitation of bioaffinity uptake into and aggregation of porous NIPAm-based (N-isopropylacrylamide) hydrogel nanoparticles specifically engineered to bind peptides and proteins, and the characterization of the negative single-nanoparticle SPRI response and log-normal Δ%RNP distributions obtained for three different types of genetically encoded gas-filled protein nanostructures derived from bacteria.
Keywords: surface plasmon polaritons, single-nanoparticle refractive index, NIPAm-based hydrogel nanoparticle, melittin, concanavalin A, gas vesicle, protein nanostructure
The rational design, synthesis, and characterization of both polymeric and protein nanoparticles (PPNPs) for applications in materials, catalysis, and biotechnology have become a significant component of the current nanoscience revolution. PPNPs have been constructed from a wide variety of polymeric materials including single-chain or cross-linked polymers, dendrimers, synthetic polypeptides, proteins, and polysaccharides.1−6 PPNPS can be designed to form compact structures, porous hydrogels, or other three-dimensional structures that can exhibit a wide variety of rheological properties, display a large number of interfacial chemical moieties with specific affinities or reactivities on the outside of the nanoparticle, or incorporate internal chemical binding sites that can be used to capture and release chemicals or smaller nanoparticles.7−10 Examples include elastin-like polypeptide nanoparticles that are biodegradable and thermally responsive,11,12 polysaccharide-based nanoparticles for medical diagnostics and therapies,13 and cross-linked N-isopropylacrylamide (NIPAm) hydrogel nanoparticles that incorporate a mixture of chemical functional groups to create specific binding sites for bioaffinity uptake.14−16 Genetically coded protein nanostructures with acoustic properties, such as gas vesicles (GVs), have been identified for use as ultrasound and magnetic resonance imaging contrast agents.17−20
The characterization of PPNPs at the single-nanoparticle level is challenging. Unlike metallic or semiconductor particles, which often exhibit a strong size-dependent optical response,21−23 PPNPs typically do not possess any convenient spectroscopic markers. Additionally, PPNPs often contain a significant amount of solvent, and their size and composition may vary with external pH, temperature, or pressure. A particularly important but difficult measurement is the quantification of bioaffinity adsorption and uptake into single PPNPs that have been designed for drug delivery or toxin neutralization applications. PPNPs are typically characterized with a combination of bulk dynamic light scattering (DLS) and multiangle light scattering (MALS) measurements,24−26 cryo-TEM,27,28 and, if the PPNPs are sufficiently rigid, scanning probe measurements.29,30 In some studies, the incorporation of fluorophores into the nanoparticle has been employed to facilitate single-nanoparticle detection and to provide some limited characterization information.31,32
The optical technique of single-nanoparticle surface plasmon resonance imaging (SPRI) microscopy has recently emerged as an excellent in situ refractive-index based method for the detection and characterization of single PPNPs. As first identified in 2010 by Zybin and Tao,33−35 an adsorbed nanoparticle can interact with traveling surface plasmon polariton waves created on a gold thin film surface to create a point diffraction pattern in the differential SPRI image. Single metallic nanoparticles, polymer nanoparticles, liposomes, cells, and viruses have been detected with SPRI microscopy.36−46 The intensity of the diffraction pattern depends on the integrated refractive index of the nanoparticle and, thus, varies with nanoparticle size and material content. Real-time SPRI measurements have been used previously for the digital biosensing of single-nanoparticle bioaffinity adsorption events at chemically modified gold thin films.41,47 In addition to nanoparticle-counting measurements, changes in the intensity of the average single-nanoparticle SPRI response (⟨Δ%RNP⟩) have been used to quantitate the bioaffinity uptake of polypeptides and proteins by hydrogel nanoparticles.42,43
Determining the distribution of Δ%RNP values obtained during a single-nanoparticle SPRI adsorption measurement in addition to the average response can provide much more detailed information about a population of PPNPs. Since the Δ%RNP response depends on the integrated refractive index of the nanoparticle, Δ%RNP frequency distributions will reflect variations in both nanoparticle size and composition. An example of the latter would be changes in a Δ%RNP distribution created by variations in molecular uptake into a population of PPNPs designed for drug delivery. While ensemble measurements such as DLS can provide limited information on the moments of a PPNP nanoparticle distribution, single-nanoparticle SPRI measurements can directly measure the detailed frequency distribution histogram of a PPNP population.
In this paper, we provide three different examples of how to obtain and use single PPNP Δ%RNP distributions from real-time SPRI adsorption measurements. As a first case, we demonstrate that Δ%RNP distributions can be used to measure nanoparticle size distributions for mixtures of solid polystyrene (PS) nanoparticles. In a second set of experiments, we demonstrate how Δ%RNP distributions obtained from porous NIPAm-based hydrogel nanoparticles (HNPs) can be used to monitor changes in PPNP structure and aggregation due to the bioaffinity uptake of peptides and proteins. In the final example, we show that the adsorption of gas-filled protein nanostructures produces an unusual negative single-nanoparticle SPRI response with a Δ%RNP distribution that depends on the shape and size of the particle. The three examples presented in this paper have been chosen to demonstrate that the single-nanoparticle SPRI measurements can be applied to three very different classes of PPNPs: solid polymer nanoparticles, highly porous, solvent-swollen polymer nanoparticles, and protein nanostructures that enclose a gas volume.
Results and Discussion
Single-Nanoparticle SPRI Adsorption Measurements
The detection and characterization of single polymer and protein nanoparticles was achieved by using real-time SPRI microscopy measurements to detect the irreversible adsorption of individual nanoparticles onto a chemically modified gold thin film surface. The optical setup of the near-infrared single-nanoparticle SPRI microscope used in these experiments is shown in Figure 1a and has been described in detail in a previous publication.41 Briefly, an 814 nm laser was expanded, collimated, and then polarized before being directed off-axis through the back of a high numerical aperture microscope objective and onto the back of a gold-coated microscope coverslip. The reflected image (56.5 μm × 56.5 μm) was captured with a CMOS camera (see the Methods section for more details). For each SPRI adsorption measurement, a 10 μL solution of nanoparticles was exposed to the chemically modified gold surface, and then a series of three-second SPRI reflectivity images (Rn, where n is the image number) were collected for 10 min as nanoparticles adsorbed to the surface. Using these images, a series of 200 frame-to-frame SPRI differential reflectivity images (ΔRn) were obtained by sequentially subtracting each image from the previous image (i.e., ΔRn = Rn – Rn–1).
Figure 1.
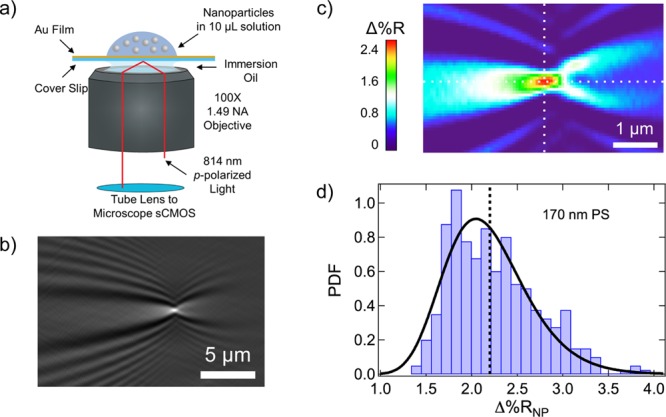
(a) Schematic diagram of the SPRI microscope. A gold-coated knife-edge mirror was used to direct collimated p-polarized light off-axis through the microscope objective and onto the back of the gold-coated glass coverslip. The reflected image was captured with a CMOS camera. A nanoparticle solution was exposed to the top of the gold-coated glass coverslip immediately preceding the image acquisition process. (b) A point diffraction pattern is observed in the SPRI differential reflectivity image when a 170 nm polystyrene (PS) nanoparticle adsorbs to the chemically modified gold surface. (c) Quantitative map displaying the Δ%R pixel intensities for the single-nanoparticle point diffraction pattern in (b). A sharp spike in Δ%R intensity is observed at the center of the diffraction pattern (the intersection of the two white dotted lines). We define Δ%RNP as the average of the Δ%R values for the nine pixels at and surrounding the pixel with the maximum Δ%R intensity. (d) Δ%RNP frequency distribution histogram obtained from the SPRI adsorption measurement of 170 nm PS nanoparticles. The average Δ%RNP value for this experiment was 2.19 ± 0.05% and is plotted in the figure as a black dotted line. The Δ%RNP distribution is also fit to a probability density function (PDF) with location (μ) and scale (σ) parameters of 0.76 and 0.21, respectively.
The adsorption of a single nanoparticle onto the chemically modified gold thin film appears in the differential reflectivity images as a point diffraction pattern. An example of a single-nanoparticle point diffraction pattern from a 170 nm diameter PS nanoparticle is shown in Figure 1b. These diffraction patterns have been observed previously in SPRI differential reflectivity images from the adsorption of metal, polymer, and lipid nanoparticles. The diffraction patterns have been modeled using a 2D Helmholtz equation, where the integrated refractive index of the adsorbed nanoparticle acts as a diffraction point for the planar surface plasmon polariton waves traveling on the gold thin film.48 Since we are using differential reflectivity images, only nanoparticles that have adsorbed within the three-second time frame of image ΔRn are observed.
The center of each nanoparticle diffraction pattern has a sharp Δ%R (change in percent reflectivity) maximum that can be used to quantitate the intensity of the single-nanoparticle SPRI response. Figure 1c shows a quantitative map of the Δ%R pixel intensities for a typical single-nanoparticle SPRI diffraction pattern. As described previously,42 the average of the nine Δ%R pixel intensities (a 3 × 3 array) at and surrounding the maximum Δ%R is used to calculate the single-nanoparticle SPRI reflectivity response that we denote as Δ%RNP. The single-nanoparticle SPRI diffraction pattern has been described previously by several researchers as the sum of a traveling plane wave and a spherical wave.48 Using the average values of the 3 × 3 array of nine pixels around the maximum is a simple, yet reliable method of calculating a reproducible Δ%RNP value for this diffraction pattern; using larger pixel arrays was also reliable, but gave lower Δ%RNP values. Fitting the entire diffraction pattern to the Helmholtz equation solution has been successfully used to determine Δ%RNP44 and has also been recently employed as a method to improve the spatial resolution of the nanoparticle location on the surface.45 For each set of differential reflectivity images associated with a 10 min SPRI adsorption measurement, several hundred point diffraction patterns are observed and analyzed to calculate both an average Δ%RNP value, denoted ⟨Δ%RNP⟩, and a frequency distribution histogram of Δ%RNP values.
An example of a Δ%RNP frequency distribution histogram obtained from an in situ real-time SPRI adsorption measurement of 170 nm PS nanoparticles onto a chemically modified gold thin film is shown in Figure 1d (details of this experiment are given in the next section). The ⟨Δ%RNP⟩ for this measurement is also plotted in Figure 1d as a black dotted line. It is evident from the distribution that the Δ%RNP values are not symmetrically distributed about ⟨Δ%RNP⟩. Therefore, in order to more precisely quantify this distribution, in addition to a standard deviation (s), we calculate a skewness (g) from the set of Δ%RNP values, where the skewness is proportional to the third central moment m3:49
| 1 |
The skewness can be either positive or negative, depending on which side of ⟨Δ%RNP⟩ the distribution is skewed; for the data in Figure 1d, g = 0.68.
The Δ%RNP distribution of 170 nm PS nanoparticles is also fitted to a log-normal probability density function (PDF), described as50
| 2 |
where μ and σ are the location and scale parameters, respectively. This log-normal fit is plotted in Figure 1d as a black solid line, and it is apparent that a log-normal probability density function gives an accurate fit of the data. Previous size measurements on PS nanoparticles have also followed a log-normal distribution.51 The values for ⟨Δ%RNP⟩, s, 95% confidence interval (95% CI), g, μ, and σ for this experiment on 170 nm PS nanoparticles are reported in Table 1.
Table 1. Hydrodynamic Size Measurements from DLS for Polystyrene and Hydrogel Nanoparticles and Statistics from Single-Nanoparticle SPRI Measurements for Polystyrene and Hydrogel Nanoparticles and Gas Vesicles.
| nanoparticle | diameter (nm) | standard deviation (nm) | ⟨Δ%RNP⟩ | standard deviation (s) | 95% CI | skewness (g) | μa | σb | no. of NPs |
|---|---|---|---|---|---|---|---|---|---|
| PS (A) | 85 | 25 | 0.34 | 0.10 | 0.01 | 0.59 | –1.13 | 0.31 | 354 |
| PS (B) | 170 | 40 | 2.19 | 0.48 | 0.05 | 0.68 | 0.76 | 0.21 | 365 |
| HNP | 271 | 55 | 1.67 | 0.43 | 0.05 | 0.60 | 0.48 | 0.27 | 324 |
| HNP + 2 μM melittin | 272 | 65 | 2.79 | 0.52 | 0.08 | 0.02 | 1.01 | 0.20 | 172 |
| HNP | 272 | 50 | 0.90 | 0.27 | 0.03 | 0.55 | –0.15 | 0.31 | 289 |
| HNP + 500 nM ConA | 357 | 75 | 3.6 | 1.3 | 0.2 | 0.79 | 1.22 | 0.37 | 307 |
| HNP + 500 nM ConA + 1 mM Man | 338 | 65 | 2.04 | 0.60 | 0.07 | 0.05 | 0.66 | 0.36 | 270 |
| HNP + 500 nM ConA + 10 mM Man | 320 | 55 | 1.74 | 0.41 | 0.05 | 0.30 | 0.53 | 0.24 | 241 |
| Mega GV | –c | – | –0.49 | 0.26 | 0.03 | –1.28 | –0.84 | 0.52 | 274 |
| Ana GV | – | – | –1.07 | 0.44 | 0.04 | –1.53 | –0.0083 | 0.38 | 395 |
| Halo GV | – | – | –3.0 | 1.5 | 0.2 | –0.74 | 0.95 | 0.58 | 345 |
Mixtures of Polystyrene Nanoparticles
As a first demonstration that single-nanoparticle SPRI measurements can provide useful information on polydisperse polymer nanoparticle samples, a series of single-nanoparticle SPRI adsorption measurements were performed on three solutions of carboxyl-terminated PS nanoparticles: 85 nm diameter PS nanoparticles, 170 nm diameter PS nanoparticles, and a one-to-one mixture of 85 and 170 nm PS nanoparticles. For each SPRI adsorption measurement, PS nanoparticle solutions were exposed to a gold surface modified with an amine-terminated (11-mercaptoundecamine, MUAM) self-assembled monolayer. SPRI reflectivity images were collected as the negatively charged carboxyl-terminated PS nanoparticles electrostatically and irreversibly adsorbed to the MUAM surface. An example SPRI differential reflectivity image from the sample of mixed size PS nanoparticles is shown in Figure 2a. As seen in the image, two PS nanoparticles irreversibly adsorbed onto the MUAM surface within the three-second time frame. The larger, more intense point diffraction pattern near the top of the image is attributed to the adsorption of a 170 nm PS nanoparticle, whereas the smaller, less intense point diffraction pattern near the bottom of the image is attributed to the adsorption of an 85 nm PS nanoparticle. The intensity of each nanoparticle point diffraction pattern is quantitated by calculating a Δ%RNP value as described in the previous section. For the two PS nanoparticles in this image, Δ%RNP was calculated to be 3.1% for the 170 nm PS nanoparticle and 0.47% for the 85 nm PS nanoparticle.
Figure 2.
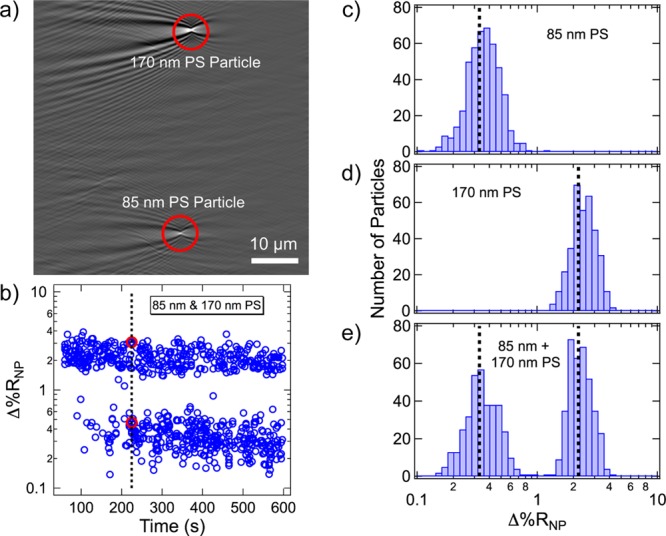
(a) Example SPRI differential reflectivity image of a mixed sample of PS nanoparticles. The larger, more intense point diffraction pattern represents a 170 nm PS nanoparticle, and the smaller, less intense point diffraction pattern represents an 85 nm PS nanoparticle. The total image area is 58.5 μm × 58.5 μm. (b) Time-dependent distribution of Δ%RNP values for the mixture of 85 and 170 nm PS nanoparticles. Each blue circle represents the Δ%RNP for a single PS nanoparticle irreversibly adsorbing to the chemically modified surface. The two red circles represent the Δ%RNP values for the point diffraction patterns in the differential reflectivity image (a) that adsorbed to the surface at the 225 s mark of the experiment (black dotted line). Δ%RNP frequency distribution histograms obtained from three different SPRI adsorption measurements of (c) 85 nm PS nanoparticles, (d) 170 nm PS nanoparticles, and (e) a one-to-one mixture of 85 and 170 nm PS nanoparticles. Average Δ%RNP values for each size of PS nanoparticle are plotted as a black dotted line. The average Δ%RNP values for 85 and 170 nm PS nanoparticles are 0.34 ± 0.01% and 2.19 ± 0.05%, respectively.
All the Δ%RNP values calculated from the series of differential reflectivity images obtained during the SPRI adsorption measurement of the mixed size PS nanoparticles are plotted in Figure 2b as a function of adsorption time. Each blue circle in Figure 2b represents a single Δ%RNP value obtained for the adsorption of a single PS nanoparticle; over 700 PS nanoparticle point diffraction patterns were quantitated over the 10 min measurement. The two Δ%RNP values calculated for the two PS nanoparticles in the differential reflectivity image shown in Figure 2a are identified in the time-dependent distribution as two red circles. The data in Figure 2b clearly indicate there are two distinct ranges of Δ%RNP values, which can be attributed to the two sizes of PS nanoparticles.
In addition to the time-dependent distribution of Δ%RNP values, we also generate a frequency distribution histogram of Δ%RNP values from the SPRI adsorption measurements. The Δ%RNP frequency distribution histograms from SPRI adsorption measurements for the three different PS nanoparticle solutions are also plotted in Figure 2: 85 nm PS nanoparticles (Figure 2c), 170 nm PS nanoparticles (Figure 2d), and a one-to-one mixture of 85 and 170 nm PS nanoparticles (Figure 2e). The black dotted lines in Figure 2c and d are the ⟨Δ%RNP⟩ values obtained for each SPRI adsorption measurement; ⟨Δ%RNP⟩ = 0.34 ± 0.01% for 85 nm PS nanoparticles and ⟨Δ%RNP⟩ = 2.19 ± 0.05% for 170 nm PS nanoparticles. Because the Δ%RNP values for PS nanoparticles are log-normally distributed, we plot the distribution histograms in logarithmically spaced bins in Figure 2 for ease of comparison. Reported in Table 1 are ⟨Δ%RNP⟩, s, 95% CI, g, μ, and σ for both the 85 and 170 nm PS nanoparticles. Even though the average ⟨Δ%RNP⟩ is more than 6 times larger for the 170 nm PS nanoparticles as compared to the 85 nm PS nanoparticles, the skewness and scale parameters are relatively similar for the two distributions.
It is apparent from the histogram in Figure 2e that the distribution obtained from the mixed size PS nanoparticle sample is simply the sum of the two single-size PS nanoparticle distributions. The ⟨Δ%RNP⟩ values obtained for each size of PS nanoparticle are plotted in Figure 2e and are the same values as those obtained from the experiments in Figure 2c and d. These results unequivocally demonstrate that the single-nanoparticle SPRI measurements can be used to study polydisperse mixtures of nanoparticles. Using the data presented in Figure 2, we estimate that we can differentiate two populations of PS nanoparticles that have a difference in diameter greater than 40 nm.
Molecular Uptake into Hydrogel Nanoparticles and Aggregation of Hydrogel Nanoparticles
In a second set of experiments, we demonstrate that Δ%RNP frequency distribution histograms from single-nanoparticle SPRI measurements can be used to characterize the bioaffinity uptake of molecules into porous PPNPs, such as NIPAm-based HNPs. HNPs are solvent-swollen nanoparticles (up to ∼65% solvent by volume as estimated from MALS measurements42) that can be engineered to incorporate chemical moieties with specific affinity for various biomolecules. We have previously shown that ⟨Δ%RNP⟩ values from single-nanoparticle SPRI measurements can be used to study the uptake of the peptide melittin and the lectin concanavalin A (ConA) into specifically designed HNPs.42,43 In this paper, we demonstrate that the analysis of Δ%RNP frequency distribution histograms can be used to provide additional information on the uptake of these molecules into HNPs.
An example of a Δ%RNP frequency distribution histogram measurement of peptide uptake by HNPs is shown in Figure 3. As depicted in Figure 3a, NIPAm-based HNPs (272 nm in diameter as measured by DLS) were synthesized with specific affinity for melittin, a small peptide composed of 26 amino acid residues.52 Single-nanoparticle SPRI measurements on these HNPs, in both the absence and presence of melittin, were used to quantitate the Δ%RNP response. Plotted in Figure 3b are two Δ%RNP frequency distribution histograms: the Δ%RNP distribution for HNPs alone (transparent blue bars) and the Δ%RNP distribution for HNPs in the presence of 2 μM melittin (solid red bars). The two distributions in Figure 3b show that there is an overall increase in the average single-nanoparticle Δ%RNP response due to the uptake of melittin into the HNPs, which is an increase in the total integrated refractive index of the HNPs. Reported in Table 1 are the values for ⟨Δ%RNP⟩, s, 95% CI, g, μ, and σ obtained from the measurements. However, although there is an increase in ⟨Δ%RNP⟩, there are no significant increases observed in the size or skewness of the Δ%RNP distributions of HNPs in the presence of melittin. Specifically, the value for σ decreases from 0.27 to 0.20 for HNPs in the presence of melittin, and the relative standard deviation (s/⟨Δ%RNP⟩) also decreases (see Table 1). These results suggest that melittin uptake does not affect the structure of the HNPs, a conclusion that is corroborated with DLS measurements that show no change in the average hydrodynamic diameter for the hydrogels in the presence of 2 μM melittin (data also reported in Table 1).
Figure 3.
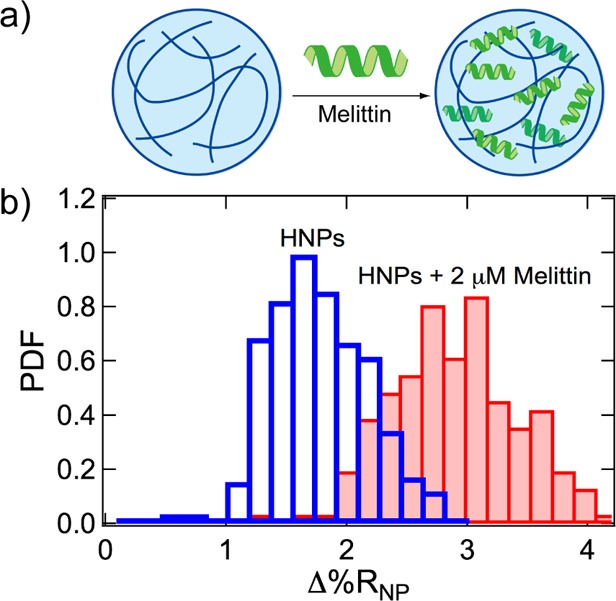
(a) Hydrogel nanoparticles (HNPs) were composed of N-isopropylacrylamide (NIPAm, 53 mol %), N-tert-butylacrylamide (TBAm, 40 mol %), acrylic acid (AAc, 5 mol %), and N,N′-methylenebis(acrylamide) (BIS, 2 mol %) and designed to uptake the peptide melittin by hydrophobic and electrostatic interactions. (b) Δ%RNP frequency distribution histograms obtained from the SPRI adsorption measurements of HNPs alone (transparent blue bars) and HNPs in the presence of 2 μM melittin (solid red bars).
In comparison, large changes in the Δ%RNP frequency distribution histograms were observed upon the uptake of the lectin ConA into HNPs modified with mannose. ConA is a large protein (MW = 104 kDa) with four subunits and a high binding specificity for mannose.53 Single-nanoparticle SPRI measurements were used to study the binding of ConA to mannose-modified HNPs as shown schematically in Figure 4a. Plotted in Figure 4b are two Δ%RNP frequency distribution histograms: mannose-modified HNPs only (transparent blue bars) and mannose-modified HNPs in the presence of 500 nM ConA (solid red bars). The ⟨Δ%RNP⟩, s, 95% CI, g, μ, and σ values for these two distributions are reported in Table 1. As evident from the data, not only is there an increase in ⟨Δ%RNP⟩ in the presence of ConA, but there is also a significant increase in the width of the Δ%RNP distribution. Specifically, there is 5-fold increase in the standard deviation of the Δ%RNP distribution for mannose-modified HNPs in the presence of ConA. Additionally, we observe an increase in the skewness and scale parameter. Because ConA has the capability to bind to multiple mannoses, ConA can induce aggregation of the mannose-modified HNPs by cross-linking. We attribute the changes in the Δ%RNP distributions to the aggregation of the mannose-modified HNPs induced by interparticle interactions of ConA that is bound to the outer regions of the HNPs. These results are also confirmed with DLS, which shows an increase in average hydrodynamic diameter of the mannose-modified HNPs from 272 to 357 nm.
Figure 4.
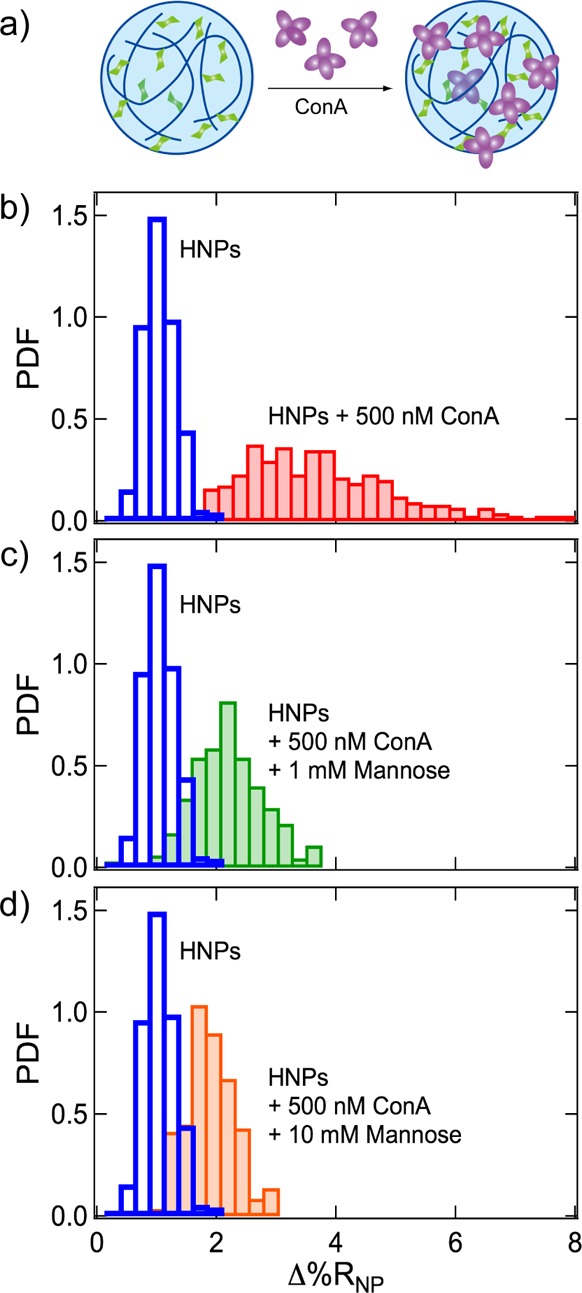
(a) Mannose-modified HNPs were composed of NIPAm (63.5 mol %), TBAm (28 mol %), AAc (5 mol %), BIS (2 mol %), and p-acrylamidophenyl-α-d-mannopyranoside (Man, 1.5 mol %). The lectin concanavalin A (ConA, purple) binds specifically to Man sugar units (green) in the mannose-modified HNPs. (b) Δ%RNP frequency distribution histograms obtained from the SPRI adsorption measurements of mannose-modified HNPs alone (transparent blue bars) and mannose-modified HNPs in the presence of 500 nM ConA (solid red bars). Δ%RNP frequency distribution histograms are also plotted for additional SPRI adsorption measurements of mixtures of mannose-modified HNPs and 500 nM ConA in the presence of (c) 1 mM mannose (solid green bars) and (d) 10 mM mannose (solid orange bars). The Δ%RNP frequency distribution histogram for mannose-modified HNPs alone is replotted in (c) and (d) for comparison.
To further study ConA binding to mannose-modified HNPs, additional single-nanoparticle SPRI measurements were made on the mixtures of mannose-modified HNPs and 500 nM ConA in the presence of free mannose in solution. By introducing free mannose into solution, we can induce competition between ConA binding to free mannose and mannose-modified HNPs and subsequently decrease the ConA-induced aggregation of mannose-modified HNPs. The Δ%RNP frequency distribution histograms for single-nanoparticle SPRI measurements of mannose-modified HNPs and 500 nM ConA with the addition of 1 mM mannose (solid green bars) and 10 mM mannose (solid orange bars) are shown in Figure 4c and d, respectively. The ⟨Δ%RNP⟩, s, 95% CI, g, μ, and σ values for these distributions are also reported in Table 1. As in Figure 4b, the Δ%RNP frequency distribution for mannose-modified HNPs without ConA is also plotted in Figure 4c and d for comparison (transparent blue bars). The distributions plotted in Figure 4c and d clearly show increases in the both ⟨Δ%RNP⟩ and the width of the distributions, compared to measurements of mannose-modified HNPs without ConA; however, these increases in ⟨Δ%RNP⟩ and the width of the distributions are less compared to measurements of mannose-modified HNPs and 500 nM ConA but without free mannose in solution (Figure 4b). This observation can also be seen quantitatively from the values listed in Table 1. For example, the standard deviation for mannose-modified HNPs increases by 480%, 220%, and 150% in the presence of 500 nM ConA and 0, 1, and 10 mM mannose, respectively. The Kd for ConA binding to monovalent mannose is on the order of 10–4–10–3 M.54 The observation that 10 mM monovalent mannose did not eliminate ConA interactions with the mannose-modified HNPs implies that there is a strong binding affinity between ConA and mannose-modified HNPs. It is well reported that the strength of interactions between sugars and lectins can be enhanced via multivalent binding,55 and various two- and three-dimensional sugar–polymer networks enhance the potency of the sugar–lectin interactions.56 It has been previously demonstrated that mannose-modified HNPs have Kd values in the micromolar to nanomolar range.57
Gas Vesicle Protein Nanostructures
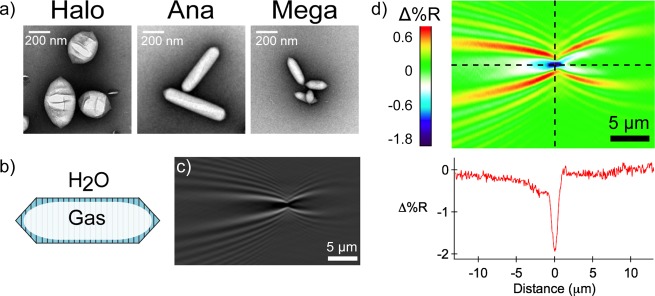
As a final example of the utility of single-nanoparticle SPRI measurements of PPNPs, we demonstrate the use of single-nanoparticle SPRI measurements to characterize gas vesicle protein nanostructures. GVs are hollow gas-filled bacterial protein nanostructures composed of a ∼2 nm protein shell that excludes water but allows gas to diffuse in and out of the particle.58,59 In this work, we characterized three genotypes of GVs encoded by the bacteria Bacillus megaterium (Mega GVs), Anabaena flos-aquae (Ana GVs), and Halobacterium salinarum (Halo GVs). TEM images of the three varieties of GVs are displayed in Figure 5a, and a schematic illustration of an Ana GV is shown in Figure 5b. The preparation of these GVs has been reported previously.17,19,20,60 Ana GVs and Mega GVs are cone-tipped cylindrical nanostructures with lengths of 519 ± 160 nm and 249 ± 99 nm, respectively, and diameters of 136 ± 21 nm and 73 ± 14 nm, respectively; Halo GVs are spindle-shaped nanostructures with lengths of 400 ± 113 nm and diameters of 251 ± 51 nm. TEM measurements of GV lengths and diameters are reported in Table 2, along with an estimate of the total volume, the molecular weight, and the gas-to-protein volume ratios for the three types of GVs.60
Figure 5.
(a) TEM images of the three genotypes of gas vesicle (GV) nanostructures: Halo GVs (left), Ana GVs (middle), and Mega GVs (right). (b) GVs are composed of a ∼2 nm protein shell that excludes water but allows gas to flow in and out of the particle. (c) A negative point diffraction pattern is observed in the SPRI differential reflectivity images when a GV electrostatically adsorbs to the chemically modified gold surface. (d) Quantitative map displaying the Δ%R pixel intensities for the single-GV point diffraction pattern. A sharp, negative spike in Δ%R intensity is observed at the center of the diffraction pattern (the intersection of the two black dotted lines). We observe negative point diffractions for GV adsorption events due to the decrease in interfacial refractive index from water to air (GV).
Table 2. Size Measurements from TEM and Volume, Molecular Weight, and Gas-to-Protein Ratio Calculations for Gas Vesicles.
| nanostructure | Mega GV | Ana GV | Halo GV |
|---|---|---|---|
| length (nm) | 249 | 519 | 400 |
| sL (nm) | 99 | 160 | 113 |
| 95% CI (nm) | 25 | 31 | 20 |
| diameter (nm) | 73 | 136 | 251 |
| sD (nm) | 14 | 21 | 51 |
| 95% CI (nm) | 4 | 4 | 9 |
| volume (nm3) | 7.4 × 105 | 6.4 × 106 | 6.6 × 106 |
| sV (nm3) | 0.8 × 105 | 0.4 × 106 | 0.4 × 106 |
| 95% CI (nm3) | 2 × 104 | 8 × 104 | 7 × 104 |
| # of GVs | 61 | 107 | 125 |
| estimated GV molecular weight (MDa) | 72 | 320 | 282 |
| estimated gas-to-protein volume ratio | 8 | 16 | 19 |
Single-nanoparticle SPRI adsorption measurements were obtained for the irreversible electrostatic adsorption of negatively charged GVs onto a gold surface modified with a positively charged amino-terminated monolayer. Figure 5c shows an example point diffraction pattern from a differential reflectivity image that was obtained for the adsorption of a single Ana GV. This diffraction pattern is similar to the diffraction pattern observed for the adsorption of a PS nanoparticle, but the signal is inverted. This can be seen most dramatically in Figure 5d, which quantifies a sharp negative spike in Δ%R that is observed at the center of the point diffraction pattern (intersection of the two black dotted lines). Calculation of Δ%RNP for an individual GV results in a negative value. Because the volumes of GVs are primarily composed of air, the displacement of water (nwater = 1.33, where n is the refractive index) with the GV (nair = 1.0) causes a decrease in the local refractive index at the location of the GV adsorption and consequently yields a negative Δ%RNP value.
We have previously observed both positive and negative diffraction patterns for PS, hydrogel, and other nanoparticles due to the transient adsorption and subsequent desorption of nanoparticles for the case where nanoparticles are not irreversibly adsorbed onto the chemically modified gold thin film.41 The observed negative diffraction pattern due to desorption always occurred after and at the same location as the previous positive diffraction pattern. For the positively charged MUAM-modified gold thin film, the GVs are irreversibly adsorbed, and the adsorption event always created a negative diffraction pattern. Occasionally, we did observe positive diffraction patterns, which we attribute to the desorption of GVs, but this occurred less than 5% of the time.
The Δ%RNP frequency distribution histograms for single-nanoparticle SPRI adsorption measurements of all three types of GVs are displayed in Figure 6: Mega GVs (Figure 6a), Ana GVs (Figure 6b), and Halo GVs (Figure 6c). Similar to the PS nanoparticles, the absolute Δ%RNP values for all three types of GVs follow log-normal distributions. The values of ⟨Δ%RNP⟩, s, 95% CI, g, μ, and σ for the GVs are all reported in Table 1. All of the GVs have larger relative standard deviations (s/⟨Δ%RNP⟩), skew factors (g), and log-normal scale factors (σ) as compared to PS nanoparticles (Table 1). We attribute these larger log-normal distributions to the heterogeneous nature of the GV biosynthesis. As expected, ⟨Δ%RNP⟩ values for the three types of GVs increase as the total volume of the GV increases (in the order Halo GV > Ana GV > Mega GV). However, a quantitative relationship of ⟨Δ%RNP⟩ to GV volume is complex; the protein component of the GV makes a positive contribution to Δ%RNP, while the gas volume makes a negative contribution. As seen in Table 2, the gas volume dominates over the protein volume in all the GVs, which is why we observe negative Δ%RNP values for all GVs. Moreover, the Ana and Mega GVs have a high length-to-width aspect ratio, which could alter the single-nanoparticle SPRI response. Because the GVs adsorbed to the surface from a quiescent 10 μL solution, we do not expect that there are any preferential orientations of the anisotropic GVs relative to the direction of the surface plasmon polaritons. The future incorporation of a microfluidic flow system for nanoparticle delivery to the gold surface could potentially be used to create oriented adsorbed GV populations. Since near-infrared surface plasmon polaritons have a decay length of approximately 200–300 nm perpendicular to the gold surface,61 Ana and Mega GVs that adsorb with their length perpendicular to the surface may fall outside the range of the surface plasmon polaritons and produce a smaller than expected Δ%RNP.
Figure 6.
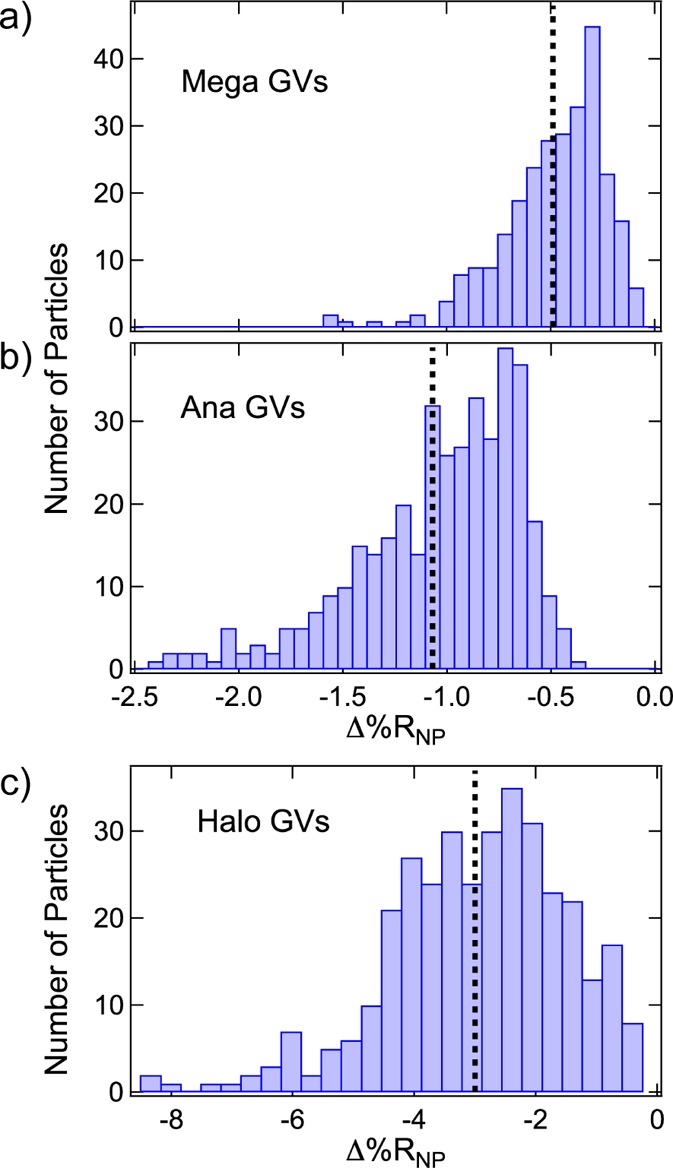
Δ%RNP frequency distribution histograms obtained from the SPRI adsorption measurements of (a) Mega GVs, (b) Ana GVs, and (c) Halo GVs. The average Δ%RNP value for each experiment is plotted as a black dotted line in each histogram. Average Δ%RNP values for Mega, Ana, and Halo GVs were respectively −0.49 ± 0.03%, −1.07 ± 0.04%, and −3.0 ± 0.2%.
Conclusions
In summary, the experiments presented in this paper have demonstrated that both the average single-nanoparticle response (⟨Δ%RNP⟩) and Δ%RNP frequency distribution measurements obtained from single-nanoparticle SPRI adsorption measurements can provide detailed characterization information for a variety of solid, porous, and gas-filled PPNPs. The Δ%RNP frequency distribution measurements of PS nanoparticles showed that Δ%RNP depends on nanoparticle volume for solid nanoparticles. The changes of ⟨Δ%RNP⟩ observed upon uptake of melittin into porous HNPs demonstrate that the single-nanoparticle SPRI measurements can also measure changes in the total material content of a nanoparticle. The ConA binding to mannose-modified HNPs indicates that both bioaffinity uptake and nanoparticle aggregation can be studied through the Δ%RNP frequency distribution histograms. Finally, the most striking evidence that single-nanoparticle SPRI experiments measure changes in interfacial refractive index due to nanoparticle adsorption is the negative point diffraction patterns and Δ%RNP values observed for the adsorption of gas vesicles, a type of gas-filled protein nanostructure.
An important parameter to ascertain for these single-nanoparticle SPRI measurements on PPNPs is how narrow of a Δ%RNP frequency distribution can be measured. Since every PPNP Δ%RNP distribution determined in this paper could be fit with a log-normal distribution, we can use the scale parameter σ to define the normal distribution. The lowest scale parameter observed in these experiments is ∼0.2, and thus this number is our current experimental lower limit for what we can measure for Δ%RNP log-normal distributions. With additional theoretical modeling of the single-nanoparticle SPRI response and the development of more accurate methods of determining Δ%RNP, we expect that this lower limit can be improved in the future.
Methods
Hydrogel Nanoparticle Synthesis
N-Isopropylacrylamide (NIPAm), acrylic acid (AAc), sodium dodecyl sulfate (SDS), V-501, and ammonium persulfate (APS) were obtained from Sigma-Aldrich, Inc. (St. Louis, MO, USA). N,N′-Methylenebis(acrylamide) (BIS) was obtained from Fluka (St. Louis, MO, USA). N-tert-Butylacrylamide (TBAm) was obtained from Acros Organics (Geel, Belgium). NIPAm was recrystallized from hexane before use. All other chemicals were used as received.
HNPs for melittin uptake experiments were synthesized following the procedure detailed in Cho etal.42 The monomers NIPAm (53 mol %), TBAm (40 mol %), AAc (5 mol %), and BIS (2 mol %) were dissolved in 50 mL of nanopure water in a round-bottom flask for a total monomer concentration of 65 mM. TBAm was dissolved in 1 mL of ethanol before addition to the monomer solution. The surfactant SDS (1.7 mg) was also added to the monomer solution to control nanoparticle size. Nitrogen gas was bubbled through the solution for 30 min. Following the addition of a 500 μL aqueous solution containing 30 mg of APS, the polymerization was carried out in an oil bath preset to 60 °C for 3 h under a nitrogen atmosphere. The polymerized solutions were purified by dialysis using 12–14 kDa molecular weight cut-off dialysis membrane against an excess amount of nanopure water (changed more than three times a day) for 4 days.
Mannose-modified HNPs for ConA uptake experiments were synthesized following the procedure detailed in Maley etal. and using a similar procedure to that described for HNP synthesis.43 The sugar unit p-acrylamidophenyl-α-d-mannopyranoside (Man) was synthesized using methods reported previously.62,63 The monomer ratio for mannose-modified HNPs was NIPAm (63.5 mol %), TBAm (28 mol %), AAc (5 mol %), BIS (2 mol %), and Man (1.5 mol %) for a total monomer concentration of 65 mM. SDS (2.5 mg) was used as a surfactant, and V-501 (131.3 μmol/0.5 mL of DMSO) was used as the radical initiator. The polymerization was carried out in an oil bath preset to 70 °C for 3 h under a nitrogen atmosphere. The polymerized solutions were purified by dialysis using 12–14 kDa molecular weight cut-off dialysis membrane against an excess amount of nanopure water (changed more than three times a day) for 4 days.
Gas Vesicles
Ana and Halo GVs were expressed and purified from their respective host bacteria, and Mega GVs were expressed and purified from E. coli, as described previously.60 Briefly, cells were cultured to confluency (and, in the case of E. coli, induced to express GVs) and lysed using hypertonic, hypotonic, or detergent lysis. GVs were isolated using centrifugally assisted buoyancy purification, and their concentration was measured using optical density at 500 nm. Mega GVs, which are natively clustered after purification from bacteria, were unclustered with a solution of 6 M urea and 20 mM Tris-HCl (pH = 8.0), followed by two rounds of centrifugally assisted buoyancy purification and overnight dialysis in 1× phosphate-buffered saline (PBS) (11.9 mM phosphates, 137 mM sodium chloride, 2.7 mM potassium chloride, pH 7.4, Fisher), before optical density quantification and use in SPRI experiments. Transmission electron microscopy was performed on a Philips Tecnai T12 LaB6 120 kV after GVs in HEPES buffer were deposited on carbon/Formvar grids stained with 2% uranyl acetate.60
Optical Setup
The detailed description of the construction of the near-infrared single-nanoparticle SPRI microscope is described in a previous publication.41 The microscope was built into the frame of an IX51 inverted microscope (Olympus, Tokyo, Japan). A 1 mW, 814 nm diode laser (Melles Griot, Carlsbad, CA, USA) was expanded and collimated using a spatial filter (Newport, Corp., Newport Beach, CA, USA). The beam was then polarized and focused with a lens (f = 200 mm) onto the back focal plane of a 100× 1.49 numerical aperture oil microscope objective (Olympus). The beam was directed upward near the edge of the objective by a gold-coated knife-edge mirror (Thorlabs, Newton, NJ, USA) that was mounted on an X–Y micrometer, in order to adjust the incident angle on the sample. The reflected image was allowed to pass out the other side of the objective and acquired by an Andor Neo sCMOS camera (South Windsor, CT, USA) by accumulating 30 11-bit, 0.1 s exposures.
Substrate Preparation
Substrates for all SPRI experiments were borosilicate No. 1.5 coverslips (Fisherbrand, Pittsburgh, PA, USA) coated with a 1 nm Cr adhesion layer and 45 nm Au. For PS nanoparticles and GV measurements, Au surfaces were immobilized with a positively charged alkanethiol monolayer (11-mercaptoundecamine, Dojindo Molecular Technologies, Inc., Gaithersburg, MD, USA) by immersing the Au substrate in a 1 mM ethanolic MUAM solution for 12 h. For HNP measurements, Au surfaces were immobilized with a hydrophobic 1-undecanethiol monolayer (C11, Sigma-Aldrich) by immersing the Au substrate in a 1 mM ethanolic C11 solution for 12 h. All Au surfaces were partitioned using adhesive silicone isolation wells (Electron Microscopy Sciences, Hatfield, PA, USA).
Polystyrene Particle SPRI Measurements
Carboxylate polystyrene spheres with mean diameters of 85 and 170 nm were purchased from Polysciences, Inc. (Warrington, PA, USA). Au slides chemically modified with MUAM were prepared and isolation wells were filled with 10 μL of nanopure water to protect the MUAM layer. Solutions of PS nanoparticles were diluted in nanopure water to concentrations of ∼109 particles/mL for all measurements. For all SPRI experiments, 10 μL of nanoparticle solution was pipetted into the isolation well immediately preceding the image acquisition process.
Hydrogel SPRI Measurements
Au slides for all hydrogel nanoparticle SPRI measurements were chemically modified with C11, and isolation wells were filled with 10 μL of 1× PBS to protect the C11 layer. For melittin uptake measurements, melittin (Sigma-Aldrich) was dissolved in 1× PBS and diluted to a concentration of 18 μM. HNPs were diluted in 1× PBS to a final concentration of 20 μg/mL, and 18 μM melittin was added with a final concentration of 2 μM. The HNP and melittin mixture was allowed to incubate at room temperature for 30 min before SPRI experiments. For ConA uptake measurements, mannose-modified HNPs were diluted in 1× PBS to a final concentration of 20 μg/mL after mixing with ConA (Sigma-Aldrich) at a final concentration of 500 nM. For mannose-modified HNP experiments with free mannose, d-(+)-mannose (Sigma-Aldrich) was also added to the solution at the specified concentration from a more concentrated solution in 1× PBS. The mannose-modified HNP and ConA mixtures were allowed to incubate at room temperature for a minimum of 30 min before SPRI experiments.
Gas Vesicle SPRI Measurements
Au slides for GV measurements were chemically modified with MUAM, and isolation wells were filled with 10 μL of 1× PBS to protect the MUAM layer. The optical density at 500 nm was measured using a NanoDrop 2000 (Thermo Scientific). All GVs were diluted in 1× PBS to the concentrations specified for SPRI experiments: Mega GVs diluted to 1 nM, Ana GVs diluted to 10 pM, and Halo GVs diluted to 5 pM.
Dynamic Light Scattering Measurements
The hydrodynamic diameters of PS nanoparticles were measured in aqueous solutions at 25 °C, and the hydrodynamic diameters of hydrogel nanoparticles were measured in 1× PBS at 25 °C by a DLS instrument equipped with Zetasizer software (Zetasizer Nano ZS, Malvern Instruments Ltd., Worcestershire, U.K.).
Acknowledgments
This work was supported by the National Science Foundation through grant CHE-1403506 (R.M.C.), the National Institutes of Health through grants R01-GM059622 (R.M.C.) and R01-EB018975 (M.G.S.), and the Heritage Medical Research Institute (M.G.S.). DLS data were acquired at the Laser Spectroscopy Facility in the Department of Chemistry at UCI. The authors acknowledge Y. Terada for the synthesis of p-acrylamidophenyl-α-d-mannopyranoside.
The authors declare no competing financial interest.
The version of this paper that was published ASAP July 12, 2017, contained an error in eq 1. The corrected version was reposted July 13, 2017.
References
- Nayak S.; Lyon L. A. Soft Nanotechnology with Soft Nanoparticles. Angew. Chem., Int. Ed. 2005, 44, 7686–7708. 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]
- Rao J. P.; Geckeler K. E. Polymer Nanoparticles: Preparation Techniques and Size-Control Parameters. Prog. Polym. Sci. 2011, 36, 887–913. 10.1016/j.progpolymsci.2011.01.001. [DOI] [Google Scholar]
- Shi X.; Thomas T. P.; Myc L. A.; Kotlyar A.; Baker J. R. Jr. Synthesis, Characterization, and Intracellular Uptake of Carboxyl-Terminated Poly(amidoamine) Dendrimer-Stabilized Iron Oxide Nanoparticles. Phys. Chem. Chem. Phys. 2007, 9, 5712–5720. 10.1039/b709147h. [DOI] [PubMed] [Google Scholar]
- Callahan D. J.; Liu W.; Li X.; Dreher M. R.; Hassouneh W.; Kim M.; Marszalek P.; Chilkoti A. Triple Stimulus-Responsive Polypeptide Nanoparticles That Enhance Intratumoral Spatial Distribution. Nano Lett. 2012, 12, 2165–2170. 10.1021/nl300630c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molino N. M.; Wang S. W. Caged Protein Nanoparticles for Drug Delivery. Curr. Opin. Biotechnol. 2014, 28, 75–82. 10.1016/j.copbio.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.; Zhang Y. S.; Pang B.; Hyun D. C.; Yang M.; Xia Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem., Int. Ed. 2014, 53, 12320–12364. 10.1002/anie.201403036. [DOI] [PubMed] [Google Scholar]
- O’Brien J.; Lee S. H.; Onogi S.; Shea K. J. Engineering the Protein Corona of a Synthetic Polymer Nanoparticle for Broad-Spectrum Sequestration and Neutralization of Venomous Biomacromolecules. J. Am. Chem. Soc. 2016, 138, 16604–16607. 10.1021/jacs.6b10950. [DOI] [PubMed] [Google Scholar]
- Appel E. A.; Tibbitt M. W.; Webber M. J.; Mattix B. A.; Veiseh O.; Langer R. Self-Assembled Hydrogels Utilizing Polymer-Nanoparticle Interactions. Nat. Commun. 2015, 6, 6295. 10.1038/ncomms7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunday D. F.; Green D. L. Thermal and Rheological Behavior of Polymer Grafted Nanoparticles. Macromolecules 2015, 48, 8651–8659. 10.1021/acs.macromol.5b00987. [DOI] [Google Scholar]
- Yonamine Y.; Yoshimatsu K.; Lee S. H.; Hoshino Y.; Okahata Y.; Shea K. J. Polymer Nanoparticle-Protein Interface. Evaluation of the Contribution of Positively Charged Functional Groups to Protein Affinity. ACS Appl. Mater. Interfaces 2013, 5, 374–379. 10.1021/am302404q. [DOI] [PubMed] [Google Scholar]
- Kracke B.; Cole J. T.; Kaiser C. J. O.; Hellenkamp B.; Krysiak S.; Ghoorchian A.; Braun G. B.; Holland N. B.; Hugel T. Thermoswitchable Nanoparticles Based on Elastin-Like Polypeptides. Macromolecules 2015, 48, 5868–5877. 10.1021/acs.macromol.5b00932. [DOI] [Google Scholar]
- Smits F. C.; Buddingh B. C.; van Eldijk M. B.; van Hest J. C. Elastin-Like Polypeptide Based Nanoparticles: Design Rationale Toward Nanomedicine. Macromol. Biosci. 2015, 15, 36–51. 10.1002/mabi.201400419. [DOI] [PubMed] [Google Scholar]
- Liu P.; Yue C.; Sheng Z.; Gao G.; Li M.; Yi H.; Zheng C.; Wang B.; Cai L. Photosensitizer-Conjugated Redox-Responsive Dextran Theranostic Nanoparticles for Near-Infrared Cancer Imaging and Photodynamic Therapy. Polym. Chem. 2014, 5, 874–881. 10.1039/C3PY01173A. [DOI] [Google Scholar]
- Li J.; Mo L.; Lu C. H.; Fu T.; Yang H. H.; Tan W. Functional Nucleic Acid-Based Hydrogels for Bioanalytical and Biomedical Applications. Chem. Soc. Rev. 2016, 45, 1410–1431. 10.1039/C5CS00586H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonamine Y.; Hoshino Y.; Shea K. J. ELISA-Mimic Screen for Synthetic Polymer Nanoparticles with High Affinity to Target Proteins. Biomacromolecules 2012, 13, 2952–2957. 10.1021/bm300986j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.; Zhu Z.; Zou Y.; Huang Y.; Liu D.; Jia S.; Xu D.; Wu M.; Zhou Y.; Zhou S.; Yang C. J. Target-Responsive ″Sweet″ Hydrogel with Glucometer Readout for Portable and Quantitative Detection of Non-Glucose Targets. J. Am. Chem. Soc. 2013, 135, 3748–3751. 10.1021/ja3114714. [DOI] [PubMed] [Google Scholar]
- Lakshmanan A.; Farhadi A.; Nety S. P.; Lee-Gosselin A.; Bourdeau R. W.; Maresca D.; Shapiro M. G. Molecular Engineering of Acoustic Protein Nanostructures. ACS Nano 2016, 10, 7314–7322. 10.1021/acsnano.6b03364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca D.; Lakshmanan A.; Lee-Gosselin A.; Melis J. M.; Ni Y. L.; Bourdeau R. W.; Kochmann D. M.; Shapiro M. G. Nonlinear Ultrasound Imaging of Nanoscale Acoustic Biomolecules. Appl. Phys. Lett. 2017, 110, 073704. 10.1063/1.4976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. G.; Goodwill P. W.; Neogy A.; Yin M.; Foster F. S.; Schaffer D. V.; Conolly S. M. Biogenic Gas Nanostructures as Ultrasonic Molecular Reporters. Nat. Nanotechnol. 2014, 9, 311–316. 10.1038/nnano.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. G.; Ramirez R. M.; Sperling L. J.; Sun G.; Sun J.; Pines A.; Schaffer D. V.; Bajaj V. S. Genetically Encoded Reporters for Hyperpolarized Xenon Magnetic Resonance Imaging. Nat. Chem. 2014, 6, 629–634. 10.1038/nchem.1934. [DOI] [PubMed] [Google Scholar]
- Kelly K. L.; Coronado E.; Zhao L. L.; Schatz G. C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. 10.1021/jp026731y. [DOI] [Google Scholar]
- Crut A.; Maioli P.; Del Fatti N.; Vallee F. Optical Absorption and Scattering Spectroscopies of Single Nano-Objects. Chem. Soc. Rev. 2014, 43, 3921–3956. 10.1039/c3cs60367a. [DOI] [PubMed] [Google Scholar]
- Crut A.; Maioli P.; Vallee F.; Del Fatti N. Linear and Ultrafast Nonlinear Plasmonics of Single Nano-Objects. J. Phys.: Condens. Matter 2017, 29, 123002. 10.1088/1361-648X/aa59cc. [DOI] [PubMed] [Google Scholar]
- Cho E. J.; Holback H.; Liu K. C.; Abouelmagd S. A.; Park J.; Yeo Y. Nanoparticle Characterization: State of the Art, Challenges, and Emerging Technologies. Mol. Pharmaceutics 2013, 10, 2093–2110. 10.1021/mp300697h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till U.; Gaucher-Delmas M.; Saint-Aguet P.; Hamon G.; Marty J. D.; Chassenieux C.; Payre B.; Goudouneche D.; Mingotaud A. F.; Violleau F. Asymmetrical Flow Field-Flow Fractionation with Multi-Angle Light Scattering and Quasi-Elastic Light Scattering for Characterization of Polymersomes: Comparison with Classical Techniques. Anal. Bioanal. Chem. 2014, 406, 7841–7853. 10.1007/s00216-014-7891-8. [DOI] [PubMed] [Google Scholar]
- Wyatt P. J. Measurement of Special Nanoparticle Structures by Light Scattering. Anal. Chem. 2014, 86, 7171–7183. 10.1021/ac500185w. [DOI] [PubMed] [Google Scholar]
- Ballauff M.; Lu Y. “Smart” Nanoparticles: Preparation, Characterization and Applications. Polymer 2007, 48, 1815–1823. 10.1016/j.polymer.2007.02.004. [DOI] [Google Scholar]
- Helvig S.; Azmi I. D. M.; Moghimi S. M.; Yaghmur A. Recent Advances in Cryo-TEM Imaging of Soft Lipid Nanoparticles. AIMS Biophysics 2015, 2, 116–130. 10.3934/biophy.2015.2.116. [DOI] [Google Scholar]
- Patterson J. P.; Robin M. P.; Chassenieux C.; Colombani O.; O’Reilly R. K. The Analysis of Solution Self-Assembled Polymeric Nanomaterials. Chem. Soc. Rev. 2014, 43, 2412–2425. 10.1039/C3CS60454C. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Hernandez J.; Babin J.; Zappone B.; Lecommandoux S. Preparation of Shell Cross-Linked Nano-Objects from Hybrid-Peptide Block Copolymers. Biomacromolecules 2005, 6, 2213–2220. 10.1021/bm0501499. [DOI] [PubMed] [Google Scholar]
- Wolfbeis O. S. An Overview of Nanoparticles Commonly Used in Fluorescent Bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. 10.1039/C4CS00392F. [DOI] [PubMed] [Google Scholar]
- Su S.; Wang H.; Liu X.; Wu Y.; Nie G. iRGD-Coupled Responsive Fluorescent Nanogel for Targeted Drug Delivery. Biomaterials 2013, 34, 3523–3533. 10.1016/j.biomaterials.2013.01.083. [DOI] [PubMed] [Google Scholar]
- Weichert F.; Gaspar M.; Timm C.; Zybin A.; Gurevich E. L.; Engel M.; Müller H.; Marwedel P. Signal Analysis and Classification for Surface Plasmon Assisted Microscopy of Nanoobjects. Sens. Actuators, B 2010, 151, 281–290. 10.1016/j.snb.2010.08.005. [DOI] [Google Scholar]
- Zybin A.; Kuritsyn Y. A.; Gurevich E. L.; Temchura V. V.; Überla K.; Niemax K. Real-Time Detection of Single Immobilized Nanoparticles by Surface Plasmon Resonance Imaging. Plasmonics 2010, 5, 31–35. 10.1007/s11468-009-9111-5. [DOI] [Google Scholar]
- Wang S.; Shan X.; Patel U.; Huang X.; Lu J.; Li J.; Tao N. Label-Free Imaging, Detection, and Mass Measurement of Single Viruses by Surface Plasmon Resonance. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16028–16032. 10.1073/pnas.1005264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel K. F.; Bechinger C.; Herminghaus S.; Riedel M.; Leiderer P.; Weiland U.; Bastmeyer M. Imaging of Cell/Substrate Contacts of Living Cells with Surface Plasmon Resonance Microscopy. Biophys. J. 1999, 76, 509–516. 10.1016/S0006-3495(99)77219-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil M. M.; Denyer M. C.; Youseffi M.; Britland S. T.; Liu S.; See C. W.; Somekh M. G.; Zhang J. Imaging of the Cell Surface Interface Using Objective Coupled Widefield Surface Plasmon Microscopy. J. Struct. Biol. 2008, 164, 75–80. 10.1016/j.jsb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Gurevich E. L.; Temchura V. V.; Überla K.; Zybin A. Analytical Features of Particle Counting Sensor Based on Plasmon Assisted Microscopy of Nano Objects. Sens. Actuators, B 2011, 160, 1210–1215. 10.1016/j.snb.2011.09.050. [DOI] [Google Scholar]
- Wang W.; Foley K.; Shan X.; Wang S.; Eaton S.; Nagaraj V. J.; Wiktor P.; Patel U.; Tao N. Single Cells and Intracellular Processes Studied by a Plasmonic-Based Electrochemical Impedance Microscopy. Nat. Chem. 2011, 3, 249–255. 10.1038/nchem.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Tao N. Detection, Counting, and Imaging of Single Nanoparticles. Anal. Chem. 2014, 86, 2–14. 10.1021/ac403890n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern A. R.; Wood J. B.; Wang Y.; Corn R. M. Single-Nanoparticle Near-Infrared Surface Plasmon Resonance Microscopy for Real-Time Measurements of DNA Hybridization Adsorption. ACS Nano 2014, 8, 1022–1030. 10.1021/nn405868e. [DOI] [PubMed] [Google Scholar]
- Cho K.; Fasoli J. B.; Yoshimatsu K.; Shea K. J.; Corn R. M. Measuring Melittin Uptake into Hydrogel Nanoparticles with Near-Infrared Single Nanoparticle Surface Plasmon Resonance Microscopy. Anal. Chem. 2015, 87, 4973–4979. 10.1021/acs.analchem.5b00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley A. M.; Terada Y.; Onogi S.; Shea K. J.; Miura Y.; Corn R. M. Measuring Protein Binding to Individual Hydrogel Nanoparticles with Single-Nanoparticle Surface Plasmon Resonance Imaging Microscopy. J. Phys. Chem. C 2016, 120, 16843–16849. 10.1021/acs.jpcc.6b05700. [DOI] [Google Scholar]
- Viitala L.; Maley A. M.; Fung H. W. M.; Corn R. M.; Viitala T.; Murtomäki L. Surface Plasmon Resonance Imaging Microscopy of Liposomes and Liposome-Encapsulated Gold Nanoparticles. J. Phys. Chem. C 2016, 120, 25958–25966. 10.1021/acs.jpcc.6b09503. [DOI] [Google Scholar]
- Yu H.; Shan X.; Wang S.; Tao N. Achieving High Spatial Resolution Surface Plasmon Resonance Microscopy with Image Reconstruction. Anal. Chem. 2017, 89, 2704–2707. 10.1021/acs.analchem.6b05049. [DOI] [PubMed] [Google Scholar]
- Jiang D.; Jiang Y.; Li Z.; Liu T.; Wo X.; Fang Y.; Tao N.; Wang W.; Chen H. Y. Optical Imaging of Phase Transition and Li-Ion Diffusion Kinetics of Single LiCoO2 Nanoparticles During Electrochemical Cycling. J. Am. Chem. Soc. 2017, 139, 186–192. 10.1021/jacs.6b08923. [DOI] [PubMed] [Google Scholar]
- Bolduc O. R.; Masson J. F. Advances in Surface Plasmon Resonance Sensing with Nanoparticles and Thin Films: Nanomaterials, Surface Chemistry, and Hybrid Plasmonic Techniques. Anal. Chem. 2011, 83, 8057–8062. 10.1021/ac2012976. [DOI] [PubMed] [Google Scholar]
- Zayats A. V.; Smolyaninov I. I.; Maradudin A. A. Nano-Optics of Surface Plasmon Polaritons. Phys. Rep. 2005, 408, 131–314. 10.1016/j.physrep.2004.11.001. [DOI] [Google Scholar]
- Joanes D. N.; Gill C. A. Comparing Measures of Sample Skewness and Kurtosis. J. R. Stat. Soc. D-Sta. 1998, 47, 183–189. 10.1111/1467-9884.00122. [DOI] [Google Scholar]
- Smith J. E.; Jordan M. L. Mathematical and Graphical Interpretation of the Log-Normal Law for Particle Size Distribution Analysis. J. Colloid Sci. 1964, 19, 549–559. 10.1016/0095-8522(64)90069-8. [DOI] [Google Scholar]
- Boyd R. D.; Pichaimuthu S. K.; Cuenat A. New Approach to Inter-Technique Comparisons for Nanoparticle Size Measurements; Using Atomic Force Microscopy, Nanoparticle Tracking Analysis and Dynamic Light Scattering. Colloids Surf., A 2011, 387, 35–42. 10.1016/j.colsurfa.2011.07.020. [DOI] [Google Scholar]
- Hoshino Y.; Koide H.; Furuya K.; Haberaecker W. W. 3rd; Lee S. H.; Kodama T.; Kanazawa H.; Oku N.; Shea K. J. The Rational Design of a Synthetic Polymer Nanoparticle that Neutralizes a Toxic Peptide In Vivo. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 33–38. 10.1073/pnas.1112828109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis H.; Sharon N. Lectins: Carbohydrate-Specific Proteins that Mediate Cellular Recognition. Chem. Rev. 1998, 98, 637–674. 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Mandal D. K.; Kishore N.; Brewer C. F. Thermodynamics of Lectin-Carbohydrate Interactions. Titration Microcalorimetry Measurements of the Binding of N-Linked Carbohydrates and Ovalbumin to Concanavalin A. Biochemistry 1994, 33, 1149–1156. 10.1021/bi00171a014. [DOI] [PubMed] [Google Scholar]
- Lee Y. C.; Lee R. T. Carbohydrate-Protein Interactions: Basis of Glycobiology. Acc. Chem. Res. 1995, 28, 321–327. 10.1021/ar00056a001. [DOI] [Google Scholar]
- Pieters R. J. Maximising Multivalency Effects in Protein-Carbohydrate Interactions. Org. Biomol. Chem. 2009, 7, 2013–2025. 10.1039/b901828j. [DOI] [PubMed] [Google Scholar]
- Terada Y.; Hashimoto W.; Endo T.; Seto H.; Murakami T.; Hisamoto H.; Hoshino Y.; Miura Y. Signal Amplified Two-Dimensional Photonic Crystal Biosensor Immobilized with Glyco-Nanoparticles. J. Mater. Chem. B 2014, 2, 3324–3332. 10.1039/C4TB00028E. [DOI] [PubMed] [Google Scholar]
- Pfeifer F. Distribution, Formation and Regulation of Gas Vesicles. Nat. Rev. Microbiol. 2012, 10, 705–715. 10.1038/nrmicro2834. [DOI] [PubMed] [Google Scholar]
- Walsby A. E. Gas Vesicles. Microbiol. Rev. 1994, 58, 94–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan A.; Lu G. J.; Farhadi A.; Nety S. P.; Kunth M.; Lee-Gosselin A.; Maresca D.; Bourdeau R. W.; Yin M.; Yan J.; Witte C.; Malounda D.; Foster F. S.; Schroder L.; Shapiro M. G.. Preparation and Noninvasive Imaging of Biogenic Gas Vesicle Nanostructures. Nat. Protoc. In press. DOI: 10.1038/nprot.2017.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. A.; Corn R. M. Surface Plasmon Resonance Imaging as a Tool to Monitor Biomolecular Interactions in an Array Based Format. Appl. Spectrosc. 2003, 57, 320A–332A. 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- Hoshino Y.; Nakamoto M.; Miura Y. Control of Protein-Binding Kinetics on Synthetic Polymer Nanoparticles by Tuning Flexibility and Inducing Conformation Changes of Polymer Chains. J. Am. Chem. Soc. 2012, 134, 15209–15212. 10.1021/ja306053s. [DOI] [PubMed] [Google Scholar]
- Toyoshima M.; Miura Y. Preparation of Glycopolymer-Substituted Gold Nanoparticles and Their Molecular Recognition. J. Polym. Sci., Part A: Polym. Chem. 2009, 47, 1412–1421. 10.1002/pola.23250. [DOI] [Google Scholar]