Abstract
The compartmentalization and association of lactate dehydrogenase (LDH) with specific cellular structures (e.g., synaptosomal, sarcoplasmic or mitochondrial) may play an important role in brain energy metabolism. Our previous research revealed that LDH in the synaptosomal fraction shifts toward the aerobic isoforms (LDH-B) among the large-brained haplorhine primates compared to strepsirrhines. Here, we further analyzed the subcellular localization of LDH in primate forebrain structures using quantitative Western blotting and ELISA. We show that, in cytosolic and mitochondrial subfractions, LDH-B expression level was relatively elevated and LDH-A declined in haplorhines compared to strepsirrhines. LDH-B expression in mitochondrial fractions of the neocortex was preferentially increased, showing a particularly significant rise in the ratio of LDH-B to LDH-A in chimpanzees and humans. We also found a significant correlation between the protein levels of LDH-B in mitochondrial fractions from haplorhine neocortex and the synaptosomal LDH-B that suggests LDH isoforms shift from a predominance of A-subunits toward B-subunits as part of a system that spatially buffers dynamic energy requirements of brain cells. Our results indicate that there is differential subcellular compartmentalization of LDH isoenzymes that evolved among different primate lineages to meet the energy requirements in neocortical and striatal cells.
Keywords: Lactate dehydrogenase, Strepsirrhine, Haplorhine, Primate forebrain, Metabolism, Cellular subfractions, Mitochondria
1. Introduction
The lactate dehydrogenase enzyme (LDH, E.C 1.1.1.27) is the protein responsible for lactate oxidation and reduction. LDH is a tetrameric enzyme that is comprised of four subunits of two types: muscle/anaerobic (LDH-A) and heart/aerobic (LDH-B). Various combinations of LDH-A sub-units and LDH-B subunits (which are produced from a different gene) can form homo- or heterotetramers, and assemble into five different combinations LDH-1 (4B), LDH-2 (3B,1A), LDH-3 (2B,2A), LDH-4 (1B,3A), and LDH-5 (4A). The direction of the LDH-mediated reaction is facilitated by the presence of functionally different isoforms. The A-prevailing isoforms act primarily in lactate formation, whereas the isoforms that show B dominance facilitate the reaction in the reverse direction. The regulation of energetic utilization by LDHs has significance for neuronal metabolism (Laughton et al., 2007; O'Brien et al., 2007; Ross et al., 2010), glutamatergic signaling (Gross et al., 1996; Magistretti, 2006; Ghosh et al., 2010; Petit et al., 2013), and brain evolution (Syner and Goodman, 1966; Goodman et al., 1969; Koen and Goodman, 1969). Furthermore, studies of LDH isoform distribution at both the mRNA and protein levels have shown variation in the expression of LDH-A and LDH-B in the central nervous system (Bittar et al., 1996; Laughton et al., 2000; Pellerin, 2003). Our previous research demonstrated that among haplorhine primates (i.e., monkeys and hominoids, including humans) with relatively large brain sizes the shift from LDH-A to LDH-B was especially pronounced in the synaptosomes of the neocortex (Duka et al., 2014). Thus, the distribution of LDH types might be related to metabolic features of their compartments. Hence, the structural, biochemical, and functional variation among subcellular compartments are likely to entail differences in metabolic demand. For example, particular cell types or subcellular compartments may show elevated concentrations of glycolytic enzymes to meet transient increases in demand for energy that occur at these unique sites and that might display changes in primate brain evolution.
At present, LDH isoenzyme composition is not known at the level of subcellular localization in the brain for different primate species. In the current study, the relative protein expression levels of cytoplasmic and mitochondrial LDH isozymes in forebrain structures were compared across a diversity of primate species by quantitative Western blotting and ELISA.
2. Experimental procedures
2.1. Brain sample collection details
In this study, a total of 25 (13 males and 12 females, all adults) primate frozen postmortem brain samples were analyzed (Lorises - Loris tardigradus, n = 1; Nycticebus pygmaeus, n = 3; Lemurs - Lemur catta, n = 2; Eulemur flavifrons, n= 1; New World monkeys -Ateles belzebuth, n = 1; Pithecia pithecia, n = 1; Old World monkeys - Papio anubis, n = 1; Macaca mulatta, n = 4; Macaca nemestrina, n = 1; Hominoids - Pan troglodytes, n = 5; Homo sapiens, n= 5). Because of the opportunistic nature of collecting frozen postmortem brain tissue samples from such a diversity of primates, it was not possible to acquire even sex and age distributions in each species. Frozen brain samples from nonhuman primates used in this study were obtained from various zoos and research facilities. All brain samples were collected at necropsy and no animals were sacrificed for the purposes of this research. Frozen human brain samples were obtained from the University of Maryland Brain and Tissue Bank. Brain samples were stored at −80 °C prior to use in this study, and tissue samples for protein extraction and further analysis were processed as previously described (Duka et al., 2014).
Tissue samples, approximately 100–150 mg each, were dissected from the left superior parietal cortex and striatum without thawing. Table A.1 shows the taxonomic composition of the individuals and full details of the age and sex distribution of the sample. Supplemental Fig. A.1 represents the phylogeny of the species used in the study. To enable statistical analysis, we divided the samples into two groups according to primate suborders: strepsirrhines (lemurs and lorises) and haplorhines (monkeys and hominoids). Also, several comparisons considered variation among three groups based on brain mass: (1) strepsirrhines, (2) the combined paraphyletic sample of New World monkeys (NWM) and Old World monkeys (OWM), and (3) hominoids (including chimpanzees and humans). Due to small sample sizes we were unable to analyze separately NWM and OWM; however, they are represented as individual phylogenetic groups in all graphs.
2.2. Reagents and chemicals
Bio-Rad protein assay reagents, NativePAGE Novex Bis-tris Gel System kit, ECL Western blotting detection reagent and other materials for SDS-PAGE were purchased from Bio-Rad (Hercules, CA, USA). Human heart whole tissue lysate (NB820-59217) or skeletal muscle whole tissue lysate (NB820-59253) were purchased from Novus Biologicals (Littleton, CO, USA).
2.3. Subcellular fractionation of proteins
For analysis of subcellular localization of LDH, isolation of a highly enriched mitochondrial and cytosolic fraction were performed using Mitochondria/Cytosol Fractionation (Cat# K-256-25) and Nuclear/Cytosol Fractionation (Cat# K-266-25) kits from BioVision (Mountain View, CA, USA) as described elsewhere (Yu et al., 2005; Guha et al., 2011), and in accordance with manufacturer's protocol, with minor modifications in the brain sample preparation step. In brief, 10 mg of tissue was resuspended in 1 ml of cytosol extraction buffer mix containing dithiothreitol and protease inhibitors. We only used a Dounce homogenizer with these kits, and avoided unnecessary excessive homogenization, as it can cause damage to the mitochondrial membrane which triggers release of mitochondrial components. To characterize the subcellular fractions obtained by differential centrifugation, the distributions of subcellular localization markers were analyzed in the same blots. In particular, β-tubulin (cytosol), cytochrome c oxidase subunit IV (COX IV) (mitochondria), and GRP75 (mitochondria) were used to verify the purity of subcellular fractions.
2.4. Western blot analysis
The following primary antibodies were used for immunoblotting: anti-LDH-A (sc-27230; Santa Cruz Biotechnology, Santa Cruz, CA), anti-LDH-B (NB100-79987; Novus Biologicals, Littleton CO), and anti-LDH (sc-133123; Santa Cruz Biotechnology, Santa Cruz, CA). The protocol for Western blot was previously described in detail (Duka et al., 2014). The blots were also probed with antibodies against cell compartment-specific marker proteins: anti-COX IV (mitochondrial control; ab16056; Abcam, Cambridge, UK), anti-GRP75 (mitochondrial marker and loading control; ab2799; Abcam, Cambridge, UK), anti-β-tubulin (cytosolic marker and loading control; ab21058; Abcam, Cambridge, UK). Supplemental Fig. A.4 shows an assessment of the purity of subcellular fractions. The signals were quantitatively evaluated with Scion Image software. Equal protein loading was confirmed with anti-β-actin antibody (sc-1616; Santa Cruz Biotechnology, Santa Cruz, CA). Protein bands were scanned using an EPSON Perfection 4870 Photo scanner and quantitatively analyzed by Scion Image software. The LDH-B/LDH-A ratio was calculated as previously described (Duka et al., 2014).
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
All ELISAs were performed using Maxisorp plates (Nunc, Roskilde, Denmark) coated for overnight incubation at 4 °C with 20 μg/ml of fractions (100 μl/well), followed by blocking with PBS/T/BSA (PBS with 0.05% Tween 20,1% BSA) for 2 h at 37 °C. ELISA assays were conducted with anti-LDH-A and anti-LDH-B antibodies as previously described for Western blot analysis (see Materials and methods). After incubation of the captured antibody during 1 h at RT, Nunc Maxisorp plates were washed three times with PBS/T. An O-phenylenediamine dihydrochloride/peroxidase solution (Thermo Scientific, Rockford, IL) was used as the chromogen to visualize the reaction product. After sufficient color development, the reaction was stopped with 40% H2SO4 and optical densities of wells were then read at 490 nm, subtracting 650 nm (background) values. The optical density of the sample was compared to a standard curve, which is a serial dilution of a known-concentration for muscle tissue and heart tissue homogenate, LDH standards in PBS. For characterization of antibody specificity and ELISA optimization see Supplemental Figs. A.2 and A.3.
2.6. Statistical analysis
The nonparametric Mann-Whitney U test was used to determine significant differences between the strepsirrhine and haplorhine groups. The Kruskal–Wallis one-way analysis of variance test was used to examine differences between LDH expression in the following primate groups: strepsirrhines, monkeys, and hominoids. PAST statistical software (Hammer et al., 2001) was used for analyses. All data in figures and tables are expressed as mean ± S.D. Pearson correlation coefficients were estimated from the optical density values from isoenzyme, Western blot, and ELISA analyses to investigate the relationship of protein expression in fractions in brain structures. Statistical significance was set at α= 0.05. Pearson correlation coefficients were estimated from the optical density values from isoenzyme.
3. Results
3.1. Levels of LDH-A and LDH-B in the cytoplasmic and mitochondrial fractions
We examined the intracellular localization of LDH isoenzymes using Western blot analysis and ELISA of subcellular fractionated samples of neocortex and striatum from a diverse group of primate species. The results of analyses of species-specific subcellular levels of LDH isoforms are shown in Figs. 1 and 2. As estimated by Western blot analysis, the expression level of cytosolic LDH-A was 1.3 and 2.6-fold lower (p ≤ 0.01) in the neocortex (Fig. 1A) and 1.3 and 1.8-fold lower (p ≤ 0.01) in the striatum (Fig. 1B) of monkeys and hominoids, respectively, compared to strepsirrhines (Table 1). In contrast, elevated cytosolic LDH-B protein expression was observed in monkey and hominoid neocortex (1.6 and 1.7-fold increase; p ≤ 0.05) (Fig. 1A) and striatum (1.6 and 1.8-fold increase; p ≤ 0.05) compared to strepsirrhines (Fig. 1B).
Fig. 1.
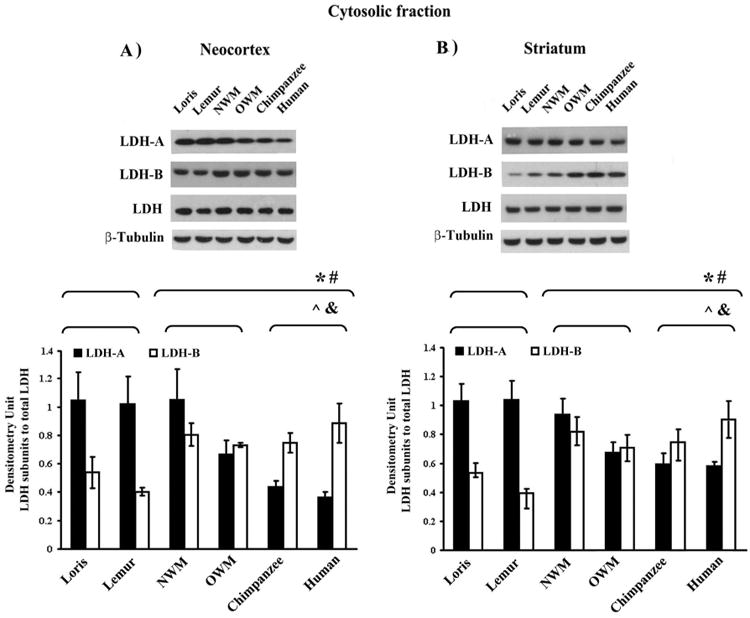
LDH isoenzyme expression levels in cytosolic fractions of primate forebrain structures. Representative Western blot showing the immunoreactive bands obtained with cytosolic LDH isoforms from neocortex (A), and striatum (B) separated by SDS-PAGE and immunoblotted with antibodies indicated from left. The amount of immunoreactive LDH isoenzymes shown were assessed by scanning densitometry of Western blots. Equal protein loading was confirmed with anti-β-tubulin antibody (see Supplemental Fig. A.4). Densitometry units were adjusted to an LDH total control. *, # p ≤ 0.05; Mann-Whitney U test for LDH-A and LDH-B, respectively. ˆ, & p ≤ 0.05; Kruskal–Wallis test for LDH-A and LDH-B, respectively.
Fig. 2.
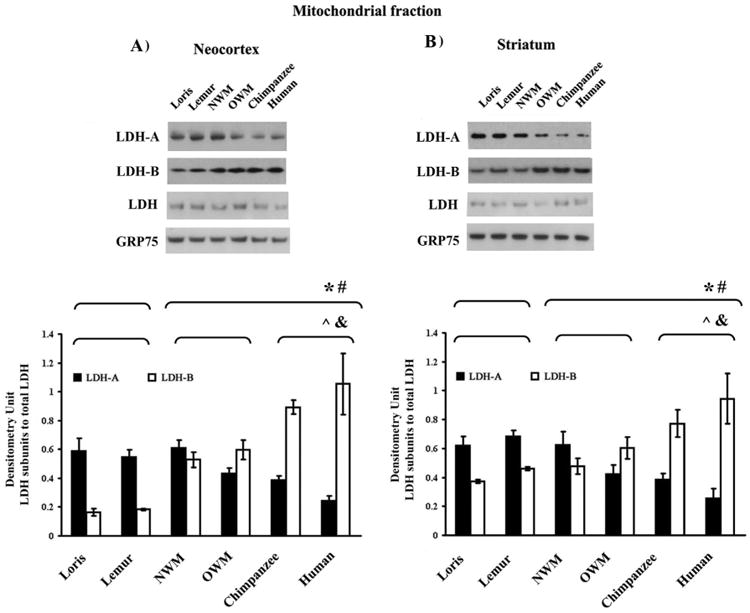
LDH isoenzyme patterns from the neocortical and striatal mitochondrial fractions. Upper panel shows representative Western blots of LDH isoenzymes from neocortex (A) and striatum (B). Quantification of the relative amount of LDH protein subunits was obtained by scanning gels. GRP75 was used as a loading control. The values obtained by densitometric analysis of the blotted LDH isoenzymes are shown in the bottom panel. *, The difference in the values of strepsirrhines vs. haplorhines was significant (Mann-Whitney U test, p ≤ 0.05). #, Kruskal-Wallis test of significant differences among the following primates groups: strepsirrhines, monkeys, and hominoids (p ≤ 0.05).
Table 1.
Quantification of the ratio LDH-B to LDH-A in the cytosolic fractions of primate brain.
| Species group | Western blot analysis | ELISA | ||
|---|---|---|---|---|
|
|
|
|||
| Neocortex | Striatum | Neocortex | Striatum | |
| Lorisidae | 0.545 ± 0.050 | 0.519 ± 0.071 | 0.808 ± 0.095 | 0.776 ± 0.091 |
| Lemuridae | 0.426 ± 0.056 | 0.379 ± 0.043 | 0.662 ± 0.094 | 0.517 ± 0.086 |
| New World monkeys | 0.793 ± 0.083 | 0.870 ± 0.134 | 1.084 ± 0.117 | 0.896 ±0.110 |
| Old World monkeys | 1.202 ± 0.361 | 1.048 ± 0. 169 | 1.519 ± 0.178 | 1.429 ± 0.193 |
| Chimpanzee | 1.759 ± 0.253 | 1.252 ± 0. 376 | 2.199 ± 0.295 | 1.708 ± 0.198 |
| Human | 2.453 ± 0.379 | 1.558 ± 0.391 | 2.780 ± 0.240 | 2.124 ± 0.249 |
We also used Western blotting and ELISA to identify phylogenetic differences in LDH isoform expression profiles in mitochondrial fractions. The mitochondrial fraction also contained LDH-A and LDH-B (Fig. 2). The two methods concordantly revealed that LDH-B levels in mitochondrial fractions from the neocortex and striatum significantly varied across species, with increased levels in monkeys and hominoids as compared to strepsirrhines. In contrast, LDH-A levels showed an opposite pattern of decreasing expression in those species where LDH-B levels were increased. Quantitative Western blot analysis demonstrated that neocortical mitochondrial LDH-B expression level was elevated 3-fold in NWM and OWM, 5-fold in chimpanzees, and 6-fold humans (p ≤ 0.01) compared to strepsirrhines (Fig. 2A). Mitochondrial LDH-B was 2- and 1.6-fold higher in the striatum (p ≤ 0.05), respectively, in monkey and hominoids compared to strepsirrhines (Fig. 2B). By contrast, the amount of mitochondrial LDH-A was 1.2- and 1.8-fold lower in the neocortex (p ≤ 0.05) (Fig. 2A) and 2.0- and 1.6-fold (p ≤ 0.05) lower in the striatum (Fig. 2B) of monkeys and hominoids compared to strepsirrhines, respectively. ELISAs for both proteins, LDH-A and LDH-B, and for both brain regions, neocortex and striatum, confirmed the results from Western blotting (Fig. A.5).
3.2. Ratios of LDH-B to LDH-A in subfractions from neocortex and striatum
The ratio of LDH-B to LDH-A monomers signifies aerobic status (Koen and Goodman, 1969; Koen and Goodman, 1971). Phylogenetic differences in the proportions of two kinds of LDH in primate brain regions were shown previously (Goodman et al., 1969). Here we calculated the polypeptide ratio of single isoforms, LDH-B/LDH-A (Western blot and ELISA) for neocortical and striatal subfractions (see Table 2). As estimated by two different methods, LDH-B/LDH-A ratios differed markedly in the neocortical mitochondrial fractions among primate taxa (Table 2). The average ratio, as measured by Western blot, increased 2- to 6-fold across taxa, from 0.47 in strepsirrhines, to 0.91 in NWM, 1.26 in OWM, and 2.99 in hominoids (p ≤ 0.05).
Table 2.
Quantification of the ratio LDH-B to LDH-A in the mitochondrial fractions of primate brain.
| Species group | Western blot analysis | ELISA | ||
|---|---|---|---|---|
|
|
|
|||
| Neocortex | Striatum | Neocortex | Striatum | |
| Lorisidae | 0.446 ± 0.048 | 0.599 ± 0.055 | 0.926 ± 0.105 | 0.714 ± 0.087 |
| Lemuridae | 0.492 ± 0.042 | 0.675 ± 0.088 | 1.125 ± 0.116 | 0.790 ± 0. 093 |
| New World monkeys | 0.914 ± 0.082 | 0.765 ± 0.094 | 1.792 ± 0.156 | 1.124 ± 0.281 |
| Old World monkeys | 1.255 ± 0.109 | 1.421 ± 0.151 | 2.655 ± 0.367 | 1.860 ± 0.095 |
| Chimpanzee | 2.208 ± 0. 237 | 1.998 ± 0.338 | 4.223 ± 0.596 | 2.059 ± 0.232 |
| Human | 3.768 ± 0.578 | 3.707 ± 0.523 | 7.549 ± 0.850 | 3.875 ± 0.476 |
The LDH-B/LDH-A ratio in the mitochondrial fraction obtained from the striatum as measured by Western blot also varied 1.7- to 2.6-fold with phylogeny, from a ratio of 0.64 in strepsirrhines, to 0.77 in NWM, 1.42 in OWM, and 3.71 (p ≤ 0.05) in hominoids. As evaluated by Western blot, values of the ratio in the cytosolic fraction obtained from the neocortex and striatum increases correspondently from an average of 0.49 and 0.45 in strepsirrhine primates to 1.00 and 0.96 in monkeys and 2.11 and 1.41 in hominoids (p ≤ 0.05).
Our analysis showed a strong positive correlation between the relative ratio of LDH-B to LDH-A in the cytosolic (r = 0.968, p = 0.002) and mitochondrial (r = 0.992, p = 0.0001) fractions, as well as between methods used to measure the LDH isoenzymes level in these two subfractions isolated from primate neocortex (r = 0.998, p = 0.0001, for both fractions) and striatum (r = 0.978, p = 0.001 and r = 0.992, p = 0.0001).
3.3. Resemblance in phylogenetic variation of LDH isoenzymes in mitochondrial and synaptosomal fractions
We have previously demonstrated the predominance of LDH-B in the synaptosomal fraction of neocortex in association with increased brain size in primates (Duka et al., 2014). Here, we examined the association between LDH isoenzymes in synaptosomal and mitochondrial fractions in our comparative sample of primates. We obtained densitometric estimates of the levels of LDH-A and LDH-B proteins in synaptosomal fractions from Duka et al. (2014). In our previous study all experiments were performed on the same primate frozen postmortem brain samples as in the current study, with the same sampling strategy, by using the same protocol for Western blot and the same data analysis. In the present study, we found a significant positive correlation (Fig. 3) in both forebrain regions in the expression level of LDH-A (for neocortex, r = 0.859, p = 0.03 and for striatum, r = 0.913, p = 0.01) and LDH-B (for neocortex, r = 0.947, p = 0.004 and for striatum, r = 0.943, p = 0.005) between synaptosomal and mitochondrial fractions, suggesting a specific parallel between mitochondrial activity and evolutionary changes in synaptic energy use.
Fig. 3.
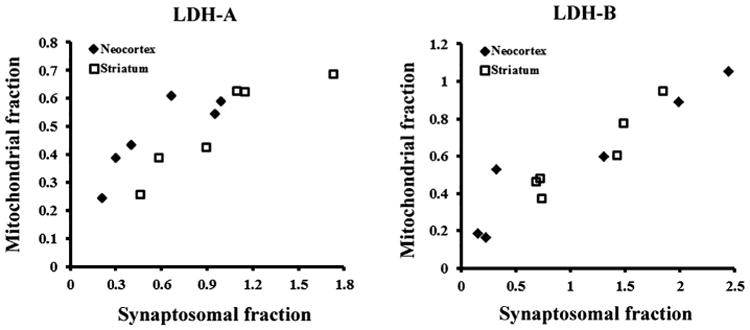
Scatterplots of mitochondria-associated LDH-A and LDH-B versus synapse-associated LDH isoform expression in primate neocortex and striatum. For this analysis, we used densitometry units for mitochondrial LDH isoenzymes based on Western blot and obtained densitometric estimates of the levels of LDH-A and LDH-B proteins in synaptosomal fractions from Duka et al. (2014). The previously published data values for the synaptosomal fractions was calculated in the same way as in the current study, and presented as the signal ratios of LDH protein subunits to the respective loading control first, and then to total LDH densities.
4. Discussion
4.1. Distribution of LDH isoenzymes between the cytosolic and mitochondrial fractions in primate neocortex and striatum
Cytosolic and mitochondrial fractions from primate forebrain structures were isolated and examined with SDS-PAGE, immunoblotting, and ELISA using antibodies against the LDH isoforms A or B (Figs. 1, 2 and Fig. A.5). The expression levels of LDH-A and LDH-B varied in the neocortex and striatum across primates in both subcellular fractions. The most pronounced elevation of LDH-B expression was observed in the mitochondrial fractions of the neocortex in chimpanzees and humans, indicating that a functional metabolic shift occurred during primate brain evolution in comparatively large-brained species to allow more pyruvate production to enter the TCA cycle and produce more ATP molecules.
Although LDH is classically considered to be a cytoplasmic marker, there are several reports, however, on the localization of LDH in other cell compartments, namely, synaptosomes, sarcoplasmic reticulum, mitochondria, and cell nuclei (Venkov et al., 1976; Vdovichenko, 1978; Fang et al., 2000; Kim and Dang, 2005; Laughton et al., 2007; Lemire et al., 2008; Brooks, 2009). Our results also confirm that LDH isoenzymes are present in isolated mitochondrial fractions, in addition to the cytosol, and demonstrate that mitochondrial LDH subunits share similarity with cytosolic forms of a 37.0 kDa molecular mass. The controversy over mitochondrial-localized LDH was a topic of discussion over many years (Baba and Sharma, 1971; Kline et al., 1986; Brandt et al., 1987). The issue was debated because of methodological limitations. Recently these problems have been resolved by studies using contemporary techniques such as electrophoretic and immunohistochemical analyses, 13C NMR, and HPLC. Additionally, it was demonstrated that mitochondria in human astrocytoma cells utilize LDH for lactate oxidation (Lemire et al., 2008). Furthermore, immunohistochemical analyses of rat brain cross-sections showed MCT1, MCT2, and LDH to colocalize with the mitochondrial inner membrane marker cytochrome oxidase in neocortical, hippocampal, and thalamic neurons (Hashimoto et al., 2006). Thus, the role of mitochondrial LDH has recently been more clearly identified (Hashimoto et al., 2006; Pagliarini et al., 2008). However, future experiments will be required to analyze possible species differences in the function of LDH isoenzymes in mitochondria fractions.
4.2. Phylogenetic differences of LDH isoenzymes in mitochondrial and synaptosomal fractions
We demonstrated previously the predominance of LDH-B in synaptosomal fractions in the neocortex and striatum across multiple primate species in correlation with increasing brain size (Duka et al., 2014). In the present study, Western blot analysis showed pronounced elevation of LDH-B expression in the mitochondrial fractions from the species with the largest brain sizes, including chimpanzees and humans, as compared to monkeys and strepsirrhines. Moreover, phylogenetic differences of LDH isoenzymes in synaptosomal and mitochondrial fractions were also correlated. A strong correlation between these two fractions can be explained by the fact that mitochondria are a principal component of the synaptosomal fraction and neurons are highly dependent on the energy provided by mitochondrial respiration. Because of their unique function and bioenergetic requirements, neurons have an uneven cellular mitochondria distribution. Areas with high energetic demand, such as synapses, contain correspondingly higher density of mitochondria than the rest of the cell (Hollenbeck, 2005; Ly and Verstreken, 2006).
4.3. Data on the polypeptide composition of LDH
Data on the polypeptide composition of LDH offer additional evolutionary insight into shifts in aerobic status of particular tissues across species (Koen and Goodman, 1969; Goodman et al., 1969; Koen and Goodman, 1971). It was shown previously that LDH-B forms are predominantly associated with haplorhine brains, whereas strepsirrhine brains displayed primarily LDH-A (Goodman et al., 1969). The current study extends these findings, showing an increase in the percentage of aerobic B forms of LDH in mitochondrial fractions of the neocortex and striatum of haplorhines as compared to strepsirrhines.
5. Conclusion
In summary our results show that (1) in cytosolic and mitochondrial subcellular fractions from haplorhines, LDH-B expression level was comparatively elevated and LDH-A declined in two forebrain structures relative to strepsirrhines; (2) mitochondrial LDH-B was increased up to 5-fold in the neocortex of chimpanzees and humans compared to strepsirrhine primates, showing a particularly elevated ratio of LDH-B to LDH-A that might reflect a selective regulation of mitochondrial LDH-B during primate brain evolution; and (3) there was a significant correlation between LDH-B in the synaptosomal and mitochondrial subcellular fractions, suggesting that as brain size enlarged in haplorrhine primates, the neocortex and striatum required a relatively higher rate of aerobic glycolysis that is intimately linked to mitochondrial metabolism.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Grant numbers (BCS-0549117, BCS-0824531, BCS-0827546, DGE-0801634), National Institutes of Health (NS-042867, RR-000165, HHSN268201100065C), and the James S. McDonnell Foundation (220020293).
Abbreviations
- LDH
lactate dehydrogenase
- NWM
New World monkeys
- OWM
Old World monkeys
- MCT
monocarboxylic acid transporter
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mcn.2017.04.007.
References
- Baba N, Sharma HM. Histochemistry of lactic dehydrogenase in heart and pectoralis muscles of rat. J Cell Biol. 1971;51:621–635. doi: 10.1083/jcb.51.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittar PG, Charnay Y, Pellerin L, Bouras C, Magistretti PJ. Selective distribution of lactate dehydrogenase isoenzymes in neurons and astrocytes of human brain. J Cereb Blood Flow Metab. 1996;16:1079–1089. doi: 10.1097/00004647-199611000-00001. [DOI] [PubMed] [Google Scholar]
- Brandt RB, Laux JE, Spainhour SE, Kline ES. Lactate dehydrogenase in rat mitochondria. Arch Biochem Biophys. 1987;259:412–422. doi: 10.1016/0003-9861(87)90507-8. [DOI] [PubMed] [Google Scholar]
- Brooks GA. Cell-cell and intracellular lactate shuttles. J Physiol. 2009;587:5591–6000. doi: 10.1113/jphysiol.2009.178350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duka T, Anderson SM, Collins Z, Raghanti MA, Ely JJ, Hof PR, Wildman DE, Goodman M, Grossman LI, Sherwood CC. Synaptosomal lactate dehydrogenase isoenzyme composition is shifted toward aerobic forms in primate brain evolution. Brain Behav Evol. 2014;83:216–230. doi: 10.1159/000358581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Li JJ, Tan W. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal Chem. 2000;72:3280–3285. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Kaushik DK, Gomes J, Nayeem S, Deep S, Basu A. Changes in cytosolic Ca2+ levels correspond to fluctuations of lactate levels in crosstalk of astrocyte-neuron cell lines. Indian J Exp Biol. 2010;48:529–537. [PubMed] [Google Scholar]
- Goodman M, Syner FN, Stimson CW, Rankin JJ. Phylogenetic changes in the proportions of two kinds of lactate dehydrogenase in primate brain regions. Brain Res. 1969;14:447–459. doi: 10.1016/0006-8993(69)90121-8. [DOI] [PubMed] [Google Scholar]
- Gross J, Ungethüm U, Andreeva N, Heldt J, Priem F, Marschhausen G, Altmann T. Glutamate-induced efflux of protein, neuron-specific enolase and lactate dehydrogenase from a mesencephalic cell culture. Eur J Clin Chem Clin Biochem. 1996;4:305–310. doi: 10.1515/cclm.1996.34.4.305. [DOI] [PubMed] [Google Scholar]
- Guha P, Dey A, Sen R, Chatterjee M, Chattopadhyay S, Bandyopadhyay SK. Intracellular GSH depletion triggered mitochondrial Bax translocation to accomplish resveratrol-induced apoptosis in the U937 cell line. J Pharmacol Exp Ther. 2011;336:206–214. doi: 10.1124/jpet.110.171983. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica. 2001;4:1–9. [Google Scholar]
- Hashimoto T, Hussien R, Brooks GA. Colocalization of MCT1, CD147 and LDH in mitochondrial inner membrane of L6 muscle cells: evidence of a mitochondrial lactate. Am J Physiol Endocrinol Metab. 2006;290:E1237–E1244. doi: 10.1152/ajpendo.00594.2005. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. Mitochondria and neurotransmission: evacuating the synapse. Neuron. 2005;47:331–333. doi: 10.1016/j.neuron.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kline ES, Brandt RB, Laux JE, Spainhour SE, Higgins ES, Rogers KS, Tinsley SB, Waters MG. Localization of l-lactate dehydrogenase in mitochondria. Arch Biochem Biophys. 1986;246:673–680. doi: 10.1016/0003-9861(86)90323-1. [DOI] [PubMed] [Google Scholar]
- Koen AL, Goodman M. Lactate dehydrogenase isozymes: qualitative and quantitative changes during primate evolution. Biochem Genet. 1969;3:457–474. doi: 10.1007/BF00485606. [DOI] [PubMed] [Google Scholar]
- Koen AL, Goodman M. Metabolic immaturity in mental retardation: quantitative relationships of lactate dehydrogenase isozymes. J Ment Defic Res. 1971;15:229–235. doi: 10.1111/j.1365-2788.1971.tb01161.x. [DOI] [PubMed] [Google Scholar]
- Laughton JD, Charnay Y, Belloir B, Pellerin L, Magistretti PJ, Bouras C. Differential messenger RNA distribution of lactate dehydrogenase LDH-1 and LDH-5 isoforms in the rat brain. Neuroscience. 2000;96:619–625. doi: 10.1016/s0306-4522(99)00580-1. [DOI] [PubMed] [Google Scholar]
- Laughton JD, Bittar P, Charnay Y, Pellerin L, Kövari E, Magistretti PJ, Bouras C. Metabolic compartmentalization in the human cortex and hippocampus: evidence for a cell- and region-specific localization of lactate dehydrogenase 5 and pyruvate dehydrogenase. BMC Neurosci. 2007;8:1–35. doi: 10.1186/1471-2202-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire J, Mailloux RJ, Appanna VD. Mitochondrial lactate dehydrogenase is involved in oxidative-energy metabolism in human astrocytoma cells (CCF-STTG1) PLoS One. 2008;3:e1550. doi: 10.1371/journal.pone.0001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly CV, Verstreken P. Mitochondria at the synapse. Neuroscientist. 2006;12:291–299. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ. Neuron-glia metabolic coupling and plasticity. J Exp Biol. 2006;209:2304–2311. doi: 10.1242/jeb.02208. [DOI] [PubMed] [Google Scholar]
- O'Brien J, Kla KM, Hopkins IB, Malecki EA, McKenna MC. Kinetic parameters and lactate dehydrogenase isozyme activities support possible lactate utilization by neurons. Neurochem Res. 2007;32:597–607. doi: 10.1007/s11064-006-9132-9. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L. Lactate as a pivotal element in neuron-glia metabolic cooperation. Neurochem Int. 2003;43:331–338. doi: 10.1016/s0197-0186(03)00020-2. [DOI] [PubMed] [Google Scholar]
- Petit JM, Gyger J, Burlet-Godinot S, Fiumelli H, Martin JL, Magistretti PJ. Genes involved in the astrocyte-neuron lactate shuttle (ANLS) are specifically regulated in cortical astrocytes following sleep deprivation in mice. Sleep. 2013;36:1445–1458. doi: 10.5665/sleep.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JM, Öberg J, Brené S, Coppotelli G, Terzioglu M, Pernold K, Goiny M, Sitnikov R, Kehr J, Trifunovic A, Larsson NG, Hoffer BJ, Olson L. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proc Natl Acad Sci U S A. 2010;107:20087–20092. doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syner FN, Goodman M. Differences in the lactic dehydrogenases of primate brains. Nature. 1966;209:426–428. doi: 10.1038/209426a0. [DOI] [PubMed] [Google Scholar]
- Vdovichenko LM. Properties of lactate dehydrogenase of brain mitochondria. Ukr Biokhim Zh. 1978;50:489–493. [PubMed] [Google Scholar]
- Venkov L, Rosental L, Manolova M. Subcellular distribution of LDH isoenzymes in neuronal- and glial-enriched fractions. Brain Res. 1976;109:323–333. doi: 10.1016/0006-8993(76)90533-3. [DOI] [PubMed] [Google Scholar]
- Yu Z, Xia X, Kone BC. Expression profile of a human inducible nitric oxide synthase promoter reporter in transgenic mice during endotoxemia. Am J Physiol Renal Physiol. 2005;288:214–220. doi: 10.1152/ajprenal.00258.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


