Abstract
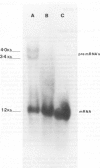
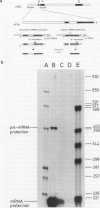
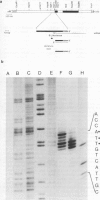
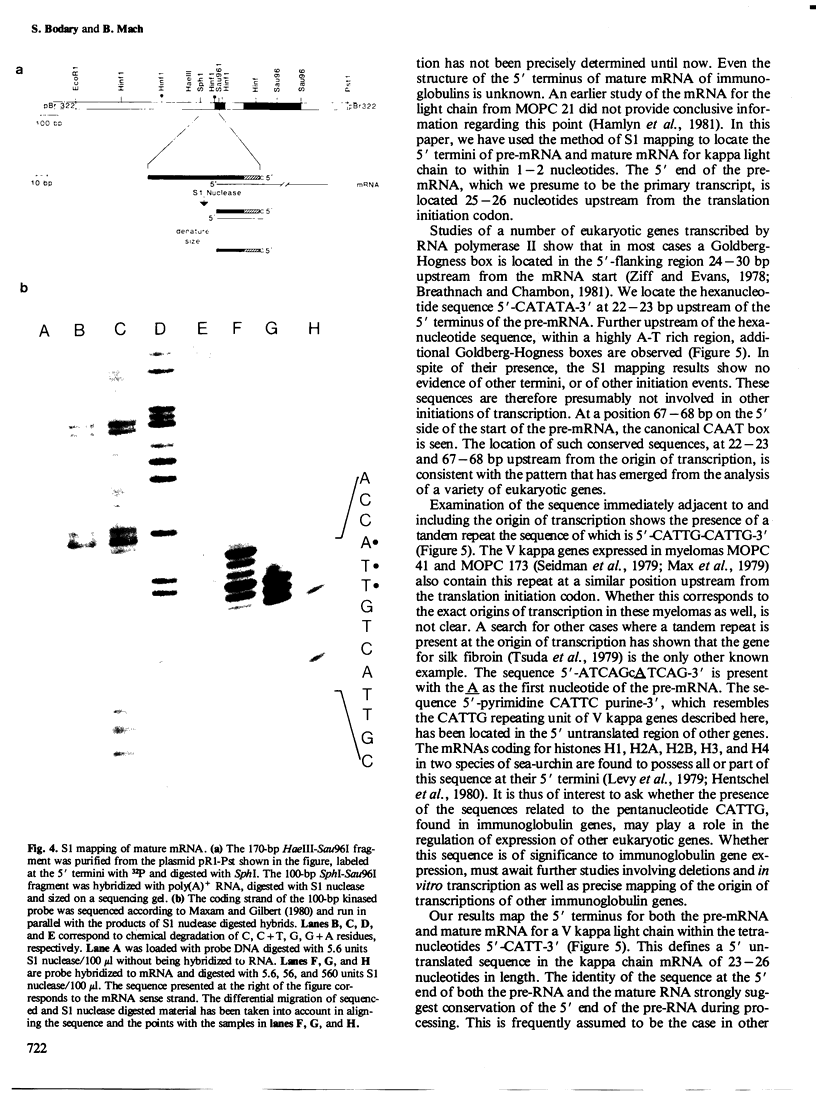
Although the organization, the structure, and the somatic rearrangement of immunoglobulin genes have been studied in detail, there is no information regarding the precise origin of transcription of immunoglobulin genes. We have analyzed the 5'-flanking region of an expressed mouse V kappa light chain gene and the pre-mRNA transcript of that light chain gene, as it is expressed in vivo. Study of the pre-mRNA and of the DNA sequence by the procedure of "S1 mapping" establishes that the 5' end of the pre-mRNA transcript is 25-26 nucleotides upstream from the initiation codon. A CATATA sequence is found 22-23 nucleotides upstream from the origin of transcription. Although several other TATA-like sequences are found further upstream, only a single origin of transcription can be documented. From the S1 protection experiments, the 5' end of the mature mRNA was found to be the same as that of the pre-mRNA, indicating conservation of that sequence during RNA processing. Finally, a conspicuous pentanucleotide repeat CATTG - CATTG has been identified at the position of transcription initiation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenburger W., Steinmetz M., Zachau H. G. Functional and non-functional joining in immunoglobulin light chain genes of a mouse myeloma. Nature. 1980 Oct 16;287(5783):603–607. doi: 10.1038/287603a0. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Baralle F. E., Brownlee G. G. AUG is the only recognisable signal sequence in the 5' non-coding regions of eukaryotic mRNA. Nature. 1978 Jul 6;274(5666):84–87. doi: 10.1038/274084a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burstein Y., Schechter I. Amino acid sequence of the NH2-terminal extra piece segments of the precursors of mouse immunoglobulin lambda1-type and kappa-type light chains. Proc Natl Acad Sci U S A. 1977 Feb;74(2):716–720. doi: 10.1073/pnas.74.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S., Webb E., Gough J., Adams J. M. Recombination events near the immunoglobulin Cmu gene join variable and constant region genes, switch heavy-chain expression, or inactivate the locus. Biochemistry. 1981 Apr 28;20(9):2662–2671. doi: 10.1021/bi00512a047. [DOI] [PubMed] [Google Scholar]
- Gilmore-Herbert M., Hercules K., Komaromy M., Wall R. Variable and constant regions are separated in the 10-kbase transcription unit coding for immunoglobulin kappa light chains. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6044–6048. doi: 10.1073/pnas.75.12.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Koster A., Flavell R. A. A transcription map for the rabbit beta-globin gene. Cell. 1981 Feb;23(2):573–584. doi: 10.1016/0092-8674(81)90153-7. [DOI] [PubMed] [Google Scholar]
- Hamlyn P. H., Gait M. J., Milstein C. Complete sequence of an immunoglobulin mRNA using specific priming and the dideoxynucleotide method of RNA sequencing. Nucleic Acids Res. 1981 Sep 25;9(18):4485–4494. doi: 10.1093/nar/9.18.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. H. Sequence of the 5'-end of Strongylocentrotus purpuratus H2b histone mRNA and its location within histone DNA. Nature. 1979 Jun 21;279(5715):737–739. doi: 10.1038/279737a0. [DOI] [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Miller H., Leder P. Variation in the crossover point of kappa immunoglobulin gene V-J recombination: evidence from a cryptic gene. Cell. 1980 Oct;21(3):793–799. doi: 10.1016/0092-8674(80)90442-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGill J. R., Herbert D. C., Hayashida T. Immunocytochemical identification of rat growth hormone cells utilizing antisera to nonmammalian growth hormones. Gen Comp Endocrinol. 1978 Jul;35(3):342–345. doi: 10.1016/0016-6480(78)90080-1. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Harvey R., Smith A. E. The size of Rous sarcoma virus mRNAs active in cell-free translation. Nature. 1977 Aug 4;268(5619):416–420. doi: 10.1038/268416a0. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Coleclough C., Seidman J. G., Leder P., Tonegawa S., Matthyssens G., Weigert M. Transcription of mouse kappa chain genes: implications for allelic exclusion. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1937–1941. doi: 10.1073/pnas.77.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Schibler U. Comparison of mRNA precursors in plasmacytomas producing closely related kappa chains. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3678–3682. doi: 10.1073/pnas.76.8.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Hüppi K., Heinrich G., Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979 Jul 26;280(5720):288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980 Aug 21;286(5775):779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Max E. E., Leder P. A kappa-immunoglobulin gene is formed by site-specific recombination without further somatic mutation. Nature. 1979 Aug 2;280(5721):370–375. doi: 10.1038/280370a0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Zachau H. G., Mach B. Cloning of immunoglobulin kappa light chain genes from mouse liver and myeloma MOPC 173. Nucleic Acids Res. 1979 Jul 25;6(10):3213–3229. doi: 10.1093/nar/6.10.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S., Hozumi N., Matthyssens G., Schuller R. Somatic changes in the content and context of immunoglobulin genes. Cold Spring Harb Symp Quant Biol. 1977;41(Pt 2):877–889. doi: 10.1101/sqb.1977.041.01.097. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Ohshima Y., Suzuki Y. Assumed initiation site of fibroin gene transcription. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4872–4876. doi: 10.1073/pnas.76.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]