Summary
The gut microbiota is implicated in numerous aspects of health and disease, but dissecting these connections is challenging because genetic tools for gut anaerobes are limited. Inducible promoters are particularly valuable tools, because these platforms allow real-time analysis of the contribution of microbiome gene products to community assembly, host physiology, and disease. We developed a panel of tunable expression platforms for the prominent genus Bacteroides in which gene expression is controlled by a synthetic inducer. In the absence of inducer, promoter activity is fully repressed; addition of inducer rapidly increases gene expression by 4 to 5 orders of magnitude. Because the inducer is absent in mice and their diets, Bacteroides gene expression inside the gut can be modulated by providing the inducer in drinking water. We use this system to measure the dynamic relationship between commensal sialidase activity and liberation of mucosal sialic acid, a receptor and nutrient for pathogens.
Keywords: Microbiome, synthetic biology, Bacteroides, gut, inducible promoter, gene regulation, sialic acid
Introduction
The human gut is populated with enormous bacterial populations that impact many aspects of host physiology. While our ability to describe these communities by high-throughput sequencing has expanded dramatically, parallel tools to manipulate or engineer the microbiota have significantly lagged behind. Such tools are critical for experimental dissection of these poorly understood microbial consortia in the short term, and for future efforts to leverage the microbiota to predict, report on, and ameliorate disease.
Synthetic biology approaches provide an attractive means to address these obstacles, but genetic parts developed and validated in model organisms such as Escherichia coli are typically nonfunctional in Bacteroidetes or Firmicutes, which together account for ~90% of the microbiota in most individuals (The Human Microbiome Project Consortium, 2012). Members of the most abundant and stable genus in healthy U.S. humans, the Bacteroides (Faith et al., 2013; The Human Microbiome Project Consortium, 2012), are typical in this regard. These Gram-negative, obligate anaerobes are associated with a range of host phenotypes in mouse models, from modulating normal physiological processes (mucus production, villus development, immune maturation) to shaping interactions with invading pathogens and even behavior (Hsiao et al., 2013; Ng et al., 2013; Wexler, 2007). However, several features of Bacteroides preclude the use of standard synthetic biology tools. First, the RNA polymerase (RNAP) σ70 holoenzyme recognizes a unique -33/-7 consensus sequence, TTTG (N19-21)TANNTTTG, in promoter DNA (Bayley et al., 2000; Mastropaolo et al., 2009). Second, standard reporters of gene expression such as green fluorescent protein or β-galactosides do not work well due to oxygen requirement or high background from native enzymes. Third, ribosome binding site (RBS) strength is correlated with a complex set of features and not with the presence of a canonical Shine-Dalgarno sequence (Wegmann et al., 2013).
While many genetic parts will likely contribute to understanding the interactions between Bacteroides and their host, highly inducible yet tightly controlled gene expression systems are arguably among the most valuable. These regulated platforms provide several features absent in constitutive expression systems. First, gene expression can be modulated with time in the same strain or experiment, permitting kinetic studies of the bacterial or host response to production, depletion, or repeated exposures of a gene product. Second, inducible systems allow mechanistic study of essential or toxic gene products. Third, precise modulation of gene expression is the foundation for many more complex devices in synthetic biology. Indeed, an E. coli inducible promoter, standardized RBS, and terminator are the three most frequently used items in the 20,000-component Registry of Standard Biological Parts (Endy, 2005).
Native Bacteroides promoters that respond to plant-, fungal-, or mammal-derived polysaccharides or sugars can provide approximately 102-fold range of activity and have been employed for regulated gene expression in B. thetaiotaomicron, B. fragilis, and B. ovatus (Hamady et al., 2008; Horn et al., 2016; Mimee et al., 2015; Parker and Smith, 2012). However, even in the absence of inducer, these promoters are significantly activated in mouse models, possibly because of background from the host or its diet (Horn et al., 2016; Mimee et al., 2015). Further, plant-, fungal- and mammal-derived polysaccharides are known to be actively consumed by the microbiota and have a large impact on microbiome composition and gene expression (Sonnenburg et al., 2005). Lastly, the performance of these native promoters in heterologous species is not known.
Tetracycline-regulated systems have become a useful tool for analyzing gene function. These systems utilize a constitutively expressed repressor, TetR, which binds the tetO operator sequence in the promoter, preventing RNAP from binding DNA and initiating transcription. TetR has a high affinity (~10−11 M) for the tetracycline analog anhydrotetracycline (aTC), and aTC binding to TetR decreases its affinity for tetO by 6–10 orders of magnitude (Lederer et al., 1995; Orth et al., 2000).
Here, we describe a panel of tunable gene expression platforms for diverse human gut Bacteroides. In the OFF state, these systems recapitulate the phenotypes of mutants that lack the target gene entirely. Upon aTC induction, these engineered promoters can individually modulate gene expression up to 9,000-fold, spanning 5 orders of magnitude across the panel. By varying the aTC concentrations provided to mice carrying responsive strains, commensal gene expression inside the gut can be modulated across 3 orders of magnitude and can be fully repressed in vivo. We use this system to measure the dynamic relationship between commensal sialidase expression and liberation of mucosal sialic acid, which serves as a receptor and nutrient for numerous viral and bacterial pathogens. These engineered systems function across diverse human gut Bacteroides and provide general design principles that are applicable across the microbiome.
Results
Design rationale
Highly active, broadly conserved Bacteroides promoters provide an ideal template for a regulated gene expression platform for these organisms. B. thetaiotaomicron RNA-seq experiments indicate that 16S rDNA promoter activity can produce 20–30% of total cellular RNA, consistent with other bacteria (Dennis and Bremer, 2008; Rey et al., 2010). Because the P1 promoter sequence upstream of the B. thetaiotaomicron 16S rDNA gene BT_r09 is known to function in other Bacteroides (Wegmann et al., 2013), this sequence provides an ideal template for a regulated gene expression platform in this genus.
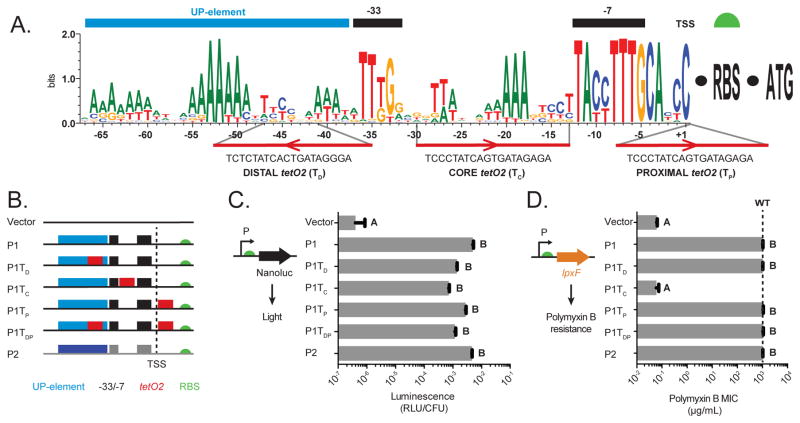
Intergenic regions upstream of Bacteroides 16S rDNAs generally contain two RNAP binding regions capable of initiating transcription, designated P1 (located farthest upstream of the 16S rDNA coding region) and P2 (Mastropaolo et al., 2009). Frequency logos generated from 182 16S rDNA promoters representing 74 different 16S rDNA loci across 19 genome-sequenced Bacteroides species reveal several highly conserved sequence elements (Figure 1A, Table S1). Consistent with other constitutive Bacteroides promoters, the 16S rDNA promoters encode strongly conserved -33 (TTTG) and -7 (TAnnTTTG) RNAP binding sites (Bayley et al., 2000; Mastropaolo et al., 2009). Upstream of the -33 element, repeated A-rich tracts are spaced 12–13 base pairs apart, consistent with these conserved sequences representing an UP-element which in E. coli stimulates transcription initiation (Gourse et al., 2000). Finally, conserved A/T-rich regions between the -33 and -7 elements may play an important role in RNAP binding and/or transcription activation. This analysis suggests permissive and restricted regions for manipulation of the promoter sequence.
Figure 1. Genetic architecture and construction of regulatable expression platforms in Bacteroides.
(A) Sequence conservation across 182 RNAP binding sites from 16S rRNA promoter regions from 19 Bacteroides genomes. Areas of conservation reflecting -33 and -7 RNAP binding sites (black bars) and the transcription-activating UP-element (blue bar) are indicated above the sequence logo. TetO2 sequences were placed and oriented as shown below the sequence logo. (B) Schematic of the P1 and P2 constitutive promoters and engineered P1 promoters containing tetO2 elements. The predicted UP-element, -33/-7 sites, and tetO2 operator sequences are designated as blue, black, and red boxes, respectively. The dashed line designates the transcription start site (+1), and the green semicircle indicates the GH023 RBS. (C) Activity of native P1, native P2, and tetO2-containing P1 promoters in B. thetaiotaomicron measured using the NanoLuc luciferase reporter and expressed as relative light units/colony forming unit (RLU/CFU). Luminescence from B. thetaiotaomicron carrying NanoLuc with no promoter is marked as “Vector”. (D) Polymyxin B (PMB) minimal inhibitory concentrations (MIC) for B. thetaiotaomicron lpxF strains expressing lpxF from each promoter. The dashed line indicates the MIC for PMB in wildtype B. thetaiotaomicron. In C and D, values represent three biological replicates performed on separate days; letters indicate significantly different groups (p < 1x10−6).
Engineered tetO2-containing promoters maintain high levels of gene expression
To transform the constitutive BT_r09 P1 promoter into an inducible system, we searched for control elements that meet three key criteria. Most importantly, the inducer should not be present in undefined growth medium used to culture Bacteroides and other human gut anaerobes, nor in the gut or tissue of mice and other mammals, and not in standard animal diets (i.e., not a poly- or monosaccharide or analogue). Second, the inducer should not be toxic to Bacteroides at concentrations that permit maximal promoter activation nor be consumed as a carbon source or other nutrient. Third, the regulatory elements should be capable of repressing strong promoters, allowing deletion mutant phenotypes to be recapitulated when regulated gene(s) are repressed and permitting a wide range of expression levels in response to varying inducer concentration. The aTC-regulated control elements from the E. coli Tn10 transposon fit these three criteria (Bertram and Hillen, 2008). Bacteroides grow at wildtype rates in aTC concentrations typically used to control gene expression from tetracycline-regulated promoters (Figure S1A) (Lutz and Bujard, 1997).
We first created several modified P1 promoters containing the tetO2 operator in each of three locations: upstream of the -33 (distal; P1TD), within the spacer region between the -33 and -7 (core; P1TC), or downstream of the transcription start site (proximal; P1TP) (Figure 1B and Table S2). In each location, tetO2 sequences were engineered as sequence insertions, partial substitutions, or complete substitutions in the orientation that best preserves the consensus promoter sequence (Figure 1A). We also created an additional promoter containing the tetO2 operator in both the distal and proximal regions (P1TDP).
To first investigate the positional effects of the tetO2 operators on basal gene expression in the absence of the TetR repressor, we fused the native P1 and P2 promoters, and each synthetic promoter, to a standard Bacteroides RBS (GH023) (Wegmann et al., 2013), followed by the NanoLuc luciferase gene as a reporter (Hall et al., 2012). Integration of these constructs into the B. thetaiotaomicron genome in single copy at a standard location revealed luminescence approximately four orders of magnitude higher than a control strain that lacks a promoter upstream of NanoLuc (Figure 1C). Placement of tetO2 in the distal, core, and/or proximal regions of P1 decreased promoter activity 1.8- to 6.8-fold (Figure 1C). Together, these results suggest that tetO2 insertion affects promoter activity, but reporter activity remains 4,200-fold to 16,000-fold above background, depending on operator placement.
To test the function of native Bacteroides genes expressed under the control of these engineered promoters, we replaced the NanoLuc reporter with BT1854 (lpxF). LpxF is a phospholipid phosphatase that determines resistance to cationic antimicrobial peptides (including polymyxin B; PMB) in Bacteroides, and deletion of lpxF in B. thetaiotaomicron reduces PMB resistance by four orders of magnitude (Cullen et al., 2015). Expression of lpxF under the control of the native P1, native P2, P1TD, P1TP, and P1TDP promoters in a B. thetaiotaomicron lpxF mutant increases PMB resistance to wildtype levels, while the P1TC-lpxF fusion does not (Figure 1D).
A synthetic inducer modulates gene expression over a broad dynamic range
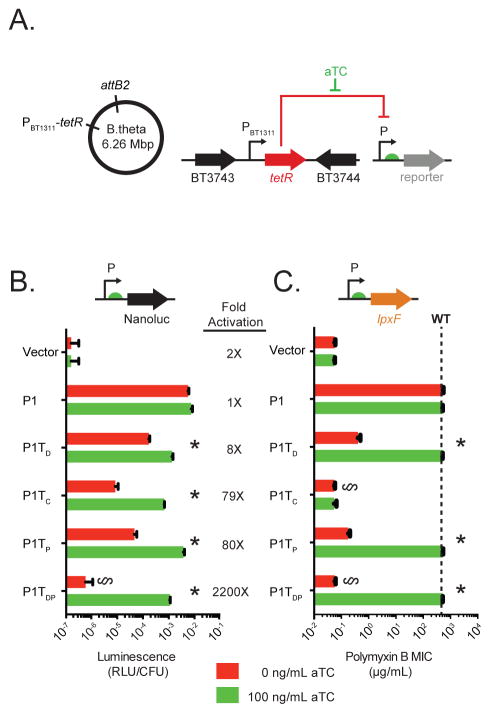
Next, we integrated a constitutive tetR cassette in a neutral (not required for fitness) location in the B. thetaiotaomicron genome to create Bt::tetR (Figure 2A, see Method Details). Introduction of the panel of promoter-NanoLuc fusions (Figure 1B) into Bt::tetR revealed that while TetR has no effect on the activity of the native P1 promoter, all of the tetO2-containing promoters exhibit a significant decrease in activity in the presence of TetR (Figure 2B, red bars). Of these promoters, P1TDP shows the greatest repression by TetR, with reporter expression 6,800-fold above background in the absence of TetR (Figure 1C) and decreasing to within 4-fold of background levels when TetR is present (Figure 2B, red bars).
Figure 2. Regulation of Bacteroides gene expression via a synthetic inducer.
(A) Strain design. (B) Activity of engineered, tetO2-containing promoters measured by luminescence (RLU/CFU) of NanoLuc-promoter fusions. (C) Addition of aTC restores lpxF-dependent PMB resistance to Bt::tetR lpxF carrying lpxF-promoter fusions. The dashed line indicates the PMB MIC for wildtype B. thetaiotaomicron (> 1024 μg/mL). In B and C, asterisks indicate significant differences in gene expression in response to aTC (p < 0.0001). § indicates no significant difference as compared to vector control (p > 0.1).
Addition of aTC (100 ng/mL) to strains constitutively expressing TetR derepresses each of the tetO2-containing promoters, producing reporter levels equivalent to those measured in strains lacking tetR (Figure 2B, green bars versus Figure 1C). This indicates that aTC readily reaches the Bacteroides cytoplasm. While the promoters with a single tetO2 exhibit at most an 80-fold induction of reporter activity in the presence of aTC, promoter P1TDP increases luminescence over 2,000-fold in response to the inducer (Figure 2B). Consistent with this result, introduction of the panel of promoter-lpxF fusions into an lpxF mutant constitutively expressing TetR (Bt::tetR lpxF) revealed that TetR represses expression of the P1TDP-lpxF fusion to levels that reduce PMB MIC by over 10,000-fold, functionally equivalent to the absence of lpxF entirely (<0.064 μg/mL; Figure 2C). In strains carrying the other promoters, TetR expression also reduces PMB MIC, although some resistance remains (Figure 2C). Except for P1TC, addition of 100 ng/mL aTC restores PMB MIC to wildtype levels in each case (Figure 2C). Additional examination of the sensitivity and temporal dynamics of the system showed that P1TDP expression can be tuned over a 100-fold range in aTC concentration and induces in response to aTC within one round of cell division (Figure S1B and S1C). Together, these results highlight the utility of the P1TDP platform for stringent regulation of both exogenous and endogenous genes in B. thetaiotaomicron.
Acquiring regulatory control of endogenous loci
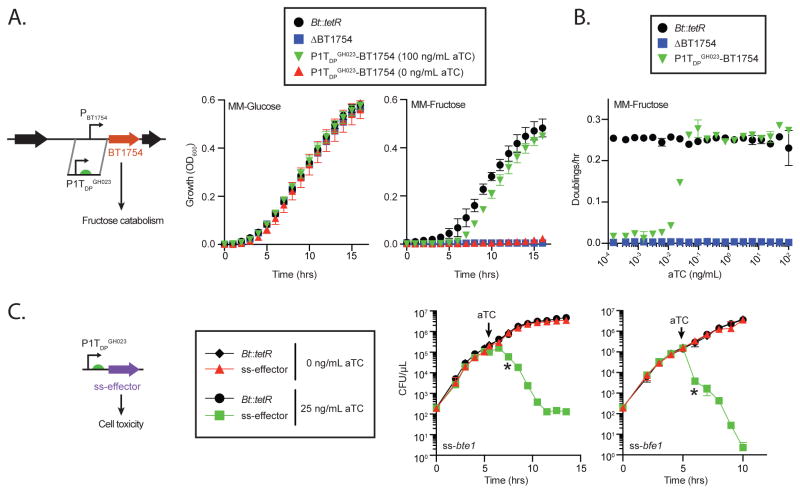
Regulatory control of a gene or locus can be acquired by deleting gene(s) of interest and then returning the gene(s) under the control of an engineered promoter as shown for lpxF above. However, direct replacement of the native promoter provides an important one-step alternative, especially if there are complications in creating a gene deletion (e.g. essential genes; deletions that impact the regulation of neighboring genes) and/or complementation (e.g. long, multiple, or undefined operons; repeating or low-complexity sequences recalcitrant to amplification). The hybrid two-component sensor BT1754 regulates a multi-gene polysaccharide utilization locus required for growth on fructose. We replaced the 200 base pairs upstream of BT1754 with the 200 base pair P1TDP promoter (Figure 3A) in Bt::tetR (designated Bt::tetR P1TDPGH023-BT1754). Growth of Bt::tetR P1TDPGH023-BT1754 is entirely dependent on the addition of aTC in fructose but not glucose (Figure 3A). Additionally, cell doubling rates in fructose can be readily tuned by varying aTC concentrations (Figure 3B). Together, these studies establish that the P1TDP system can be used to assume control of endogenous loci in Bacteroides.
Figure 3. Attaining regulatory control of endogenous loci and highly toxic gene products.
(A and B) Promoter design and aTC-dependent growth of Bt::tetR P1TDPGH023-BT1754 in fructose and glucose. Error bars (for A and B) represent the standard deviation of three biological replicates on separate days. (C) Bt::tetR encoding either of two highly toxic antibacterial effectors from B. fragilis under the control of P1TDPGH023 grow at wildtype rates unless expression is induced with aTC. Error bars represent the standard deviation of two biological replicates on separate days; asterisks indicate the earliest timepoint with significant (p < 0.001) differences compared to uninduced controls.
Gene expression is tightly repressed in the absence of inducer
Tight control in the OFF state is a critical goal for engineered gene regulatory platforms. In theory, genes that encode highly deleterious proteins should be nonlethal under repressing conditions if tight control is achieved. The Type VI secretion system (T6SS)-delivered antibacterial effectors Bte1 and Bfe1, encoded by B. fragilis strains NCTC 9343 and 638R, respectively, allow these strains to efficiently kill B. thetaiotaomicron upon transient contact (Chatzidaki-Livanis et al., 2016; Wexler et al., 2016). We constructed translational fusions of each effector with an N-terminal periplasmic localization signal sequence (“ss”) and placed these fusions under the control of P1TDP in Bt::tetR (Figure 3C). Expression of ss-Bte1 or ss-Bfe1 in B. thetaiotaomicron is highly toxic, because addition of aTC results in immediate decreases in viability and culture density (Figure 3C and Figure S1D). Toxicity resulting from aTC-induced effector expression is likely a consequence of their underlying mechanism of action and not from overproduction of a heterologous protein, as expression of either effector without a periplasmic signal sequence does not affect B. thetaiotaomicron viability (data not shown). In the OFF state (without aTC), growth of strains carrying P1TDP-ss-bte1, P1TDP-ss-bfe1, and the Bt::tetR parental strain is equivalent (Figure 3C and Figure S1D). Together, these studies establish that the P1TDP platform provides strict repression of gene expression in the OFF state.
An RBS panel extends the dynamic range of the Bacteroides inducible promoter to capture promoter activity levels observed across the B. thetaiotaomicron transcriptome
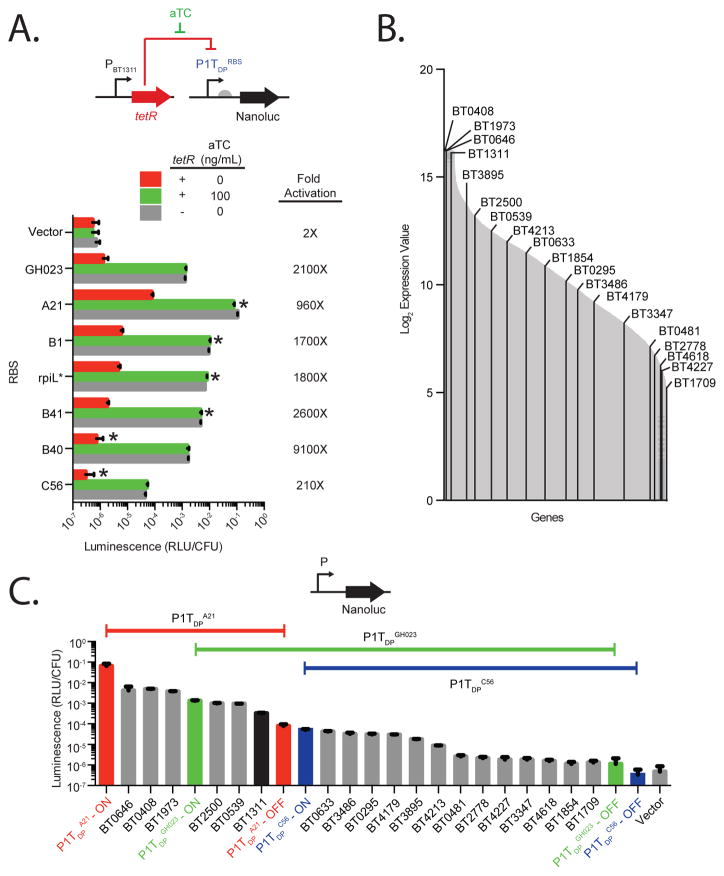
Ribosome binding site (RBS) variation provides an additional layer of gene expression control to engineered promoters. We replaced the original RBS (GH023) in P1TDP with 6 alternate RBSs (designated P1TDPRBS) (Mimee et al., 2015). The 6 RBSs modulate P1TDP promoter expression by three orders of magnitude, in a pattern generally similar to that previously reported for a native B. thetaiotaomicron promoter (Figure 4A and Figure S2A–B) (Mimee et al., 2015). RBS variation impacts expression in the induced and uninduced state proportionally (Figure S2A-B) and can extend the dynamic response to aTC: for example, P1TDPB40 provides negligible expression in the uninduced state (1.4-fold over background) which increases 9,100-fold when aTC is provided, a 3.3-fold increase over P1TDPGH023. Luminescence production from P1TDP-NanoLuc carrying each RBS is comparable between wildtype B. thetaiotaomicron and Bt::tetR in the ON state, suggesting that varying the RBS does not disrupt promoter activation and that TetR repression is fully relieved in all RBS variants with the addition of aTC (Figure 4A; green bar vs. grey bar). Together, this panel of RBS variants coupled to the P1TDP promoter further extends the range of expression to 320,000-fold (e.g., between P1TDPC56 in the OFF state and P1TDPA21 in the ON state).
Figure 4. A panel of ribosome binding sites extends the dynamic range of P1TDP to over 105-fold, spanning the complete range of native gene expression in B. thetaiotaomicron.
(A) Promoter activity of the P1TDP promoter fused to various RBSs (Mimee et al., 2015) was measured using the NanoLuc reporter in wildtype B. thetaiotaomicron (grey) and Bt::tetR in the absence (red) or presence (green) of aTC. Luminescence from B. thetaiotaomicron carrying NanoLuc with no promoter is marked as “Vector”. Error bars represent the standard deviation of three biological replicates on separate days. Asterisks above red bars indicate significant (p < 0.02) reduction in activity compared to P1TDPGH023 in the OFF state; asterisks above green bars indicate significant (p < 1x10−5) increase in activity compared to P1TDPGH023 in the ON state. (B) B. thetaiotaomicron gene expression levels as previously measured by genome-wide transcriptional profiling (Sonnenburg et al., 2005) are marked with grey lines. Promoters tested were selected from the designated genes labeled as black lines. (C) Luminescence from NanoLuc fusions to three engineered promoters can be modulated by aTC to span the entire expression range of the 18 native promoters selected to span the B. thetaiotaomicron transcriptome.
To compare this range of expression to native promoter strengths in B. thetaiotaomicron, we fused the NanoLuc reporter to the promoters of 18 B. thetaiotaomicron ORFs whose mRNA levels span the complete expression range of this organism in rich medium based on transcriptome studies (Sonnenburg et al., 2005) (Figure 4B). Luminescence production from these 18 promoter fusions is largely proportional to the previously reported mRNA levels of the corresponding ORFs (Figure S2C). The P1TDP platform fully spans the range of native promoter activities, producing luminescence levels ranging from 14-fold higher than the strongest native promoter tested (P1TDPA21, ON state) to 6.7-fold lower than the weakest native promoter tested (P1TDPC56, OFF state) (Figure 4C). Notably, the expression range of P1TDPGH023 alone captures ~86% of the 18 native B. thetaiotaomicron promoters tested, which corresponds to ~97% of the transcriptome (Figure 4C). These results establish that the P1TDP platform provides the capacity to express Bacteroides proteins to their native levels.
Evaluation and performance of the P1TDP platform in different Bacteroides species
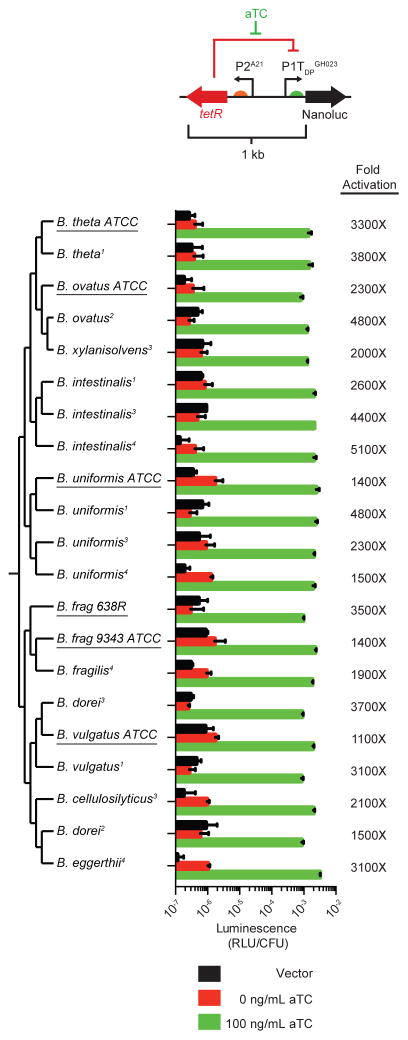
To facilitate gene expression control across species, we built a self-contained, ~1kb inducible expression cassette (designated TetR-P1TDP) in which tetR is expressed from the P2A21 promoter and oriented in the opposite direction from P1TDPGH023, thereby providing the ability to artificially regulate gene expression in unmodified strains of multiple species in a single step (Figure 5). TetR-P1TDPGH023-NanoLuc reporter fusions displayed robust induction over 3 orders of magnitude in an aTC-dependent manner in 6 different Bacteroides type strains representing 5 species (Figure 5, underlined). We also introduced the cassette into 15 additional Bacteroides isolates (representing 11 different species) cultured directly from 4 unrelated human donors. These isolates encode diverse 16S rRNA gene sequences and exhibit doubling times ranging from 38 to 137 minutes in rich medium, consistent with a broad range of phylogeny and physiology (Figure 5). The TetR-P1TDPGH023-NanoLuc cassette could be readily introduced into all isolates, and each exhibited robust induction of luminescence in an aTC-dependent manner (Figure 5).
Figure 5. A self-contained, inducible expression cassette functions across diverse Bacteroides species.
Strains are sorted by 16S rDNA phylogeny, with type strains (Table S3) underlined and novel isolates (isolated directly from human donors) noted with superscript indicating donor number. Activity of TetR-P1TDPGH023-NanoLuc in each strain is shown. Error bars represent the standard deviation of three biological replicates on separate days.
To further test the cross-species performance of TetR-P1TDP, we used this platform to acquire regulatory control of an endogenous gene in wildtype B. ovatus. To this end, we identified the lpxF homolog in B. ovatus (BACOVA_04598) and replaced the 62 base pairs upstream of this gene with the TetR-P1TDPB1 cassette (Figure S3A). With this one-step promoter replacement, PMB resistance becomes entirely dependent on aTC (Figure S3B). Together, these results indicate that the key requirements of the P1TDP regulatory system – comprising the P1 and P2 promoters, tetO2 operator sequences, TetR repressor, and responsiveness to aTC – are widely functional across this genus, in both type strains and novel isolates.
Control of B. thetaiotaomicron gene expression in the mouse gut
A central design element of the P1TDP platform is the use of a synthetic inducer that should be absent from mammalian intestinal tissue and diet. To test this system in vivo, we first introduced aTC at a concentration that does not adversely affect mouse health (100 μg/mL) (Kotula et al., 2014) for 4 days into the drinking water of gnotobiotic mice carrying wildtype B. thetaiotaomicron. Liquid chromatography-mass spectrometry (LC-MS)-based quantification of aTC in fecal pellets reveals that the inducer is undetectable prior to exogenous addition, is readily detected within 24 hours of its addition to drinking water, and is again undetectable 6 days after its removal from the water, exhibiting a half-life of 19 hours in vivo (Figure S4). Germfree mice given the same regime of aTC administration rapidly show comparable aTC levels in fecal pellets (data not shown), indicating that B. thetaiotaomicron does not degrade aTC in vivo.
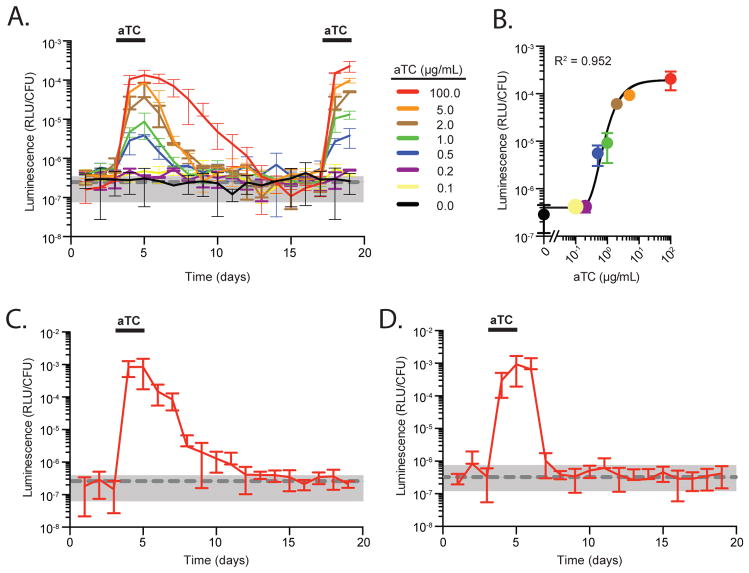
To directly test the function of the P1TDP platform in the context of the mammalian gut, we colonized groups of germfree mice with Bt::tetR P1TDPGH023-NanoLuc. On days 3–5 and 17–19, aTC was added to the drinking water and promoter activity quantified by measuring luminescence in fecal pellets over time and along the length of the gut at day 19. Within 24 hours of aTC addition (the first timepoint after induction), luminescence in fecal pellets increased ~1,000-fold (Figure 6A, red line). When aTC was removed, luminescence returned to levels observed in fecal pellets of mice colonized with wildtype B. thetaiotaomicron (Figure 6A, grey dashed line and shading), indicating that the promoter returns to a tight OFF state upon inducer removal in vivo (NanoLuc half-life of 3–4 days; complete repression 9 days after aTC removal). The longer half-life of luminescence versus aTC is likely due to the high stability of the engineered Nanoluc protein (Hall et al., 2012). By contrast, mice colonized with Bt::tetR P1TDPGH023-NanoLuc and maintained for the same time period without aTC exhibit no induction of luminescence (Figure 6A, black line). This engineered system and aTC exposure do not impact B. thetaiotaomicron stability in the gut (Figure S5A), and the kinetics and magnitude of induction are identical each time aTC is provided and are highly reproducible across mice (Figure 6A). Administration of lower aTC concentrations to mice carrying the reporter strain resulted in the induction of intermediate luminescence measurements within 24 hours, with detectable responses starting at 0.2–0.5 μg/mL aTC (Figure 6A). The response of P1TDP to aTC inside the gut can be accurately described using a sigmoidal function (R2 = 0.952) (Figure 6B), suggesting that 100 μg/mL aTC provides maximal gene induction in vivo. Modulation of gene expression by varying aTC concentrations is recapitulated in the distal small intestine, cecum and throughout the large intestine (Figure S5B and S5C).
Figure 6. Exogenous control of Bacteroides gene expression in mice via a synthetic inducer.
(A) Groups of germfree mice were colonized with Bt::tetR carrying the P1TDPGH023-NanoLuc, and promoter activity measured in feces over time. Mice were provided aTC as indicated. The grey dashed line and shading represent the average and standard deviation of fecal luminescence measured over time from mice colonized with wildtype B. thetaiotaomicron under the same regime of aTC exposure (n = 6 mice for groups treated with 100 μg/mL aTC; n = 2 mice for groups administered lower aTC concentrations). (B) Dose response of the P1TDPGH023 promoter to varying aTC concentrations in mice. (C) Luminescence production from Bt::tetR P1TDPGH023-NanoLuc in mice co-colonized with 13 other prominent human gut microbes (Figure S5D). Mice (n = 6) were given aTC as indicated. The grey dashed line and shading represents the average and standard deviation of fecal luminescence measured over time from mice colonized with wildtype B. thetaiotaomicron from Figure 6A. (D) Inducer-dependent fecal luminescence production by Bt::tetR P1TDPGH023-NanoLuc in specific pathogen-free Rag−/− mice carrying a complete microbiota. The grey dashed line and shading represents the average and standard deviation of fecal luminescence on day −1; n = 7 mice.
To test the function of P1TDP platform in the context of a complex microbiota, we first colonized germ-free mice with a community consisting of Bt::tetR P1TDPGH023-NanoLuc and 13 additional microbial species representative of the 3 dominant phyla found in the human gut. In this community context, Bt::tetR P1TDPGH023-NanoLuc represented ~11% of the total community (~3x1011 CFU/mL) based on strain- and species-specific qPCR analysis on DNA isolated from fecal pellets and selective culturing (Figure S5A and S5D). Prior to aTC addition (days 1–3), fecal pellets exhibited no induction of luminescence (compared to luminescence measurements from mice monocolonized with wildtype B. thetaiotaomicron; Figure 6C, grey dashed line and shading), indicating that this human community does not produce compounds that activate P1TDP. Within 24 hours of administering 100 μg/mL aTC (day 4), luminescence in fecal pellets increased ~4,500-fold, and was stable throughout aTC administration (days 4–5). Fecal luminescence returned to baseline levels after removing the inducer, with complete repression 7–9 days after removal. Alpha and beta diversity analyses of these communities over the 19-day experiment indicate that the presence of aTC did not impact community structure (Figure S5D-S5F). Additionally, CFU measurements of Bt::tetR P1TDPGH023-NanoLuc were unchanged throughout the experiment (Figure S5A), indicating that increases in luminescence are due to in vivo gene modulation in the target organism, not changes in the relative or absolute abundance of this species in the community.
We further interrogated the performance of the P1TDP platform within a complete microbiota. Like most human gut Bacteroides, B. thetaiotaomicron is rapidly outcompeted in conventional, specific pathogen-free (SPF) wildtype mice that harbor a native murine microbiota. However, this species stably colonizes conventional, SPF Rag−/− animals at ~0.2–0.5% of the total community, similar to its natural abundance in humans (Cullen et al., 2015; Lee et al., 2014). We gavaged conventional Rag−/− mice with Bt::tetR P1TDPGH023-NanoLuc and administered aTC for 48 hours beginning 3 days after gavage. Luminescence in fecal pellets prior to aTC administration (days 1–3) was similar to that observed in fecal pellets before Bt::tetR P1TDPGH023-NanoLuc was introduced, demonstrating tight repression in the absence of aTC (Figure 6D, grey dashed line and shading). Addition of aTC to the drinking water resulted in a ~4,800-fold increase in fecal luminescence which was sustained for the duration of aTC administration and returned to baseline levels 3 days after aTC removal, suggesting that a complete murine microbiota does not produce any inducing molecules, degrade aTC, nor inhibit the ability of the P1TDP platform to activate and repress optimally. Further, aTC does not affect the stability of B. thetaiotaomicron in a complete murine microbiota (Figure S5A). These results demonstrate that the P1TDP platform enables predictable tuning of gene expression within the microbiota in vivo.
Inducible expression platforms reveal new dynamics of host-microbiome interactions
To demonstrate the utility of this system, we focused on sialic acid (N-acetylneuraminic acid), a sugar liberated from the gut mucosa by sialidases expressed by certain commensal bacteria lacking the necessary enzymes for sialic acid catabolism. Free sialic acid is a nutrient source for the antibiotic-associated enteric pathogens Clostridium difficile and Salmonella typhimurium, and can determine pathogen burden in mouse models (McDonald et al., 2016; Ng et al., 2013). Mucosal glycoproteins carrying sialic acid also serve as viral receptors (Wasik et al., 2016). Despite this broad importance, how commensal sialidase activity determines sialic acid levels in the gut lumen is unexplored. While a healthy microbiota has been reported to produce low levels of free sialic acid, at least one antibiotic formulation alters the community to increase luminal sialic acid concentrations, creating a pathogen-sensitive state (Ng et al., 2013). It is unknown whether antibiotic-induced sialic acid release reflects a specific reconfiguration of the microbiota dependent on the initial community composition and antibiotics used, or instead is a result of fundamental properties of this microbiome-host interaction.
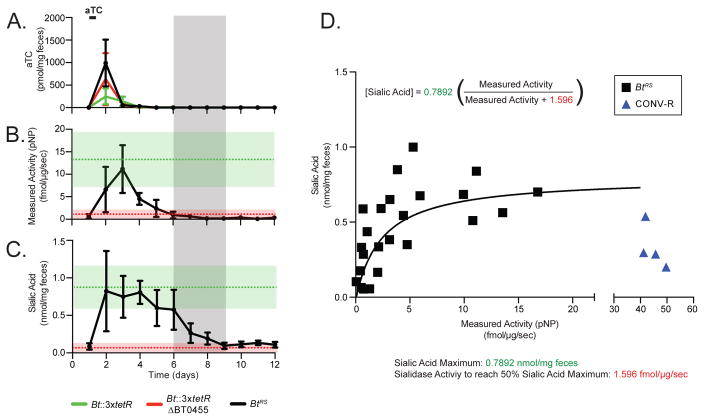
As previously reported (Ng et al., 2013), B. thetaiotaomicron efficiently liberates sialic acid from the gut mucosa of monoassociated gnotobiotic mice; this activity is dependent on the B. thetaiotaomicron sialidase BT0455 (Figure S6A). To understand how commensal sialidase activity modifies the gut environment, we placed BT0455 under the control of the P1TDPGH023 promoter (Bt::3xtetR BT0455 P1TDPGH023-BT0455, abbreviated BtRS for regulated sialidase; see Method Details and Figure S6B). We colonized germfree mice with BtRS (or Bt::3xtetR or Bt::3xtetR BT0455 controls) and collected fecal samples daily before and after 12 hours of aTC induction via drinking water. Quantification of aTC in fecal pellets by LC-MS revealed that inducer levels spike within 12 hours of administration and become undetectable 24 hours later (Figure 7A).
Figure 7. Modulating commensal sialidase expression in mice reveals that sialic acid persists in the gut after microbial enzyme activity is repressed and uncovers a non-linear relationship between enzyme activity and luminal sialic acid.
(A) aTC concentrations, (B) sialidase activity, and (C) free sialic acid levels in fecal samples collected over time from gnotobiotic mice monocolonized with Bt::3xtetR (green line), Bt::3xtetR BT0455 (red line), or BtRS (black line). Mice were given aTC as indicated (n = 6 mice per group in panels A and C; n = 4 per group in panel B). In B and C, green and red dashed lines and shadings represent the average and standard deviation of sialidase activity (from the first 4 days; shown in Figure S6D) and free sialic acid (from the full experiment; shown in Figure S6E) in mice carrying Bt::3xtetR and Bt::3xtetR BT0455, respectively. The grey shading represents the time window when sialidase activity is no longer detected in mice carrying BtRS yet free sialic acid remains. (D) Sialic acid levels are positively correlated with sialidase activity at low levels of enzyme activity but remain constant at higher levels of enzyme activity, suggesting that the reaction is substrate-limited. A best fit line based on Michaelis-Menten kinetics is shown in black. Conventional mice are shown in blue (n = 4 mice).
To measure sialidase enzyme activity, cell-free lysates from fecal samples were incubated with the artificial substrate 2-O-(p-Nitrophenyl)-tropheacetylneuraminic acid (pNP-SA), which is hydrolyzed by sialidases, and the subsequent release of the pNP moiety was monitored over time by LC-MS. pNP-SA was provided in 10-fold molar excess to its Km (Km = 0.11 mM) (Park et al., 2013) in order to ensure direct proportionality between the calculated initial reaction velocity and the sialidase concentration in each sample (Figure S6C). Prior to aTC administration, fecal samples from mice carrying BtRS show no detectable sialidase activity, indicating tight repression of the enzyme and the absence of pNP in the gut (Figure 7B and Figure S6E). After induction, sialidase activity of BtRS-associated mice became equivalent to the control strain expressing BT0455 from its native promoter, suggesting that the P1TDP platform expressed the gene to wildtype levels (Figure 7B). Sialidase activity was again undetectable 4 days after aTC removal (Figure 7B). By contrast, fecal samples from control mice monocolonized with wildtype or BT0455 B. thetaiotaomicron strains exhibit constitutive or no sialidase activity, respectively (Figure S6D).
We next used LC-MS/MS to quantify free sialic acid liberated from the host mucosa in the same fecal pellets. Consistent with the ex vivo sialidase activity results, prior to aTC treatment, mice colonized with BtRS had low levels of fecal sialic acid, equivalent to germfree animals or those colonized with the BT0455 deletion mutant (Figure 7C, S6A, and S6E). After aTC was provided in drinking water, fecal sialic acid levels in BtRS mice were equivalent to mice colonized with wildtype B. thetaiotaomicron (Figure 7C, S6A, and S6E). The inducer was not directly responsible for sialic acid release, because mice colonized with the BT0455 deletion mutant had no increase in fecal sialic acid upon aTC administration (Figure S6A and S6E). Strikingly, significant levels of sialic acid persisted in BtRS animals for multiple days after both inducer and sialidase activity were no longer detectable (Figure 7C, grey shading). Consistent with this result, exogenous sialic acid delivered by oral gavage to gnotobiotic mice monocolonized with B. thetaiotaomicron BT0455 exhibits a half-life of ~ 9 hours (Figure S6F). Together, these results indicate that the levels of this microbiome-dependent metabolite reflect not only the current activity of the microbiota, but also its activity from days before.
BtRS dynamically modulates sialidase expression in response to changing inducer levels, providing the opportunity to compare sialidase activity and fecal sialic acid concentration in vivo as aTC is eliminated from the gut and sialidase gene expression changes accordingly (Figure 7D). At low levels of sialidase activity, fecal sialic acid scales in proportion to enzyme activity. However, this relationship becomes non-linear once sialidase activity exceeds 25% of the maximum: regardless of sialidase levels, free sialic acid concentration remains constant (Figure 7D). This suggests that in microbial communities exhibiting at least 25% of the enzyme activity of B. thetaiotaomicron, substrate availability (i.e., the host), not community enzyme activity, is the limiting factor that determines free sialic acid levels. Indeed, activity measured in fecal samples from wildtype SPF (conventional) mice reveals high levels of sialidase activity and intermediate levels of free sialic acid, suggesting that the liberation of sialic acid from the mucosa of these animals is substrate (host)-limited and not enzyme-limited. By contrast, catabolism of free sialic acid is likely enzyme (microbe)-limited, as the substrate of this reaction (sialic acid) is readily detectable in SPF animals (Figure 7D). In this way, these results using precise modulation of a single microbiome-encoded activity in vivo suggest that it is not necessary for antibiotics to reconfigure the microbiota in any specific manner in order to increase free sialic acid levels: instead, even general antibacterial activity can reduce sialic acid catabolism without a proportional impact on the sialic liberation from the host, shifting the community into an increasingly pathogen-sensitive state.
Discussion
The lack of genetic tools for gut anaerobes and the incompatibility of genetic parts from model organisms are significant obstacles in the microbiome field. We focused on inducible promoters, a fundamental genetic part which allows real-time analysis of the contribution of microbiome-encoded gene products over the course of microbiome establishment, host development, or disease progression (Rogers et al., 2015). For example, regulated control of microbiome-host signals (secondary bile acids, short chain fatty acids) would allow new insight into the temporal dynamics of these interactions, and modulation of microbiome-encoded small molecules that reach systemic circulation would enable study of the pharmacokinetics of these compounds. Further, these systems could enable on-demand delivery of therapeutics or enzymes from the gut microbiota. Tightly regulated, highly inducible expression platforms also provide the basis for more complex synthetic architectures.
Here, we developed a panel of tunable expression platforms that allow Bacteroides gene expression to be strictly controlled by the presence of a synthetic inducer not found in bacterial growth media, mouse intestines or diets, or the human and mouse microbial communities tested in this study. In the absence of inducer, gene expression is repressed to levels that recapitulate the phenotypes of strains lacking the target gene. When aTC is provided, individual promoters within this panel exhibit up to a 104-fold dynamic range of expression, and promoter activity across the panel ranges 3 x 105-fold. This dynamic control of gene repression and expression are recapitulated or even exceeded in mice, and investigation of sialic acid provides a first example of insights gained from manipulating a host-microbe interaction by tuning expression of a single gene in the microbiome.
Variations in TetR-regulated promoters expand the versatility, range, and regulatory properties of this system (Bertram and Hillen, 2008; Hillen and Berens, 1994). For example, modifications of TetR have been identified that alter its specificity for tetracycline analogs (Henssler et al., 2004; Scholz et al., 2003), change its allosteric interactions so that aTC acts as a corepressor (Kamionka et al., 2004; Scholz et al., 2004), or change its operator specificity to selectively recognize distinct tetO variants (Helbl and Hillen, 1998; Helbl et al., 1998; Krueger et al., 2007). These modifications considerably expand the range of conceivable applications of TetR-regulated systems and increase the possibilities for multi-gene regulation within Bacteroides.
The absence of aTC or related molecules in mice, their diets, or the microbial community allows precise control of Bacteroides gene expression in vivo, potentially enabling on-demand delivery of therapeutic compounds. The tetracycline analog doxycycline is highly stable and binds tightly to TetR, and has been administered to mice for several months at a time (Bohl et al., 1998; Manfredsson et al., 2009). The use of tetracycline analogs that differ in stability and sensitivity to host metabolism, bioavailability and excretion will increase the options for fine temporal control in microbiome studies. Additionally, administering inducers through different routes (orally via water, diet, or time- or pH-dependent delayed release capsules (Cetin et al., 2011); via enema; or surgically through catheterization of specific locations in the GI tract) will allow additional means to precisely tune gene expression in a spatially restricted manner. Lastly, the use of a synthetic inducer to alter gene expression will enable functional and kinetic studies of the microbiome in a matter that does not itself perturb the dynamics of microbial metabolism, growth, and community composition as could be anticipated from the administration of dietary polysaccharides or sugars. Further, changes in host diet and its component polysaccharides are unlikely to impact reporter function.
This regulated expression platform was designed using general principles that should apply across the microbiome (Figure S7), and provides a route for regulated gene expression in other prominent genera from the human gut microbiome and in microbes that inhabit other body regions or habitats. These genetic platforms could potentially be combined with important recent developments (Kotula et al., 2014; Mimee et al., 2015), and will enable progress in understanding the biochemical and ecological processes that contribute to the homeostasis of the intestinal microbiome and its impact on host health and disease.
STAR Methods
Contact for Reagent and Resource Sharing
Requests for further information and resources may be directed to and will be fulfilled by Lead Contact Andrew Goodman (andrew.goodman@yale.edu).
Experimental Model and Subject Details
Microbial Strains and Growth Conditions
All strains used are listed in Table S3. All Bacteroides thetaiotaomicron and Bacteroides ovatus strains used were derivatives of B. thetaiotaomicron VPI-5482 or B. ovatus ATCC 8483 (NCBI Taxonomy ID: 411476), respectively, carrying a deletion in the tdk gene (Larsbrink et al., 2014; Martens et al., 2008), except for Figure 5, which used wildtype strains. Bacteroides strains were cultured anaerobically at 37°C in liquid TYG media [10g/L tryptone, 5g/L yeast extract, 10mM glucose, 100mM potassium phosphate buffer (pH 7.2), 20mg/L MgSO4-7H2O, 400mg/L NaHCO3, 80mg/L NaCl, 0.0008% CaCl2, and 4 μg/mL FeSO4-7H20, 10mg/L hemin (Sigma), 0.5g/L cysteine and 1mg/L vitamin K3], Gut Microbiota Medium (for Figure 5) [10 g/L tryptone, 5g/L yeast extract, 2.2 mM glucose, 2.9 mM cellobiose, 2.8 mM maltose, 2.2 mM fructose, 5g/L meat extract, 100 mM potassium phosphate buffer (pH 7.2), 20mg/L MgSO4-7H2O, 400mg/L NaHCO3, 80mg/L NaCl, 0.0008% CaCl2, and 4 μg/mL FeSO4-7H20, 10mg/L hemin (Sigma), 0.5g/L cysteine and 1mg/L vitamin K3, 0.05% Tween 80, 10 mL/L ATCC Vitamin Mix, 10mL/L ATCC Trace Mineral Mix, 30 mM acetic acid, 1 mM isovaleric acid, 8 mM propionic acid, 4 mM butyric acid] (Goodman et al., 2011a), or on brain heart infusion (BHI; Becton Dickinson) agar supplemented with 10% horse blood (Quad Five). Cultures were grown and manipulated in an anaerobic chamber (Coy Laboratory Products) with an atmosphere of 20% CO2, 10% H2, and 70% N2 at 37°C. E. coli strains EC100D pir-116 and S17-1 lambda pir were used for propagation and transfer, respectively, of R6K pir plasmids. E. coli strains were grown aerobically in LB medium at 37°C. When required, antibiotics were added to the medium as follows: carbenicillin 100 μg/mL, gentamicin 200 μg/mL, and erythromycin 25 μg/mL. Anhydrotetracycline (aTC; Cayman Chemicals) was dissolved in 100% ethanol at 2 mg/mL as a master stock. Working stock solutions of aTC were made at 1,000X in 100% ethanol.
Human Studies
All human studies were conducted with the permission of the Yale Human Investigation Committee. Recruitment of healthy human volunteers, sample collection, anaerobic processing, and −80°C storage of fecal samples in cryoprotectant under anaerobic conditions was previously described (Cullen et al., 2015).
Animal Experiments
All experiments using mice were performed using protocols approved by the Yale University Institutional Animal Care and Use Committee. Germ-free Swiss Webster mice were maintained in flexible plastic gnotobiotic isolators with a 12-hr light/dark cycle and provided a standard, autoclaved mouse chow (5K67 LabDiet, Purina) ad libitum and autoclaved water for the duration of the experiment. Filter-sterilized aTC was added to drinking water stored in red-tinted bottles (Ancare, #PC9RH8.5RD) to inhibit degradation by light. Mice were co-housed for the duration of the experiments. Cage changes were always carried out when aTC was withdrawn from drinking water. Fecal and luminal samples were collected and processed in 2.0 mL screw cap tubes (Axygen Scientific; #SCT-200-C). Germ free status was monitored by 16S rDNA-targeted PCR of fecal DNA and anaerobic and aerobic culture of fecal samples.
Method Details
Sequence Conservation in 16S Promoters
Genomic sequences of 19 different Bacteroides strains (Table S1) were searched using BLAST (Altschul et al., 1990) for 16S rRNA loci using the B. thetaiotaomicron VPI-5482 (NBCI Taxonomy ID: 226186) 16S rRNA sequence as a query. For each 16S rRNA locus identified, upstream regions (up to 1000 base pairs) were manually searched for the RNA polymerase recognition sequence, TTTG(N19-21)TANNTTTG (Mastropaolo et al., 2009). The fifth nucleotide downstream of the -7 element is a conserved cytosine residue and shown to be the transcription start site (Mastropaolo et al., 2009). This cytosine residue and the upstream 67 nucleotides were designated as a 16S rRNA promoter. In total, 182 promoters were identified and frequency logos generated for each nucleotide position using WebLogo3 (Crooks et al., 2004).
Genetic Parts
Genetic parts (promoters, ribosome binding sites (RBSs), localization signal) developed in this study are listed in Table S2 and will be deposited in Addgene upon publication. RBSs strength correlates with a complex set of features, including enrichment in adenine and thymine, secondary structure, and potential interactions with the ribosomal protein S1 (Accetto and Avguštin, 2011; Mimee et al., 2015; Wegmann et al., 2013). Alternate RBSs of varying strengths (Mimee et al., 2015; Wegmann et al., 2013), were used in this study.
Molecular Cloning
The plasmids used in this study are listed in Table S3. The template P1 and P2 promoters containing the ribosome binding site GH023 (see Table 23) were synthesized by Integrated DNA Technologies (IDT). All primers used in this study were obtained from the Keck Biotechnology Resource Laboratory (Yale University). DNA amplification for cloning procedures were carried out using Q5 High Fidelity DNA Polymerase (New England Biolabs), and colony PCR was carried out using OneTaq DNA Polymerase (New England Biolabs). Splicing by Overlap Extension (SOE) PCR (Bryksin and Matsumura, 2010) was utilized to insert or replace genetic parts (tetO2 or RBSs) of the parental P1 and P2 promoters, followed by Gibson cloning (HiFi DNA Assembly Master Mix, New England Biolabs). All vectors containing P1 and P2 promoters and their derivatives were built to contain an NcoI restriction site at the ATG site and a SalI restriction site further downstream. For insertion of reporters downstream of the modified promoters, vectors were digested with both NcoI and SalI, and reporters were inserted using Gibson cloning. Genetic modifications generated on plasmids and Bacteroides strains were verified by sequencing at the Keck Biotechnology Resource Laboratory (Yale University).
Integration of pNBU2 Vectors into Bacteroides
Single-copy introduction of pNBU2 vectors into Bacteroides sp. genomes and determination of genomic insertion site location was carried out as previously described (Koropatkin et al., 2008; Larsbrink et al., 2014; Martens et al., 2008), with minor modifications described below.
For Bacteroides type strains, overnight cultures of E. coli S17-1 donor strains were diluted 1,000-fold in LB medium containing carbenicillin and Bacteroides recipients diluted 250-fold in TYG medium and each grown to early exponential phase (OD600 0.2–0.3). Donor and recipient strains were combined at a 1:10 donor:recipient culture volume ratio, centrifuged at 4,000 rpm for 20 minutes, resuspended in 100 μL of BHI liquid medium and plated as a dime-sized puddle on non-selective BHI-blood agar plates for 20 hours at 37°C under aerobic conditions to allow for conjugation. Mating lawns were resuspended in 1mL 1X phosphate buffered saline (PBS) and transconjugants selected by plating serial dilutions on BHI-blood agar plates containing gentamicin and erythromycin. Insertion site location was verified by PCR (primers provided in Table S3).
For Bacteroides isolates cultured directly from humans, the same procedure was used except equal volumes of donor (E. coli S17-1 pNBU2[Erm]TetR-P1TDPGH023-NanoLuc) and recipient overnight cultures were combined and 100 μL plated on BHI-blood agar plates for the conjugation step. After 20 hours of aerobic incubation at 37°C, transconjugants were selected on gentamicin and erythromycin as above. For each human isolate, 4 separate clones were colony purified on the same media and directly inoculated into Gut Microbiota Medium to test the performance of the P1TDP platform.
Construction of Bt::tetR and BtRS
A constitutively-expressing tetR construct was built by splicing 300 base pairs of the BT1311 (rpoD; σ70) promoter (Table S2) to the tetR gene. To identify a neutral location in the B. thetaiotaomicron genome for stable integration of the tetR cassette, we searched for an intergenic region between the 3′ ends of two genes, where disruption of either gene or the intergenic region itself by transposon insertion confers no fitness defect or benefit in rich or minimal medium, or in gnotobiotic mice mono-associated with B. thetaiotaomicron or colonized with multi-species communities (Goodman et al., 2009). Constructs were integrated into the B. thetaiotaomicron genome by conjugation as described below. For strain Bt::tetR, the tetR cassette was placed between genomic position 4861701 and 4861702 (between BT3743 and BT3744).
For strain BtRS, repression of sialidase activity likely confers a fitness cost to the organism (Ng et al., 2013). Consequently, three constitutive tetR constructs were placed in the chromosome to reduce the risk of selecting spontaneous mutants in tetR. In addition to the construct located between BT3743 and BT3744, two additional exact copies of the construct were placed between genomic position 2660693 and 2660694 (between BT2113 and BT2114), and between genomic position 6193956 and 6193957 (between BT4719 and BT4720), generating strain Bt::3xtetR.
Unmarked Chromosomal Modifications
Unmarked chromosomal modifications, including the insertion of the constitutively-expressing tetR cassette(s), in-frame deletions of BT1754 and BT0455, replacement of the BT1754 promoter with P1TDPGH023, and replacement of the BACOVA_04598 promoter with TetR-P1TDPB1, were generated using a counterselectable allelic exchange procedure (Koropatkin et al., 2008). Approximately 1,000-basepair regions flanking the genomic region to be modified were amplified by PCR, joined using Gibson cloning or SOE PCR, and cloned into pExchange-tdk. Sequence-verified plasmids were introduced into Bacteroides as above, except a 1:1 donor:recipient culture volume ratio was used for conjugation. Bacteroides transconjugants (merodiploids) were selected after conjugation by dilution plating on BHI-blood agar plates containing gentamicin and erythromycin. Insertion of the pExchange-tdk vector upstream or downstream of the genomic region of interest was verified by colony PCR. Merodiploids harboring appropriate integration of the pExchange-tdk vector were grown in liquid TYG overnight, and plated onto BHI-blood agar containing 200 μg/mL 5-fluoro-2-deoxy-uridine (FUdR) (Abcam) to select for the loss of the pExchange-tdk vector. Individual clones were further verified for erythromycin sensitivity and FUdR resistance. Erythromycin-sensitive, FUdR-resistant clones were screened for the appropriate genetic manipulation by PCR and verified by sequencing.
Luciferase Assay from In Vitro Samples
The NanoLuc luciferase assay was performed as described (Mimee et al., 2015), with the following modifications. All strains were first grown for 18 hours in TYG medium or Gut Microbiota Medium and diluted 250-fold into fresh medium in appropriate conditions (with or without aTC). Cultures were grown to mid exponential phase (OD600 0.3–0.4) and colony forming units (CFU) were measured by dilution plating. 500 μL of the culture was centrifuged at 21,130 x g for 3 minutes, resuspended in 50 μL 1X BugBuster (Novagen) in 1X PBS and lysed by one cycle of freeze-thaw and nutation at 26°C for 10 minutes. Lysates were cleared of cellular debris by centrifugation at 21,130 x g for 10 minutes at 4°C. 10 μL of the supernatant was mixed with an equal volume of NanoLuc Reaction Buffer using the Nano-Glo Luciferase Assay System (Promega) and incubated for 5 minutes at 26°C to induce luminescence production. Relative Light Units (RLU) were measured using a fluorescence plate reader (BioTek Synergy H1) with an integration time of 1 second at a gain setting of 100. Luminescence values were normalized to colony forming units (CFUs) because experimental conditions (e.g., growth medium, antibiotics, aTC, heterologous gene expression) affect the correlation between CFUs and OD600 in Bacteroides (data not shown).
Polymyxin B Susceptibility
Minimum inhibitory concentrations (MICs) were determined on solid medium using E-test strips (Biomerieux) as previously described (Cullen et al., 2015), except that cultures were initially grown for 18 hours in liquid medium. Cultures were adjusted to OD600 ~1.0 and 100 μL of cell suspension was then spread onto BHI-blood agar plates. Surface liquid was allowed to dry in a fume hood for 10 minutes, followed by application of the E-test strip to the agar surface. Plates were incubated anaerobically at 37°C for 24 hours before scoring the MIC.
Isolation of Bacteroides from Humans
Bacteroides isolates were enriched by plating dilutions of the fecal material on Brucella Laked Blood Agar containing Kanamycin and Vancomycin (BD #297840). After 48 hours anaerobic incubation at 37°C, candidate Bacteroides isolates were selected and cultured in Gut Microbiota Medium for DNA isolation by the cetyltrimethylammonium bromide (CTAB) method (Wilson, 2001) and −80°C storage in cryoprotectant. 16S rDNA was amplified and sequenced from each isolate using primers 8F and 1492Rm (Tables S3). The taxonomy of the isolate was identified by BLAST (Altschul et al., 1990) using the sequenced 16S rDNA and designated as the most closely related species. In each case, isolate 16S sequences were within 97% identity of a known species. A phylogenetic tree of sequenced 16S rDNA from the isolates and type strains was generated using Phlogeny.fr (Dereeper et al., 2008; 2010).
BT1754-Dependent Growth Assay
Minimal media was made as previously described (Martens et al., 2008), but with the addition of 1 g/L Na2CO3, 10 mg/L MnCl2-4H2O, and 10 mg/L CoCl2-6H2O. Glucose or fructose was added as the sole carbon source at a final concentration of 0.5%. Cultures were grown in minimal medium containing glucose for 18 hours and diluted 1:1000 in minimal medium containing either glucose or fructose with or without 100 ng/mL aTC. Cultures were incubated anaerobically at 37°C in clear flat bottom 96-well plates (Costar #3595) and growth was monitored (OD600) every 15 minutes in a plate reader (BioTek Eon). Cell doublings per hour were calculated as (log(OD600final) - log(OD600initial)) / (duration x log(2)) from cultures during exponential phase of growth (OD600 ~0.15 to ~0.4).
Periplasmic Targeting of T6SS Effectors
To direct Bte1 and Bfe1 to the periplasm upon expression in B. thetaiotaomicron, the genes encoding each effector were first PCR amplified from genomic DNA purified from B. fragilis NCTC 9343 and B. fragilis 638R, respectively, using the CTAB method. We searched for a periplasmic localization signal that would be cleaved after translocation to prevent steric hindrance of the effector. To identify proteins that encode N-terminal B. thetaiotaomicron periplasmic localization signals that are post-translationally cleaved after protein translocation, the proteome of the Bacteroides thetaiotaomicron VPI-5482 chromosome was searched for signal peptides (PTM/Processing; Molecule Processing, Signal Peptide) using UniProt (UniProt Consortium, 2015). To exclude membrane proteins that potentially encode an uncleaved signal peptide, the search was refined by an additional query for the term “periplasmic” under Protein name. To identify a short periplasmic signal, the maximum length for the signal peptide was set at 20 amino acids. BT4676, which encodes a hypothetical protein, gave the highest confidence value in predicting both a periplasmic-localized signal peptide and the position of cleavage (between amino acid 20 and 21) according to SignalP (Petersen et al., 2011). Therefore, the B. fragilis effectors were fused at the 5′ end (minus the ATG start codon) to the first 63 base pairs (encoding the first 21 amino acids) of the BT4676 open reading frame (see Table S2 for sequence) in order to localize them to the periplasm after expression in B. thetaiotaomicron.
Monocolonized Gnotobiotic Mice
Germ-free Swiss Webster mice were orally gavaged with 1 x 109 CFU of Bt::tetR P1TDPGH023-NanoLuc and colonization was allowed to stabilize for at least 7 days prior to the start of the experiment. Fecal pellets were collected every day, and groups of mice were given aTC in the drinking water at various concentrations for 48 hours after stool collection on Day 3 and Day 17. Mice belonging to the same treatment group were co-housed for the duration of the experiment. To obtain samples for luminescence production, fecal pellets and intestinal lumen (~100–200 mg) were homogenized in 1 mL of 1X PBS with a single 5 mm stainless steel bead using a bead beater (BioSpec Products) at 26°C for 1 minute on the high setting, followed by vortexing at top speed for 8 minutes. Samples were centrifuged at 500 x g for 1 minute to pellet fibrous and insoluble matter. The supernatant was used for CFU determination by plating dilutions on BHI-blood agar plates containing gentamicin and erythromycin, and 500 μL was used for luminescence measurements (described below).
Defined Human Gut Microbiota Gnotobiotic Mice
Germ-free Swiss Webster mice were orally gavaged with a mixture of 1 x 108 CFU of each of 14 human gut microbes shown in Figure S5D and Table S3. With the exception of the reporter strain, all species are sensitive to gentamicin and/or erythromycin (data not shown). Colonization was allowed to stabilize for at least 7 days prior to the start of the experiment. Fecal pellets were collected daily, and mice were given aTC (100 μg/mL) in the drinking water for 48 hours after stool collection on Day 3. Fecal samples were homogenized, and fibrous/insoluble matter removed as described above. From the supernatant, 200 μL was stored for DNA purification as previously described (Goodman et al., 2011b) and subsequent qPCR analysis (described below), 10 μL was used for CFU determination of the reporter strain by plating dilutions on BHI-blood agar plates containing gentamicin and erythromycin, and 500 μL was used for luminescence measurements (described below).
At each time point, abundance of each species was assessed by qPCR from fecal DNA using strain specific primers described previously (Cullen et al., 2015). qPCR was performed as described (Goodman et al., 2009; Martens et al., 2008) using a CFX96 instrument (BioRad) and SYBR FAST universal master mix (KAPA Biosystems). Mean strain quantities were calculated using a standard curve.
SPF Rag−/− Conventional Mice
8–10 week old SPF Rag−/− mice (Jackson Laboratories) (n = 7) were colonized by oral gavage with 1 x 109 CFU of B. thetaiotaomicron TetR-P1TDPGH023-NanoLuc. Fecal pellets were collected daily after gavage and processed as described above. The supernatant was used for CFU determination by plating dilutions on BHI-blood agar plates containing gentamicin and erythromycin, and 500 μL was used for luminescence measurements (described below). Culturing of bacteria from fecal pellets obtained from these animals prior to gavage confirms that they do not harbor native species resistant to gentamicin and erythromycin (data not shown).
Luciferase Assay from Murine Samples
The 500 μL collected from the resuspended fecal supernatant was centrifuged at 21,130 x g for 3 minutes to pellet bacterial cells, which were then resuspended in 50 μL 1X BugBuster (Novagen) in 1X PBS and lysed by one round of freeze-thaw and 10 minutes of nutation at 26°C. Samples were centrifuged at 21,130 x g for 10 minutes at 4°C, and 10 μL of the supernatant was mixed with an equal volume of NanoLuc Reaction Buffer to induce luminescence production. Luminescence was measured after a 5 minute incubation at 26°C on a plate reader (BioTek Synergy H1) with an integration time of 1 second at a gain setting of 100. Luminescence values were normalized to colony forming units (CFUs) determined by dilution plating of the resuspended fecal supernatant on BHI-blood agar plates containing gentamicin and erythromycin.
Sialic Acid and aTC in Murine Samples
Groups of 6 germ-free Swiss Webster mice were orally gavaged with 1 x 109 CFU of the desired B. thetaiotaomicron strain. Fecal samples (~100–200 mg) were collected daily for 12 days. aTC (100 μg/mL) was administered in drinking water for 12 hours after fecal collection on day 1. Fecal samples were homogenized in 1 mL of 1X tris-buffered saline (TBS) with a single 5 mm stainless steel bead using a bead beater (BioSpec Products) at 26°C for 1 minute on the high setting, followed by vortexing at top speed for 8 minutes. Samples were then centrifuged at 500 x g for 1 minute to pellet fibrous and insoluble matter. Bacterial cells from 600 μL of the supernatant was isolated through centrifugation at 21,130 x g for 3 minutes, resuspended in 50 μL of Sialidase Reaction buffer (50 mM Tris-HCl pH 7.0, 5 mM EDTA) containing 1X BugBuster, and stored at −80°C for ex vivo sialidase measurements (described below).
To the remaining material, 400 mg of 0.1 mm glass beads was added and further homogenized by bead beating for 2 minutes. Insoluble material was pelleted by centrifuging the sample for 3 minutes at 21,130 x g, and 20 μL of the supernatant was used for mass spectrometry measurements of sialic acid and aTC by liquid chromatography coupled tandem mass spectrometry (QqQ) and mass spectrometry (qTOF), respectively.
For sialic acid measurements, 5 μL of internal standard solution (50 μM 1,2,3-13C3 sialic acid in H2O) were added to each sample (20 μL) and proteins were precipitated through addition of 100 μL organic solvents at −20°C (acetonitrile : methanol, 1:1). After incubation at −20°C for 1 h, samples were centrifuged at 4,000 x g for 15 min and 100 μL supernatant was collected, dried under vacuum at 22°C, and re-suspended in 32 μL H2O. LC-MS/MS analysis was carried out using an Agilent 1290 UHPLC system coupled to an Agilent 6490A QqQ mass spectrometer. Chromatographic separation was performed on a BEH Amide column (Waters, 150 mm × 2.1 mm, 1.7 μm particle size) using mobile phase A: H2O, 0.1% formic acid and B: acetonitrile, 0.1% formic acid at 45°C. 2 μL of processed sample were injected at 99% B and 0.4 mL/min flow followed by a linear gradient to 70% B over 10 min and 0.4 mL/min flow, leading to sialic acid elution at 6.5 min. The column was re-equilibrated at starting conditions for 8 min. The QqQ was operated in negative ionization mode using dynamic MRM scans with the following source parameters: VCap: 3500 V, nozzle voltage: 2000 V, gas temp: 275°C; drying gas 12 L/min; nebulizer: 35 psig; sheath gas temp 275°C; sheath gas flow 12 L/min. MRM parameters were optimized following the manufacturer’s recommendation: Sialic acid 308.1 m/z to 58.8, 87, 97,9, 118.9, and 169.9 m/z with respective collision energies 33, 13, 29, 9, 13 (au). Corresponding settings were used for the 1,2,3-13C3 sialic acid standard. The MassHunter Quantitative Analysis Software (Agilent, version 7.0) was used for peak integration with the most abundant ion fragment as quantifier and the others as qualifiers of the peak. Quantification was based on calibration curves using commercially available sialic acid and the signal intensity of the internal standard spiked into each sample.
For aTC measurements, 5 μL of internal standard solution (0.25 μM sulfamethoxazole and caffeine in H2O) were added to each sample (20 μL) and further processed as described above. Dried samples were re-suspended in 4 μL methanol and diluted in 12 μL H2O. Chromatographic separation was performed on a C18 Kinetex Evo column (Phenomenex, 100 mm × 2.1 mm, 1.7 μm particle size) using mobile phase A: H2O, 0.1% formic acid and B: methanol, 0.1% formic acid at 45°C. 10 μL of sample were injected at 100% A and 0.4 mL/min flow followed by a linear gradient to 95% B over 5.5 min and 0.4 mL/min flow leading to aTC, caffeine and sulfamethoxazole elution at 3.0, 1.9, 2.1 min, respectively. The qTOF was operated in positive scanning mode (50 – 1000 m/z) and the following source parameters: VCap: 3500 V, nozzle voltage: 2000 V, gas temp: 225°C; drying gas 13 L/min; nebulizer: 20 psig; sheath gas temp 225°C; sheath gas flow 12 L/min. Online mass calibration was performed using a second ionization source and a constant flow (5 μL /min) of reference solution (121.0509 and 922.0098 m/z). Compounds were identified based on the retention time of chemical standards and their accurate mass (tolerance 20 ppm). The MassHunter Quantitative Analysis Software (Agilent, version 7.0) was used for peak integration and quantification was based on calibration curves using chemical standards and the signal intensity of the internal standard spiked into each sample.
Sialidase Activity in Murine Samples
Bacterial cells (collected and stored as described above) were lysed through one round of freeze-thaw followed by nutation for 10 minutes at 26°C. Cell lysates, which contained sialidase activity, were obtained from the supernatant after centrifugation at 21,130 x g for 10 minutes at 4°C. The reaction to determine sialidase activity was performed in a total volume of 100 μL consisting of 72 μL Sialidase Reaction Buffer, 10 μL of 100 mM of substrate 2-O-(p-Nitrophenyl)-t-D-N-acetylneuraminic acid (pNP-SA; Sigma-Aldrich) resuspended in Sialidase Reaction Buffer, and 18 μL of cell lysate. The reaction mixture was incubated at 37°C and 10 μL of the reaction was transferred to an equal volume of ice-cold acetonitrile at 0, 5, 10, 15, 20, 25, 30, and 60 minutes to stop the reaction. 5 μL of internal standard solution (1 μM sulfamethoxazole and caffeine in H2O) were added to each sample (20 μL) and further processed as described above. Dried samples were re-suspended in 6 μL methanol followed by the addition of 26 μL H2O and further dilution (1:4) in H2O. Quantification of the enzyme reaction product, p-nitrophenol (pNP), was identical to the LC-MS analysis of aTC described above, but with reduced injection volume (5 μL) and methanol replacing acetonitrile as solvent B for the reversed phase chromatography. Retention time of pNP was 2.1 min and a commercially available standard was used for the calibration curve. Linear fits of pNP were performed for at least the first 10 min of the enzyme reaction using the ‘polyfit’ function of Matlab R2016a (Mathworks). Linear fit values were normalized to amount of protein in the cellular lysates added to each reaction, which was determined by Bradford.
A best fit line (Figure 7D) based on Michaelis-Menten kinetics was created (Prism 6) on data plotting sialic acid levels (Figure 7C) against fecal sialidase activity (Figure 7B) from fecal pellets collected daily between Day 0 (prior to aTC administration) and Day 6 (when sialidase activity becomes undetectable) from mice carrying the BtRS strain (n = 4 mice).
Quantification and Statistical Analysis
Datasets were analyzed within the GraphPad Prism 6 software. Pairwise comparisons were generated with two-tailed t tests. P values and n values are indicated in the associated figure legends for each figure.
Analysis of Defined Microbial Communities
Relative abundance calculations based on qPCR were used to construct an OTU table for alpha and beta diversity analyses in QIIME v1.8 (Caporaso et al., 2010). Alpha diversity (Simpson, Shannon, and Simpson_e metrics) was compared between day 3 (immediately prior to aTC administration) and each other day using a paired Student’s t-test and a p-value cutoff of 0.05; no significant differences were found regardless of multiple hypothesis testing correction. Beta diversity analysis was conducted using Hellinger distances.
Supplementary Material
Table S1. Related to Figure 1. Name and strain IDs of species used for identifying promoter regions upstream of 16S rRNA.
Contains the list of Bacteroides strains whose 16S rDNA promoters were used for determining conserved nucleotides.
Table S2. Related to STAR Methods. Genetic Parts used in this study.
Contains information regarding the genetic parts used and created in this study – see Method Details.
Table S3. Related to STAR Methods. Bacterial strains, plasmids, and primers used in this study.
Contains information regarding the strains, plasmids, and primers used and created in this study – see Method Details.
(A) Wildtype B. thetaiotaomicron grows normally in 100 ng/mL aTC. Culture densities over time of wildtype B. thetaiotaomicron diluted into TYG media with varying concentrations of aTC are shown. (B, C) Expression from the P1TDPGH023 promoter can be modulated over a 100-fold range of aTC concentration from 0.01 ng/mL to 1 ng/mL and activates within minutes, achieving a 395-fold increase in expression within 45 minutes and maximal induction at ~120 minutes after aTC addition. Because the B. thetaiotaomicron doubling rate is 47.1 minutes in the conditions tested, this synthetic promoter therefore activates gene expression in response to aTC within one round of cell division. Dashed lines represents the average luminescence value from the vector control at mid-exponential phase. Error bars represent the standard deviation of three biological replicates on separate days. (D) Culture densities of Bt::tetR harboring antibacterial effectors under the control of P1TDPGH023. Bt::tetR encoding either of two highly toxic antibacterial effectors fused to periplasmic signal sequences (“ss”) from B. fragilis under the control of P1TDPGH023 grow at wildtype rates unless expression is induced with aTC. Error bars represent the standard deviation of two biological replicates on separate days. Asterisks indicate the earliest timepoint with significant (p < 0.002) differences compared to uninduced controls.
(A and B) The impact of RBS variation on promoter activity is generally conserved between constitutive and inducible promoters. Luminescence values and standard deviations from the P1TDPRBS-NanoLuc constructs (Figure 4A), in the absence (A) and presence (B) of aTC, were plotted against previously published luminescence values and standard deviations of the same RBSs fused to the constitutive BT1311 promoter (Mimee et al., 2015). (C) mRNA levels generally correspond to the strength of their promoters based on promoter-NanoLuc fusions. Luminescence values and standard deviations from the promoter-NanoLuc constructs (Figure 4C), were plotted against previously published values and standard deviations of the mRNA levels generated by the corresponding promoter as determined by microarray (Sonnenburg et al., 2005).
Promoter replacement of the lpxF homologue (BACOVA_04598) in B. ovatus ATCC 8483 with the self-contained, ~1kb inducible expression cassette, TetR-P1TDPB1, provides aTC-dependent regulatory control of PMB resistance in a one-step procedure. (A) Genomic position and replacement of the native promoter (upstream 62 basepairs) of BACOVA_04598, with TetR-P1TDPB1. (B) PMB MICs for the wildtype (tdk), lpxF deletion (BACOVA_04598), and aTC-regulated lpxF (Bo::P1TDPB1-BACOVA_04598) strains were determined with E-test strips.
Gnotobiotic mice monocolonized with B. thetaiotaomicron were given aTC (100 μg/mL in drinking water) beginning after collection of fecal samples on day 4 and continuing for 4 days (n = 6 mice/group). Mice were placed on water without inducer immediately after the fecal collection on day 8. Fecal aTC concentrations reach 22 μM at the highest point, about an order of magnitude below the lowest toxic concentration observed in vitro (216 μM).
(A) Addition of aTC to drinking water does not impact colonization of B. thetaiotaomicron in monocolonized gnotobiotic mice (green, black, and red lines), in gnotobiotic mice carrying defined human communities (orange line), or in the presence of a complete murine microbiota (blue line). Colony forming units (CFU) were determined per gram of feces collected each day from mice in Figure 6. For gnotobiotic mice monocolonized with B. thetaiotaomicron or Bt::tetR P1TDP-NanoLuc, aTC was given for 48 hours in drinking water on days 3–5 and days 17–19, while mice carrying a human or murine microbiota were given aTC on days 3–5 (n ≥ 6 mice per data point). Grey shading designates days when mice were administered aTC. (B) P1TDPGH023 activity was measured in luminal contents collected along the length of the gastrointestinal tract (upper, middle, and lower small intestine; cecum; upper and lower large intestine) on day 19 from mice in Figure 6A. Asterisks indicate significant induction (p < 0.05) as compared to the same GI location in mice colonized with Bt::tetR P1TDPGH023-NanoLuc but not provided aTC in the drinking water. (C) Addition of aTC to drinking water does not impact colonization of B. thetaiotaomicron along the length of the gut. Day 19 CFU measurements from each gastrointestinal section from mice from Figure 6A are shown (n ≥ 4 mice per data point). (D) Community composition of gnotobiotic mice (n = 6) colonized with Bt::tetR P1TDP GH023-NanoLuc and 13 prominent human gut microbes (Figure 6C) does not significantly change in response to aTC. Inducer was provided in drinking water for 48 hours from days 3–5 and community composition monitored by species-specific quantitative polymerase chain reaction (qPCR) and reported as mean percentage of the total. (E). Alpha diversity of defined human microbial communities in mice (n = 6) from Figure 6C before, during, and after aTC administration. Using each of the described alpha diversity metrics, no significant differences exist between the diversity at day 3 (the last day before aTC administration) and any other timepoint. (F) Beta diversity of defined human microbial communities from mice described in Figure 6C, as calculated from Hellinger distances.
(A) BT0455 in B. thetaiotaomicron is responsible for liberating sialic acid from the host mucosa. Free sialic acid concentrations were measured by LC-MS in fecal pellets of germfree mice and gnotobiotic mice monocolonized with wildtype B. thetaiotaomicron or Bt::tetR BT0455 (n = 4 mice per group). (B) Because sialidase activity likely provides a benefit to B. thetaiotaomicron by exposing mucosal nutrients, BtRS was engineered with multiple copies of tetR to avoid loss of repressor activity by spontaneous mutation. (C) Ex vivo sialidase assay for determination of enzyme activity in fecal pellets. Sialidase activity kinetics were measured with the indicated dilutions of lysed bacterial cells from fecal pellets of gnotobiotic mice monocolonized with wildtype B. thetaiotaomicron. Enzyme reactions contained the artificial substrate 2-O-(p-Nitrophenyl)-tropheacetylneuraminic acid (pNP-SA), which is cleaved by BT0455 to release sialic acid and p-Nitrophenyl (pNP), the latter of which was measured by LC-MS. The initial velocities of this reaction were measured every 5 minutes at 37°C for 30 minutes, and a best-fit line and its slope was determined to ensure that enzyme concentrations in the lysate were directly proportional to the initial velocities. (D) Sialidase activity and (E) free sialic acid in feces was measured for Bt::3xtetR (green line), Bt::3xtetR BT0455 (red line) for the first four days (for sialidase activity) or for the full experiment (for free sialic acid) shown in Figure 7. The average values and standard deviations were then used for shaded boxes shown in Figure 7B and 7C, respectively. (F) Transit time of exogenous sialic acid through the intestinal tract. Gnotobiotic mice monocolonized with Bt::tetR BT0455 were gavaged with 5 mg of sialic acid and sialic acid in fecal pellets were measured every 3 hours for 48 hours (n = 2 – 6 mice per timepoint).
Highly active promoters were identified from RNA-seq data, and conserved regions determined by alignment of homologous promoters collected from genome sequences. Next, a systematic approach identified regions permissible for tetO2 operator placement, and experimental validation confirmed that these engineered promoters achieve high promoter activity and tight repression in the presence of the repressor. Further, additional performance tests were carried out determine whether functional and regulatory integrity was maintained across diverse Bacteroides species. For organisms that lack phage attachment sites (used by the pNBU2 integrating vector) or replicating plasmids, single-unit systems (Figure 5) could potentially be encoded in a transposon for delivery into the genome. Lastly, performance of the regulatable platform was determined in vivo to confirm that the gut environment does not hinder the functionality of the engineered promoter.
Acknowledgments
We thank the Goodman and Groisman labs for helpful discussions and L. Valle and D. Lazo for technical assistance. This study was funded by grants from the NIH (GM105456, GM118159), the Burroughs Wellcome Fund, and the DuPont Young Professors, Pew Scholars, and HHMI Faculty Scholars Programs (to A.L.G.). M.Z. received a Long-Term Fellowship (ALTF 670-2016) from the European Molecular Biology Organization and an Early Postdoc Mobility Fellowship from the Swiss National Science Foundation.
Footnotes
Author Contributions
Conceptualization, B.L., M.Z., and A.L.G.; Methodology, B.L., M.Z., and A.L.G.; Investigation, B.L., M.Z., and N.A.B.; Writing – Original Draft, B.L. and A.L.G.; Writing –Review & Editing, B.L., M.Z., N.A.B., and A.L.G.
References
- Accetto T, Avguštin G. Inability of Prevotella bryantii to form a functional Shine-Dalgarno interaction reflects unique evolution of ribosome binding sites in Bacteroidetes. PLoS ONE. 2011;6:e22914–e22919. doi: 10.1371/journal.pone.0022914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bayley DP, Rocha ER, Smith CJ. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol. 2008;1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl D, Salvetti A, Moullier P, Heard JM. Control of erythropoietin delivery by doxycycline in mice after intramuscular injection of adeno-associated vector. Blood. 1998;92:1512–1517. [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotech. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin M, Atila A, Sahin S, Vural I. Preparation and characterization of metformin hydrochloride loaded-Eudragit ®RSPO and Eudragit ®RSPO/PLGA nanoparticles. Pharmaceutical Development and Technology. 2011;18:570–576. doi: 10.3109/10837450.2011.604783. [DOI] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Bremer H. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3:1–49. doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439–1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011a;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host and Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Wu M, Gordon JI. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc. 2011b;6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady ZZR, Farrar MD, Whitehead TR, Holland KT, Lodge JPA, Carding SR. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology. 2008;154:3165–3174. doi: 10.1099/mic.0.2008/019109-0. [DOI] [PubMed] [Google Scholar]
- Helbl V, Hillen W. Stepwise selection of TetR variants recognizing tet operator 4C with high affinity and specificity. Journal of Molecular Biology. 1998;276:313–318. doi: 10.1006/jmbi.1997.1540. [DOI] [PubMed] [Google Scholar]
- Helbl V, Tiebel B, Hillen W. Stepwise selection of TetR variants recognizing tet operator 6C with high affinity and specificity. Journal of Molecular Biology. 1998;276:319–324. doi: 10.1006/jmbi.1997.1539. [DOI] [PubMed] [Google Scholar]
- Henssler EM, Scholz O, Lochner S, Gmeiner P, Hillen W. Structure-based design of tet repressor to optimize a new inducer specificity. Biochemistry. 2004;43:9512–9518. doi: 10.1021/bi049682j. [DOI] [PubMed] [Google Scholar]
- Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- Horn N, Carvalho AL, Overweg K, Wegmann U, Carding SR, Stentz R. A novel tightly regulated gene expression system for the human intestinal symbiont Bacteroides thetaiotaomicron. Front Microbiol. 2016;7:207–209. doi: 10.3389/fmicb.2016.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Research. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci USA. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger M, Scholz O, Wisshak S, Hillen W. Engineered Tet repressors with recognition specificity for the tetO-4C5G operator variant. Gene. 2007;404:93–100. doi: 10.1016/j.gene.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer T, Takahashi M, Hillen W. Thermodynamic analysis of tetracycline-mediated induction of Tet repressor by a quantitative methylation protection assay. Analytical Biochemistry. 1995;232:190–196. doi: 10.1006/abio.1995.0006. [DOI] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2014;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Research. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredsson FP, Burger C, Rising AC, Zuobi-Hasona K, Sullivan LF, Lewin AS, Huang J, Piercefield E, Muzyczka N, Mandel RJ. Tight long-term dynamic doxycycline responsive nigrostriatal GDNF using a single rAAV vector. Molecular Therapy. 2009;17:1857–1867. doi: 10.1038/mt.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host and Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastropaolo MD, Thorson ML, Stevens AM. Comparison of Bacteroides thetaiotaomicron and Escherichia coli 16S rRNA gene expression signals. Microbiology. 2009;155:2683–2693. doi: 10.1099/mic.0.027748-0. [DOI] [PubMed] [Google Scholar]
- McDonald ND, Lubin JB, Chowdhury N, Boyd EF. Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo. mBio. 2016;7:e02237–15. doi: 10.1128/mBio.02237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth P, Schnappinger D, Hillen W, Saenger W. Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- Park KH, Kim MG, Ahn HJ, Lee DH, Kim JH, Kim YW, Woo EJ. Structural and biochemical characterization of the broad substrate specificity of Bacteroides thetaiotaomicron commensal sialidase. BBA - Proteins and Proteomics. 2013;1834:1510–1519. doi: 10.1016/j.bbapap.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Parker AC, Smith CJ. Development of an IPTG inducible expression vector adapted for Bacteroides fragilis. Plasmid. 2012;68:86–92. doi: 10.1016/j.plasmid.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Heijne von G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JK, Guzman CD, Taylor ND, Raman S, Anderson K, Church GM. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Research. 2015;43:7648–7660. doi: 10.1093/nar/gkv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz O, Henssler EM, Bail J, Schubert P, Bogdanska-Urbaniak J, Sopp S, Reich M, Wisshak S, Köstner M, Bertram R, et al. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- Scholz O, Köstner M, Reich M, Gastiger S, Hillen W. Teaching TetR to recognize a new inducer. Journal of Molecular Biology. 2003;329:217–227. doi: 10.1016/s0022-2836(03)00427-3. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Barnard KN, Parrish CR. Effects of sialic acid modifications on virus binding and infection. Trends in Microbiology. 2016;24:991–1001. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, Horn N, Carding SR. Defining the Bacteroides ribosomal binding site. Appl Environ Microbiol. 2013;79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler HM. Bacteroides: the Good, the Bad, and the Nitty-Gritty. Clinical Microbiology Reviews. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 2001;Chapter 2:2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 1. Name and strain IDs of species used for identifying promoter regions upstream of 16S rRNA.
Contains the list of Bacteroides strains whose 16S rDNA promoters were used for determining conserved nucleotides.
Table S2. Related to STAR Methods. Genetic Parts used in this study.
Contains information regarding the genetic parts used and created in this study – see Method Details.
Table S3. Related to STAR Methods. Bacterial strains, plasmids, and primers used in this study.
Contains information regarding the strains, plasmids, and primers used and created in this study – see Method Details.
(A) Wildtype B. thetaiotaomicron grows normally in 100 ng/mL aTC. Culture densities over time of wildtype B. thetaiotaomicron diluted into TYG media with varying concentrations of aTC are shown. (B, C) Expression from the P1TDPGH023 promoter can be modulated over a 100-fold range of aTC concentration from 0.01 ng/mL to 1 ng/mL and activates within minutes, achieving a 395-fold increase in expression within 45 minutes and maximal induction at ~120 minutes after aTC addition. Because the B. thetaiotaomicron doubling rate is 47.1 minutes in the conditions tested, this synthetic promoter therefore activates gene expression in response to aTC within one round of cell division. Dashed lines represents the average luminescence value from the vector control at mid-exponential phase. Error bars represent the standard deviation of three biological replicates on separate days. (D) Culture densities of Bt::tetR harboring antibacterial effectors under the control of P1TDPGH023. Bt::tetR encoding either of two highly toxic antibacterial effectors fused to periplasmic signal sequences (“ss”) from B. fragilis under the control of P1TDPGH023 grow at wildtype rates unless expression is induced with aTC. Error bars represent the standard deviation of two biological replicates on separate days. Asterisks indicate the earliest timepoint with significant (p < 0.002) differences compared to uninduced controls.
(A and B) The impact of RBS variation on promoter activity is generally conserved between constitutive and inducible promoters. Luminescence values and standard deviations from the P1TDPRBS-NanoLuc constructs (Figure 4A), in the absence (A) and presence (B) of aTC, were plotted against previously published luminescence values and standard deviations of the same RBSs fused to the constitutive BT1311 promoter (Mimee et al., 2015). (C) mRNA levels generally correspond to the strength of their promoters based on promoter-NanoLuc fusions. Luminescence values and standard deviations from the promoter-NanoLuc constructs (Figure 4C), were plotted against previously published values and standard deviations of the mRNA levels generated by the corresponding promoter as determined by microarray (Sonnenburg et al., 2005).
Promoter replacement of the lpxF homologue (BACOVA_04598) in B. ovatus ATCC 8483 with the self-contained, ~1kb inducible expression cassette, TetR-P1TDPB1, provides aTC-dependent regulatory control of PMB resistance in a one-step procedure. (A) Genomic position and replacement of the native promoter (upstream 62 basepairs) of BACOVA_04598, with TetR-P1TDPB1. (B) PMB MICs for the wildtype (tdk), lpxF deletion (BACOVA_04598), and aTC-regulated lpxF (Bo::P1TDPB1-BACOVA_04598) strains were determined with E-test strips.
Gnotobiotic mice monocolonized with B. thetaiotaomicron were given aTC (100 μg/mL in drinking water) beginning after collection of fecal samples on day 4 and continuing for 4 days (n = 6 mice/group). Mice were placed on water without inducer immediately after the fecal collection on day 8. Fecal aTC concentrations reach 22 μM at the highest point, about an order of magnitude below the lowest toxic concentration observed in vitro (216 μM).
(A) Addition of aTC to drinking water does not impact colonization of B. thetaiotaomicron in monocolonized gnotobiotic mice (green, black, and red lines), in gnotobiotic mice carrying defined human communities (orange line), or in the presence of a complete murine microbiota (blue line). Colony forming units (CFU) were determined per gram of feces collected each day from mice in Figure 6. For gnotobiotic mice monocolonized with B. thetaiotaomicron or Bt::tetR P1TDP-NanoLuc, aTC was given for 48 hours in drinking water on days 3–5 and days 17–19, while mice carrying a human or murine microbiota were given aTC on days 3–5 (n ≥ 6 mice per data point). Grey shading designates days when mice were administered aTC. (B) P1TDPGH023 activity was measured in luminal contents collected along the length of the gastrointestinal tract (upper, middle, and lower small intestine; cecum; upper and lower large intestine) on day 19 from mice in Figure 6A. Asterisks indicate significant induction (p < 0.05) as compared to the same GI location in mice colonized with Bt::tetR P1TDPGH023-NanoLuc but not provided aTC in the drinking water. (C) Addition of aTC to drinking water does not impact colonization of B. thetaiotaomicron along the length of the gut. Day 19 CFU measurements from each gastrointestinal section from mice from Figure 6A are shown (n ≥ 4 mice per data point). (D) Community composition of gnotobiotic mice (n = 6) colonized with Bt::tetR P1TDP GH023-NanoLuc and 13 prominent human gut microbes (Figure 6C) does not significantly change in response to aTC. Inducer was provided in drinking water for 48 hours from days 3–5 and community composition monitored by species-specific quantitative polymerase chain reaction (qPCR) and reported as mean percentage of the total. (E). Alpha diversity of defined human microbial communities in mice (n = 6) from Figure 6C before, during, and after aTC administration. Using each of the described alpha diversity metrics, no significant differences exist between the diversity at day 3 (the last day before aTC administration) and any other timepoint. (F) Beta diversity of defined human microbial communities from mice described in Figure 6C, as calculated from Hellinger distances.
(A) BT0455 in B. thetaiotaomicron is responsible for liberating sialic acid from the host mucosa. Free sialic acid concentrations were measured by LC-MS in fecal pellets of germfree mice and gnotobiotic mice monocolonized with wildtype B. thetaiotaomicron or Bt::tetR BT0455 (n = 4 mice per group). (B) Because sialidase activity likely provides a benefit to B. thetaiotaomicron by exposing mucosal nutrients, BtRS was engineered with multiple copies of tetR to avoid loss of repressor activity by spontaneous mutation. (C) Ex vivo sialidase assay for determination of enzyme activity in fecal pellets. Sialidase activity kinetics were measured with the indicated dilutions of lysed bacterial cells from fecal pellets of gnotobiotic mice monocolonized with wildtype B. thetaiotaomicron. Enzyme reactions contained the artificial substrate 2-O-(p-Nitrophenyl)-tropheacetylneuraminic acid (pNP-SA), which is cleaved by BT0455 to release sialic acid and p-Nitrophenyl (pNP), the latter of which was measured by LC-MS. The initial velocities of this reaction were measured every 5 minutes at 37°C for 30 minutes, and a best-fit line and its slope was determined to ensure that enzyme concentrations in the lysate were directly proportional to the initial velocities. (D) Sialidase activity and (E) free sialic acid in feces was measured for Bt::3xtetR (green line), Bt::3xtetR BT0455 (red line) for the first four days (for sialidase activity) or for the full experiment (for free sialic acid) shown in Figure 7. The average values and standard deviations were then used for shaded boxes shown in Figure 7B and 7C, respectively. (F) Transit time of exogenous sialic acid through the intestinal tract. Gnotobiotic mice monocolonized with Bt::tetR BT0455 were gavaged with 5 mg of sialic acid and sialic acid in fecal pellets were measured every 3 hours for 48 hours (n = 2 – 6 mice per timepoint).
Highly active promoters were identified from RNA-seq data, and conserved regions determined by alignment of homologous promoters collected from genome sequences. Next, a systematic approach identified regions permissible for tetO2 operator placement, and experimental validation confirmed that these engineered promoters achieve high promoter activity and tight repression in the presence of the repressor. Further, additional performance tests were carried out determine whether functional and regulatory integrity was maintained across diverse Bacteroides species. For organisms that lack phage attachment sites (used by the pNBU2 integrating vector) or replicating plasmids, single-unit systems (Figure 5) could potentially be encoded in a transposon for delivery into the genome. Lastly, performance of the regulatable platform was determined in vivo to confirm that the gut environment does not hinder the functionality of the engineered promoter.