Abstract
Background
Delayed cerebral infarction (DCI) is a major cause of morbidities after aneurysmal subarachnoid hemorrhage (SAH) and typically starts at day 4 to 7 after initial hemorrhage. MicroRNAs (miRNAs) play an important role in posttranscriptional gene expression control, and distinctive patterns of circulating miRNA changes have been identified for some diseases. We aimed to investigate miRNAs that characterize SAH patients with DCI compared with those without DCI.
Methods and Results
Circulating miRNAs were collected on day 7 after SAH in healthy, SAH‐free controls (n=20), SAH patients with DCI (n=20), and SAH patients without DCI (n=20). We used the LASSO (least absolute shrinkage and selection operator) method of regression analysis to characterize miRNAs associated with SAH patients with DCI compared with those without DCI. In the 28 dysregulated miRNAs associated with DCI and SAH, we found that a combination of 4 miRNAs (miR‐4532, miR‐4463, miR‐1290, and miR‐4793) could differentiate SAH patients with DCI from those without DCI with an area under the curve of 100% (95% CI 1.000–1.000, P<0.001). This 4‐miRNA combination could also distinguish SAH patients with or without DCI from healthy controls with areas under the curve of 99.3% (95% CI 0.977–1.000, P<0.001) and 82.0% (95% CI 0.685–0.955, P<0.001), respectively.
Conclusions
We found a 4‐miRNA combination that characterized SAH patients with DCI. The findings could guide future mechanistic study to develop therapeutic targets.
Keywords: biomarker, delayed cerebral infarction, miRNA, stroke, subarachnoid hemorrhage
Subject Categories: Intracranial Hemorrhage
Introduction
Aneurysmal subarachnoid hemorrhage (SAH) accounts for about 3% to 5% of stroke and is an important cause of stroke in young populations, causing significant socioeconomic burden worldwide.1 Up to two thirds of SAH patients experience cognitive impairment and impaired quality of life and may not be able to return to their previous work.2, 3, 4, 5, 6, 7, 8
Delayed cerebral infarction (DCI) occurs in up to 44% of SAH patients and typically starts at day 4 to 7 after the initial hemorrhage.9, 10, 11 DCI is a well‐established and relevant clinical surrogate marker for neurological outcome after SAH.12 Numerous factors including age, initial neurological impairment, intraventricular hemorrhage, SAH load, and aneurysm size are reported to be associated with the development of DCI.13, 14, 15, 16
MicroRNAs (miRNAs) are a family of small (19–23 base pairs), noncoding, and deeply conserved RNA molecules that regulate target gene expression at a posttranscriptional level by inhibiting mRNA translation17 or destabilizing mRNA molecules.18 There are 1881 miRNAs in human genomes, 296 of which are currently annotated as high confidence, according to miRBase 21.0.19 Circulating miRNAs have been demonstrated to be promising diagnostic or prognostic biomarkers for cerebrovascular conditions such as myocardial infarction,20 atherosclerotic diseases,21 stroke,22 cerebral infarction,23 hypertension,24 intracranial aneurysms,25, 26, 27, 28 and SAH.29, 30
In the current study, we aimed to investigate miRNAs that characterized SAH patients with DCI compared with those without DCI.
Methods
Patient Recruitment and Sample Collection
The study was approved by the Joint NTEC‐CUHK (New Territories East Cluster‐Chinese University of Hong Kong) Clinical Research Ethics Committee, and written informed consent was obtained from all participants or their next of kins. Circulating miRNAs were collected from healthy controls (n=20), SAH patients with DCI (n=20) at day 7 after SAH, and SAH patients without DCI (n=20) at day 7 after SAH. SAH patients were recruited from Prince of Wales Hospital, Chinese University of Hong Kong, Hong Kong Special Administrative Region, People's Republic of China, between 2012 and 2013. Ruptured cerebral aneurysms were diagnosed by computer tomography angiography. For SAH patients, the healthy controls (n=20) were recruited from the family members of SAH patients with the no major medical problem (including no smoking history and hypertension). Participant characteristics are shown in Table 1.
Table 1.
Participant Characteristics
| Control (n=20) | SAH With DCI (n=20) | SAH Without DCI (n=20) | P Value | |
|---|---|---|---|---|
| Demographics | ||||
| Age, y | 50±17 | 59±12 | 59±11 | |
| Female, % | 65 (13) | 45 (9) | 65 (13) | |
| WFNS grade on admission, % | ||||
| 1–2 | ··· | 60 (12) | 75 (15) | 0.311 |
| 3–5 | ··· | 40 (8) | 25 (5) | |
| CT feature on admission | ||||
| Fisher grade 3/4, % | ··· | 100 (20) | 100 (20) | 1.000 |
| Risk factors, % | ||||
| Hypertension | ··· | 55 (11) | 30 (6) | 0.200 |
| Smokers | ··· | 10 (2) | 0 (0) | 0.487 |
| Outcome | ||||
| Infarction, % | ··· | 65 (13) | 0 (0) | <0.001a |
| mRS 3 mo, median | ··· | 2 | 1 | 0.034a |
| mRS 3 mo, >2 | ··· | 45 (9) | 15 (3) | 0.082 |
| mRS 3 mo, ≤2 | ··· | 55 (11) | 85 (17) | |
Data are % (N), mean±SD, or median. CT indicates computed tomography; DCI, delayed cerebral infarction; mRS 3 mo, modified Rankin Scale 3‐month; SAH, subarachnoid hemorrhage; WFNS, World Federation of Neurosurgical Societies.
P<0.05.
Delayed Cerebral Infarction
DCI is defined as a new cerebral infarction identified on computed tomography after exclusion of procedure‐related infarctions.3, 12 Procedure‐related infarction was defined as new hypodensity appearing on the posttreatment computed tomography at 12 to 24 hours after aneurysm treatment. All recruited patients had delayed computed tomography of the brain at 2 to 3 weeks after presentation available for assessment. The diagnoses of DCI were made by consensus of 2 neuroradiologists.
Quantitative Polymerase Chain Reaction
Peripheral blood samples were obtained using EDTA tubes with standard procedures. Samples were placed on ice immediately and centrifuged at 1000g for 15 minutes at 4°C. Plasma fraction was aliquoted and stored at −80°C. RNA isolation and quantitative real‐time polymerase chain reaction were performed, as described previously.29 Briefly, serum was prepared by adding 20% wt/vol CaCl2 (Sigma‐Aldrich) into the plasma samples at a ratio of 1:100, followed by clotting overnight and centrifugation. Total RNA was isolated from pooled serum samples using the miRNeasy Serum/Plasma Kit following the manufacturer's instructions (Qiagen). The quantity of the extracted RNA was determined using a Nanodrop 2000 UV‐Vis spectrophotometer (Thermo Scientific). The results were analyzed with the Applied Biosystems SDS software for Ct values and melting curve analyses. Of the 99 possible deregulated miRNAs associated with SAH,29 we selected the top 20 differentially regulated miRNAs between SAH patients with and without DCI; the top 8 miRNAs between SAH patients with DCI and healthy controls (2 overlapped); and miR‐132‐3p and miR‐324‐3p, the 2 previously reported circulating miRNAs in SAH patients29 from a total of 28 unique miRNAs for expression analysis. Table S1 is the list of quantitative polymerase chain reaction primers for miRNA expression profiling used in this study.
Statistical Analyses
Heat maps with hierarchical clustering and principal component analysis (PCA) were computed and visualized using the functions pHeatmap and prcomp, respectively, from the stats package for R version 3.0.1 (R Foundation for Statistical Computing). The R package pROC was used to plot and visualize receiver operating characteristic (ROC) curves to compute the area under the curve (AUC) and confidence intervals to evaluate the performance of the miRNA‐based classifier.31 Different R packages and functions were used to construct the 4 classification models to identify the miRNA‐based classifiers that could differentiate SAH patients with or without DCI after SAH—the linear support vector machine (R package e1071, function svm, kernel=“linear”), nonlinear support vector machine (R package e1071, function svm, kernel=“radial”), linear discriminant analysis (R package MASS, function lda), and logistic regression (R package stats, function glm, family=“binomial”)—as previously described.32 The R package glmnet was used to fit the LASSO (least absolute shrinkage and selection operator) regression model via penalized maximum likelihood to define the miRNA‐based classifier.33 The value λ=0.07765104 based on cross‐validation, and t tests were used to compute the P value (Tables 1 and 2). A value of P<0.05 was regarded as statistically significant.
Table 2.
Performance of Individual MicroRNAs in Subarachnoid Hemorrhage Subtypes Classification
| miRNA | AUC (95% CI) | P Value |
|---|---|---|
| miR‐4532 | 0.9475 (0.873–1.000) | 1.80E‐08 |
| miR‐4793 | 0.93 (0.584–1.000) | 2.14E‐07 |
| miR‐1290 | 0.9525 (0.897–1.000) | 3.54E‐07 |
| miR‐421 | 0.9175 (0.836–0.999) | 3.24E‐06 |
| miR‐4492 | 0.895 (0.798–0.992) | 7.26E‐06 |
| miR‐574 | 0.8625 (0.753–0.972) | 2.70E‐05 |
| miR‐4689 | 0.86 (0.748–0.972) | 2.87E‐05 |
| miR‐4449 | 0.855 (0.741–0.969) | 3.16E‐05 |
| miR‐93‐5p | 0.835 (0.713–0.957) | 9.31E‐05 |
| miR‐1268 | 0.795 (0.648–0.942) | 2.69E‐04 |
| miR‐4497 | 0.8175 (0.685–0.950) | 3.77E‐04 |
| miR‐3178 | 0.7875 (0.640–0.935) | 6.71E‐04 |
| miR‐4674 | 0.765 (0.602–0.928) | 7.94E‐04 |
| miR‐297 | 0.81 (0.671–0.949) | 9.15E‐04 |
| miR‐4463 | 0.815 (0.677–0.953) | 1.03E‐03 |
| miR‐4690 | 0.7325 (0.568–0.897) | 5.38E‐03 |
| miR‐4459 | 0.73 (0.560–0.900) | 9.75E‐03 |
| miR‐3651 | 0.705 (534–0.876) | 1.99E‐02 |
| miR‐132 | 0.7125 (0.547–0.878) | 2.37E‐02 |
| miR‐4433 | 0.67 (0.497–0.843) | 7.86E‐02 |
| miR‐339‐5p | 0.6625 (0.490–0.835) | 7.95E‐02 |
| miR‐125a | 0.66 (0.483–0.837) | 1.58E‐01 |
| miR‐324 | 0.59875 (0.419–0.779) | 2.29E‐01 |
| miR‐4440 | 0.5675 (0.385–0.750) | 4.16E‐01 |
| miR‐4454 | 0.5075 (0.321–0.694) | 7.66E‐01 |
| miR‐27b | 0.49 (0.304–0.676) | 8.62E‐01 |
| miR‐27a | 0.485 (0.299–0.671) | 9.24E‐01 |
| miR‐15a | 0.525 (0.339–0.711) | 9.52E‐01 |
AUC indicates area under the receiver operating characteristic curve.
Results
Hierarchical Clustering of miRNAs Associated With SAH With and Without DCI
We performed unsupervised hierarchical clustering to graphically represent the averaged microarray expressions of the 28 miRNAs derived from the 3 cohorts for exploratory analysis: healthy controls, SAH with DCI, and SAH without DCI. The dendrogram represented with the heat map showing the entire data matrix of the 3 cohorts indicated that the expression profiles of the 28 miRNAs among the 3 cohorts are distinctive, with clear cluster dissimilarity (results not shown), and suggested that a subset of the 28 miRNAs could characterize SAH patients with DCI from those without DCI.
We next performed quantitative polymerase chain reaction array analyses to quantify the expression levels of the 28 SAH‐associated miRNAs (Figure 1A) and used PCA for pattern recognition to evaluate the distance connectivity of the 2 SAH subtypes (Figure 1B). PCA generated 2 distinct clusters with minimal overlap of the 2 SAH subtypes, highlighting the possibility of characterizing the disease subtypes using a miRNA combination.
Figure 1.
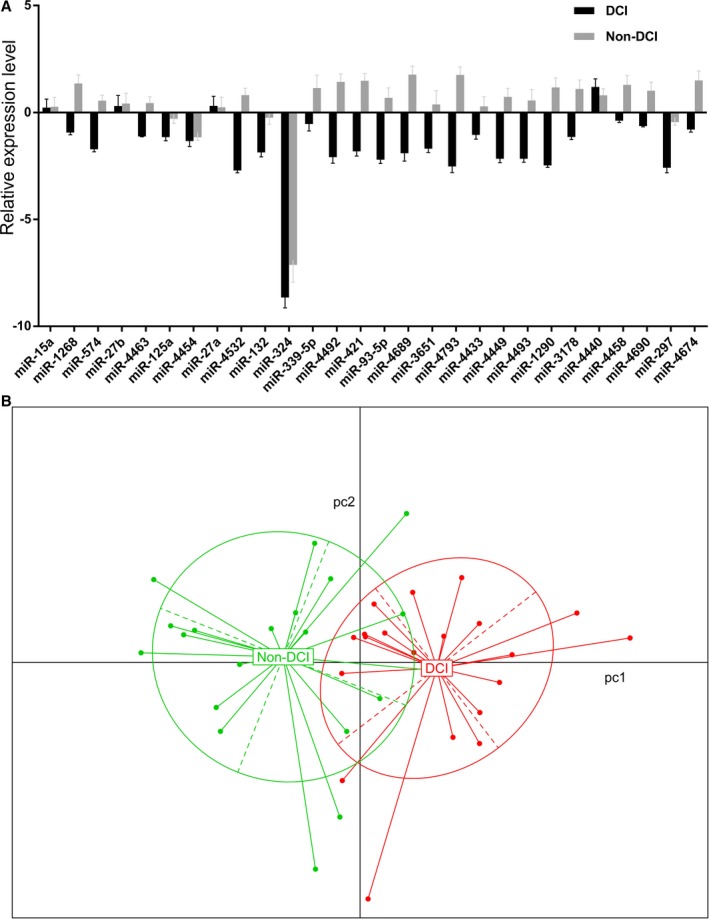
A, Quantitative polymerase chain reaction array of the 28 subarachnoid hemorrhage (SAH)‐associated microRNAs. The expression levels were normalized to the control and expressed as ΔΔCt. B, Principal component analysis of the 2 SAH subtypes with or without delayed cerebral infarction (DCI).
miRNAs That Characterized SAH With DCI
Given the distinctive pattern of miRNA expression profile between the 2 SAH subtypes, we initially evaluated the performance of individual miRNAs to characterize SAH patients with and without DCI using ROC analysis.34 Based on the sensitivity (true‐positive rate) and specificity (false‐positive rate) at varied threshold levels, the ROC analysis allowed us to select possibly optimal models for DCI discrimination.
We plotted the ROC curves for individual miRNAs from a total of 28 miRNAs according to their quantitative polymerase chain reaction–measured expression levels in the SAH groups with DCI and without DCI. The results are shown in Table 2. The AUCs of 8 miRNAs can achieve >85% (P<0.0005), and 4 of them—miR‐4532, miR‐4793, miR‐1290, and miR‐421—are >90%. For miR‐1290, the AUC, specificity, and sensitivity were 95.3% (95% CI 0.897–1), 0.900, and 0.850, respectively, at a cutoff level of 8.653.
We postulated that a combination of miRNAs would show characteristics for SAH with versus without DCI. We examined each miRNA combination of the 28 SAH‐associated miRNAs, based on their sensitivity, specificity, and accuracy, using the 4 different classifier algorithms: linear support vector machine, nonlinear support vector machine, linear discriminant analysis, and logistic regression. The results are shown in Figure 2. All 3 readouts improved while increasing the number of miRNAs contained in each classifier.
Figure 2.
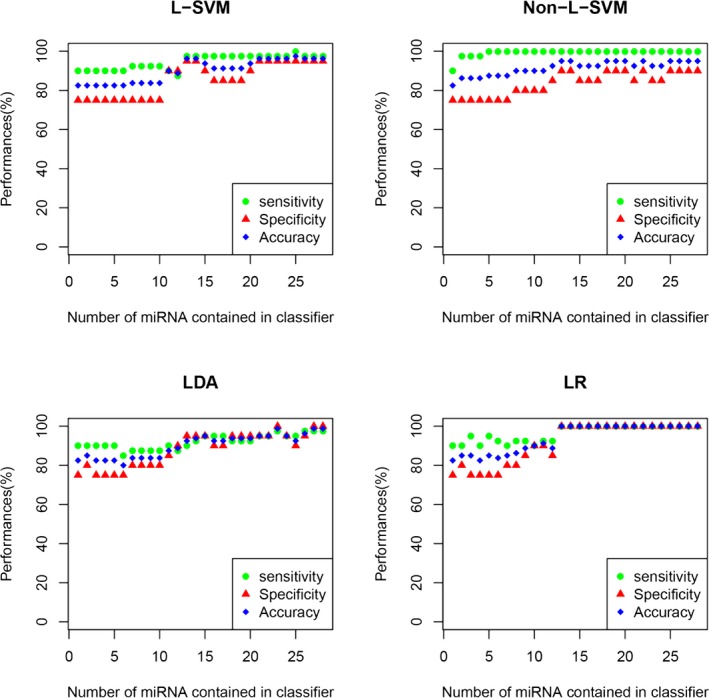
Four classification models, namely, linear support vector machine (L‐SVM), nonlinear support vector machine (non‐L‐SVM), linear discriminant analysis (LDA), and logistic regression (LR) models, were applied to construct microRNA (miRNA) classifiers. The performance of the miRNA‐based classifiers was determined by leave‐one‐out cross‐validation. The x‐axis denotes the number of miRNAs contained in the classifier, whereas the y‐axis indicates the performance of the classifiers including sensitivity (green circle), specificity (blue square), and accuracy (red triangle). In each group with the same miRNA number, only the classifier displaying the best performance is presented.
We next performed the LASSO regression analysis involving penalizing the absolute size of the regression coefficients to determine the accuracy using a combination of miRNAs to distinguish SAH with and without DCI (Figure 3A). The results are shown in Figure 3B. ROC analysis indicated that the AUC achieved 100% (95% CI 1–1, P<0.0001) using 4 miRNAs, namely, miR‐4463, miR‐4532, miR‐4793, and miR‐1290, according to the LASSO regression model and the ROC analysis using the following formula: ln(Y/1−Y)=−4.29759355+miR‐4463×0.16589606+miR‐4532×0.43352951+miR‐4793×0.09569081+miR‐1290×0.2602013. PCA analysis indicated that the 4‐miRNA combination could produce 2 nonoverlapping clusters to unambiguously differentiate SAH with and without DCI.
Figure 3.
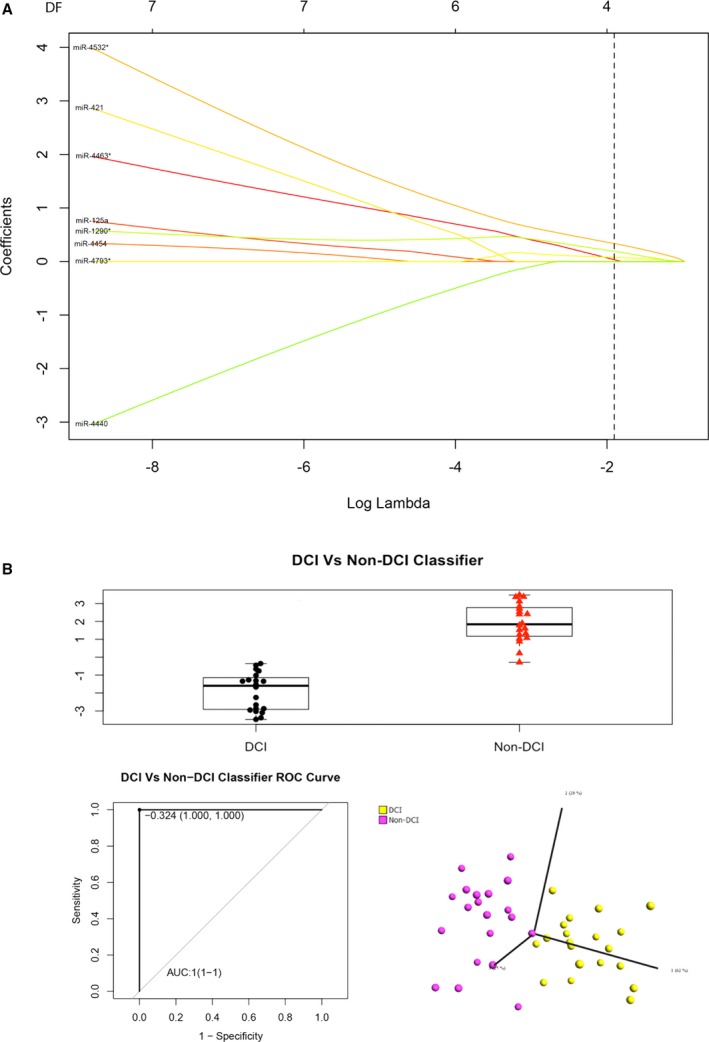
A, LASSO (least absolute shrinkage and selection operator) regression analysis, (B) the box‐and‐whisker plot (upper panel), receiver operating characteristic (ROC) curve (bottom left panel), and principal component analysis (bottom right panel) of the 4 microRNAs (miR‐4532, miR‐4463, miR‐1290, and miR‐4793‐3p) in the subarachnoid hemorrhage groups with and without delayed cerebral infarction (DCI) in the classification algorithm. AUC indicates area under the curve.
Interestingly, the 4 miRNAs were strongly repressed in SAH blood samples with DCI but activated in the non‐DCI samples compared with healthy controls (Figure 4). The biphasic expressions of these 4 miRNAs in the SAH patients with or without DCI also provided discrimination with an AUC >80% by ROC analyses to differentiate the SAH patients with or without DCI from healthy controls with AUCs of 99.3% (95% CI 0.977–1, P<0.0001) and 82.0% (95% CI 0.685–0.955, P<0.0005), respectively. The results supported the disease‐specific nature of the 4 miRNAs.
Figure 4.
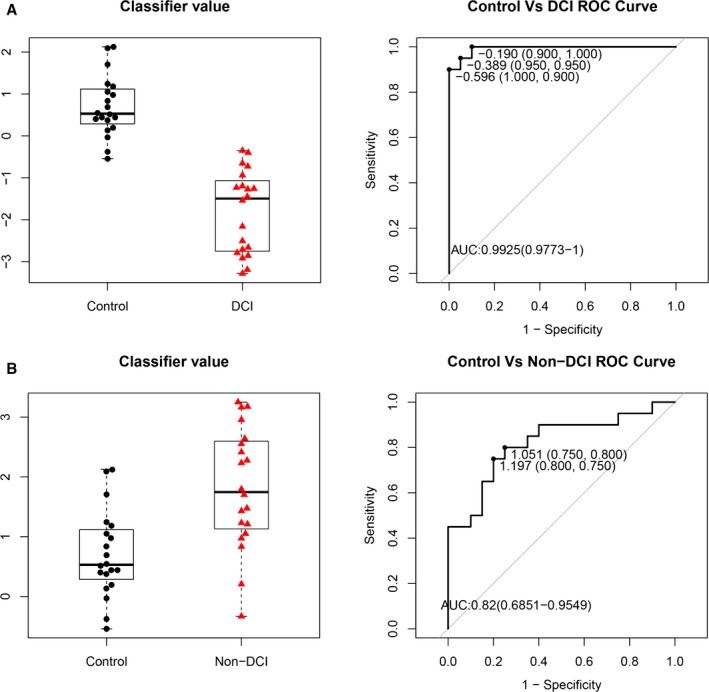
Box‐and‐whisker plots (left panel) and receiver operating characteristic curves (right panel) of the 4 microRNAs (miR‐4532, miR‐4463, miR‐1290, and miR‐4793‐3p) in (A) subarachnoid hemorrhage (SAH) patients with delayed cerebral infarction (DCI) vs control and (B) SAH patients without DCI (non‐DCI) vs control, using the R package. AUC indicates area under the curve.
Discussion
In this study, both the heat map and PCA analysis suggested that a combination of miRNAs could characterize SAH patients with DCI from those without DCI (Figure 1). We demonstrated that a number of miRNAs could offer good to excellent discriminatory power between SAH with and without DCI (Table 2). LASSO regression analysis and PCA analysis further demonstrated that a 4‐miRNA combination (miR‐4463, miR‐4532, miR‐4793, and miR‐1290) could produce 2 nonoverlapping clusters to unambiguously differentiate SAH with and without DCI, with an AUC of 100% (95% CI 1–1, P<0.0001) (Figure 3). In addition, the classifier can also distinguish the healthy controls from each SAH subtype with or without DCI (Figure 4), highlighting its disease specificity.
Recently, studies of miRNAs in serum and cerebrospinal fluid identified biomarkers associated with occurrence of SAH, including miR‐92a, let‐7b, miR‐204‐5p, miR‐223‐3p, miR‐337‐5p, miR‐451a, miR‐489, miR‐508‐3p, miR‐514‐3p, miR‐516‐5p, miR‐548, miR‐599, miR‐937, miR‐1224‐3p and miR‐1301.29, 35, 36 Our previous genomewide serum miRNA expression profiling also suggested that miR‐132‐3p and miR‐324‐3p could be potential biomarkers for SAH.29, 35, 36 Styllis and others investigated cerebrospinal fluid miRNAs in 10 SAH patients with and 10 without cerebral vasospasm and identified miR‐27a‐3p, miR‐516a‐5p, miR‐566, and miR‐1197 as potential biomarkers for cerebral vasospasm but not DCI.36 The identification of miRNAs associated with DCI after SAH has been understudied, hindering understanding and development of therapeutic strategies for DCI after SAH.
Given that the downstream targets and functional specificities of the 4 miRNAs (miR‐4463, miR‐4532, miR‐4793 and miR‐1290) are uncharacterized, we performed gene‐centric analysis to identify their common and unique potential targets using 3 algorithms: TargetScan, miRanda, and PITA. Considerable overlaps of their downstream targets were shared by these 4 miRNAs, inferring that these miRNAs may act on common regulatory pathways mediated by their shared targets for the development of DCI after SAH (Figure 5). KEGG (Kyoto Encyclopedia of Genes and Genomes) and gene ontology analyses also consistently revealed that the targets of these 4 miRNAs associated with multiple developmental pathways including Wnt signaling pathway, hedgehog and oxytocin signaling pathways. These pathways are broadly involved in neurogenesis and nervous system development, suggesting that these 4 miRNAs may function as causative factors in DCI pathology associated with SAH (Table S2).
Figure 5.
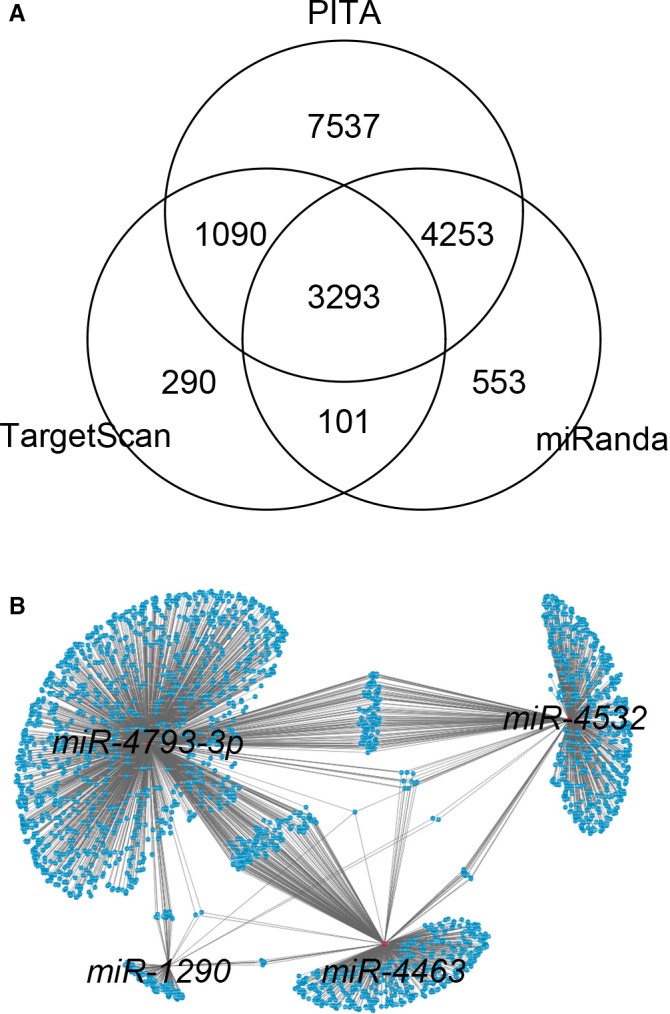
A, Gene‐centric analysis of the putative targets of the 4 microRNAs (miRNAs; miR‐4532, miR‐4463, miR‐1290, and miR‐4793‐3p) using TargetScan, miRanda, and PITA. B, Topological network view of the downstream targets shared by the 4 miRNAs, constructed by Cytoscape.
The 4 novel miRNAs—miR‐4463, miR‐4532, miR‐4793, and miR‐1290—have not been associated with SAH pathology. miR‐4532 has been associated with breast cancer cell chemotherapy resistance.37 miR‐4793 expression was elevated in liver metastases of sporadic colorectal cancer patients.38 miR‐1290 has been widely associated with different types of cancers including colorectal cancer, cervical cancer, non–small cell lung cancer, breast cancer, hepatocellular carcinoma, gastric cancer, laryngeal carcinoma, lymphoblastic leukemia, esophageal squamous cell carcinoma, lung adenocarcinoma, prostate cancer, pancreatic cancer, and bladder carcinoma, as well as oral submucosal fibrosis, nonalcoholic fatty liver disease, and chronic rhinosinusitis.39, 40, 41 miR‐4463 has been shown to serve as a biomarker for a common female reproductive disease called polycystic ovary syndrome and arteriosclerosis obliterans.42, 43
Our current study had several limitations. First, our SAH‐associated miRNA pools were obtained from patients at day 7 after SAH, which is a commonly reported time point for etiological study of DCI. Time‐course expression profiling of serum miRNAs in SAH patients at multiple time points could investigate stage/time‐specific miRNAs for SAH with or without DCI in future. Second, the current study did not investigate the pathophysiological roles of these miRNAs in DCI after SAH and propose therapeutic targets. Third, the sample size was small, and subgroup analyses such as admission neurological grade and the mode of aneurysm treatment were not performed. In addition, the sample size did not allow meaningful matched group analyses. Fourth, 28 miRNAs were selected to find the important miRNAs involved to guide further mechanistic study to find therapeutic targets. Fifth, there could be a limitation in diagnosing DCI with computed tomography hypodensity, as magnetic resonance imaging was not routinely performed in our patients. Sixth, validation with another patient cohort would be warranted to confirm our findings. Seventh, cerebrospinal fluid samples were not collected for miRNA analysis and should be considered in future study.
Our work was important as it provides an understanding of the characteristics of circulating miRNAs associated with DCI after SAH. The 4 miRNAs might play distinct roles in DCI after SAH. Further investigations into their roles might bring new therapeutic targets for DCI after SAH. Future time‐course studies of miRNAs might also identify SAH patients at risk of subsequent DCI development for stringent monitoring in neurocritical care units and timely treatment.
Conclusions
We found a 4‐miRNA combination (miR‐4532, miR‐4463, miR‐1290, and miR‐4793) that characterized SAH patients with DCI. The findings could guide future mechanistic study to develop therapeutic targets.
Sources of Funding
This work was supported by the Chinese University of Hong Kong Direct Grant for Research (MD11782) and a collaborative funding from SDIVF R&D Centre.
Disclosures
None.
Supporting information
Table S1. MicroRNA Primer Sets for Quantitative Polymerase Chain Reaction
Table S2. Gene Ontology and KEGG Enrichment Analyses of the Targets of miR‐4532, miR‐4463, miR‐1290 and miR‐4793‐3p
(J Am Heart Assoc. 2017;6:e005363 DOI: 10.1161/JAHA.116.005363.)28442458
References
- 1. Lovelock CE, Rinkel GJ, Rothwell PM. Time trends in outcome of subarachnoid hemorrhage: population‐based study and systematic review. Neurology. 2010;74:1494–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al‐Khindi T, Macdonald RL, Schweizer TA. Cognitive and functional outcome after aneurysmal subarachnoid hemorrhage. Stroke. 2010;41:e519–e536. [DOI] [PubMed] [Google Scholar]
- 3. Wong GK, Lam S, Ngai K, Wong A, Mok V, Poon WS; Cognitive Dysfunction after Aneurysmal Subarachnoid Haemorrhage I . Evaluation of cognitive impairment by the montreal cognitive assessment in patients with aneurysmal subarachnoid haemorrhage: prevalence, risk factors and correlations with 3 month outcomes. J Neurol Neurosurg Psychiatry. 2012;83:1112–1117. [DOI] [PubMed] [Google Scholar]
- 4. Wong GK, Poon WS, Boet R, Chan MT, Gin T, Ng SC, Zee BC. Health‐related quality of life after aneurysmal subarachnoid hemorrhage: profile and clinical factors. Neurosurgery. 2011;68:1556–1561; discussion 1561. [DOI] [PubMed] [Google Scholar]
- 5. Scott RB, Eccles F, Molyneux AJ, Kerr RS, Rothwell PM, Carpenter K. Improved cognitive outcomes with endovascular coiling of ruptured intracranial aneurysms: neuropsychological outcomes from the International Subarachnoid Aneurysm Trial (ISAT). Stroke. 2010;41:1743–1747. [DOI] [PubMed] [Google Scholar]
- 6. Wong GK, Wun Tam YY, Zhu XL, Poon WS. Incidence and mortality of spontaneous subarachnoid hemorrhage in Hong Kong from 2002 to 2010: a Hong Kong hospital authority clinical management system database analysis. World Neurosurg. 2014;81:552–556. [DOI] [PubMed] [Google Scholar]
- 7. de Rooij NK, Linn FH, van der Plas JA, Algra A, Rinkel GJ. Incidence of subarachnoid haemorrhage: a systematic review with emphasis on region, age, gender and time trends. J Neurol Neurosurg Psychiatry. 2007;78:1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Passier PE, Visser‐Meily JM, Rinkel GJ, Lindeman E, Post MW. Life satisfaction and return to work after aneurysmal subarachnoid hemorrhage. J Stroke Cerebrovasc Dis. 2011;20:324–329. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt JM, Rincon F, Fernandez A, Resor C, Kowalski RG, Claassen J, Connolly ES, Fitzsimmons BF, Mayer SA. Cerebral infarction associated with acute subarachnoid hemorrhage. Neurocrit Care. 2007;7:10–17. [DOI] [PubMed] [Google Scholar]
- 10. Dorsch NW, King MT. A review of cerebral vasospasm in aneurysmal subarachnoid haemorrhage part I: incidence and effects. J Clin Neurosci. 1994;1:19–26. [DOI] [PubMed] [Google Scholar]
- 11. Ionita CC, Baker J, Graffagnino C, Alexander MJ, Friedman AH, Zaidat OO. Timing of symptomatic vasospasm in aneurysmal subarachnoid hemorrhage: the effect of treatment modality and clinical implications. J Stroke Cerebrovasc Dis. 2010;19:110–115. [DOI] [PubMed] [Google Scholar]
- 12. Vergouwen MD, Vermeulen M, van Gijn J, Rinkel GJ, Wijdicks EF, Muizelaar JP, Mendelow AD, Juvela S, Yonas H, Terbrugge KG, Macdonald RL, Diringer MN, Broderick JP, Dreier JP, Roos YB. Definition of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage as an outcome event in clinical trials and observational studies: proposal of a multidisciplinary research group. Stroke. 2010;41:2391–2395. [DOI] [PubMed] [Google Scholar]
- 13. Sundt TM Jr, Piepgras DG, Fode NC, Meyer FB. Giant intracranial aneurysms. Clin Neurosurg. 1991;37:116–154. [PubMed] [Google Scholar]
- 14. Kassell NF, Torner JC, Jane JA, Haley EC Jr, Adams HP. The international cooperative study on the timing of aneurysm surgery. Part 2: surgical results. J Neurosurg. 1990;73:37–47. [DOI] [PubMed] [Google Scholar]
- 15. Rabinstein AA, Friedman JA, Weigand SD, McClelland RL, Fulgham JR, Manno EM, Atkinson JL, Wijdicks EF. Predictors of cerebral infarction in aneurysmal subarachnoid hemorrhage. Stroke. 2004;35:1862–1866. [DOI] [PubMed] [Google Scholar]
- 16. Dority JS, Oldham JS. Subarachnoid hemorrhage: an update. Anesthesiol Clin. 2016;34:577–600. [DOI] [PubMed] [Google Scholar]
- 17. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozomara A, Griffiths‐Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C, Chen X, Huang J, Sun Q, Wang L. Clinical impact of circulating miR‐26a, miR‐191, and miR‐208b in plasma of patients with acute myocardial infarction. Eur J Med Res. 2015;20:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Araldi E, Chamorro‐Jorganes A, van Solingen C, Fernandez‐Hernando C, Suarez Y. Therapeutic potential of modulating microRNAs in atherosclerotic vascular disease. Curr Vasc Pharmacol. 2015;13:291–304. [PubMed] [Google Scholar]
- 22. Leung LY, Chan CP, Leung YK, Jiang HL, Abrigo JM, Wang de F, Chung JS, Rainer TH, Graham CA. Comparison of miR‐124‐3p and miR‐16 for early diagnosis of hemorrhagic and ischemic stroke. Clin Chim Acta. 2014;433:139–144. [DOI] [PubMed] [Google Scholar]
- 23. Zhou J, Zhang J. Identification of miRNA‐21 and miRNA‐24 in plasma as potential early stage markers of acute cerebral infarction. Mol Med Rep. 2014;10:971–976. [DOI] [PubMed] [Google Scholar]
- 24. Heggermont WA, Heymans S. MicroRNAs are involved in end‐organ damage during hypertension. Hypertension. 2012;60:1088–1093. [DOI] [PubMed] [Google Scholar]
- 25. Jin H, Li C, Ge H, Jiang Y, Li Y. Circulating microRNA: a novel potential biomarker for early diagnosis of intracranial aneurysm rupture a case control study. J Transl Med. 2013;11:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li P, Zhang Q, Wu X, Yang X, Zhang Y, Li Y, Jiang F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J Am Heart Assoc. 2014;3:e000972 DOI: 10.1161/JAHA.114.000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu D, Han L, Wu X, Yang X, Zhang Q, Jiang F. Genome‐wide microRNA changes in human intracranial aneurysms. BMC Neurol. 2014;14:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y, Zhang M, He H, Chen J, Zeng H, Li J, Duan R. microRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med Genomics. 2013;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su XW, Chan AH, Lu G, Lin M, Sze J, Zhou JY, Poon WS, Liu Q, Zheng VZ, Wong GK. Circulating microRNA 132‐3p and 324‐3p profiles in patients after acute aneurysmal subarachnoid hemorrhage. PLoS One. 2015;10:e0144724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muller AH, Povlsen GK, Bang‐Berthelsen CH, Kruse LS, Nielsen J, Warfvinge K, Edvinsson L. Regulation of microRNAs miR‐30a and miR‐143 in cerebral vasculature after experimental subarachnoid hemorrhage in rats. BMC Genomics. 2015;16:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, Li SP, Xiong Y, Yuan Y, Min J, Jia WH, Jie Y, Chen MS, Chen MX, Fang JH, Zeng C, Zhang Y, Guo RP, Wu Y, Lin G, Zheng L, Zhuang SM. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case‐control study. Lancet Oncol. 2015;16:804–815. [DOI] [PubMed] [Google Scholar]
- 33. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 34. Hajian‐Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 35. Powers CJ, Dickerson R, Zhang SW, Rink C, Roy S, Sen CK. Human cerebrospinal fluid microRNA: temporal changes following subarachnoid hemorrhage. Physiol Genomics. 2016;48:361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, Kaye AH. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. 2017;126:1131–1139. [DOI] [PubMed] [Google Scholar]
- 37. Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan KG, Yin WF, Satharasinghe DA, Liew WC, Tan SW, Ong HK, Cheong SK. MiRNA transcriptome profiling of spheroid‐enriched cells with cancer stem cell properties in human breast MCF‐7 cell line. Int J Biol Sci. 2016;12:427–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sayagues JM, Corchete LA, Gutierrez ML, Sarasquete ME, Del Mar Abad M, Bengoechea O, Ferminan E, Anduaga MF, Del Carmen S, Iglesias M, Esteban C, Angoso M, Alcazar JA, Garcia J, Orfao A, Munoz‐Bellvis L. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget. 2016;7:72908–72922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, Tan W, Nandy D, Bevan GH, Longenbach S, Sun Z, Lu Y, Wang T, Thibodeau SN, Boardman L, Kohli M, Wang L. Exosomal miR‐1290 and miR‐375 as prognostic markers in castration‐resistant prostate cancer. Eur Urol. 2015;67:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mao Y, Liu J, Zhang D, Li B. MiR‐1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother. 2015;76:82–93. [DOI] [PubMed] [Google Scholar]
- 41. Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, Lim PX, Lee JZ, Tan BJ, Shyh‐Chang N, Lim EH, Lim WT, Tan DS, Tan EH, Tai BC, Soo RA, Tam WL, Lim B. Tumour‐initiating cell‐specific miR‐1246 and miR‐1290 expression converge to promote non‐small cell lung cancer progression. Nat Commun. 2016;7:11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He XM, Zheng YQ, Liu SZ, Liu Y, He YZ, Zhou XY. Altered plasma microRNAs as novel biomarkers for arteriosclerosis obliterans. J Atheroscler Thromb. 2016;23:196–206. [DOI] [PubMed] [Google Scholar]
- 43. Ding CF, Chen WQ, Zhu YT, Bo YL, Hu HM, Zheng RH. Circulating microRNAs in patients with polycystic ovary syndrome. Hum Fertil (Camb). 2015;18:22–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. MicroRNA Primer Sets for Quantitative Polymerase Chain Reaction
Table S2. Gene Ontology and KEGG Enrichment Analyses of the Targets of miR‐4532, miR‐4463, miR‐1290 and miR‐4793‐3p


