Abstract
Almost all the reported capillary convective polymerase chain reaction (CCPCR) systems to date are still limited to research use stemming from unresolved issues related to repeatability, reliability, convenience, and sensitivity. To move CCPCR technology forward toward commercialization, a couple of critical strategies and innovations are discussed here. First, single- and dual-end heating strategies are analyzed and compared between each other. Especially, different solutions for dual-end heating are proposed and discussed, and the heat transfer and fluid flow inside the capillary tube with an optimized dual-end heating strategy are analyzed and modeled. Second, real-time CCPCR is implemented with light-emitting diode and photodiode, and the real-time fluorescence detection method is compared with the post-amplification end-point detection method based on a dipstick assay. Thirdly, to reduce the system complexity, e.g., to simplify parameter tuning of the feedback control, an internal-model-control-based proportional-integral-derivative controller is adopted for accurate temperature control. Fourth, as a proof of concept, CCPCR with pre-loaded dry storage of reagent inside the capillary PCR tube is evaluated to better accommodate to point-of-care diagnosis. The critical performances of improved CCPCR, especially with sensitivity, repeatability, and reliability, have been thoroughly analyzed with different experiments using influenza A (H1N1) virus as the detection sample.
I. INTRODUCTION
While microfluidic-based polymerase chain reaction (PCR) offers significant advantages, there are nevertheless several limitations of current implementations where their convenience, cost, and performance could be considerably improved.1–3 First, microfluidic PCR amplification has to be performed on really sophisticated and costly thermal cyclers that provide repeated and precise (within ±1 °C or better) temperatures, respectively, for different reaction stages, including denaturing, annealing, and extension.4 Second, every PCR cycle has some “wasted” time in transitioning between two or three reaction stages as well as additional dwell time for each stage, both of which contribute over the 30–40 cycles to significantly longer time than theoretically needed for a specified amount of amplification. Further, time spent in transition tends to degrade specificity due to false priming and extension at non-optimal temperatures.
Many efforts have been made to improve the microfluidic PCR, ranging from simplification of instrumentation, reduction in sample or reaction volume, and decrease in amplification time needed to produce significant amplicon for detection or quantification. To simplify thermal cycling, one alternative to temporal thermal cycling PCR is continuous-flow PCR, where the reaction volume is pushed through a long (often serpentine) channel with distinct zones maintained at temperatures needed for melting, annealing, and priming.5,6 However, such continuous-flow PCR does not appear to offer substantially shorter amplification times, nor simpler supporting instrumentation, partly due to requirements of active pumping. To reduce amplification time, small size PCR reactors as well as high energy power based heating strategies have been developed to speed up the ramping rate of thermal cycling.7,8 However, detection sensitivity may be decreased by small size reactors with low quantity of templates.9 Device complexity and cost will be increased with high energy power based heating because of high cost components and elaborate control strategy.10
A promising solution to simplify thermal cycling as well as to reduce amplification time is so-called convective PCR, which was first reported in 2002.11 In convective PCR, an effective thermal cycling of the reaction mixture is realized in a pseudo-isothermal manner by imposing a steady-state vertical temperature gradient on the reaction volume.12–14 This temperature gradient induces a continuous, sustained circulation of reactants between hot and cool zones of the reaction, thus stratifying the reaction into spatially separate and stable melting, annealing, and extension zones.15–19 With thermal convection, convective PCR thus eliminates the need for controlled time-domain temperature cycling and external pumps or other fluid actuation or flow control.20–24 However, most of existed capillary convective PCR (CCPCR) systems are unavailable for commercial application because of those unresolved issues related to repeatability, reliability, convenience, and sensitivity.
To move forward microfluidic convective PCR toward commercial application, several essential issues, including heating strategy, detection method, system design, and reagent form, need to be addressed. In this paper, critical innovations and strategies are proposed and discussed to direct CCPCR to commercial application. First, different heating strategies including both single- and dual-end heating are compared between each other. A fully coupled three-dimensional mathematical model for the heat transfer inside both fluid and wall of the capillary tube as well as the fluid flow has been developed and numerically simulated to evaluate the performance of the optimized dual-end heating strategy. Second, a real-time detection method with light-emitting diode (LED) and photodiode, which can be easily incorporated into the integrated system, is developed and its performance is compared with that of the post-amplification detection with a dipstick assay. Third, to reduce the system complexity, for example, to simplify parameter tuning of the feedback control, an internal-model-control-based PID (IMC-PID) controller with good disturbance rejection is adopted here. Fourth, as a proof of concept, an in-situ storage method through pre-loading dry reagents inside the capillary PCR tube is demonstrated and evaluated. Systematic evaluation and characterization of the improved CCPCR system was performed based on experiments with influenza A (H1N1) virus as the detection sample.
II. RT-CCPCR REACTION TUBE AND PLATFORM
A. Convective PCR in a capillary tube with different heating strategies
Microfluidic diagnostics systems are typically comprised of a single-use chip in which the sample is processed, and which mates with a portable instrument that provides actuation, heating, and detection of the analyte or amplicon such as by fluorescence measurements.25,26 Here, we use a newly designed capped tube vial for CCPCR, which significantly improves the convenience of lab operation compared to existing capillary PCR reactors. The capillary tube reactor is made of polycarbonate by injection molding for low-cost, smooth surface finishes for low binding of reactants, biocompatibility, low autofluorescence, and high optical transparency in the fluorescence wavelength ranges.27–30 In most of the reported CCPCR, instruments with a single-end heating strategy have been adopted to heat the capillary tube from the bottom to introduce thermal convection. It has been demonstrated that satisfactory convective amplification can be achieved with the dual-end heating strategy when additional heating from the top of the capillary tube is incorporated.31 Here, specific properties between different heating strategies have been analyzed and compared to achieve more robust and repeatable CCPCR amplification toward commercialization.
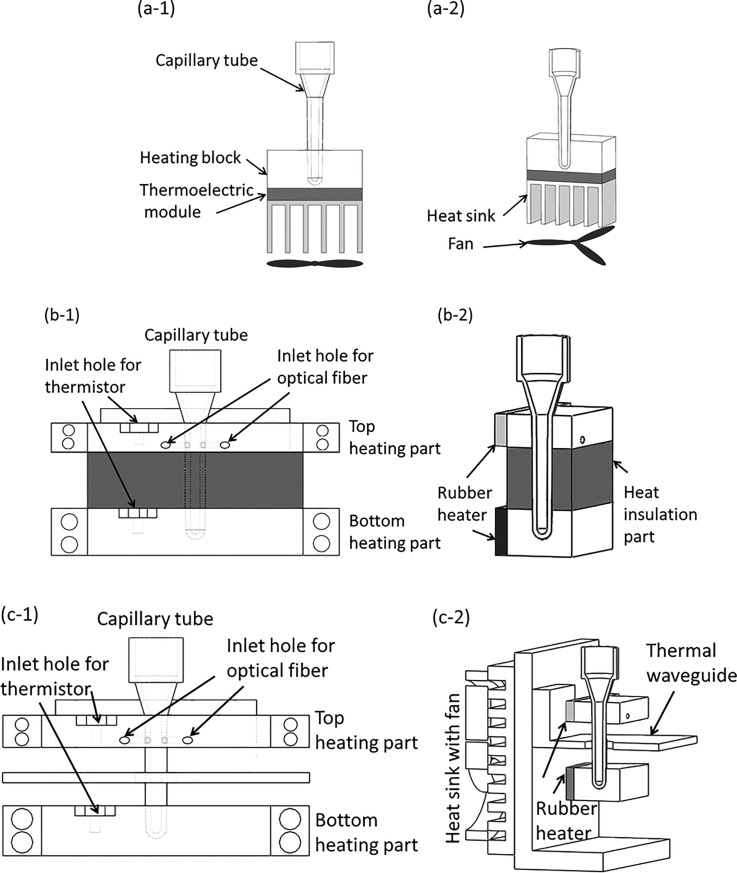
As shown in Fig. 1(a), for the single-end heating strategy, the capillary tube is heated from the bottom through a heating block by a thermoelectric module or other heater, which could cause nonuniform heating because of the temperature gradient along the heated portion of the capillary tube. The performance of convective PCR will be deteriorated not only by the fluctuation of environment temperature, but also by the nonuniform heating. As shown in Fig. 1(b), the first version of dual-end heating strategy is developed to improve the performance of convective PCR. To prevent inadvertent overheating of the top heating block with a low temperature, e.g., 55 °C, due to its close proximity (3.5 mm) to the bottom heating block with a high temperature, e.g., 95 °C, a heat insulation part made from material with low heat conduction is inserted in between the top and bottom heating parts. However, without the capacity of heat dissipation, its effectiveness will be significantly weakened when the system runs for a long time. Therefore, as shown in Fig. 1(c), an optimized dual-end heating strategy is developed. The dimensions of the top and bottom heating blocks are 50 mm × 20 mm × 8 mm and 50 mm × 20 mm × 12 mm, respectively. A thermal waveguide part, which is made from a cantilevered aluminum (1.5 mm thick) sheet with a 1 mm air gap to both the top and bottom heating parts, is adopted to shield the top block from thermal radiation from the bottom heater by preventing convective heat transfer in the air gap between blocks. To ensure robustness and stability of long time running, the thermal waveguide is attached to a fan-cooled heat sink at the backside of the frame to dissipate the accumulated heat in time. The waveguide is essential for precise maintenance of the 55–95 °C temperature difference over the length of the capillary since neither block can efficiently dissipate heat. As shown in Fig. 1(c-2), to ensure temperature uniformity along the heated portion of the capillary tube, for each end, the capillary tube is heated from its side wall through an aluminum heated block whose interior side is attached with a rubberized resistive heater element (12 V/6 W, Beijing Youpusi Science and Technology Ctr., Ltd.).
FIG. 1.
Schematic depiction of three version CCPCR platforms: (a-1) and (a-2), respectively, the front view of the schematics, and the 3D view of the single-end heating strategy, (b-1) and (b-2), respectively, the front view of the schematics, and the 3D view of the first version dual-end heating strategy, (c-1) and (c-2), respectively, the front view of the schematics, and the 3D view of the optimized dual-end heating strategy.
As shown in Fig. 1(c), with the developed dual-end heating strategy, the accessibility and convenience of lab operation has been significantly improved by just inserting the capillary tube into the well of the bottom heated block by following the alignment from the top heated block. It has been found that with a single rubber heater fixed on one side of the aluminum heating block (one type of aluminum material with good thermal conductivity), desired temperature uniformity can be achieved within a tiny CCPCR tube (inner diameter less than 2 mm). The proposed method is helpful to simplify the system design.
B. Capillary tube-based real-time convective PCR reactor
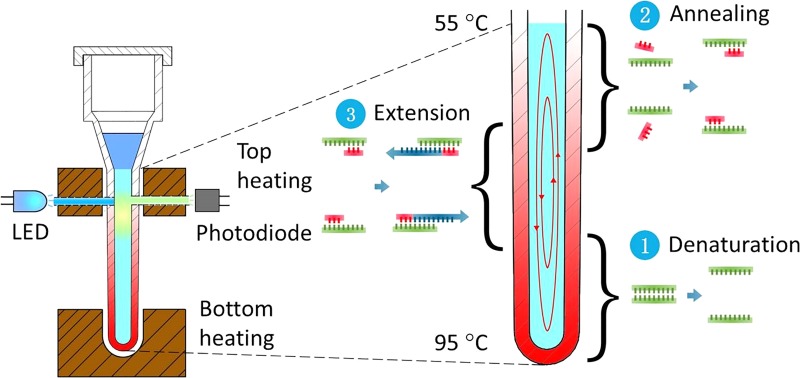
The geometry of the capillary tube for convective PCR was optimized by multiphysics (thermal and fluidic) modeling and experimentation. The capillary tube reactor is comprised of a top head volume with cap for loading, and a PCR reaction chamber (40 μl volume) in the form of a vertical capillary tube with an outside diameter of 3.3 mm, an inside diameter of 1.7 mm, and a length of 19 mm. As above discussed, a dual-end heating strategy with two independently controlled heaters (top and bottom) is adopted to move CCPCR forward toward commercialization, as shown in Fig. 2.
FIG. 2.
Schematic depiction of real-time CCPCR with dual-end heating.
In particular, we have found that for this geometry, a bottom temperature around 95 °C and a top temperature around 55 °C will induce convective currents that effectively cycle template and reagents through the required melting, annealing, and extension steps. Further, this configuration can accommodate the two optical fibers for real-time fluorescence monitoring as well as offer flexibility to other assays requiring different cycling temperatures.
Because the strongest fluorescence signal occurs during the extension step of each cycle when using Taqman probes, the optical probes are inserted through wells in the top heating block in closer proximity to the zone of extension. The fluorescence module of the system has an LED (light-emitting diode) excitation source coupled through an optical filter to a multimode optical fiber (plastic, diameter = 1 mm, numerical aperture = 0.5) which abuts one side of the capillary tube. The fluorescence emission is collected with a similar optical fiber on the side of the capillary tube opposite the excitation fiber, and then focused by a glass lens (8 mm diameter, focal length = 10 mm) before it is coupled through an optical filter (to block the excitation) to a high-sensitive silicon photodiode (Hamamatsu S2386–5 K, spectral range 320 to 110 nm) with an amplifier (OPA602, Texas Instruments). A microcontroller (STM32F407 32-bit RISC core @ 168 MHz with 1 M bytes of flash memory), also used for temperature control (see below), is used to control the LED and read and process the photodiode amplifier output. According to the particular fluorescent dye used as a reporter, e.g., Taqman probes labeled with FAM™ (fluorescein, λabs = 494 nm, λem = 518 nm), a LED (e.g., Lumileds, LXML-PB01-0040, λpeak = 495 nm), an excitation optical filter (λcut-off = 495 nm), and an emission optical filter (λcut-off = 515 nm) are selected based on their excitation, emission, and cut-off wavelengths.
C. Control algorithm
In principle, the dynamics of thermal convection inside the capillary tube is primarily determined by its heating geometry, which directly affects the state and efficiency of thermal cycling of reagents. Further, the accuracy and stability of temperature control is important to convective PCR, and especially with regard to good repeatability, consistent and stable heating has to be obtained. Therefore, a temperature control algorithm with good robustness and stability needs to be implemented to accurately maintain the temperature gradient to induce steady thermal convection with the required temperature profiles. The control algorithm is programmed with C and implemented on the same STM32F407 microcontroller as the fluorescence module. Proportional-integral-derivative (PID) control with negative feedback is widely used because of its relative simplicity, effectiveness, and wide latitude for “tuning” (adjustment of control parameters for optimization).32 However, because PCR involves relatively fast temperature cycling (abrupt changes of setpoints) and overshoot and undershoot must be minimal (<1 °C), more complicated control schemes are often necessary to achieve close temperature tracking of the temperature-time program used to affect optimal thermal cycling.33,34 Further, active heating and cooling using Peltier (thermoelectrics) modules are needed, rather than simple electrical resistance heaters, in order to achieve sufficiently rapid cooling ramps. For example, we previously reported a modified feedforward-feedback combined control scheme to speed up cooling and heating ramps and reduce overshoots in a microfluidic, large-reaction-volume (>100 μl) microfluidic PCR chip.1 Since convective PCR requires maintaining only one or two constant temperatures, the burden of the control is considerably reduced.
One drawback to PID control is that the tuning can be time consuming since the three parameter gains that need to be optimized. This is often done by trial and error, following general rules, or sometimes with auto-tuning algorithms. To simplify tuning, a PID control with an internal model control feature (IMC-PID) was adopted for this work. In addition to automatic gain parameter optimization (“auto-tuning”), IMC-PID has excellent robustness and stability35,36 even when the plant model changes with the aged rubberized resistive heater. The schematic diagram of IMC-PID negative feedback temperature control is depicted in Fig. S1 (see supplementary material). To implement the “top-bottom” heater scheme, a dual-loop control scheme was developed. Both control loops have their own thermistors to provide the process temperature subtracted from the corresponding setpoint as the error signal, which is inputted to the filter.
III. EXPERIMENTAL RESULTS AND DISCUSSION
A. Analysis of thermal convection inside the capillary tube
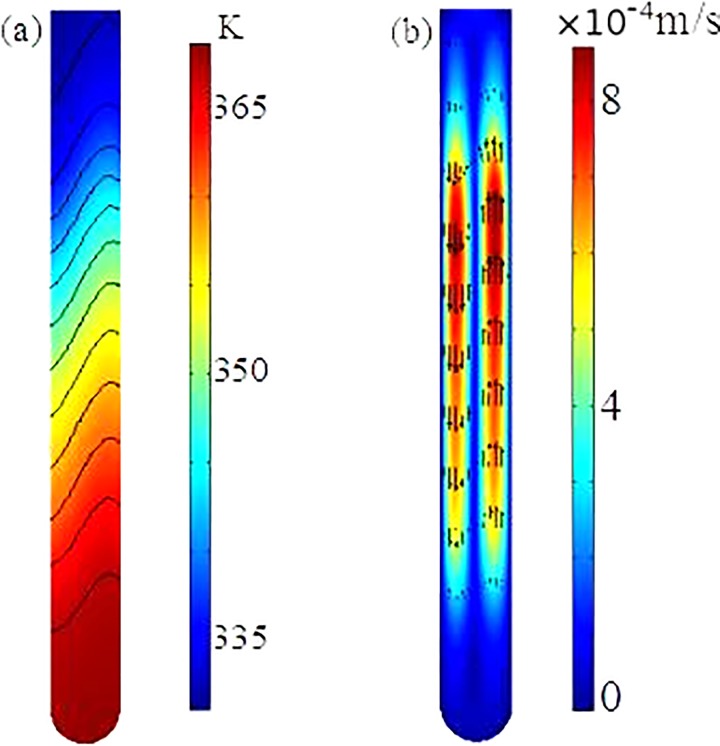
A coupled three-dimensional mathematical model, which consists of the momentum transfer inside the fluid, convection and conduction inside the fluid, and heat conduction within the wall of the capillary tube, is utilized to investigate the flow and heat transfer processes within the reactor with the dual-end heating strategy. The details of the mathematical model are described in Chen et al.14 and is omitted here for simplicity. The same as the experimental setup, the outer boundary of the capillary tube is divided into three parts: the top heater part maintained at , the bottom heater part maintained at , and the middle part using convective heat transfer boundary condition. The geometry and conditions in the numerical simulation are the same as those in the experiments. The fully coupled 3D model is numerically solved with the commercial finite element package, COMSOL (version 5.1), installed in a high performance cluster.
Fig. 3 depicts the temperature distribution and fluid flow inside the fluid in one cross-section of the reactor. Due to the use of dual-end heating, the temperatures in the top and bottom regimes are very close to the pre-specified values, and slightly non-uniform temperatures in the two zones arise from the induced convection inside the tube. In the middle non-heated part, due to convection temperature along one side of the tube is higher than that along the other side. Due to the dependence of the fluid density on the temperature, a counter-clockwise circulation is induced in the tube, as shown in Fig. 3(b). Maximum fluid velocity is on the order of 1 mm/s and occurs in the middle regime of the reactor. The existing of the one significant circulation through the entire tube is essential for the success of the PCR in the experiments, which is further confirmed in our PCR results. The mathematical model and numerical simulation would allow us to optimize our design.
FIG. 3.
Temperature distribution (a) and flow field (b) within the fluid in the CCPCR reactor.
B. Assessment of temperature control
To assess the performance of the CCPCR system with respect to temperature control, we use capillary tubes with embedded temperature sensors: (1) a 0.51 mm diameter Type K thermocouple (Omega Engineering, Stamford CT, 5TC-GG-K-24-36) at the top-end of the capillary reactor, and (2) a 0.76 mm diameter thermistor (Amphenol Thermometrics, Inc., Changzhou, China, SC30F103V) at the bottom end of the capillary. The capillary tube is inserted in between the heaters as shown in Fig. 2(c-2). The experiments were performed with an ambient room temperature of 24 °C. Setpoint offset temperatures were determined (in the range of 1 to 2 °C) to achieve precise target temperatures of 95.0 and 55.0 °C inside the capillary tube for the bottom and top temperatures, respectively.
The thermal response of the closed-loop system was tested in a separate set of experiments. The ramping rate is determined by both the thermal response of the heaters with the thermal mass of the filled capillary tube, and the controller response. The average ramp rates for the bottom and top heating blocks are 0.28 °C/s and 0.23 °C/s, respectively. With proper tuning, overshoot was negligible (<0.1 °C), and the steady-state temperatures were maintained within ±0.1 °C.
C. Analysis of heating strategy
First, evaluation and comparison between the single- and dual-end heating strategies have been performed. All the experiments were completed on a CCPCR system with dual-end heating. When the single-end heating strategy was evaluated, the top heater was shut down. The utility and performance of single or dual-end heating strategy was analyzed by detecting viral pathogens, e.g., H1N1 influenza virus, a subtype of influenza A. H1N1 is an RNA virus and one of the most common causes of human influenza. The H1N1 designation of this orthomyxovirus specifies its heamaglutinin and neuroaminadase glycoproteins.37 As some strains (which can be distinguished through selection of primers in PCR) are endemic in human populations and cause serious illness,38,39 there has been previous work on developing microfluidics-based point-of-care (POC) diagnostics platforms.40
Viral RNA was isolated from viral cultures using a magnetic-bead based kit for blood samples (Beijing Wantai Biological Pharmacy Enterprise, Ltd.). The viral samples (200 μl) were influenza A (H1N1) virus culture stock (10 TCID50/ml). Purified viral RNA was amplified in a single-tube reaction combining reverse transcription and complementary DNA (cDNA) amplification. The CCPCR reaction mix is comprised of 3 μl mixture of primers (Beijing Wantai Biological Pharmacy Enterprise, Ltd.) and a Taqman probe which is labeled with FAM at 5′end and Eclipse at the 3′end, 4 mM dNTP and 4 μl of Fast Buffer I (Mg ion buffer) (all Takara Bio Inc., Shiga, Japan), 0.4 U of Avian Myeloblastosis Virus (AMV) reverse transcriptase (Promega, USA), 1 U of SpeedSTAR HS DNA polymerase (Takara), 10 μl of the purified influenza A (H1N1) RNA as per above protocol, and molecular-biology grade water to a total reaction volume of 40 μl. After loading PCR reaction mix in the top, the reaction is sealed with a layer of 10 μl of sterile mineral oil (Sigma-Aldrich, St. Louis, MO, USA).
For the single- or dual-end heating strategy, following the reagent suppliers recommended temperatures, the bottom heater is first set 56.3 °C to incubate the reaction for 5 min at 55.0 °C for reverse transcription, then for amplification, the heater is set for a bottom melting temperature 95 °C (setpoint 95.8 °C). For the dual-end heating strategy, the top heater is set for an annealing/extension temperature of 55 °C (setpoint 56.3 °C). The primer sequence used with influenza H1N1 template is expected to yield a 105-bp amplicon. The production of amplicon was monitored in real time by measuring fluorescence via optical fiber probes as described above.
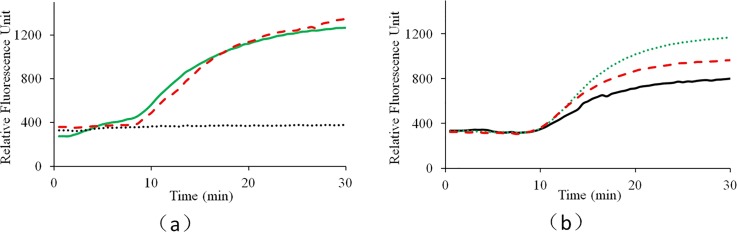
To evaluate the performance for both the single- and dual- end heating strategies, CCPCR experiments have been repeated for 3 batches, and comparable results have been achieved among different batches. For each batch, three runs have been repeated with each heating strategy. For one typical batch test, as shown in Fig. 4, for the single-end heating strategy, the successful amplification rate is less than 100%, while for the dual-end heating strategy, it is 100%. It is found that the successful amplification rate with the single-end heating strategy is around 66%, which proves that more consistent amplification is achieved with the dual-end heating strategy. As shown in Figure 4, even in an environment with a stable room temperature, we have demonstrated that dual-end heating strategy is superior to the single-end heating strategy. We suspect that with dual-end heating strategy, more stable temperature gradient can be established compared with the single-end heating strategy which just relies on the bottom heating. As suggested by the reviewer, especially in an environment with a fluctuated room temperature, the performance of single-end heating strategy will be further deteriorated. Even part of the capillary tube is still exposed to air during reaction, with dual-end heating strategy, the inside temperature gradient is mainly decided by both the bottom and top heating temperatures, which accordingly ensure the robustness of CCPCR.
FIG. 4.
Real-time CCPCR with H1N1 influenza virus RNA template, the detected fluorescent intensity is indicated as a function of the amplification time: (a) experiment with the single-end heating strategy, (b) experiment with the dual-end heating strategy, for both (a) and (b), the solid line, dotted line, and dashed line correspond, respectively, to three typical repeated tests with the same target sample concentration of 10 TCID50/ml.
The reaction temperature range allowing successful CCPCR amplification is justified with experiments. It is found that 55.0 ± 2 °C for the top reaction and 95.0 ± 2 °C for the bottom reaction are, respectively, the two appropriate temperature domains for consistent amplification. Although CCPCR amplification works within a relatively large temperature region, because the dynamics of thermal cycling of reagents inside the capillary tube is primarily determined by its heating, consistent and accurate heating should be ensured to achieve highly repeatable convective amplification, especially for quantitative detection.
D. Analysis of real-time fluorescence detection
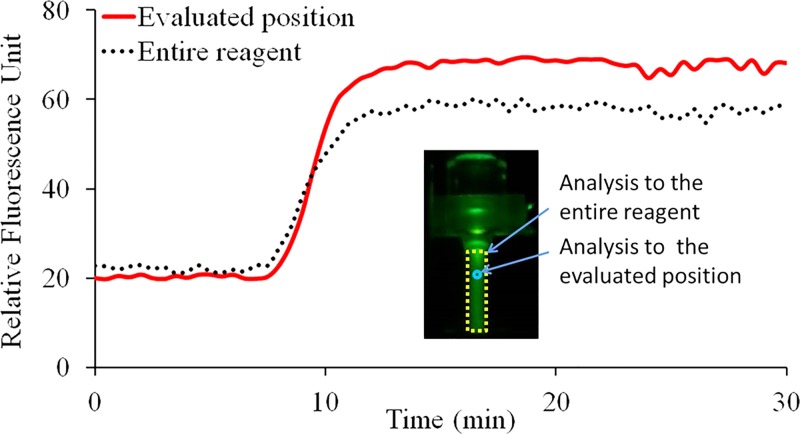
To monitor the state and distribution of fluorescence signal intensity of circulated CCPCR reagent inside the capillary tube in real-time using camera, the top heater was removed temporarily and the capillary tube was heated by just the bottom heater. The fluorescence signal images were analyzed and characterized to evaluate the proper scheme for fluorescence detection with photodiode and LED. A LED excitation source positioned on the top of the capillary tube is used to shine the reagent through a short pass optical filter when the light passes through the capillary tube from top to bottom. At the same time, the fluorescent image of the reagent inside the capillary tube is taken by a camera after it is coupled through an optical filter (to block the excitation). The fluorescent images are further processed with imaging software (Wright Cell Imaging Facility (WCIF) ImageJ 1.43 m, National Institutes of Health, USA). First, the average grey value of the entire reaction volume inside the capillary tube is obtained to represent the relative fluorescence signal intensity after it is subtracted by the average grey value of the background. Next, another average grey value of the CCPCR reagent subtracted by the background is obtained from the specifically evaluated or probed position where the fluorescence signal of the capillary tube is supposed to be detected with photodiode and LED via optical fibers. As shown in Fig. 5, two real-time curves represent the extracted fluorescence signal from the entire reagent and the specifically probed position, respectively.
FIG. 5.
Real-time monitoring of CCPCR with H1N1 template (1.0 TCID50/ml) with a camera, the extracted fluorescent signal intensity from image is indicated as a function of the amplification time: the solid and dotted lines correspond, respectively, to the average relative fluorescence signal intensity from the specifically probed position and the entire reagent. The inset features a typical fluorescence signal image of the capillary tube with two marked analyzing zones, respectively, for the probed position and the entire reagent.
As shown in Fig. 5, following amplification, the real-time curve of the probed position on the top of capillary tube with low reaction temperature gradually surpasses that of the entire reaction mainly for two reasons: (1) the melting temperature (Tm) of Taqman probe is 60–70 °C, and (2) the fluorescence signal intensity of Taqman probe is attenuated with increased temperature. On the whole, two real-time curves are comparable, which justify the feasibility of real-time monitoring of reagent through wells in the top heating block with photodiode and LED via optical fibers.
For high-throughput CCPCR, an alternative way for real-time fluorescence detection is to perform on-site monitoring of reactions from the top of the multiple capillary tubes with a single camera, which is helpful to reduce the system cost. With the fluorescence image based detection, the dynamic range of measurement will be narrowed when photodiode is replaced by camera.
The performance of real-time fluorescence detection was compared with that of dipstick assay based on lateral flow strip. For dipstick assay, the CCPCR reaction was performed in a 40 μl total reaction mixture containing 0.2 μM of each primer labeled with biotin at the 5′ end (Sangon, Shanghai, P. R. China), 0.2 μM of a probe labeled with fluorescein isothiocyanate (FITC) at the 3′ end (Sangon), 3.2 mM of dNTP (Takara Bio Inc., Shiga, Japan), 4 μl of 10× Fast Buffer I (Mg2+ plus), 1 U of SpeedSTAR HS DNA polymerase (Takara Bio Inc.), and 5 μl of the template. After loading PCR reaction mix in the top, the reaction is sealed with a layer of 10 μl of sterile mineral oil (Sigma-Aldrich, St. Louis, MO, USA). To compare the sensitivity of both methods, the amplification time of CCPCR for both methods is shortened from normal 25–30 min to less than 20 min when amplification is not saturated.
In dipstick assay, after amplification, the product is applied onto the sample pad of the lateral flow strip, and the result could be observed with the naked eye after a couple of min. The conjugate pad of the lateral flow strip is made from a fiberglass membrane pre-stored with latex particles conjugated with the anti-FITC antibody. In a positive test, two red bands are observed, of which the first red band (test line, immobilized with streptavidin) indicates a positive result and the second red band (control line, immobilized with goat anti-mouse (GAM)-IgG antibody) justifies the validity of the dipstick assay. Only the second red band appears in negative test. Failed appearance of the second band indicates an invalid detection.
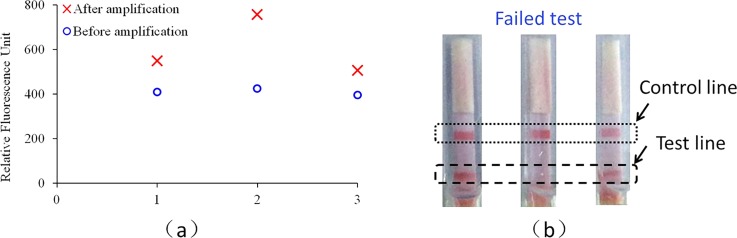
To show the difference of sensitivity between two methods, for real-time detection, the two fluorescence signal intensities before and after amplification were provided. As shown in Fig. 6, when the amplification time is set for 16 min, 100% successful detection is achieved with real-time detection, while for the dipstick assay, 2 successful detections of 3 repeated tests is achieved.
FIG. 6.
Comparison between the real-time fluorescence detection and the dipstick assay with the detection to H1N1 influenza virus (10 TCID50/ml): (a) fluorescence based detection and (b) dipstick based detection.
As shown in Fig. 6(a), for the real-time fluorescence based detection, the signal ratios between the end and the initial point fluorescence intensities are 1.34, 1.77, and 1.28, respectively, for three repeated tests. As shown in Fig. 6(b), for two successful tests, both the test and control lines appear, while for one failed test, only the control line appears. For 12 runs of real-time CCPCR, all shows positive, while for another 12 runs of dipstick with the amplicons, only 2/3 show positive. It is shown that higher sensitivity can be achieved with the real-time fluorescence based detection than the dipstick based detection, which is helpful to further decrease the required detection time.
E. Detection of influenza virus
The performance of the CCPCR reactor for detecting viral pathogens is demonstrated with H1N1 influenza virus testing. The viral samples (200 μl) were 10-fold serially diluted from influenza A (H1N1) virus culture stock (1000 to 1.0 TCID50/ml) using a semi-automated instrument (Taiwan Nanotechnology Corp., SLA-32). For repeatability test, H1N1 influenza virus with the same concentration (100 TCID50/ml) was used. Currently, for H1N1 clinical diagnostics applications, a limit of detection of 1.0 TCID50/ml or better (101.29 TCID50/ml) is feasible with commercial kits (Influenza A virus real-time PCR kit, Guangzho Hauyin Medicine Science, Ltd.). Purified viral RNA was amplified in a single-tube reaction combining reverse transcription and cDNA amplification. Parallel experiments using a conventional benchtop thermal cycler and the real-time CCPCR platform were performed for benchmarking, controls, and comparisons.
Preliminary experiments with gel electrophoresis test have confirmed that the convective PCR amplification products are the desired specific size without primer dimmer. Although the reagent is circulated by the up and down streams, the properly designed primers ensure the specific amplification based on the dual-end heating strategy. Instead of using normal DNA-intercalating fluorescent dye, such as SYTO-9 Green™, a Taqman probe is used for specific fluorescence labeling, which ensures the accuracy and specification of pathogen detection.
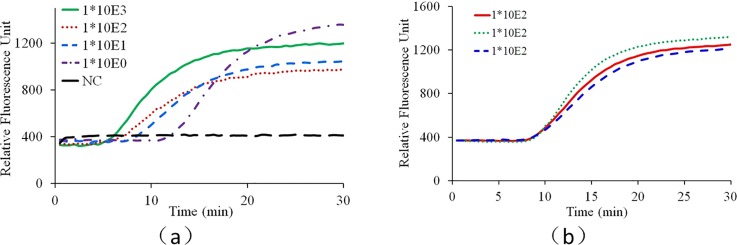
As shown in Fig. 7(a), from the real-time fluorescence curves, all the positive tests can be readily differentiated from the negative control. Ideally, for quantitative detection with isothermal amplification such as loop-mediated isothermal amplification (LAMP), the amplification time of one test needed to reach the threshold fluorescence intensity is inversely related to the initial target template concentration in the sample. However, because a much better understanding of convectional reactor and reproducible circulation among different capillary reactors is needed to realize quantitative CCPCR, currently the developed system aims to provide qualitative detection. Nevertheless, as shown in Fig. 7(b), for this prototype stage, the repeatability of the CCPCR system is noteworthy, as the equivalent CT (cycle threshold) values of the real-time fluorescence curves of repeated tests with the same target concentration (100 TCID50/ml) are quite close, which is important to the further improvement of CCPCR toward quantitative assays.
FIG. 7.
Real-time CCPCR with H1N1 influenza virus RNA template. The detected fluorescent intensity is indicated as a function of the amplification time: (a) experiment with diluted samples, the solid line, dotted line, short dashed line, dashed and dotted line, and long dashed line correspond, respectively, to samples with concentrations of 1000, 100, 10, 1.0 TCID50/ml and negative control (NC), (b) experiment with samples with one concentration (100 TCID50/ml), the solid line, dotted line, and dashed line correspond, respectively, to three repeated tests.
As shown in Fig. 7(a), the real-time fluorescence intensity curves saturate after approx. 25 min, and thus, the amplification time can be reduced accordingly without loss in sensitivity. The shortened amplification time, compared to conventional temperature cycling (typical times of 60 min or more) is a decisive advantage of the CCPCR method for POC diagnostics. In normal thermal cycling PCR, the fluorescence detection must be synchronized with the extension step and therefore with the cycling. In real-time quantitative PCR, template quantification can be done by calibration studies with serial dilutions where the number of cycles needed to reach the threshold fluorescence intensity can be related to initial target template concentration in the sample. In CCPCR, there is no periodic cycling, and thus analogous quantification of template based on number of cycles is not feasible. However, as with isothermal amplification techniques (where a cycle number is not relevant), the amplification time to reach a threshold fluorescence intensity (referred to as Tt) is expected to be inversely related to the log of the template concentration. As shown in Fig. 7(b), currently, the reproducibility of CCPCR is not as good as that of conventional PCR. In principle, the repeatability of the convection induced thermal cycling of reagents inside the capillary tube is primarily determined by the heating condition. Beside the accuracy of temperature control, the situation of heat conduction between the heating block and the capillary tube is another critical issue deciding the reproducibility of CCPCR. Currently, due to the limited machining precision, the heat condition of the capillary tube may vary among different tests because the rigid surface of the capillary tube meet with another rigid surface from the inner side of the aluminum heating block. A couple of efforts can be made to improve the consistence and repeatability of heating condition to improve the reproducibility of CCPCR. For example, to apply a downward force on the top of the capillary tube or to integrate an interfacing layer made of soft materials into the inner side of the heating block to achieve satisfied heat contact between the capillary tube and the heating block.
F. Real-time convective PCR with pre-loaded, dry-storage reagents
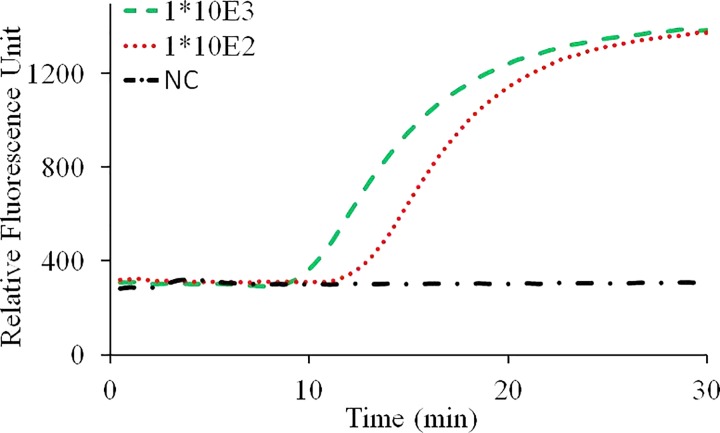
A mixture of 4 mM of deoxy-ribonucleoside triphosphate (dNTP) (Takara Bio Inc., Japan), 4 μl of Fast Buffer (Mg ion buffer) (Takara Bio Inc., Shiga, Japan), 1 μM of Betaine (sigma-aldrich, USA), 0.4 U of AMV Reverse Transcriptase (Promega, USA), 1 U of SpeedSTAR HS DNA polymerase (Takara Bio, Japan), 3 μl mixture of primers (Beijing Wantai Biological Pharmacy Enterprise, Ltd.), and a Taqman probe which is labeled with FAM at 5′end and Eclipse at the 3′ end were loaded into the capillary tube, then pre-frozen in an −80 °C ultra-low temperature freezer (ULT1386–3-V41, thermo, USA) for 2 h before lyophilization in a vacuum freeze dryer (6KBTES-55, Virtis, USA) at 300 mTorr for 4 h. Then, the capillary tube containing freeze-dried powder was sealed using a rubber stopper on top and an aluminum foil from outside before experiment. Before CCPCR experiment, 10 μl of the purified influenza A (H1N1) RNA and molecular-biology grade water were added into the capillary tube with pre-loaded dry-storage reagent to a total reaction volume of 40 μl, and then, the reaction is sealed with a layer of 10 μl of sterile mineral oil (Sigma-Aldrich, St. Louis, MO, USA), experiment result as shown in Fig. 8.
FIG. 8.
Real-time CCPCR with H1N1 influenza virus RNA template. The detected fluorescent intensity is indicated as a function of the amplification time: experiment with diluted samples, the dashed line, dotted line, and dashed and dotted line correspond, respectively, to samples with concentrations of 1000, 100 TCID50/ml and negative control.
From the preliminary experimental result, as a proof of concept, it was demonstrated that successful CCPCR amplification can be achieved with the pre-loaded dry-stored PCR reagent inside the capillary tube. Next step, more experiment will be performed to systematically evaluate the effectiveness of long-term storage of dry reagent, as well as the critical performance, such as sensitivity, stability, and repeatability of CCPCR with dry-storage reagent. As a rapid nucleic acid based diagnostics method, long-term pre-storage of the reagent inside the tube will extend CCPCR's application into more application areas, environments, for example, in resource-limited areas of the world in the of point-of-care testing.
IV. CONCLUSIONS AND OUTLOOK
As a high efficient nucleic acid amplification method, convective PCR can substantially shorten the test time compared to conventional temperature-cycling PCR (by 50%, from about 1 h to less than 30 min), which is important in many POC diagnostics applications. The test time can be further shortened by incorporating real-time fluorescence detection of the amplification in place of post-amplification end-point detection with, e.g., a lateral flow strip or gel electrophoresis. We have demonstrated here for the first time about how to solve essential issues in order to improve the performance of CCPCR toward commercialization. Based on the comparison between different heating strategies, an optimized dual-end heating strategy with two different setpoint temperatures is developed to improve system's repeatability and reliability. Thermal convection inside the capillary tube is studied with a three-dimensional model based on finite element analysis, and one big circulation developing across the capillary tube is generated, which is essential for the success of the PCR. For real-time detection, it is demonstrated that fiber optic probes provides good optical coupling of the reaction to an LED light source and photodiode detector for monitoring of increased fluorescence amplicon production, which can be easily incorporated into the diagnosis system. Instead of using traditional PID control, the system complexity is further reduced by introducing IMC-PID control to simplify parameter tuning of the feedback controller. To better accommodate to the application in point-of-care settings, as a proof of concept, CCPCR with pre-loaded dry storage of reagent inside the capillary PCR tube is evaluated. Experimental result with the detection of H1N1 influenza virus demonstrates that the critical performances of the optimized real-time CCPCR reactor, e.g., repeatability, sensitivity, and convenience, have been significantly improved. In the next step, effort will be made to build the first commercial real-time CCPCR nucleic acid analysis system based on the capillary tube reactor for point-of-care diagnostics.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the discussion on the internal-model-control-based PID (IMC-PID) controller with block diagrams and transfer functions, proportional-integral (PI) control, and tuning of control parameters.
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Foundation of China (Grant Nos. 81371711 and 81501835) and the Fundamental Research Funds for the Central Universities (Grant Nos. ZZ1329 and YS1404).
The authors declare that they have no conflicts of interest.
References
- 1. Qiu X., Mauk M. G., Chen D., Liu C., and Bau H. H., Lab Chip 10, 3170–3177 (2010). 10.1039/c0lc00038h [DOI] [PubMed] [Google Scholar]
- 2. Foudeh A. M., Didar T. F., Veres T., and Tabrizian M., Lab Chip 12, 3249–3266 (2012). 10.1039/c2lc40630f [DOI] [PubMed] [Google Scholar]
- 3. Khandurina J., McKnight T. E., Jacobson S. C., Waters L. C., Foote R. S., and Ramsey J. M., Anal. Chem. 72, 2995–3000 (2000). 10.1021/ac991471a [DOI] [PubMed] [Google Scholar]
- 4. Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., and Arnheim N., Science 230, 1350–1354 (1985). 10.1126/science.2999980 [DOI] [PubMed] [Google Scholar]
- 5. West J., Karamata B., Lillis B., Gleeson J. P., Alderman J., Collins J. K., Lane W., Mathewson A., and Berney H., Lab Chip 2, 224–230 (2002). 10.1039/b206756k [DOI] [PubMed] [Google Scholar]
- 6. Shu B., Zhang C., and Xing D., Lab Chip 15, 2597–2605 (2015). 10.1039/C5LC00139K [DOI] [PubMed] [Google Scholar]
- 7. Marchiarullo D. J., Sklavounos A. H., Oh K., Poe B. L., Barker N. S., and Landers J. P., Lab Chip 13, 3417–3425 (2013). 10.1039/c3lc50461a [DOI] [PubMed] [Google Scholar]
- 8. Ahrberg C. D., Ilic B. R., Manza A., and Neužil P., Lab Chip 16, 586–592 (2016). 10.1039/C5LC01415H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neuzil P., Pipper J., and Hsieh T. M., Mol. BioSyst. 2, 292–298 (2006). 10.1039/b605957k [DOI] [PubMed] [Google Scholar]
- 10. Shaw K. J., Docker P. T., Yelland J. V., Dyer C. E., Greenman J., Greenway G. M., and Haswell S. J., Lab Chip 10, 1725–1728 (2010). 10.1039/c000357n [DOI] [PubMed] [Google Scholar]
- 11. Krishnan M., Victor M. U., and Burns M. A., Science 298, 793–793 (2002). 10.1126/science.298.5594.793 [DOI] [PubMed] [Google Scholar]
- 12. Braun D., Goddard N. L., and Libchaber A., Phys. Rev. Lett. 91, 158103 (2003). 10.1103/PhysRevLett.91.158103 [DOI] [PubMed] [Google Scholar]
- 13. Wheeler E. K., Benett B., Stratton P., Richards J., Christian A., Chen A., Weisgraber T., Ness K., Ortega J., and Milanovich F., in Proceedings of μTAS 2003 Seventh International Conference on Micro Total Analysis Systems, Squaw Valley, CA, October 2003. [Google Scholar]
- 14. Chen Z., Qian S., Abrams W. R., Malamud D., and Bau H. H., Anal. Chem. 76, 3707–3715 (2004). 10.1021/ac049914k [DOI] [PubMed] [Google Scholar]
- 15. Muddu R., Hassan Y. A., and Ugaz V. M., J. Vis. Exp. 49, e2366 (2011) 10.3791/2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang H. G., Tsai Y., Tsai C., Lin C., Lee P., Teng P., Su C., and Jeng C., Biotechnol. J. 7, 662–666 (2012). 10.1002/biot.201100453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chung K. H., Park S. H., and Choi Y. H., Lab Chip 10, 202–210 (2010). 10.1039/B915022F [DOI] [PubMed] [Google Scholar]
- 18. Agrawal N. and Ugaz V. M., J. Lab Autom. 11, 217–221 (2006). 10.1016/j.jala.2006.06.004 [DOI] [Google Scholar]
- 19. Priye A., Hassanbc Y. A., and Ugaz V. M., Lab Chip 12, 4946–4954 (2012). 10.1039/c2lc40760d [DOI] [PubMed] [Google Scholar]
- 20. Li Z., Zhao Y., Zhang D., Zhuang S., and Yamaguchi Y., Sens. Actuators, B 230, 779–784 (2016). 10.1016/j.snb.2016.02.143 [DOI] [Google Scholar]
- 21. Zhang S., Lin Y., Wang J., Wang P., Chen J., Xue M., He S., Zhou W., Xu F., Liu P., Chen P., Ge S., and Xia N., J. Mol. Diagn. 16, 452–458 (2014). 10.1016/j.jmoldx.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 22. Zhang S., Xue M., Zhang J., Chen Q., Chen J., Wang Z., Zhou W., Chen P., Xia N., and Ge S., Clin. Biochem. 46, 1852–1856 (2013). 10.1016/j.clinbiochem.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 23. Priye A., Wong S., Bi Y., Carpio M., Chang J., Coen M., Cope D., Harris J., Johnson J., Keller A., Lim R., Lu S., Millard A., Pangelinan A., Patel N., Smith L., Chan K., and Ugaz V. M., Anal. Chem. 88, 4651−4660 (2016). 10.1021/acs.analchem.5b04153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu X., Zhang S., Xiang F., Wu D., Guo M., Ge S., Li K., Ye X., Xia N., and Qian S., Sens. Actuators, B 243, 738–744 (2017). 10.1016/j.snb.2016.12.058 [DOI] [Google Scholar]
- 25. Qiu X., Thompson J. A., Chen Z., Liu C., Chen D., Ramprasad S., Mauk M. G., Ongagna S., Barber C., Abrams W. R., Malamud D., Corstjens P. L. A. M., and Bau H. H., Biomed. Microdevices 11, 1175–1186 (2009). 10.1007/s10544-009-9334-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen D., Mauk M. G., Qiu X., Liu C., Kim J., Ramprasad S., Ongagna S., Abrams W. R., Malamud D., Corstjens P. L. A. M., and Bau H. H., Biomed. Microdevices 12, 705–719 (2010). 10.1007/s10544-010-9423-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu X., Liu C., Mauk M. G., Hart R. W., Chen D., Qiu J., Kientz T., Fiene J., and Bau H. H., Sens. Actuators, B 160, 1529–1535 (2011). 10.1016/j.snb.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu X., Chen D., Liu C., Mauk M. G., Kientz T., and Bau H. H., Biomed. Microdevices 13, 809–817 (2011). 10.1007/s10544-011-9551-5 [DOI] [PubMed] [Google Scholar]
- 29. Chin C. D., Linder V., and Sia S. K., Lab Chip 12, 2118–2134 (2012). 10.1039/c2lc21204h [DOI] [PubMed] [Google Scholar]
- 30. Piruska A., Nikcevic I., Lee S. H., Ahn C., Heineman W. R., Limbacha P. A., and Seliskar C. J., Lab Chip 5, 1348–1354 (2005). 10.1039/b508288a [DOI] [PubMed] [Google Scholar]
- 31. Qiu X., Ge S., Gao P., Li K., Yang Y., Zhang S., Ye X., Xia N., and Qian S., SLAS Technology (published online 2017) 10.1177/2211068216652847. [DOI] [PubMed] [Google Scholar]
- 32. Lee D., Allan J., Thompson H. A., and Bennett S., Control Eng. Pract. 9, 1235–1244 (2001). 10.1016/S0967-0661(01)00069-7 [DOI] [Google Scholar]
- 33. Alfaro V. M., ISA Trans. 46, 555–559 (2007). 10.1016/j.isatra.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 34. Dinca M. P., Gheorghe M., Aherne M., and Galvin P., J. Micromech. Microeng. 19, 1–15 (2009). 10.1088/0960-1317/19/6/065009 [DOI] [Google Scholar]
- 35. Jin Q. and Liu Q., J. Process Control 24, 22–32 (2014). 10.1016/j.jprocont.2013.12.011 [DOI] [Google Scholar]
- 36. Zhao Z., Liu Z., and Zhang J., J. Cent. South Univ. Technol. 18, 1153–1160 (2011). 10.1007/s11771-011-0817-0 [DOI] [Google Scholar]
- 37. Lim B. H. and Mahmood T. A., J. Obstet. Gynecol. India 61, 386–393 (2011). 10.1007/s13224-011-0055-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lessler J., Reich N. G., Cummings D. A., New York City Department of Health and Mental Hygiene Swine Influenza Investigation Team, Nair H. P., and Jordan H. T., N. Engl. J. Med. 361, 2628–2636 (2009) 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- 39. Cao B., Li X., Mao Y., Wang J., Lu H., Chen Y., Liang Z., Liang L., Zhang S., Zhang B., Gu L., Lu L., Wang D., Wang C., and National Influenza A Pandemic (H1N1) 2009 Clinical Investigation Group of China, N. Engl. J. Med. 361, 2507–2517 (2009). 10.1056/NEJMoa0906612 [DOI] [PubMed] [Google Scholar]
- 40. Pal R., Yang M., Lin R., Johnson B. N., Srivastava N., Razzacki S. Z., Chomistek K. J., Heldsinger D. C., Haque R. M., Ugaz V. M., Thwar P. K., Chen Z., Alfano K., Yim M. B., Krishnan M., Fuller A. O., Larson R. G., Burked D. T., and Burns M. A., Lab Chip 5, 1024–1032 (2005). 10.1039/b505994a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the discussion on the internal-model-control-based PID (IMC-PID) controller with block diagrams and transfer functions, proportional-integral (PI) control, and tuning of control parameters.