Abstract
Protein synthesis stands at the last stage of the central dogma of molecular biology, providing a final regulatory layer for gene expression. Reacting to environmental cues and internal signals, the translation machinery can quickly tune the translatome from a pre-existing pool of RNAs, before the transcriptome changes. Although the translation reaction itself has been known since the 1950s, the quantitative or even qualitative measurement of its efficacy in cells has posed experimental and analytic hurdles. This review outlines the array of state-of-art methods that have emerged to tackle the hidden aspects of translational control.
Translation as conductor to orchestrate gene expression
Translation is the fundamental biosynthetic reaction whose regulation determines cell fate temporally and spatially. Since protein synthesis and cell growth are tightly coupled, the dysregulation of translation is a common mechanism underlying unrestrained growth in cellular transformation and tumor development [1]. Indeed, re-tuning of the aberrant translation status by translation inhibitors is an attractive strategy for tumor treatment [2]. Translational control also underlies many other cellular and organismal processes. Perhaps most notably, regulated synaptic translation is linked to long-term potentiation and thus to learning and memory [3].
Whereas it has been historically well accepted that mRNA and protein levels are highly correlated, recent quantitative and genome-wide analyses revealed a larger contribution of translational regulation to the final output of proteins in cells than previously thought [4,5]. Indeed, although the biochemical fundamentals of protein synthesis have been well studied in great detail in vitro, monitoring protein synthesis in vivo has been a demanding task. Thus, diverse approaches have been developed to explore the variety of translation status in cells. Here we review the current advances of those methodologies, which are answering questions that were inaccessible to earlier methods and are creating new puzzles in our understanding of translation.
Luminescent labeling of newly synthesized protein
Conceptually, the most straightforward way to measure translation is detecting its output – protein. However, the huge amount of pre-existing protein in cells usually poses a challenge for analyzing the relatively subtle amount of newly synthesized protein accumulating in a short time-frame. Researchers thus need sufficiently sensitive and specific methods for capturing newly synthesized polypeptides.
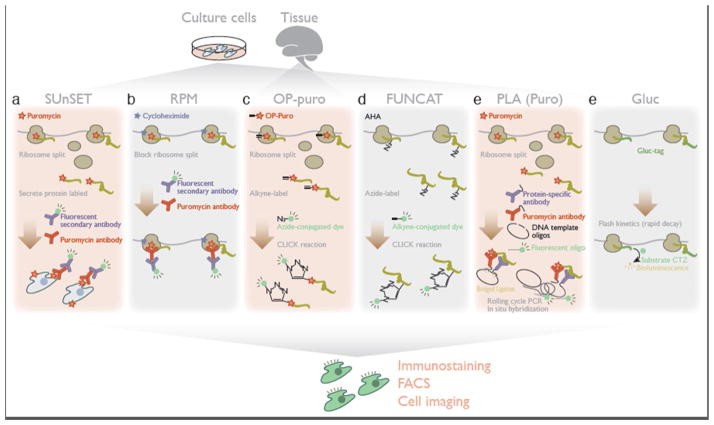
Specific fluorophore labeling of newly synthesized protein allows visualization and quantitation of the overall translation activity in cells. Conjugating the fluorescent signal directly with the peptide is highly advantageous, because this labeling is amenable to conventional immunochemical methods, microscopic analysis, and fluorescence-activated cell sorting (FACS) (Figure 1).
Figure 1. Methods for translation measurement based on luminescent labeling of newly produced peptides.
a. SUnSET (surface sensing of translation) harnesses puromycylated secreted proteins localized on the cell surface for detection by anti-puromycin antibody.
b. Combined with cycloheximide, RPM (ribo-puromycylation) traps puromycylated peptides on the ribosome and visualizes the subcellular sites of translation by staining with an anti-puromycin antibody.
c. Alkyne-bearing puromycin[O-propargyl-puromycin (OP-puro)] allows conjugation of a fluorescent dye to puromycylated proteins by CLICK chemistry.
d. FUNCAT (fluorescent non-canonical amino acid tagging) uses bio-orthogonal amino acids containing a CLICK-reactive alkyne or azide, enabling subsequent fluorophore attachment.
e. PLA (proximity ligation assay) detects newly-synthesized molecules of one target protein by the coincidental localization of a protein-specific antibody with a nascent-protein antibody recognizing puromycin or bio-orthogonal amino acids.
f. Gluc (Gussia luciferase) tagging on the protein of interests monitors its synthesis in real-time with subcellular resolution, by Gluc’s rapid maturation and flash kinetics.
The natural translation inhibitor puromycin from the bacterium Streptomyces alboniger has long been used as a key tool for studying translation [6]. Since puromycin functions as an analog of aminoacyl tRNAs, this drug is incorporated into the C-terminal end of a nascent peptide during the ribosomal elongation cycle and induces premature termination and subsequent drop-off of the ribosome from the mRNA. This “puromycylation” reaction places a “tag” into nascent peptides undergoing active translation, but not into completed protein. Although, in principle, puromycylation produces a prematurely terminated, shorter protein, a fraction of the proteins destined for cell secretion does reach the cell surface even after puromycin-mediated truncation. The surface sensing of translation (SUnSET) method detects this population of puromycylated secreted proteins on the cell surface, using an anti-puromycin antibody, as an indicator of bulk translation activity [7] (Figure 1a). In a further elaboration of this approach, puromycin is combined with cycloheximide, which blocks the dissociation of ribosomes and their nascent peptides from mRNA [8]. By trapping nascent peptides at the site of translation, this ribo-puromycylation (RPM) method focuses specifically the subcellular locus of translation [9,10] (Figure 1b). Photo-caged puromycin (NVOC-puromycin), which is activated upon UV exposure after cellular uptake, enables control over puromycylation-mediated nascent protein labeling with higher spatiotemporal resolution [11]. To facilitate the detection of the puromycin label, a derivative bearing a terminal alkyne [O-propargyl-puromycin (OP-puro)] has been synthesized; this enables the puromycylated nascent peptides to further be labeled with a fluorophore by CLICK chemistry in situ [12,13] (Figure 1c).
Nascent peptides can also be labeled by the metabolic incorporation of amino acid analogues bearing CLICK-reactive moieties. As in puromycylation, this fluorescent non-canonical amino acid tagging (FUNCAT) technique incorporates the targeting site for a fluorophore only in actively synthesized polypeptides. Using methionine-free media, culturing cells with either of two methionine surrogates – azide-containing azidohomoalanine (AHA) or alkyne-bearing homopropargylglycine (HPG) – leads to incorporation of these amino acids into translated protein, enabling CLICK conjugation with a fluorophore [14] (Figure 1d). One limitation of this approach, shared by the traditional metabolic labeling strategy with radioactive 35S-methionine, is the fact that labeling density is controlled by the methionine content of proteins.
In addition to bulk measurements, it is possible to monitor fluorescently the translation of a specific target protein by combining puromycylation and bio-orthogonal amino acids with protein-specific antibodies. The in situ proximity ligation assay (PLA) is based on the coincidental localization of two antibodies, which are connected to DNA oligonucleotides and guide the circularization of hybridized linker oligonucleotide [15]. Rolling cycle PCR of the circular DNA then amplifies the template for in situ hybridization, detecting the locus of the two target epitopes close together in cells. Applying this method with one of the antibodies targeting puromycin or bio-orthogonal amino acids and a second antibody against the protein of interest restricts proximity ligation to specifically monitor the new synthesis of the target protein [16] (Figure 1e).
Alternatively, synthetic reporters harboring the rapidly maturing Gaussia luciferase (Gluc) can be used to monitor protein synthesis in real time with sub-cellular resolution [17]. Gluc requires only the substrate coelenterazine (CTZ), and no other cofactors (e.g. ATP/magnesium ion), to emit the light, and this emission is rapidly quenched as the enzyme displays “flash” kinetics. These characters of Gluc enable measurements that focus solely on the fraction of reporter protein synthesized de novo (Figure 1f).
These methods based on fluorescence or bioluminescence are compatible with conventional FACS, microscopic imaging, and immunochemistry, which enable quantitative measurements of translational activity in single cells and often with sub-cellular resolution. In general, these techniques do rely on the delivery of chemical reagents, and in many cases require fixation before measurement as well. Nonetheless, they can reveal the variations in translatability among cell populations [7,12,13] and animal tissue [12], and of local translation [9] such as in the dendrites of neurons [10,14–17].
Mass spectrometry-based approaches
Whereas the methods mentioned above monitor the broad sweep of translation activity, understanding mRNA-specific translation regulation is an important but distinct issue. Indeed response to extracellular stimuli, intracellular stress, and in developmental programs are known to drive the translational control for a subset of mRNAs. Mass spectrometry has emerged as the preeminent approach for global and unbiased proteomics analyses. Applying proteomic mass spectrometry to study translation again hinges on distinguishing the new protein from the pre-existing population of proteins in the cell (Figure 2).
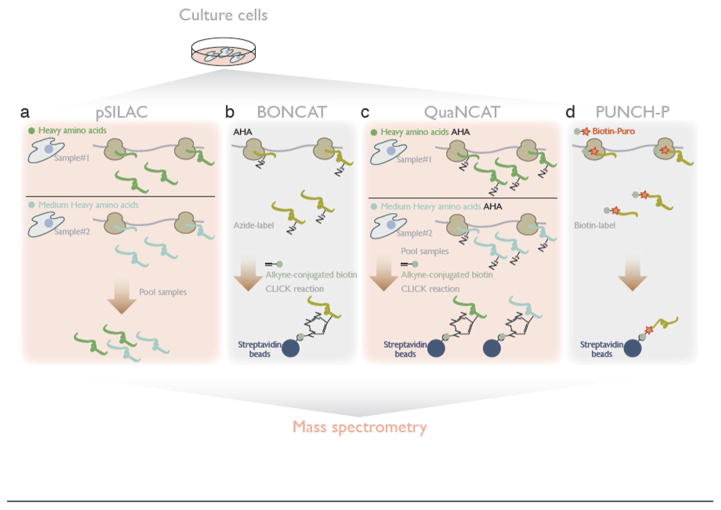
Figure 2. Mass spectrometry-based methods for measuring differential translation changes across proteome.
a. Pooling proteins from two samples, in the pSILAC (pulsed stable isotope labeling by amino acid in cell culture) method, provides quantitative mass spectrometry of protein produced during the labeling period.
b. In BONCAT (bio-orthogonal non-canonical amino acid tagging), CLICK-reactive bio-orthogonal amino acids [e.g. AHA (azidohomoalanine)] are incorporated into protein and subsequent biotin-tagging allows affinity purification of newly synthesized protein, for wider coverage of the proteome.
c. QuaNCAT (quantitative noncanonical amino acid tagging) combines pSILAC and BONCAT for quantitative mass spectrometry with broader coverage.
d. PUNCH-P captures polypeptides from active translation through biotin-conjugated puromycin and subsequent purification by streptavidin beads.
Metabolic pulse labeling of newly synthesized proteins with stable isotopes offers a solution to this problem, enabling quantitative analysis of protein synthesis and decay. In pulsed stable isotope labeling by amino acid in cell culture (pSILAC) [5,18], arginine and lysine composed of heavy nitrogen, carbon, or hydrogen isotopes, are placed into tissue culture media to mark the protein translated during the given time. Two different compositions of labeled amino acids enable the labeling of proteins with “heavy” and “medium-heavy” molecular weight. Pooling those two differentially labeled samples together and performing mass spectrometry on the mixture yields a quantitative comparison of the translation status in two different conditions (i.e. “heavy” vs. “medium-heavy”), distinguishing new proteins in each sample from pre-existing “light” proteins (Figure 2a).
To achieve broader proteome coverage, isolation and enrichment of newly synthesized protein is highly advantageous for enabling deeper mass spectrometry analysis, which can be limited by interference from more abundant pre-existing proteins and other detectability biases. Here again, CLICK chemistry offers a solution – after bio-orthogonal non-canonical amino acid tagging (BONCAT), a CLICK reaction with alkyne-conjugated biotin allows purification with streptavidin/neutravidin-beads [19,20] (Figure 2b). BONCAT can be combined with pSILAC, in an approach termed quantitative noncanonical amino acid tagging (QuaNCAT), that allows quantitative, ratiometric, and deep proteomic analysis of newly-synthesized protein [21] (Figure 2c). Moreover, cell-selective BONCAT/QuanCAT can be performed using cell-type-specific expression of an engineered aminoacyl-tRNA synthetase (tRS) that will charge azide-bearing bio-orthogonal amino acids, which are originally poor substrate for non-engineered tRS [22–26]. Alternatively, in vitro puromycylation with a biotin conjugated puromycin derivative can label nascent proteins in cell extract, which can then be enriched by streptavidin affinity purification. Proteomic analysis of these tagged translation intermediates is termed puromycin-associated nascent chain proteomics (PUNCH-P) [27] (Figure 2d).
mRNA sequencing-based approaches
The process of translation sits at the interface between nucleic acids and proteins. The advent of powerful and sensitive techniques for mRNA profiling – first microarrays, and then deep sequencing – spurred an interest in analyzing translation from the perspective of the mRNA template rather than the protein product. The number of ribosomes loaded on a single mRNA indicates how much it is being translated; this can be discerned by fractionation of poly-ribosomes, or polysomes, by ultracentrifugation through a sucrose density gradient. RNA profiles across the polysome fractions, compared to the total RNA pool in cells, can identify translation status of mRNAs across the transcriptome [28–30] (Figure 3a).
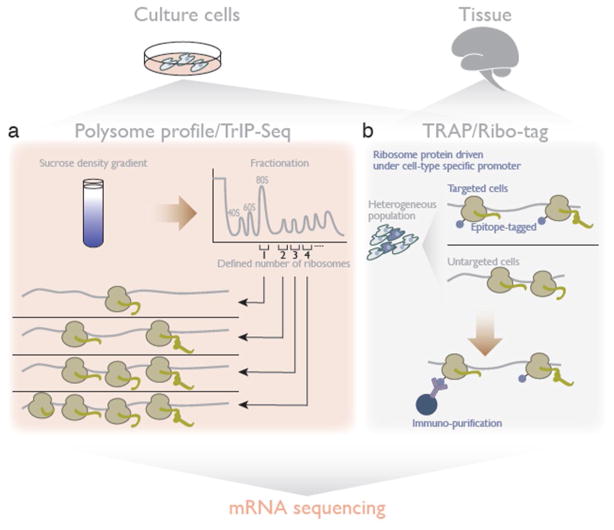
Figure 3. Analyzing translation by mRNA sequencing.
a. In polysome profile/TrIP-Seq (transcript isoforms in polysome sequencing), defined fractionation according to the number of ribosomes on an mRNA dissects the mRNA pools under active translation and monitors differential translation status among mRNA isoforms.
b. TRAP (translating ribosome affinity purification) and Ribo-tag isolate ribosomes – including constituent mRNAs – from specific cells and lineages.
One major factor affecting the diverse translation efficiencies among mRNAs in cells is their 5′ and 3′ untranslated region (UTR) [31,32], which can differ between transcript isoforms. Defining the number of ribosomes on mRNAs by polysome fractionation coupled with mRNA-seq can deconvolve the distribution of isoforms and thereby reveal differences in their translation efficiency. This approach for measuring translational differences among mRNA isoforms is termed transcript isoforms in polysome sequencing (TrIP-Seq)] [30] (Figure 3a).
Like all of the proteomic approaches discussed above, polysome profiling requires a large biological sample and yields an average across all cells comprising that sample. This poses challenges for translational profiling in complex tissues, and especially in the brain, where it could reveal new insights into synaptic translation and many other exciting open questions. In order to measure translation in specific cells and lineages, it is necessary to first isolate enough cells of the desired type from the complex mixture of diverse cells in an animal to perform polysome profiling. The translating ribosome affinity purification (TRAP) and Ribo-Tag approaches both circumvent the challenge of isolating specific cells by instead expressing an epitope-tagged ribosomal protein driven under a tissue-specific promoter and subsequently sequencing the mRNA that co-purifies with the tagged ribosome [33–35] (Figure 3b). Although the resolution is still qualitative, with these approaches it may now be easier to profile translated mRNA than total mRNA in a lineage-specific manner.
Surveying translation genome-wide with ribosome profiling
Although mRNA-Seq-based methods can provide a genome-wide view of the translatome, the data cannot easily address questions such as where the ribosome starts and ends translation, and how fast elongation proceeds. The emergence of ribosome profiling promised to unravel such hidden aspects of translation, comprehensively and in vivo [4,36]. The essence of ribosome profiling is deep sequencing of ribosome-protected mRNA fragments generated by RNase digestion (Figure 4a). Since the position of the sequenced “ribosome footprint” along an mRNAs directly represents the codons physically enclosed by the ribosome and being decoded, the reads aligned back to genome represent the number and position of translating ribosomes. Quantifying these footprints can measure translation efficiency, which is normally evaluated by the over- or under-representation of a gene in ribosome footprinting relative to total mRNA-Seq, and can also delineate the coding region on an mRNA and estimate elongation speed at every codon. This versatile technique has been applied to variety of species, including model and non-model organisms, and even viruses. The tissue-specific translatome can be addressed by immuno-purification of epitope tagged ribosomes, which enables tissue-specific ribosome profiling [37–39], similar to TRAP/Ribo-tag, which involves mRNA-seq of affinity-purified polysomes [33,34].
Figure 4. Ribosome footprinting-based methods for genome-wide translatome surveys.
a. Ribosome profiling is based on deep sequencing of RNA fragment protected by the ribosome during RNase digestion.
b. Ribosome footprints from translation initiation sites are highly enriched by the use of translation inhibitors LTM or harringtonine.
c. Artificially localized BirA (ex. on the ER or mitochondria) specifically biotinylates Avi-tags on ribosome proteins, only when those two molecules are coincidentally localized. Purification and footprinting of biotinylated ribosomes thereby surveys the translatome at a specific place within cells.
d. Co-translational events can be monitored transcriptome-wide by profiling of ribosomes selected through interacting factors.
e. In TCP-Seq (translation complex profiling sequencing), the scanning 40S ribosome and its footprint are crosslinked, purified, and sequenced.
Translation initiation site profiling
Translated reading frames can be inferred from the presence of elongating ribosomes in profiling data. The combination of ribosome profiling with a variety of translation drugs can further expand our understanding of the in vivo translatome [37,40–50] by providing a direct, genome-wide survey of translation initiation sites.
Puromycin treatment terminates translation and releases actively elongating ribosomes, but does not stop new initiation, so ribosome footprints from puromycin-treated cells are enriched at and just after the translation initiation site [40]. Even greater sensitivity can be achieved by pre-treatment of cells with translation inhibitors harringtonine and lactimidomycin (LTM), which specifically capture the ribosome footprints at the start codon [41–43]. Since these drugs only trap the ribosomes that do not have tRNA in their E-site [51] – a situation that arises only for the initiating ribosome – elongating ribosomes run off from the mRNA. Brief treatment with those inhibitors, followed by ribosome profiling, specifically enriches ribosome footprints from initiating ribosomes and thereby defines translation initiation codons, in an approach sometimes termed global translation initiation site sequencing, or GTI-Seq (Figure 4b). These analyses not only rediscover AUG start codons from canonical coding DNA sequences (CDSes), but also assign novel translation initiation codons with AUG as well as near-cognate non-AUG codons. Sequential treatment with LTM and puromycin allows an even more quantitative survey of differential selection between translation initiation sites on an mRNA [a method termed quantitative translation initiation sequencing (QTI-seq)] [37]. Much as harringtonine and LTM capture initiating ribosomes in mammalian cells, tetracycline treatment causes a similar pattern of footprint accumulation on start codons in bacteria and uncovers initiation codons comprehensively [44,52].
Elongation speed survey by ribosome profiling
Because of its mechanism of action, harringtonine (or LTM) also allows a ribosome run-off assay to profile the speed of elongation directly [41]. When harringtonine is added, it blocks new ribosomes from entering the CDS body; a short time later, cycloheximide addition freezes elongating ribosomes, and ribosome profiling can then be used to assess the amount of ribosome movement occurring between the two drug treatments. This analysis revealed the average ribosome elongation speed is 5.6 amino acids per second [41].
While the overall speed of elongation seemed similar between different genes, individual codons have different decoding rates. Ribosome footprint accumulation represents the time that the ribosome spends decoding the A-site codon, and thus identifies these differences in elongation speed by codons. Although many studies have sought to understand the factors that determine elongation speed, and reached disparate and sometimes contradictory conclusions [53–59], it does seem that amino acid availability, which in turn reflects charged tRNA abundance, is a key determinant of the decoding rate of corresponding codons [49,60]. Conversely, the ribosome occupancy over codons can be harnessed as a proxy for sensing the amino acid pool in cells [61].
Moreover, ribosome profiling data can reveal kinetic aspects of conformational changes of the ribosome during the elongation cycle. Because the ribosome adopts distinct conformations at different stages of the elongation cycle, and these conformations vary in their accessibility to RNase, ribosome footprints can differ in length. Indeed, footprints occur in two distinct populations, one shorter (~21 nt) and one longer (~29 nt) [49], with differential characteristics of accumulation over codons, presumably reflecting disparate effects of tRNAs or amino acids on the kinetics of individual steps of the elongation cycle.
Survey of co-translational processing by ribosome profiling
The ribosome functions not only as the decoding machinery, but also as the hub of co-translational events: folding of the nascent peptide, its export to organelles, protein complex assembly, and mRNA decay [62]. Selective profiling of ribosomes engaging those processes provides a new, unbiased way to address which mRNAs are targeted and where within the mRNAs those processes start.
Localization of the ribosome can restrict the production of proteins to specific places inside the cell where the protein plays its role. For example, ribosomes on the endoplasmic reticulum (ER), known as the rough ER, are well known as the locus of local translation for secretory proteins. Although it is possible to learn about the rough ER translatome by fractionation of cytosolic and membrane-bound ribosomes and the subsequent ribosome profiling on these fractions [63,64], this approach is challenging because of incomplete fractionation, which is exacerbated by the collapse of the organelle during cell lysis. Proximity-specific ribosome profiling circumvents these challenges by exploiting in vivo labeling of localized ribosomes. The bacterial biotin ligase BirA and its substrate peptide, called Avi-tag, provide a binary system for biotin tagging a protein of interest, which is fused to the Avi-tag. Localized BirA on sub-cellular organelle restricts the biotin tagging to the specific place where both BirA and Avi-tag coincide. Placing the Avi-tag on a ribosome protein and localizing BirA on the ER surface allows the specific biotinylation of ribosomes at the ER, which can then be purified and footprinted to detect the co-translational localization of nascent peptides [65] (Figure 4c). This approach also revealed the translatome on the mitochondria membrane, where localized ribosomes are not tightly coupled to the membrane and are thus less amenable to co-fractionation [66].
Immuno-purification of targeted ribosomes via their association with ribosome/nascent peptide binding factors leads to the simple but powerful adaptation of ribosome profiling to look at interesting sub-populations with high resolution (Figure 4d). This analysis has been applied to a variety of co-translational events: folding assisted by trigger factor chaperone [67–69], membrane targeting by signal recognition particle (SRP) [64,69], protein complex assembly [70,71], and Ski complex-mediated on-ribosome mRNA decay [72]. In addition to ribosome profiling, unexpected coupling of translation with mRNA decay was revealed by mRNA degradome analysis of decapped intermediates, since 5′-3′ transcript degradation is so processive that it reaches the translating ribosome and stops at its trailing edge [73].
In organello protein synthesis survey
In addition to the cytosolic translation system, eukaryotes have another translational machinery in their mitochondria, which specifically drives translation of mRNAs encoded in the mitochondrial genome. Isolation of the mitochondrial ribosome by sucrose density gradient fractionation [74], sucrose cushion purification [45,75], or epitope tagging of a mitochondrial ribosome protein [76] allows access to the translatome in organello as well.
Profiling of scanning ribosome
In eukaryotic translation initiation, the 40S small ribosomal subunit is first loaded onto the mRNA and then moves along the 5′ UTR to scan the cognate start codon prior to the assembly of mature 80S ribosome [31,77]. However, ribosome profiling does not provide information about scanning ribosomes, since the interaction between the 40S ribosome and the mRNA is weak, and so any footprint would be susceptible to dissociation from the 40S [78]. Formaldehyde cross-linking of the scanning 40S and mRNA freezes this fragile complex, and subsequent 40S isolation allows profiling of the scanning pre-initiation complex – a methodology called translation complex profiling sequencing, or TCP-Seq [79] (Figure 4e). Three distinct populations of 40S footprints sizes (19, 29, 37 nt) reflect the sequential changes in conformation and/or assembly/dissociation of factors on the translation initiation complex. This method even captures the 40S ribosome on the stop codon, after 60S dissociation but before recycling.
Single-molecule analysis in vivo
Although techniques based on mass spectrometry and deep sequencing provide comprehensive measurements of translation in vivo, the data are combined and averaged from every copy of an mRNA across millions of cells, masking the variance of translation of an individual mRNA molecule. The recent development of single molecule-based technologies reveals translation molecule-by-molecule and allows versatile tools for detailed observation of protein synthesis.
To observe translation directly, in real time and in vivo, it is first necessary to visualize mRNA at the single molecule level. Such single-molecule mRNA tracking is enabled by the combination of MS2 or PP7 short hairpin RNAs (typically placed into 3′ UTR) with fluorescently tagged versions of their cognate binding proteins [MS2 coat protein (MCP) or PP7 coat protein (PCP)]. In its simplest form, this approach allowed the visualization and co-localization of the mRNA with ribosomes [80] (Figure 5a). Moreover, placing the phage-derived RNA hairpins in the coding region, rather than the 3’ UTR, enables one to see the displacement of fluorescent hairpin-binding proteins by elongating ribosomes [termed as translating RNA imaging by coat protein knock-off (TRICK)], and thereby measure the location and the kinetics of first-round translation on an mRNA after its export from nucleus [81] (Figure 5b).
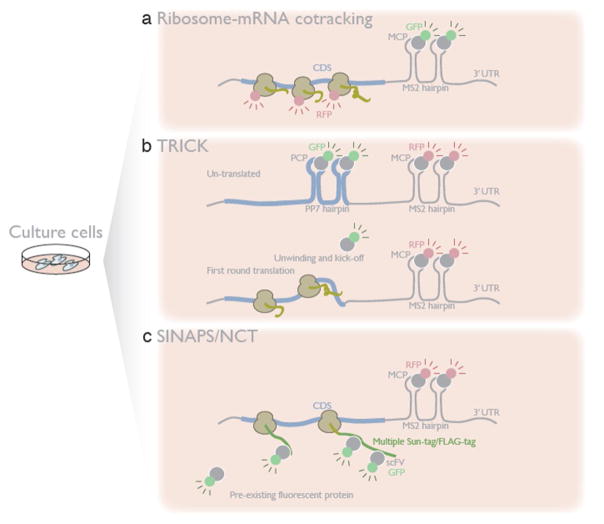
Figure 5. In vivo single-molecule translation.
Single mRNAs are visualized with a hairpin RNA together with its cognate binding protein fused to a fluorescent reporter.
a. Simultaneous visualization of an mRNA along with fluorescently tagged ribosomes reveals the population of mRNAs under translation.
b. In TRICK (translating RNA imaging by coat protein knock-off), by placing a second pair of RNA hairpins within the CDS, bound by a different coat protein with a distinct fluorescent label, untranslated mRNAs are monitored by the overlap of two colors. As the elongating ribosome melts the CDS hairpins and kicks off their binding protein, loss of the second color reflects the first round of translation.
c. Multimerized Sun-tags in SINAPS (single-molecule imaging of nascent peptides) or spaghetti monster protein tags in NCT (nascent chain tracking), encoded in the reporter transcript can bind quickly with pre-existing fluorescent antibody, as soon as nascent peptides emerge from ribosome. Co-localization of mRNA and nascent peptide directly measures the kinetics and dynamics of active protein synthesis.
In addition to tracking mRNA, fluorescent labeling of the nascent peptide from a translating mRNA offers a more direct and versatile method to visualize translation in vivo. Although simply monitoring a fluorescent protein as a nascent peptide on the ribosome seems straightforward, the dim signal of a individual fluorescent protein and its slow maturation pose fundamental technical challenges. Since SunTag [82] and spaghetti monster (multimerized FLAG-tags), used in single-molecule imaging of nascent peptides (SINAPS) and nascent chain tracking (NCT) respectively, can be rapidly recognized by a pre-existing pool of fluorescently-conjugated antibody fragments (single-chain variable fragment (scFV) and anti-FLAG), placing such tandem epitopes in the coding region enables the detection of the nascent peptide as a multi-fluorophore spot immediately upon its emergence from the ribosome [83–86] (Figure 5c). These nascent peptide visualization experiments have been performed with varying experimental and analytical approaches, including different cell types and reporters. Despite those differences, the four studies yielded similar results, representing the fundamental characteristics of translation: elongation at 3–10 amino acids per second, ribosomes spaced out every 200–300 nt on the mRNA, and translation initiation every 30–40 seconds, all of which agree well with bulk measurement of translation by other approaches [4,87]. Furthermore, these analyses uncover fluctuations in protein synthesis across individual mRNA molecules: translation bursts (switching between on- and off-states) [85,86] as observed in transcription [88], occasional ribosome stalling [84], and formation of di-polysomes [83] comprising two physically associated mRNAs, probably reflecting co-translational complex assembly [70].
Through their resolution of single-molecule translation in time and space, the results of these in vivo, single-molecule analyses make us reconsider the paradigm of established translational control mechanisms. For example, the integrated stress response represses overall translation while actually activating translation from a subset of mRNAs. Remarkably, single-molecule analysis showed that this stress-induced activation is transient and lasts only ~150 seconds [84]. In neurons, restricted translation at the synapse is necessary for long-term potentiation, and thus memory. The analysis of this localized and regulated synaptic translation reflects one of the most exciting and technically challenging applications for single-molecule studies of translation. It has been long thought that translational repression during transport of the mRNA along the dendrite, followed by release of this repression at the synapse, is the basis of local translation. However, in vivo single-molecule analysis actually showed that silencing of translation is not complete and ~20% of mRNAs are actively translated during transport [84,85]. Future application of these approaches will doubtlessly provide greater insight into this key feature of neuronal physiology.
Concluding remarks
We have a growing appreciation for the depth and complexity of translation control. Recent developments include the appreciation of diverse non-AUG start codons and their use in a cell-context-dependent manner [37,41,42], the elucidation of the ribosome quality control (RQC) system for resolving aberrantly stalled ribosomes [89], the emergence of codon optimality as additional code for mRNA half-life [90], the role of RNA modifications in translational control [91], and the global analysis of cellular internal ribosome entry sites (IRESes) [92].
The wealth of methods reviewed here is providing new ways to address such biological phenomena (see Outstanding Questions), which have previously been much less accessible. Systematic analysis by mass spectrometry of protein and deep sequencing of RNA allows us to perform unbiased surveys of translation and deduce the underlying regulatory rules. In vivo single-molecule analysis is currently limited to artificial reporter transcripts, but allows detailed kinetic analysis of translation with great resolution in time and space. The single-molecule study of translation is still in its infancy, and as it matures we expect a rich body of new insights.
Outstanding Questions Box.
How different is the protein synthesis rate among tissues across development and disease states? How does it link to cell physiology?
What kind of characteristics (e.g., RNA/protein motifs) determine the kinetics of ribosome elongation in cells? How does it affect function of the synthesized proteins?
How variable are ribosomes in cells? How do modifications of ribosome protein/rRNA and interacting factors specialize translation?
How does the epi-transcriptome affect translation?
What is the underlying mechanism of translation bursts?
Covering many endogenous transcripts by single-molecule translational analysis will be warranted.
Single-cell application of deep-sequencing based approaches, including ribosome profiling, TCP-seq, and others, may also be the future direction.
Although each individual approach discussed above has clear strengths, the combination of multiple approaches should provide even deeper and more robust analysis of translation, as the distinct techniques complement each other well. We look forward to the possibility of high-throughput, single-molecule translational analysis covering many endogenous transcripts, and single-cell application of deep-sequencing based approaches like those now performed in RNA sequencing and chromatin immunoprecipitation [93]. Moreover, applications of these techniques to complicated tissues in animals – an area of active development – will provide new insights into the contribution of translation control in development and diseases.
Trends Box.
Fluorescent labeling of newly synthesized protein measures overall translation activity by conventional microscopic analysis and fluorescence-activated cell sorting (FACS).
Mass spectrometry analysis with specific isolation of newly synthesized protein monitors the differential changes of proteome caused by translation alteration.
Ribosome profiling and its derivative methods globally survey in vivo kinetics of translation, local translatome, and co-translational events.
In vivo single-molecule analysis addresses the kinetic and dynamic fluctuation of translation, molecule-by-molecule, in real time.
Acknowledgments
We thank Hiro-oki Iwakawa, Yuichiro Mishima, and members of Iwasaki laboratory for critical comments on this manuscript. This work was supported in part by the Damon Runyon Cancer Research Foundation (DRR-37-15), the Searle Scholars Program (11-SSP-229), the National Institute of General Medical Sciences of the National Institutes of Health (P50GM102706) to N.T.I and Grant-in-Aid for Scientific Research on Innovative Areas “nascent chain biology” (JP17H05679) and Grant-in-Aid for Young Scientists (A) (JP17H04998) from Japan Society for the Promotion of Science (JSPS) to S.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat M, et al. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 3.Buxbaum AR, et al. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 6.NATHANS D. Inhibition of protein synthesis by puromycin. Fed Proc. 1964;23:984–989. [PubMed] [Google Scholar]
- 7.Schmidt EK, et al. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 8.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David A, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graber TE, et al. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci U S A. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buhr F, et al. Design of photocaged puromycin for nascent polypeptide release and spatiotemporal monitoring of translation. Angew Chem Int Ed Engl. 2015;54:3717–3721. doi: 10.1002/anie.201410940. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, et al. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signer RA, et al. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 16.tom Dieck S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods. 2015;12:411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na Y, et al. Real-time imaging reveals properties of glutamate-induced Arc/Arg 3.1 translation in neuronal dendrites. Neuron. 2016;91:561–573. doi: 10.1016/j.neuron.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwanhäusser B, et al. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 19.Dieterich DC, et al. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieterich DC, et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 21.Howden AJ, et al. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuet KP, et al. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2015;112:2705–2710. doi: 10.1073/pnas.1421567112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erdmann I, et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat Commun. 2015;6:7521. doi: 10.1038/ncomms8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdavi A, et al. Engineered aminoacyl-tRNA synthetase for cell-selective analysis of mammalian protein synthesis. J Am Chem Soc. 2016;138:4278–4281. doi: 10.1021/jacs.5b08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aviner R, et al. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 2013;27:1834–1844. doi: 10.1101/gad.219105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016:5. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinnebusch AG, et al. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia J, et al. Regulation and dysregulation of 3’UTR-mediated translational control. Curr Opin Genet Dev. 2013;23:29–34. doi: 10.1016/j.gde.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekstrand MI, et al. Molecular profiling of neurons based on connectivity. Cell. 2014;157:1230–1242. doi: 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao X, et al. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 2015;12:147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez C, et al. Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J Neurosci. 2014;34:10924–10936. doi: 10.1523/JNEUROSCI.0084-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornstein N, et al. Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biol. 2016;17:149. doi: 10.1186/s13059-016-1005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritsch C, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingolia NT, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–32. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern-Ginossar N, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakahigashi K, et al. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 2016;23:193–201. doi: 10.1093/dnares/dsw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwasaki S, et al. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popa A, et al. Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics. 2016;17:52. doi: 10.1186/s12864-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubio CA, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lareau LF, et al. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sidrauski C, et al. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015:4. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garreau de Loubresse N, et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 52.Nakahigashi K, et al. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics. 2014;15:1115. doi: 10.1186/1471-2164-15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Artieri CG, Fraser HB. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gritsenko AA, et al. Unbiased quantitative models of protein translation derived from ribosome profiling data. PLoS Comput Biol. 2015;11:e1004336. doi: 10.1371/journal.pcbi.1004336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pop C, et al. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reuveni S, et al. Genome-scale analysis of translation elongation with a ribosome flow model. PLoS Comput Biol. 2011;7:e1002127. doi: 10.1371/journal.pcbi.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian W, et al. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hussmann JA, et al. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guydosh NR, Green R. Dom34 rescues ribosomes in 3’ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loayza-Puch F, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- 62.Pechmann S, et al. The ribosome as a hub for protein quality control. Mol Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chartron JW, et al. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jan CH, et al. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams CC, et al. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker AH, et al. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc. 2013;8:2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schibich D, et al. Global profiling of SRP interaction with nascent polypeptides. Nature. 2016;536:219–223. doi: 10.1038/nature19070. [DOI] [PubMed] [Google Scholar]
- 70.Shieh YW, et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 71.Han Y, et al. Monitoring cotranslational protein folding in mammalian cells at codon resolution. Proc Natl Acad Sci U S A. 2012;109:12467–12472. doi: 10.1073/pnas.1208138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt C, et al. The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science. 2016;354:1431–1433. doi: 10.1126/science.aaf7520. [DOI] [PubMed] [Google Scholar]
- 73.Pelechano V, et al. Widespread co-translational RNA decay reveals ribosome dynamics. Cell. 2015;161:1400–1412. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rooijers K, et al. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat Commun. 2013;4:2886. doi: 10.1038/ncomms3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Couvillion MT, et al. Synchronized mitochondrial and cytosolic translation programs. Nature. 2016;533:499–503. doi: 10.1038/nature18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valásek L, et al. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 79.Archer SK, et al. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- 80.Katz ZB, et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. Elife. 2016:5. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halstead JM, et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanenbaum ME, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morisaki T, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–1429. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]
- 84.Wang C, et al. Real-time imaging of translation on single mRNA transcripts in live cells. Cell. 2016;165:990–1001. doi: 10.1016/j.cell.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu B, et al. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352:1430–1435. doi: 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan X, et al. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165:976–989. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shah P, et al. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukaya T, et al. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inada T. The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem Sci. 2017;42:5–15. doi: 10.1016/j.tibs.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 90.Chen YH, Coller J. A universal code for mRNA stability. Trends Genet. 2016;32:687–688. doi: 10.1016/j.tig.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li X, et al. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- 92.Weingarten-Gabbay S, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016:351. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 93.Grün D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]