Abstract
Background & Aims
Little is known about the incidence of celiac disease in the general population of children in the United States. We aimed to estimate the cumulative incidence of celiac disease in adolescents born in the Denver metropolitan area.
Methods
We collected data on HLA-DR, DQ genotypes of 31,766 infants, born from 1993 through 2004 at St. Joseph’s Hospital in Denver, from the Diabetes Autoimmunity Study in the Young. Subjects with susceptibility genotypes for celiac disease and type 1 diabetes were followed for up to 20 years for development of tissue transglutaminase autoantibodies (tTGA). Outcomes were the development of celiac disease autoimmunity (CDA) or celiac disease. CDA was defined as persistence of tTGA for at least 3 months or development of celiac disease. Celiac disease was defined based on detection of Marsh 2 or greater lesions in biopsies or persistent high levels of tTGA. For each genotype, the cumulative incidence of CDA and celiac disease were determined. To estimate the cumulative incidence in the Denver general population, outcomes by each genotype were weighted according to the frequency of each of these genotypes in the general population.
Results
Of 1339 subjects followed, 66 developed CDA and met criteria for celiac disease and 46 developed only CDA. Seropositivity for tTGA resolved spontaneously, without treatment, in 21 of the 46 subjects with only CDA (46%). The estimated cumulative incidence for CDA in the Denver general population at 5, 10, and 15 years of age was 2.4%, 4.3%, and 5.1% respectively; incidence values for celiac disease were 1.6%, 2.8%, and 3.1%, respectively.
Conclusions
In a 20-year prospective study of 1339 children with genetic risk factors for celiac disease, we found the cumulative incidence of CDA and celiac disease to be high within the first 10 years. Although more than 5% of children may experience a period of CDA, not all develop celiac disease or require gluten-free diets.
Keywords: transglutaminase, disease progression, DR3-DQ2, DR4-DQ8, celiac disease, HLA, birth cohort, incidence, celiac disease autoimmunity
Introduction
Recent trials to prevent celiac disease (CD) by modifying early infant experiences have not proven successful [1 2]. Meanwhile, CD continues to increase with current childhood incidence estimates increasing in the US [3 4] and Europe [5 6] with a reported prevalence of 1–3%. A gluten-free diet is an effective treatment and strategies for universal screening have been suggested, since the low diagnostic rates imply many cases of CD remain unrecognized and untreated [7] [8 9]. In addition to the lack of a clear cost-benefit to screening, other major questions remain including the optimal age to screen, the need for repeated testing, and the relationship between serological markers and long-term outcomes. Prospective population-based studies are needed to clarify the natural history of celiac disease autoimmunity (CDA) and its evolution to CD.
We have previously reported a prospective cohort study in which the cumulative incidence of CD by 5 years of age was estimated to be 0.9% [10]. However, it is not known how long this high rate of development of new onset CD continues throughout childhood. Here we report up to 15-year follow-up of this cohort. The cumulative incidence of CD is presented by each celiac-risk HLA genotype along with an estimate for the general population by adjusting for the frequencies by which these genotypes occur in the population.
Methods
Study subjects and design
Between 1993 and 2004, the Diabetes Autoimmunity Study in the Young (DAISY), completed HLA-DR, DQ genotyping of 31,766 newborns (Table 1)[11]. This typing was designed to detect DRB1*03 that is uniquely linked to DQ2.5. All newborns born at St. Joseph’s Hospital in Denver were eligible for screening; only newborns with severe congenital abnormalities or extreme prematurity were excluded. Approximately 75% of the newborns were members of Kaiser Permanente Colorado while the others represented a mix of Medicaid, uninsured and privately insured populations. Informed consent and cord blood samples were obtained for 87% of the eligible newborns, while 7% were discharged prior to being asked for consent and 6% of parents declined participation. Of those who declined, 71% did not want to be involved in any kind of research and 9% did not want to know the results. The reasons for nonparticipation in the remaining 20% were a mix of passive refusals, being too busy to participate, a variety of family reasons and plans to relocate. The refusal rate was similar among Kaiser Permanente members (5.8%) and non-members (6.3%) and did not differ by ethnicity. Blood samples of newborns for which consent could not be obtained were destroyed.
Table 1.
Ethnicity and HLA frequency, N (%) for 31,766 subjects undergoing newborn screening.
| Asian- American |
African- American |
Hispanic | Non- Hispanic White |
Biracial & others |
Total | |
|---|---|---|---|---|---|---|
| DR3-DQ2/DR3-DQ2 | 1 (0.2) | 28 (1.2) | 58 (0.6) | 250 (1.4) | 22 (1.4) | 359 (1.1) |
| DR3-DQ2/DR4-DQ8 | 4 (0.7) | 30 (1.3) | 235 (2.4) | 415 (2.4) | 19 (1.2) | 703 (2.2) |
| DR3-DQ2/X | 68 (11.8) | 472 (20.6) | 1234 (12.7) | 3376 (19.2) | 296 (19.0) | 5446 (17.1) |
| DR4-DQ8/DR4-DQ8 | 5 (0.9) | 6 (0.3) | 280 (2.9) | 385 (2.2) | 27 (1.7) | 703 (2.2) |
| DR4-DQ8/X | 51 (8.9) | 172 (7.5) | 2388 (24.6) | 2803 (15.9) | 225 (14.4) | 5639 (17.8) |
| DRX/X | 447 (77.6) | 1582 (69.1) | 5530 (56.9) | 10386 (59.0) | 971 (62.2) | 18916 (59.5) |
| Total | 576 | 2290 | 9725 | 17615 | 1560 | 31766 |
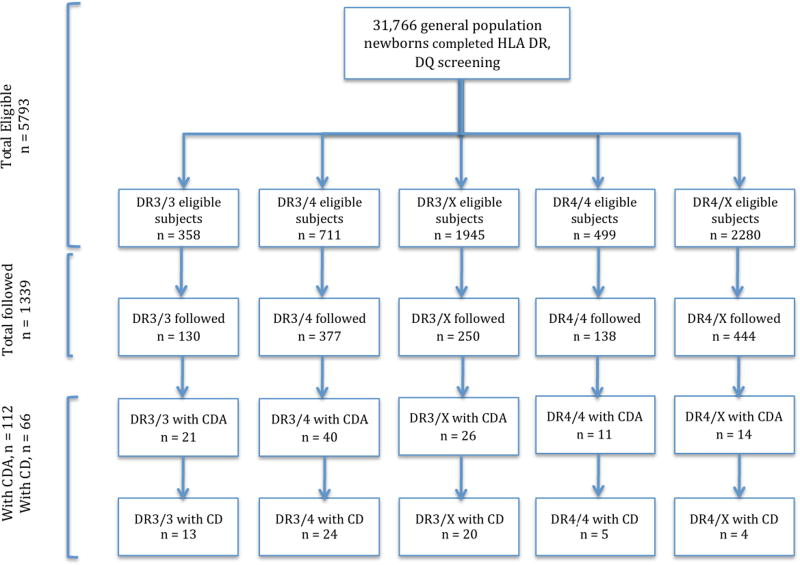
Those with HLA genotypes associated with CD, as well as a small number (54 subjects) without an identified genetic risk for type 1 diabetes or CD were recruited soon after birth. Subjects with additional high-risk celiac genotypes, DR3-DQ2/X were enrolled from the general population at age 2 to supplement the group at risk for CD. A total of 1339 enrolled into prospective follow-up and all had complete ethnicity and HLA information (Table 2). A flowchart of all screened and eligible subjects for recruitment is shown in Figure 1. Information regarding family history (having a first-degree relative) of type 1 diabetes or CD was obtained through questionnaires and updated during study visits. Information about the presence of symptoms and whether or not the subjects were on a gluten-free diet was recorded during study visits.
Table 2.
Demographic characteristics of the 1339 subjects followed prospectively for outcome of celiac disease autoimmunity or celiac disease.
| Celiac disease autoimmunity | Celiac disease | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| No N=1227 |
Yes N=112 |
p value | No N=1273 |
Yes N=66 |
p value | ||
| Sex N (%) | Female | 569 (46.4) | 64 (57.1) | 0.03 | 595 (46.7) | 38 (57.6) | 0.09 |
|
| |||||||
| HLA genotype N (%) | DR3-DQ2/DR3-DQ2 | 109 (8.9) | 21 (18.8) | <0.0001 | 117 (9.2) | 13 (19.7) | <0.0001 |
| DR3-DQ2/DR4-DQ8 | 337 (27.5) | 40 (35.7) | 353 (27.7) | 24 (36.4) | |||
| DR3-DQ2/X | 224 (18.3) | 26 (23.2) | 230 (18.1) | 20 (30.3) | |||
| DR4-DQ8/DR4-DQ8 | 127 (10.4) | 11 (9.8) | 133 (10.4) | 5 (7.6) | |||
| DR4-DQ8/X | 430 (35.0) | 14 (12.5) | 440 (34.6) | 4 (6.1) | |||
|
| |||||||
| Ethnicity N (%) | Black | 33 (2.7) | 0 (0) | 0.002 | 33 (2.6) | 0 (0) | 0.003* |
| Hispanic | 333 (27.1) | 15 (13.4) | 341 (26.8) | 7 (10.6) | |||
| NHW | 800 (65.2) | 92 (82.1) | 834 (65.5) | 58 (87.9) | |||
| Other | 61 (5.0) | 5 (4.5) | 65 (5.1) | 1 (1.5) | |||
|
| |||||||
| Family history of CD N (%) | Yes | 14 (1.1) | 17 (15.2) | <0.0001 | 16 (1.3) | 15 (22.7) | <0.0001 |
|
| |||||||
| Family history of T1D N (%) | Yes | 75 (6.1) | 10 (8.9) | 0.24 | 80 (6.3) | 5 (7.6) | 0.6 |
SD - standard deviation; T1D – type 1 diabetes
NHW - non-Hispanic white; “other” ethnicity: Asian, biracial or “other” self-reported ethnicities
X = not DR3-DQ2 or DR4-DQ8
Indicates Fisher exact test p value instead of chi-square test p value.
Figure 1.
Flow chart for subject screening and recruitment
Children with the following celiac-risk HLA genotypes were enrolled in follow-up:
DR3-DQ2/DR3-DQ2 (DR3-DQA1*05:01-DQB1*02:01/DR3-DQA1*05:01-DQB1*02:01)
DR3-DQ2/X (DR3-DQA1*05:01-DQB1*02:01/X)
DR3-DQ2/DR4-DQ8 (DR3-DQA1*05:01-DQB1*02:01/ DR4-DQA1*03:0X–DQB1*03:02)
DR4-DQ8/DR4-DQ8 (DR4-DQA1*03-DQB1*03:02/ DR4-DQA1*03-DQB1*03:02)
DR4-DQ8/X (DR4-DQA1*03-DQB1*03:02/X),
where X is neither DR3-DQA1*05:01-DQB1*02:01 or DR4-DQA1*03-DQB1*03:02.
The self-reported ethnic distribution was representative of the entire Denver metro area based on U.S. census data and on data for live births in the seven county Denver metropolitan area during the study period – the population base for this study that accounts for 54% of the Colorado population. At least one copy of either DR3-DQ2 or DR4-DQ8 was present in 40% of the screened population. Only 1.1% expressed the highest-risk HLA DR3-DQ2/DR3-DQ2 genotype. The most common risk genotypes, a single copy of either DR3-DQ2 or DR4-DQ8, were each present in about 17%.
Subjects had blood draws for tissue transglutaminase autoantibodies (tTGA) at 9, 15, and 24 months of age, and yearly thereafter. Any positive test was repeated in 3 to 6 months. After two positive blood samples, the family was referred to a pediatric gastroenterologist. A standard letter was provided to the physician that encouraged further evaluation particularly when the tTGA was >0.5. However, the decision as to whether or not to perform a biopsy was outside the protocol of this study, but if done, results were obtained and graded using the system described by Marsh[12] by a non-blinded clinical pathologist.
This study was approved by the Colorado Multiple Institutional Review Board. Written informed consent was obtained.
Autoantibody Assay
A well-described radiobinding assay using human recombinant tissue transglutaminase protein was used to detect tTGA[13 14]. A level of 0.05 was the cutoff for a positive test.
Definitions of celiac disease autoimmunity and celiac disease
The primary outcome was celiac disease autoimmunity (CDA), defined as either positive tTGA persistent on 2 consecutive blood draws at least 3 months apart, or a single positive tTGA test plus intestinal biopsy confirming CD. The secondary outcome was celiac disease, defined as tTGA plus a Marsh 2 or 3 lesion on intestinal biopsy or CDA with high tTGA levels (>0.5) on at least 2 visits. The cutoff of >0.5 was chosen because at 10x the normal cutoff of 0.05, the positive predictive value for finding villous atrophy on biopsy was 96% for children identified by screening[15]. All subjects with CD met criteria for CDA.
Statistical analysis
The dataset reflects follow-up of the cohort as of August 10, 2015. Statistical analyses were performed using the SAS software version 9.2 (SAS Institute, Cary, North Carolina). Two group t-tests were used to compare cases to controls for age at last visit. Chi-square test or Fisher exact test were used for the comparisons of categorical variables. Two outcomes were analyzed: time to development of CDA and time to CD. Ongoing recruitment since 1994 and continuing follow-up have resulted in variable lengths of follow-up, producing right-censored data. Finally we have interval-censored data in that we know only the time of the last negative and first positive autoantibody blood draw, rather than the actual time of conversion to autoantibody positivity.
Cumulative risk of CDA and CD by HLA group was estimated for each HLA group using nonparametric survival analysis of interval-censored data (SAS PROC ICLIFETEST) with Turnbull’s algorithm[16]. The cumulative risk for CD and CDA in the Denver population was estimated by using genotype-specific risk weighting for the population frequencies of HLA[17].
In order to examine the HLA effects on CD and CDA, survival analyses (SAS PROC LIFEREG) were carried out using a parametric model with different underlying time to event distributions accounting for right, left and interval censoring. Probability plots from models with Weibull, exponential, log-logistic, lognormal and generalized gamma distribution were compared. The model based on the generalized gamma distribution was the best fit. Calculations of follow-up time began at birth. HLA genotype, family history of CD and sex were included in the model because they are known to affect the risk factors for CD.
Results are presented as acceleration factors that summarize the relative time to event compared to a reference group. The acceleration factor γ allows evaluation of the effect of predictor variables on the disease-free survival time. When two groups are compared for development of celiac disease, e.g., females vs. males, γ < 1 indicates that female sex is harmful to survival free of CD, while γ >1 would indicate a benefit to disease-free survival. A 2-sided p < 0.05 was considered significant.
Results
Among the 1339 study subjects, 112 developed the outcome of CDA, including 66 subjects that met the criteria for CD: 38 by biopsy criteria and 28 by persistent and high tTGA levels. None of the 54 control subjects without an HLA genotype risk developed celiac disease during this time period. Of the 38 with CD confirmed by biopsy, 31 (81%) were known to be symptomatic, and all but 6 had reported 2 or more symptoms. Of the 28 with CD based on persistent and high tTGA levels, 15 (54%) were symptomatic, and 8 of them had reported 2 or more symptoms. Overall, 30% of the CD subjects were asymptomatic.
Predictors of celiac disease
Univariate comparisons (Table 2) confirmed that older age, female sex, HLA genotype, non-Hispanic white ethnicity and having a first-degree relative with CD were associated with both CDA and CD. Having a first-degree relative with type 1 diabetes was not associated with either outcome.
The gamma accelerated failure time model for progression to CDA and CD (Table 3) included HLA genotype, family history of CD, and sex as covariates. Ethnicity was not included because it did not allow the model to converge when analyzed. After adjusting for covariates, subjects with DR3-DQ2 homozygosity had a median progression time of 42% (acceleration factor 0.42) and 14% (acceleration factor 0.14) for CDA and CD respectively, when compared to the progression time of the reference group with the lower-risk genotype of DR4-DQ8/X.
Table 3.
Adjusted Survival Analysis of Time to Progression for Celiac Disease Autoimmunity (CDA) and Celiac Disease (CD). Acceleration factor summarizes the relative time to event compared to a reference group.
| Celiac Disease Autoimmunity | Celiac Disease | |||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Acceleration Factor |
95% CI | P value | Acceleration Factor |
95% CI | P value | ||
| Sex | Female | 0.83 | 0.66 – 1.04 | 0.11 | Female | 0.65 | 0.42 – 1.01 | 0.06 |
| Male | reference group | Male | reference group | |||||
| HLA genotype | DR3-DQ2/DR3-DQ2/ | 0.42 | 0.25 – 0.73 | 0.002 | DR3-DQ2/DR3-DQ2/ | 0.14 | 0.06–0.33 | <0.0001 |
| DR3-DQ2/DR4-DQ8 | 0.60 | 0.42 – 0.86 | 0.006 | DR3-DQ2/DR4-DQ8 | 0.24 | 0.12 – 0.48 | <0.0001 | |
| DR3-DQ2/X | 0.61 | 0.41 – 0.91 | 0.01 | DR3-DQ2/X | 0.21 | 0.10 – 0.44 | <0.0001 | |
| DR4-DQ8/DR4-DQ8 | 0.81 | 0.50 – 1.31 | 0.39 | DR4-DQ8/DR4-DQ8 | 0.40 | 0.16 – 1.00 | 0.05 | |
| DR4-DQ8/X | reference group | DR4-DQ8/X | reference group | |||||
| First-degree relative with CD | yes | 0.64 | 0.31 1.32 | 0.23 | Yes | 0.13 | 0.05 – 0.36 | <0.0001 |
| no | reference group | no | reference group | |||||
CI – confidence interval
Cumulative incidence by HLA genotype
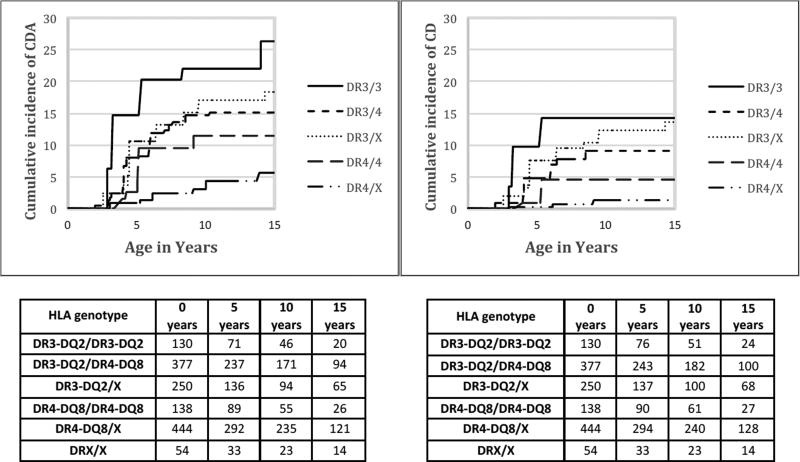
The cumulative incidence by each HLA genotype for the development of CDA and celiac disease is shown in Figure 2. By 5, 10 and 15 years, the cumulative incidence of CDA in those with DR3/3 was 14.8%, 22.1%, and 26.4% respectively (Supplementary Table S1). In those with a single DR3-DQ2 (as DR3-DQ2/X or DR3-DQ2/DR4-DQ8), the cumulative incidence of CDA was 8.6%, 15.1%, and 16.3%.
Figure 2.
Cumulative incidence (%) of celiac disease autoimmunity (CDA ) and celiac disease (CD) for each HLA genotype through the first 15 years of life. Number of subjects in study grouped by 5-year intervals.
The cumulative incidence of CD in those with DR3/3 was 9.8%, 14.2%, and 14.2% by 5, 10 and 15 years respectively (Supplementary Table S2). In those with a single DR3 (as DR3-DQ2/X or DR3-DQ2/DR4-DQ8), the combined cumulative incidence of CD was 5.6%, 10.3%, and 10.8%.
Estimated population cumulative incidence
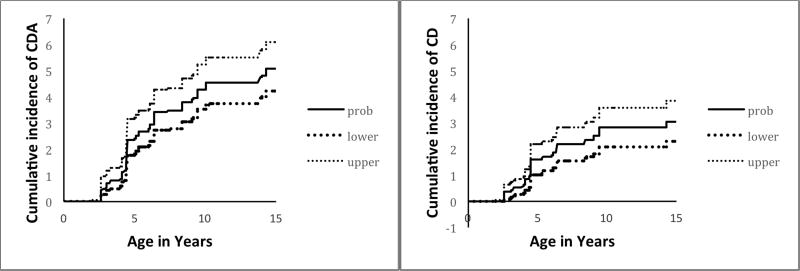
After weighting for the general population frequencies of each HLA genotype, the estimated cumulative incidence of CDA and CD in the Denver general population is shown in Figure 3. The estimated cumulative incidence in the Denver general population for CDA was 2.4% (95% CI 1.8–3.2) by age 5 years, 4.3% (3.5–5.3) by 10 years, and 5.1% (4.2–6.1) by 15 years. The estimated cumulative incidence of CD in the Denver general population was 1.6% (95% CI 1.0–2.2) by age 5 years. 2.8% (95% CI 2.1–3.6) by 10 years, and 3.1% (2.3–3.8) by 15 years. As a subgroup analysis, we considered only those with biopsy-proven CD. The estimated cumulative incidence in this group at 5, 10 and 15 years of age was 1.2%, 1.9% and 1.9%. However, it should be noted that this would be an underestimation of CD in the general population.
Figure 3.
Estimated cumulative incidence (%) of celiac disease autoimmunity (CDA) and celiac disease (CD) in the Denver, Colorado general population.
Transient or fluctuating CDA
Of the 46 study subjects who developed CDA but not CD, 21 (46%) had transient tTGA that self-resolved and did not reappear and an additional 6 (13%) had tTGA that fluctuated between positive and negative before becoming positive again despite remaining on a gluten-containing diet. All of these subjects either had tTGA levels < 0.5 (n = 23) or had a tTGA measurement >0.5 only once, followed immediately by a declining tTGA level 3–6 months later, with levels that eventually became negative (2 transients, 2 fluctuating). None of the 4 subjects in this group with biopsies had Marsh 2 or 3 lesions. In most subjects, duration of tTGA positivity was between 6–18 months. However, there were 4 subjects with tTGA positivity lasting for 2, 3, 5 years and 9 years before spontaneously becoming negative while continuing on a gluten-containing diet. There was no association between those with transient/fluctuating tTGA and having type 1 diabetes or islet autoimmunity, nor was there an association with a specific HLA genotype.
Discussion
Forty percent of the general population carries HLA-DQ2 or DQ8, and is therefore at risk for CDA and CD. We have previously estimated the risk of CDA and CD in the first 5 years of life[10]. We now show that children continue to develop CDA and CD throughout at least the first 10 years of life, with a relative decrease in incidence by 10–15 years of age. The estimated cumulative incidence of CDA of 5.1% and of CD of 3.1% by age 15 years suggests a significant increase from historical estimates in the US. However, this assessment of rising incidence is consistent with recent studies using different approaches in North America [3 4 18]and Europe[5 19].
The 3.1% cumulative incidence of CD in Denver by age 15 is the highest to date in North America and is consistent with the 3% prevalence reported in Sweden for 12 year olds born during an “epidemic” thought to be due to early introduction of large amounts of gluten in the infant diet6. Although timing of gluten introduction is likely not a factor in the increased incidence, it is possible that the amount of gluten at the time of introduction could be a factor for both countries [20].
While it is possible that this high incidence of CD could be an isolated cohort effect limited to Colorado children, the findings are consistent with the observed trend of increasing incidence noted worldwide. Prior estimates of prevalence in the US are much lower: 0.4% of US blood donors in 1998[21], and then 0.71% in 2010[22] followed by 0.79% in 2012[23] according to the NHANES study. However, these numbers are based on screening of both adults and children, as opposed to our study, which was in only children and adolescents. Therefore, the marked increase in incidence in our study may be a new phenomenon that has affected this current generation of youth.
The reasons for the dramatic increase are unknown, but in the absence of identified genetic differences and rapid changes in CD frequency over the past two decades, environmental causes seem likely. The role of early infant factors related to timing of gluten introduction and breast feeding duration has been placed into serious doubt based on two recent clinical trials[1 2] and a prospective cohort study[24]. Other proposed causes include the amount of gluten consumed during infancy[20 25] and early infections[26 27] such as rotavirus[28] possibly causing alterations of the gut microbiome[29 30].
According to the 2010 US Census data, the Denver area has a higher percentage of children of Hispanic ethnicity (32% vs 16%), and a lower percentage with African American ethnicity when compared to the US general population. These ethnicity differences are unlikely to substantially bias extrapolation of the estimates of CD and CDA, although additional studies are needed to determine if our data accurately reflects the US general population.
It is also important to note that while all subjects were recruited from the general population, there was a higher likelihood that a subject having a first degree relative with type 1 diabetes would consent to enrollment and have better study retention. However, our data and a prior study demonstrates that unlike those with a first degree relative for CD, having a first degree relative for type 1 diabetes does not impact the risk of developing celiac disease when adjusted for HLA and sex[31].
The results from this study provide unique data on the natural history of CD that has implications for public health issues such as universal screening. From this study, it might be argued that more aggressive screening during early childhood would be indicated for the first 5–10 years, followed by less frequent testing thereafter given the decrease in the rate of seroconversion during this time.
Our results emphasize that the presence of CDA does not predict universal progression to CD. Such transient autoantibodies may limit public acceptance of universal screening[15][38]. Others have also observed fluctuating tTGA[15] and transient tTGA antibodies in children with serial testing[39–41]. Continued long term follow-up will identify whether the autoimmunity in these subjects truly abates and tolerance develops, or if CDA will recur in time, possibly in response to additional stimulating events. At present, low positive tTGA results should be interpreted with caution, and do not necessarily indicate need for biopsy or for treatment.
One of the limitations is that our study did not account for the DR5/DR7 risk genotype present in 1.4–5.3% the general population[42 43] and accounting for <5% of all patients with CD[44]. These individuals express a single copy of the DQ2 heterodimer as DQA1*05:01,DQB1*02:02 in trans. An additional 5% are negative for DQ2 and DQ8 heterodimer in both cis and trans position; most have only half of the DQ2 heterodimer (DQA1*05 or DQB1*02)[45] and many have the DQA1*02:01, DQB1*02:02 haplotype (also known as DQ2.2)[46]. It also doesn’t take in to account those in the general population with DR3/DR7 who can be considered high risk and present in about 2.6% of the general population[42]. Therefore, this report should be seen as conservative since we may underestimate the true cumulative incidence of CD and CDA by not accounting for subjects with these genotypes.
Another potential limitation involves the definitions of CDA and CD. We considered persistent seropositivity (CDA) with a well-characterized assay as sufficient to identify cases since many subjects did not pursue biopsy. Of those undergoing biopsy, 30% had normal villous architecture similar to other reports[47 48]. These children were not considered to have CD unless the tTGA levels were very high and persistent (greater than 10x the normal cutoff). Therefore, only those with biopsy-proven CD or very high tTGA levels were included, while subjects with persistent low tTGA levels and no biopsy were considered to not have CD. Symptoms were not considered in the categorization of CD in this study since half of all screening-identified children with CD are known to be asymptomatic [49].
In summary, this study of a population-based birth cohort in Denver, CO shows continuing development of CD throughout early childhood. By 15 years of age, an estimated 3.1% of the Denver population develops CD and an additional 2% develops a lesser degree of CDA. Although approximately 5% of children may experience a period of time with CDA, a high proportion will remit without treatment. It seems likely that the majority of celiac autoimmunity develops in childhood, and mostly before age 10, which could inform future efforts for universal screening.
Supplementary Material
Acknowledgments
Funding source: This research was supported by NIH grants R01 DK32493, R01 DK50979 and R01 DK32083, Diabetes Endocrinology Research Center P30 DK57516, and M01RR00069 General Clinical Research Centers Program, NCRR, NIH.
Abbreviations
- tTGA
tissue transglutaminase autoantibody
- HLA
Human Leukocyte Antigen
- CD
celiac disease
- CDA
celiac disease autoimmunity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
E.L. contributed to the initial study design, performed analysis, wrote the initial draft of the manuscript
F.D. performed data analysis, statistical assistance
A.B. performed data analysis, statistical assistance
I.T. was responsible for gathering and managing the data
J.N. contributed to the initial study design, helped write and review the manuscript
B.F. helped write and review the manuscript
E.H. contributed to the initial study design, helped write and review the manuscript
M.R. contributed to the initial study design, helped write and review the manuscript
There are no conflicts of interest to declare for any of the authors of this manuscript.
References
- 1.Lionetti E, Castellaneta S, Francavilla R, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. 2014;371(14):1295–303. doi: 10.1056/NEJMoa1400697. [DOI] [PubMed] [Google Scholar]
- 2.Vriezinga SL, Auricchio R, Bravi E, et al. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. 2014;371(14):1304–15. doi: 10.1056/NEJMoa1404172. [DOI] [PubMed] [Google Scholar]
- 3.Catassi C, Kryszak D, Bhatti B, et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann Med. 2010;42(7):530–8. doi: 10.3109/07853890.2010.514285. [DOI] [PubMed] [Google Scholar]
- 4.Ludvigsson JF, Rubio-Tapia A, van Dyke CT, et al. Increasing incidence of celiac disease in a North American population. Am J Gastroenterol. 2013;108(5):818–24. doi: 10.1038/ajg.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohi S, Mustalahti K, Kaukinen K, et al. Increasing prevalence of coeliac disease over time. Aliment Pharmacol Ther. 2007;26(9):1217–25. doi: 10.1111/j.1365-2036.2007.03502.x. [DOI] [PubMed] [Google Scholar]
- 6.Myleus A, Ivarsson A, Webb C, et al. Celiac disease revealed in 3% of Swedish 12-year-olds born during an epidemic. J Pediatr Gastroenterol Nutr. 2009;49(2):170–6. doi: 10.1097/MPG.0b013e31818c52cc. [DOI] [PubMed] [Google Scholar]
- 7.Hill ID, Dirks MH, Liptak GS, et al. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2005;40(1):1–19. doi: 10.1097/00005176-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Husby S, Koletzko S, Korponay-Szabo IR, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54(1):136–60. doi: 10.1097/MPG.0b013e31821a23d0. [DOI] [PubMed] [Google Scholar]
- 9.Webb C, Norstrom F, Myleus A, et al. Celiac disease can be predicted by high levels of anti-tissue transglutaminase antibodies in population-based screening. J Pediatr Gastroenterol Nutr. 2015;60(6):787–91. doi: 10.1097/MPG.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 10.Hoffenberg EJ, MacKenzie T, Barriga KJ, et al. A prospective study of the incidence of childhood celiac disease. J Pediatr. 2003;143(3):308–14. doi: 10.1067/s0022-3476(03)00282-8. [DOI] [PubMed] [Google Scholar]
- 11.Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY) Diabetologia. 1996;39(7):807–12. doi: 10.1007/s001250050514. [DOI] [PubMed] [Google Scholar]
- 12.Marsh MN. Gluten major histocompatibility complex the small intestine A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’) Gastroenterology. 1992;102(1):330–54. [PubMed] [Google Scholar]
- 13.Bao F, Yu L, Babu S, et al. One third of HLA DQ2 homozygous patients with type 1 diabetes express celiac disease-associated transglutaminase autoantibodies. J Autoimmun. 1999;13(1):143–8. doi: 10.1006/jaut.1999.0303. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Yu L, Tiberti C, et al. A report on the International Transglutaminase Autoantibody Workshop for Celiac Disease. Am J Gastroenterol. 2009;104(1):154–63. doi: 10.1038/ajg.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu E, Bao F, Barriga K, et al. Fluctuating transglutaminase autoantibodies are related to histologic features of celiac disease. Clin Gastroenterol Hepatol. 2003;1(5):356–62. doi: 10.1053/s1542-3565(03)00180-0. [DOI] [PubMed] [Google Scholar]
- 16.Turnbull B. The Empirical Distribution Function with Arbitrarily Grouped, Censored and Truncated Data. Journal of the Royal Statistical Society Series B (Methodological) 1976;(38):6. [Google Scholar]
- 17.Norris JM, Barriga K, Hoffenberg EJ, et al. Risk of celiac disease autoimmunity and timing of gluten introduction in the diet of infants at increased risk of disease. JAMA. 2005;293(19):2343–51. doi: 10.1001/jama.293.19.2343. [DOI] [PubMed] [Google Scholar]
- 18.Riddle MS, Murray JA, Porter CK. Percieved rise in the incidence of celiac diseases in the US military may be due to more than one factor--response to Dosanjh et al. Am J Gastroenterol. 2013;108(1):144–5. doi: 10.1038/ajg.2012.378. [DOI] [PubMed] [Google Scholar]
- 19.Ress K, Luts K, Rago T, Pisarev H, Uibo O. Nationwide study of childhood celiac disease incidence over a 35-year period in Estonia. Eur J Pediatr. 2012;171(12):1823–8. doi: 10.1007/s00431-012-1835-0. [DOI] [PubMed] [Google Scholar]
- 20.Andren Aronsson C, Lee HS, Koletzko S, et al. Effects of Gluten Intake on Risk of Celiac Disease: A Case-Control Study on a Swedish Birth Cohort. Clin Gastroenterol Hepatol. 2016;14(3):403–09. doi: 10.1016/j.cgh.2015.09.030. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Not T, Horvath K, Hill ID, et al. Celiac disease risk in the USA: high prevalence of antiendomysium antibodies in healthy blood donors. Scand J Gastroenterol. 1998;33(5):494–8. doi: 10.1080/00365529850172052. [DOI] [PubMed] [Google Scholar]
- 22.Rubio-Tapia A, Ludvigsson JF, Brantner TL, Murray JA, Everhart JE. The prevalence of celiac disease in the United States. Am J Gastroenterol. 2012;107(10):1538–44. doi: 10.1038/ajg.2012.219. quiz 37, 45. [DOI] [PubMed] [Google Scholar]
- 23.Mardini HE, Westgate P, Grigorian AY. Racial Differences in the Prevalence of Celiac Disease in the US Population: National Health and Nutrition Examination Survey (NHANES) 2009–2012. Dig Dis Sci. 2015;60(6):1738–42. doi: 10.1007/s10620-014-3514-7. [DOI] [PubMed] [Google Scholar]
- 24.Aronsson CA, Lee HS, Liu E, et al. Age at gluten introduction and risk of celiac disease. Pediatrics. 2015;135(2):239–45. doi: 10.1542/peds.2014-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivarsson A, Persson LA, Nystrom L, et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000;89(2):165–71. doi: 10.1080/080352500750028771. [DOI] [PubMed] [Google Scholar]
- 26.Myleus A, Hernell O, Gothefors L, et al. Early infections are associated with increased risk for celiac disease: an incident case-referent study. BMC Pediatr. 2012;12:194. doi: 10.1186/1471-2431-12-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemppainen KM, Lynch KF, Liu E, et al. Factors that Increase Risk of Celiac Disease Autoimmunity Following a Gastrointestinal Infection in Early Life. Clin Gastroenterol Hepatol. 2016 doi: 10.1016/j.cgh.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stene LC, Honeyman MC, Hoffenberg EJ, et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am J Gastroenterol. 2006;101(10):2333–40. doi: 10.1111/j.1572-0241.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 29.Wacklin P, Kaukinen K, Tuovinen E, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19(5):934–41. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 30.Sellitto M, Bai G, Serena G, et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS One. 2012;7(3):e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu E, Lee HS, Aronsson CA, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371(1):42–9. doi: 10.1056/NEJMoa1313977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen A, Sandstrom O, Carlsson A, et al. Usefulness of symptoms to screen for celiac disease. Pediatrics. 2014;133(2):211–8. doi: 10.1542/peds.2012-3765. [DOI] [PubMed] [Google Scholar]
- 33.Marine M, Farre C, Alsina M, et al. The prevalence of coeliac disease is significantly higher in children compared with adults. Aliment Pharmacol Ther. 2011;33(4):477–86. doi: 10.1111/j.1365-2036.2010.04543.x. [DOI] [PubMed] [Google Scholar]
- 34.Pratesi R, Gandolfi L, Garcia SG, et al. Prevalence of coeliac disease: unexplained age-related variation in the same population. Scand J Gastroenterol. 2003;38(7):747–50. doi: 10.1080/00365520310003255. [DOI] [PubMed] [Google Scholar]
- 35.Volta U, Bellentani S, Bianchi FB, et al. High prevalence of celiac disease in Italian general population. Dig Dis Sci. 2001;46(7):1500–5. doi: 10.1023/a:1010648122797. [DOI] [PubMed] [Google Scholar]
- 36.Evans KE, Hadjivassiliou M, Sanders DS. Is it time to screen for adult coeliac disease? Eur J Gastroenterol Hepatol. 2011;23(10):833–8. doi: 10.1097/MEG.0b013e328348f9aa. [DOI] [PubMed] [Google Scholar]
- 37.Hershcovici T, Leshno M, Goldin E, Shamir R, Israeli E. Cost effectiveness of mass screening for coeliac disease is determined by time-delay to diagnosis and quality of life on a gluten-free diet. Aliment Pharmacol Ther. 2010;31(8):901–10. doi: 10.1111/j.1365-2036.2010.04242.x. [DOI] [PubMed] [Google Scholar]
- 38.Lionetti E, Castellaneta S, Pulvirenti A, et al. Prevalence and natural history of potential celiac disease in at-family-risk infants prospectively investigated from birth. J Pediatr. 2012;161(5):908–14. doi: 10.1016/j.jpeds.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Simell S, Hoppu S, Simell T, et al. Age at development of type 1 diabetes- and celiac disease-associated antibodies and clinical disease in genetically susceptible children observed from birth. Diabetes Care. 2010;33(4):774–9. doi: 10.2337/dc09-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjorck S, Brundin C, Lorinc E, Lynch KF, Agardh D. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J Pediatr Gastroenterol Nutr. 2010;50(1):49–53. doi: 10.1097/MPG.0b013e3181b477a6. [DOI] [PubMed] [Google Scholar]
- 41.Castellaneta S, Piccinno E, Oliva M, et al. High rate of spontaneous normalization of celiac serology in a cohort of 446 children with type 1 diabetes: a prospective study. Diabetes Care. 2015;38(5):760–6. doi: 10.2337/dc14-2890. [DOI] [PubMed] [Google Scholar]
- 42.Meuli R, Pichler WJ, Gaze H, Lentze MJ. Genetic difference in HLA-DR phenotypes between coeliac disease and transitory gluten intolerance. Arch Dis Child. 1995;72(1):29–32. doi: 10.1136/adc.72.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall MAMM, Satz ML, Barboni F, Brunier G. Coeliac Disease Study. XIth Workshop Joint Report. Oxford: Oxford University Press; 1991. [Google Scholar]
- 44.Monsuur AJ, de Bakker PI, Zhernakova A, et al. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. PLoS One. 2008;3(5):e2270. doi: 10.1371/journal.pone.0002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karell K, Louka AS, Moodie SJ, et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol. 2003;64(4):469–77. doi: 10.1016/s0198-8859(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 46.Pietzak MM, Schofield TC, McGinniss MJ, Nakamura RM. Stratifying risk for celiac disease in a large at-risk United States population by using HLA alleles. Clin Gastroenterol Hepatol. 2009;7(9):966–71. doi: 10.1016/j.cgh.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 47.Auricchio R, Tosco A, Piccolo E, et al. Potential celiac children: 9-year follow-up on a gluten-containing diet. Am J Gastroenterol. 2014;109(6):913–21. doi: 10.1038/ajg.2014.77. [DOI] [PubMed] [Google Scholar]
- 48.Kurppa K, Ashorn M, Iltanen S, et al. Celiac disease without villous atrophy in children: a prospective study. J Pediatr. 2010;157(3):373–80. doi: 10.1016/j.jpeds.2010.02.070. 80 e1. [DOI] [PubMed] [Google Scholar]
- 49.Agardh D, Lee HS, Kurppa K, et al. Clinical features of celiac disease: a prospective birth cohort. Pediatrics. 2015;135(4):627–34. doi: 10.1542/peds.2014-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.