Abstract
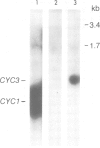
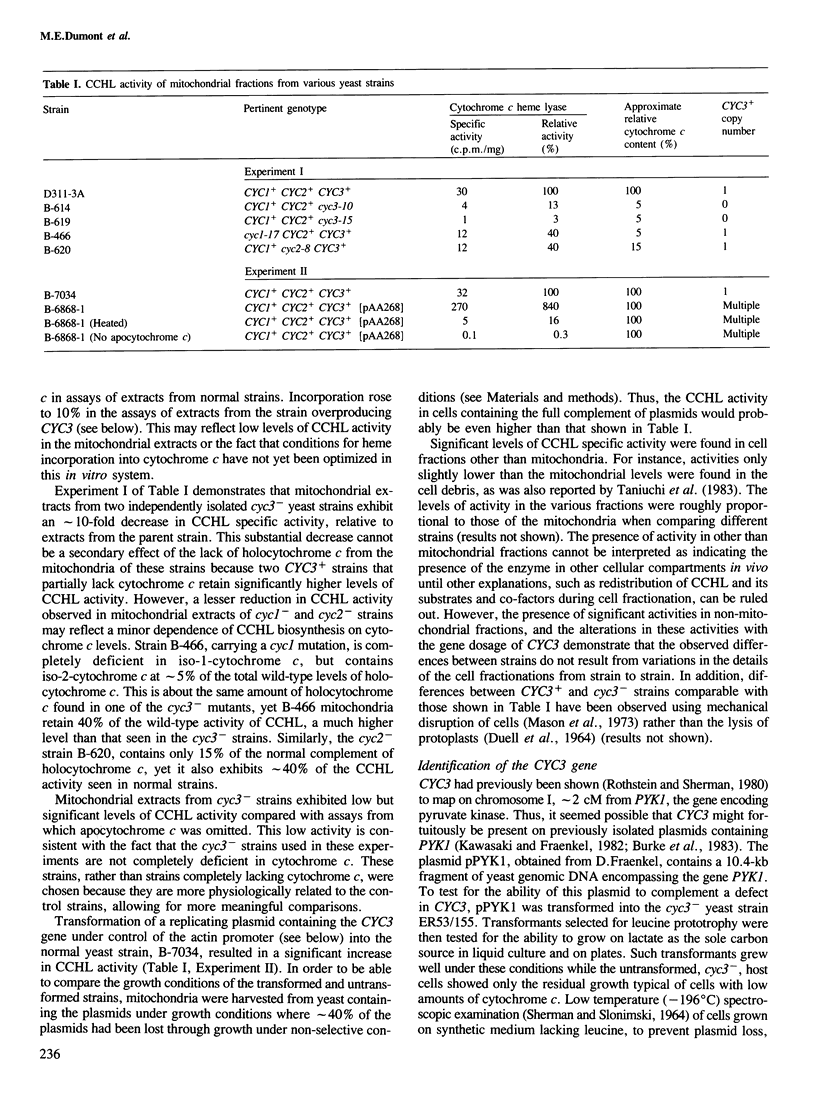
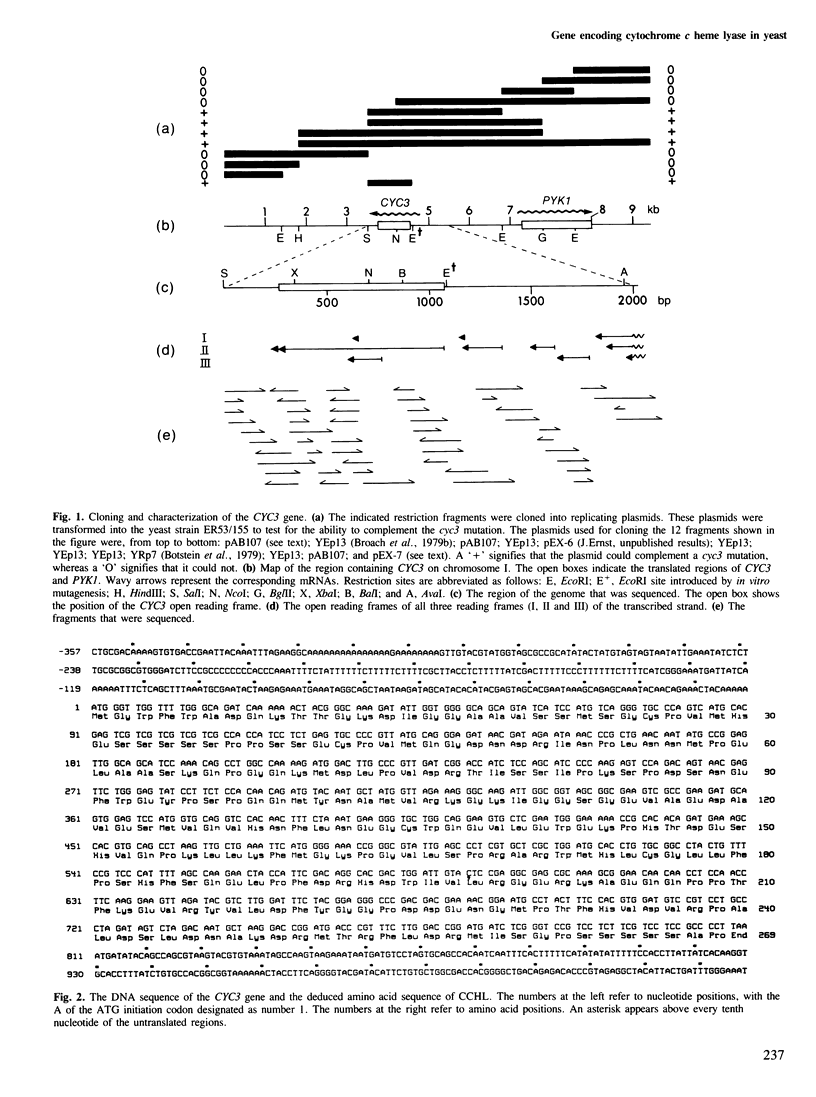
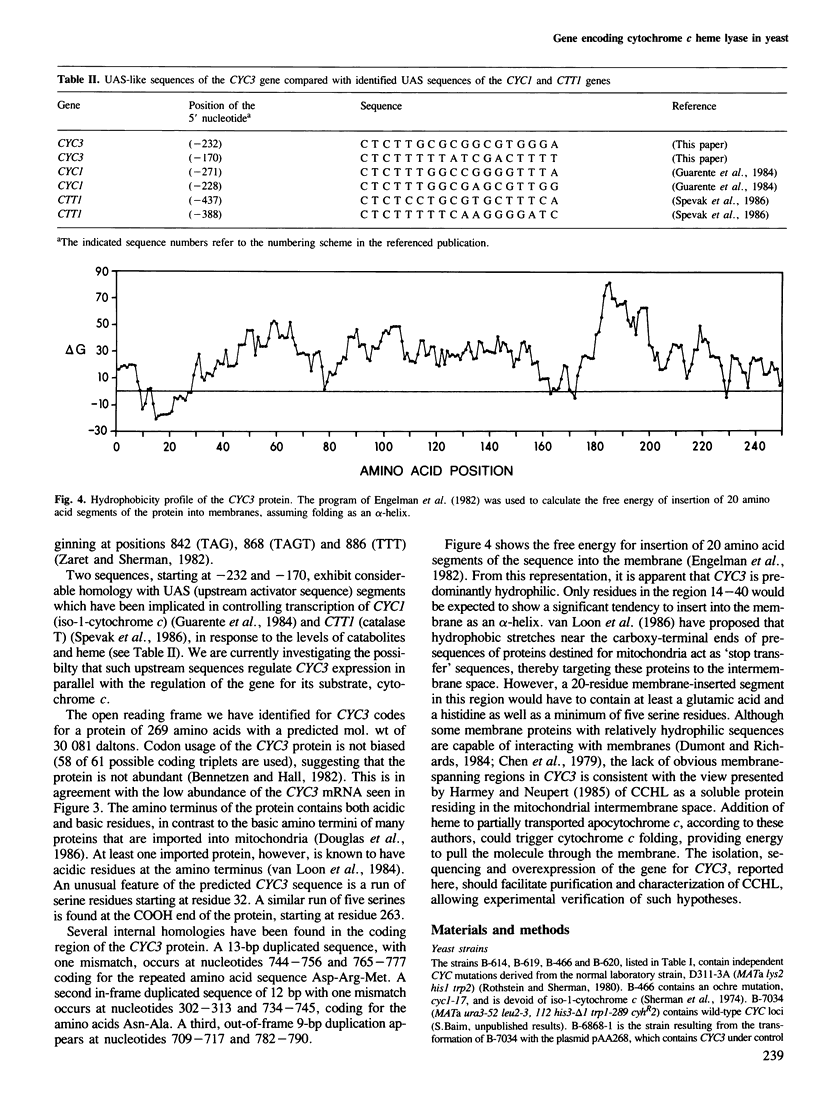
Mitochondrial cytochrome c contains a heme group covalently attached through thioether linkages to two cysteinyl residues of the protein. We demonstrate here that the nuclear gene, CYC3, in the yeast Saccharomyces cerevisiae, encodes cytochrome c heme lyase (CCHL), the enzyme catalyzing the attachment of heme to apocytochrome c. Mitochondrial extracts from cyc3- mutants are deficient in CCHL activity compared with extracts from normal strains, whereas strains carrying multiple copies of the CYC3 gene exhibit high levels of the activity. The CYC3 gene was cloned by functional complementation of a cyc3- mutant using a previously isolated plasmid containing the gene PYK1, which is tightly linked to CYC3. An open reading frame encoding a protein of 269 amino acids was identified from the DNA sequence of a fragment encompassing the CYC3 gene, and the corresponding transcript shown to be approximately 0.9 kb in length. CCHL appears to be a single polypeptide chain which acts specifically on the two forms of cytochrome c, but not on cytochrome c1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basile G., Di Bello C., Taniuchi H. Formation of an iso-1-cytochrome c-like species containing a covalently bonded heme group from the apoprotein by a yeast cell-free system in the presence of hemin. J Biol Chem. 1980 Aug 10;255(15):7181–7191. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Atkins J. F., McGill C., Chow L. Identification and mapping of the transcriptional and translational products of the yeast plasmid, 2mu circle. Cell. 1979 Apr;16(4):827–839. doi: 10.1016/0092-8674(79)90098-9. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Chen R., Krämer C., Schmidmayr W., Henning U. Primary structure of major outer membrane protein I of Escherichia coli B/r. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5014–5017. doi: 10.1073/pnas.76.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleran E. M., Jones O. T. Studies on the biosynthesis of cytochrome c. Biochem J. 1973 May;134(1):89–96. doi: 10.1042/bj1340089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUELL E. A., INOUE S., UTTER M. F. ISOLATION AND PROPERTIES OF INTACT MITOCHONDRIA FROM SPHEROPLASTS OF YEAST. J Bacteriol. 1964 Dec;88:1762–1773. doi: 10.1128/jb.88.6.1762-1773.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M. G., McCammon M. T., Vassarotti A. Targeting proteins into mitochondria. Microbiol Rev. 1986 Jun;50(2):166–178. doi: 10.1128/mr.50.2.166-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M. E., Richards F. M. Insertion of apocytochrome c into lipid vesicles. J Biol Chem. 1984 Apr 10;259(7):4147–4156. [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Ernst J. F., Chan R. K. Characterization of Saccharomyces cerevisiae mutants supersensitive to aminoglycoside antibiotics. J Bacteriol. 1985 Jul;163(1):8–14. doi: 10.1128/jb.163.1.8-14.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher W. R., Taniuchi H., Anfinsen C. B. On the role of heme in the formation of the structure of cytochrome c. J Biol Chem. 1973 May 10;248(9):3188–3195. [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig B., Neupert W. Assembly of cytochrome c. Apocytochrome c is bound to specific sites on mitochondria before its conversion to holocytochrome c. Eur J Biochem. 1981 Dec;121(1):203–212. doi: 10.1111/j.1432-1033.1981.tb06450.x. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Kawasaki G., Fraenkel D. G. Cloning of yeast glycolysis genes by complementation. Biochem Biophys Res Commun. 1982 Oct 15;108(3):1107–1122. doi: 10.1016/0006-291x(82)92114-3. [DOI] [PubMed] [Google Scholar]
- Korb H., Neupert W. Biogenesis of cytochrome c in Neurospora crassa. Synthesis of apocytochrome c, transfer to mitochondria and conversion to Holocytochrome c. Eur J Biochem. 1978 Nov 15;91(2):609–620. doi: 10.1111/j.1432-1033.1978.tb12714.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz T. M., Pietras D. F., Sherman F. Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4475–4479. doi: 10.1073/pnas.81.14.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H. N., Sherman F. Yeast cytochrome c-specific protein-lysine methyltransferase: coordinate regulation with cytochrome c and activities in cyc mutants. J Bacteriol. 1979 Jun;138(3):853–860. doi: 10.1128/jb.138.3.853-860.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason T. L., Poyton R. O., Wharton D. C., Schatz G. Cytochrome c oxidase from bakers' yeast. I. Isolation and properties. J Biol Chem. 1973 Feb 25;248(4):1346–1354. [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- Matsuura S., Arpin M., Hannum C., Margoliash E., Sabatini D. D., Morimoto T. In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4368–4372. doi: 10.1073/pnas.78.7.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Oostra B. A., Harvey R., Ely B. K., Markham A. F., Smith A. E. Transforming activity of polyoma virus middle-T antigen probed by site-directed mutagenesis. Nature. 1983 Aug 4;304(5925):456–459. doi: 10.1038/304456a0. [DOI] [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980 Apr;94(4):871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELS A. A., FUKUHARA H., PERE G., SLONIMSKI P. P. CIN'ETIQUE DE LA BIOSYNTH'ESE INDUITE DE L'ISO-1-CYTOCHROME C ET DE L'ISO-2-CYTOCHROME C AU COURS DE L'ADAPTATION 'A L'OXYG'ENE. Biochim Biophys Acta. 1965 Mar 15;95:486–502. [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- STICKLAND L. H. The determination of small quantities of bacteria by means of the biuret reaction. J Gen Microbiol. 1951 Oct;5(4):698–703. doi: 10.1099/00221287-5-4-698. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak W., Hartig A., Meindl P., Ruis H. Heme control region of the catalase T gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1986 Apr;203(1):73–78. doi: 10.1007/BF00330386. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi H., Basile G., Taniuchi M., Veloso D. Evidence for formation of two thioether bonds to link heme to apocytochrome c by partially purified cytochrome c synthetase. J Biol Chem. 1983 Sep 25;258(18):10963–10966. [PubMed] [Google Scholar]
- Van Loon A. P., De Groot R. J., De Haan M., Dekker A., Grivell L. A. The DNA sequence of the nuclear gene coding for the 17-kd subunit VI of the yeast ubiquinol-cytochrome c reductase: a protein with an extremely high content of acidic amino acids. EMBO J. 1984 May;3(5):1039–1043. doi: 10.1002/j.1460-2075.1984.tb01924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso D., Basile G., Taniuchi H. Formation of a cytochrome c-like species from horse apoprotein and hemin catalyzed by yeast mitochondrial cytochrome c synthetase. J Biol Chem. 1981 Aug 25;256(16):8646–8651. [PubMed] [Google Scholar]
- Visco C., Taniuchi H., Berlett B. S. On the specificity of cytochrome c synthetase in recognition of the amino acid sequence of apocytochrome c. J Biol Chem. 1985 May 25;260(10):6133–6138. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- van Loon A. P., Brändli A. W., Schatz G. The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell. 1986 Mar 14;44(5):801–812. doi: 10.1016/0092-8674(86)90846-9. [DOI] [PubMed] [Google Scholar]