ABSTRACT
The stimulator of interferon (IFN) genes (STING) is a broad antimicrobial factor that restricts herpes simplex virus (HSV) by activating type I interferon and proinflammatory responses upon sensing of foreign DNA. UL46 is one of the most abundant tegument proteins of HSV-1, but a well-established function has yet to be found. We found that the HSV-1 UL46 protein interacts with and colocalizes with STING. A ΔUL46 virus displayed growth defects and activated innate immunity, but both effects were alleviated in STING knockdown cells. UL46 was also required for the inhibition of the 2′,3′-cyclic GMP-AMP (cGAMP)-dependent immune responses during infection. In cells expressing UL46, out of the context of the infection, innate immunity to a ΔICP0 virus was largely compromised, and that permitted ICP0-deficient mutants to replicate. The UL46-expressing cell lines also rescued the defects of the ΔUL46 virus and enhanced wild-type virus infection. The UL46-expressing cell lines did not activate interferon-stimulated gene (ISG) transcription following treatment with the noncanonical cyclic dinucleotide 2′,3′-cGAMP, suggesting that the STING pathway may be compromised. Indeed, we found that both proteins STING and IFI16 were eliminated in cells constitutively expressing UL46 and that the accumulation of their transcripts was blocked. Finally, we demonstrated that UL46 via its N terminus binds to STING and, via its C terminus, to TBK1. These interactions appear to modulate the functions of STING during HSV-1 infection. Taken together, our studies describe a novel function for one of the least-studied proteins of HSV, the tegument protein UL46, and that function involves the evasion of foreign DNA-sensing pathways.
IMPORTANCE Herpes simplex virus 1 (HSV-1) afflicts 80% of the population worldwide, causing various diseases. After initial infection, the virus establishes latent reservoirs in sensory neurons and persists for life. Here we describe novel interactions between HSV-1 and the DNA sensor STING. We found that (i) HSV-1 tegument protein UL46 interacts with and colocalizes with STING; (ii) UL46 expressed out of the context of the infection blocks type I interferon triggered by STING stimuli, through the elimination of STING and of interferon-inducible protein 16 (IFI16); (iii) a ΔUL46 virus displayed growth defects, which were rescued in STING knockdown cells; (iv) the ΔUL46 virus failed to block innate immunity triggered by ligands of STING such as 2′,3′-cGAMP and also activated IFN-β and ISG expression; and (v) UL46 binds to both STING and TBK1 through different domains. We conclude that UL46 counteracts the actions of STING during HSV-1 infection.
KEYWORDS: UL46 (VP11/12), STING, IFI16, herpes simplex virus, DNA sensors, innate immunity, VP11/12 (UL46)
INTRODUCTION
Herpes simplex virus (HSV) is a burden for individuals worldwide (1). Following primary infection of epithelial cells, the virus establishes latent infections in sensory neurons, where it persists for the life of the individual (1). Reactivation of the viral genome upon stress, weakened immune response, or immunosuppression results in replication of the virus, causing recurrent disease (1).
Previous studies identified the DNA sensor STING as a broad antimicrobial factor that restricts HSV by activating type I interferon (IFN) and proinflammatory responses upon sensing of foreign DNA, or noncanonical cyclic dinucleotides, which are synthesized by the cyclic GMP-AMP synthase (cGAS or cGAMP synthase) (2–4). STING knockout mice succumb to HSV infection due to uncontrollable spread of the virus to the central nervous system and subsequent development of encephalitis (2, 3, 5). How STING senses the HSV DNA has remained elusive. STING associates with another DNA sensor, interferon-inducible protein 16 (IFI16), which is involved in interferon regulatory factor 3 (IRF3)-mediated signaling (6). IFI16 localizes predominantly in the nucleus, but under certain conditions, a significant amount of the protein relocalizes to the cytoplasm to interact with STING and trigger its activation (6). Depletion of p204, the mouse functional ortholog of IFI16, from bone marrow-derived macrophages resulted in decreased IRF3 and NF-κB responses to HSV infection, while depletion of p204 expression from mouse cornea resulted in increased HSV-1 replication in the cornea tissue (6, 7). HSV targets for elimination the IFI16 protein early after infection to combat its antiviral responses (8, 9). Another connection between STING and IFI16 has emerged through studies on the stability of the two proteins. We found that depletion of STING in the cancer cell line HEp-2 resulted in elimination of IFI16 as well (10). This phenomenon was not observed in immortalized HEL cells. These data imply that the two proteins might share common regulators or partners that determine their stability and maybe activity.
While the aforementioned paradigms suggest that the actions of STING and IFI16 are hostile to the virus, we have found that HSV-1 stabilizes STING, suggesting that this protein may be utilized by the pathogen to its advantage (10). Indeed, during HSV infection, STING is released from cells in extracellular vesicles (EVs) and can be delivered to uninfected cells. The excreted STING most likely controls the dissemination of the virus in the host (10, 11). These data imply that viral proteins may interact with STING. The goal of this study was to identify viral proteins with potential effects in the stability and activity of STING.
UL46 is one of the most abundant tegument proteins of HSV-1, with approximately 1,000 to 2,000 copies per virion, although a well-established function has not been described (12–14). The protein accumulates late in infection, and its expression is dependent on DNA synthesis. Earlier studies proposed that UL46, in conjunction with VP16, modulates the VP16-dependent transcriptional induction of α genes (13–15). During infection UL46 localizes in the cytoplasm as multiple punctate structures, a phenotype that resembles the localization of the membrane-associated protein VP22, suggesting that UL46 may also associate with membranes (14, 16–20). Consistent with the proposed membrane localization, several studies show that UL46 binds members of the Src family tyrosine kinases (SFKs) (21–23). This interaction results in tyrosine phosphorylation of UL46 and recruitment of the p85 subunit of the phosphatidylinositol 3 (PI3)-kinase in an SFK-dependent fashion, resulting in HSV-induced phosphorylation of AKT on its activating residues (21–23). Several downstream targets of AKT are phosphorylated during HSV-1 infection but the contribution of UL46 remains unclear, as other viral proteins influence the AKT pathway. The viral kinase Us3 has garnered particular attention because it directly phosphorylates some AKT substrates and also mediates the disappearance of phosphorylated species of AKT (24–27). Thus, in the context of the infection it becomes more complex to understand how discrete viral functions are coordinated and implemented.
Here we show that the HSV-1 tegument protein UL46 interacts with and colocalizes with STING. A ΔUL46 mutant virus, at a low multiplicity of infection, displayed growth defects and activated innate immunity, but these defects were rescued in STING knockdown cells. UL46 was required for the inhibition of the 2′,3′-cGAMP-dependent immune responses during HSV-1 infection. In cells expressing UL46, out of the context of the infection, innate immunity to the ΔICP0 virus was largely compromised, and that permitted ICP0-deficient mutants to replicate. UL46-expressing cell lines also rescued ΔUL46 virus growth, and after infection with the wild-type virus, they yielded higher titers of progeny viruses. Moreover, UL46-expressing cell lines did not activate transcription of interferon-stimulated genes (ISGs) following treatment with the noncanonical cyclic dinucleotide 2′,3′-cGAMP, suggesting that the STING pathway may be compromised. Indeed, we found that both proteins STING and IFI16 were eliminated in cells constitutively expressing UL46 and that the accumulation of their transcripts was blocked. Finally, we demonstrated that UL46 via its N terminus binds to STING and via its C terminus binds to TBK1. These interactions appear to modulate the functions of STING during HSV-1 infection.
RESULTS
Interaction of the HSV-1 UL46 with STING.
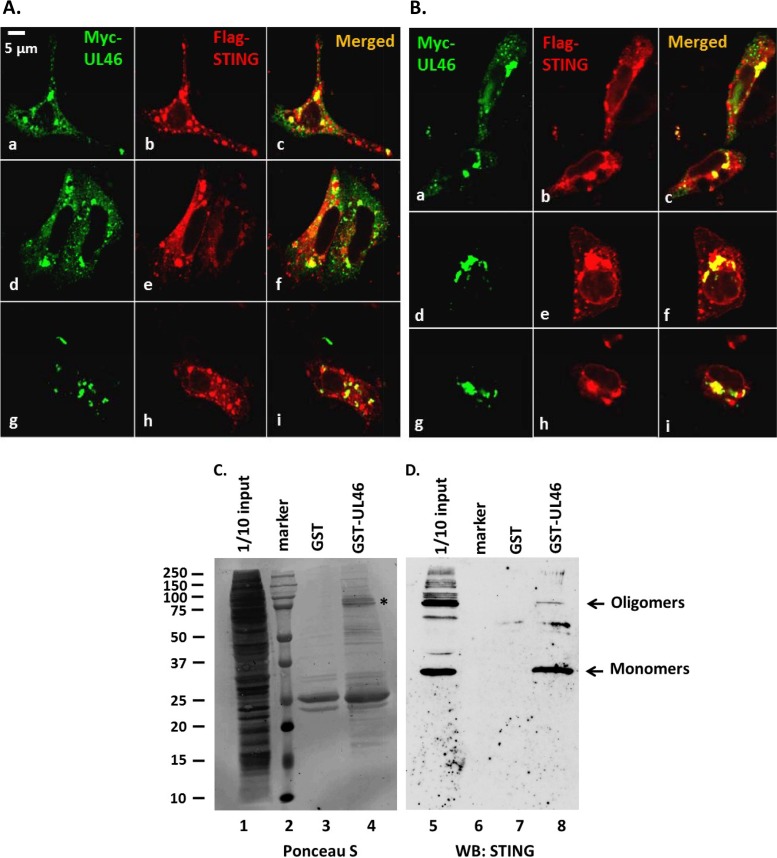
The subcellular distribution of STING and UL46 proteins was monitored in two cell lines expressing the human STING and the HSV-1 protein UL46. Vero (Fig. 1A) or HEp-2 (Fig. 1B) cells were cotransfected with plasmids encoding Flag-tagged STING and Myc-tagged UL46. At 24 h posttransfection, the cells were fixed and the localization of the proteins was monitored by immunofluorescence. The STING protein (Fig. 1A and B, red) accumulated in the perinuclear area and in globular structures in the cytoplasm. A similar pattern was observed for UL46 (Fig. 1A and B, green). Colocalization of STING with UL46 in some of these structures (Fig. 1A and B, yellow) was observed both in the perinuclear area and in distinct structures in the cytoplasm. Next we investigated whether HSV-1 UL46 and STING interact. Bacterially purified glutathione S-transferase (GST) fusions of the full-length UL46 (FL) or GST alone was immobilized on glutathione beads and reacted with lysates derived from HEp-2 cells. The bead-bound protein complexes were analyzed in a denaturing polyacrylamide gel, and immunoblotting was done with an antibody against STING. As shown in Fig. 1D, GST-UL46 pulled down monomers and, less efficiently, the oligomers of STING (∼75 kDa) (lane 8). GST alone did not pull down STING (lane 7). Depicted in lane 5 is 1/10 of the input of STING present in HEp-2 cell lysates. Ponceau S staining of the purified proteins and their quantities are depicted in Fig. 1C. We conclude that UL46 preferentially associates with the monomeric forms of STING.
FIG 1.
Association of UL46 with STING. (A and B) Vero (A) or HEp-2 (B) cells were cotransfected with plasmids encoding Myc-UL46 and Flag-STING. At 24 h posttransfection, the cells were fixed and doubly reacted with antibodies to c-Myc and to STING. Multiple fields from each cell line are depicted. All pictures were taken at the same settings using a Zeiss confocal microscope. (C) Ponceau S staining of the purified proteins and their quantities used in the pulldown assay. The asterisk indicates purified GST-UL46. (D) Purified GST or GST-UL46 proteins were incubated with equal amounts of lysates derived from HEp-2 cells. The electrophoretically separated protein complexes bound to the beads were probed with antibody to STING. STING protein was present in 1/10 of the input of HEp-2 cell lysates used for pulldown. The arrows indicate the monomers and oligomers of STING. WB, Western blotting.
UL46 blocks innate immune responses triggered by 2′,3′-cGAMP or after exposure to the ΔICP0 virus.
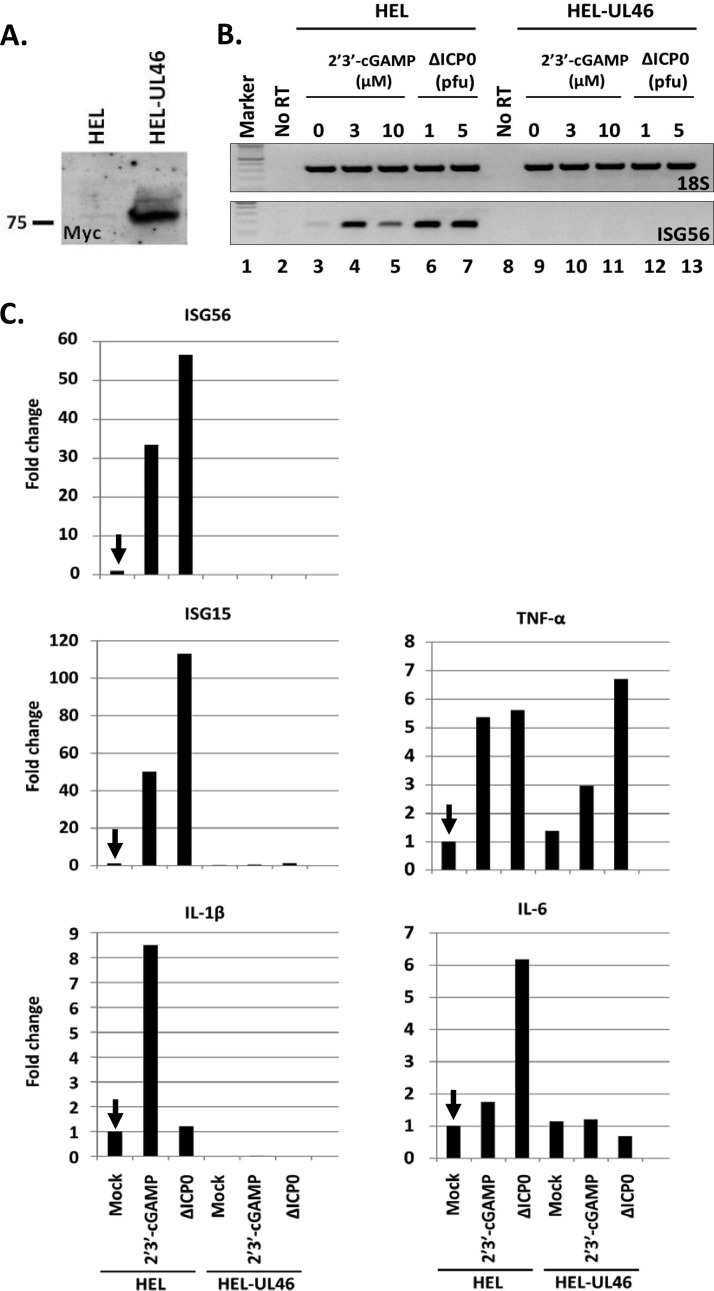
To address the effect of UL46 on the activity of STING, an HEL cell line constitutively expressing UL46 was established using lentiviral vectors. The expression of UL46 was verified by immunoblot analysis as depicted in Fig. 2A. The immortalized HEL cells and their derivatives expressing UL46 either were treated with different concentrations (3 and 10 μM) of 2′,3′-cGAMP or were infected with the ΔICP0 mutant virus, which cannot block innate immunity, at various multiplicities of infection (1 or 5 PFU/cell). At 8 h posttreatment the cells were harvested, total RNA was extracted, cDNA was synthesized, and innate immunity and inflammatory gene transcription were semiquantified or quantified by real-time PCR analysis. A semiquantification analysis, depicted in Fig. 2B, demonstrated that the HEL-UL46 cells either treated with 2′,3′-cGAMP (3 or 10 μM) (lanes 10 and 11) or exposed to the ΔICP0 mutant virus (1 or 5 PFU/cell) (lanes 12 and 13) did not activate transcription of interferon-stimulated gene 56 (ISG56). The parental HEL cells exposed to the ΔICP0 virus (lanes 6 and 7) or treated with 2′,3′-cGAMP as before (lanes 4 and 5) strongly activated transcription of ISG56. In addition to ISG56, the transcription of other ISGs and inflammatory genes was quantified by real-time PCR analysis in samples that were treated with 3 μM 2′,3′-cGAMP or infected with the ΔICP0 virus (5 PFU/cell). The results shown in Fig. 2C can be summarized in the following statements. First, in HEL-UL46 cells, activation of ISG15, ISG56, and interleukin 1β (IL-1β) transcription by 2′,3′-cGAMP or after infection with the ΔICP0 virus was negligible (Fig. 2C). In contrast, in the parental HEL cells, a robust activation of ISG56, ISG15, and IL-1β transcription compared to that in untreated cells was recorded following exposure to 3 μM 2′,3′-cGAMP (Fig. 2C). Infection with the ΔICP0 mutant virus also induced robust transcription of ISG56 and ISG15 but not IL-1β (Fig. 2C). Second, some inflammatory genes, e.g., the tumor necrosis factor alpha (TNF-α) gene but not the IL-6 gene, were only slightly induced after treatment with 3 μM 2′,3′-cGAMP or after infection with the ΔICP0 mutant at 5 PFU/cell, but there were no substantial differences between the two cell lines. All the experiments described above were done at least three independent times, and the pattern was reproducible. We conclude that expression of UL46 protein alone suppresses innate immune responses to nucleic acids or to the ΔICP0 virus.
FIG 2.
Inhibition of innate immune responses triggered by 2′,3′-cGAMP or the ΔICP0 virus in cells expressing the HSV-1 UL46 protein. (A) HEL cells constitutively expressing the Myc-UL46 protein. (B) HEL or HEL-UL46 cells were either treated with 2′,3′-cGAMP (3 or 10 μM) (lanes 4, 5, 10, and 11), exposed to the ΔICP0 virus (1 or 5 PFU/cell) (lanes 6, 7, 12, and 13), or left untreated (lanes 3 and 9). At 8 h posttreatment, the cells were harvested and the ISG56 transcripts were semiquantified. 18S rRNA served as a control. (C) HEL or HEL-UL46 cells were treated with 2′,3′-cGAMP or exposed to the ΔICP0 virus, similar to the description for panel B. The ISG56, IL-6, ISG15, IL-1β, and TNF-α transcripts were quantified in duplicate assays by real-time PCR analysis relative to their transcripts in untreated HEL cells, indicated by the arrow. 18S rRNA was used for normalization. Each experiment was repeated three times. Results of a representative experiment are depicted.
The growth defects of the ΔICP0 virus were reversed in UL46-expressing cell lines.
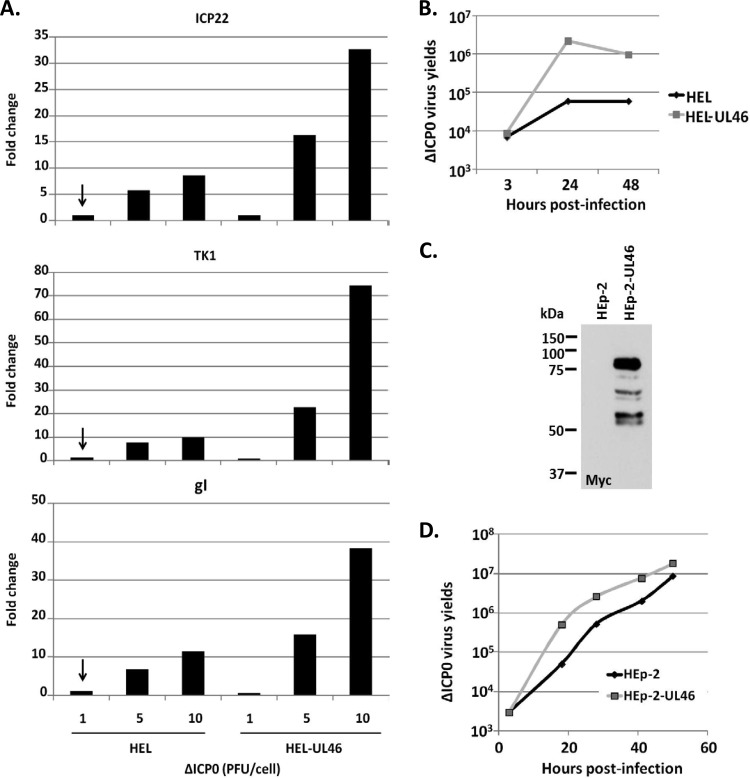
We sought to determine whether the growth defects of the ΔICP0 virus could be reversed in cells expressing the UL46 protein. We performed a series of three experiments. In the first experiment, the HEL-UL46 and the parental HEL cells were exposed to different doses of the ΔICP0 virus (1, 5, and 10 PFU/cell). The cells were harvested at 8 h after infection, and ΔICP0 virus gene transcription was analyzed by real-time PCR analysis, using primer pairs against the immediate early gene Us1, which encodes ICP22, the early gene UL23, which encodes thymidine kinase 1 (TK1), and the late gene Us7, which encodes glycoprotein I (gI). The experiment was repeated three times, and representative results are shown in Fig. 3. As shown in Fig. 3A, at a low multiplicity of infection (1 PFU/cell), the levels of transcription of ICP22, TK1, and gI were comparable between HEL and HEL-UL46 cells exposed to the ΔICP0 virus. At higher doses of the ΔICP0 virus (5 and 10 PFU/cell), transcription of all classes of viral genes was 2- to 8-fold higher in the HEL-UL46 cells than in the parental HEL cells. The gene silencing and innate immune machineries both block ΔICP0 infection at a low multiplicity of infection. At a high multiplicity of infection, some copies of the viral genome escape the gene silencing machinery, and in the presence of UL46, which impairs innate immunity, ΔICP0 virus gene expression is enhanced. In the second experiment, the titers of the ΔICP0 virus were compared in HEL and HEL-UL46 cells infected with the virus at 0.05 PFU/cell. The cells were harvested at 3, 24, and 48 h postinfection, and titrations were done in the U2OS cell line. As shown in Fig. 3B, the yields of the ΔICP0 virus in the HEL-UL46 cells were approximately 20-fold higher at 24 h postinfection than those obtained in the parental HEL cells. In the third experiment, we established a HEp-2 cell line constitutively expressing UL46 (Fig. 3C). Similar to the case with the HEL-UL46 cell line, the yields of the ΔICP0 virus in the HEp-2-UL46-expressing cell line were almost 10-fold higher between 18 and 28 h postinfection than in the parental HEp-2 cells (Fig. 3D). These results were reproducible in two more independent experiments. We conclude that expression of UL46 protein in cells, in the absence of other viral proteins, can rescue ΔICP0 virus growth.
FIG 3.
HSV-1 UL46 rescues the growth of the ΔICP0 mutant virus. (A) HEL or HEL-UL46 cells were exposed to ΔICP0 virus at 1, 5, or 10 PFU/cell. Quantification of ICP22, TK1, and gI viral transcripts was done by quantitative PCR (qPCR) at 8 h postinfection. Normalization was done against the 18S rRNA. The results represent the fold change of the viral transcripts relative to the amounts of mRNAs present in HEL cells exposed to 1 PFU/cell, indicated by the arrow. (B) HEL or HEL-UL46 cells were exposed to ΔICP0 virus at 0.05 PFU/cell. The cells were harvested at 3 h, 24 h, and 48 h after infection, and titrations of progeny viruses were done in U2OS. (C) HEp-2 cell line expressing Myc-UL46. (D) HEp-2 and the HEp-2-UL46 derivatives were exposed to the ΔICP0 mutant at 0.1 PFU/cell. The cells were harvested at 3, 18, 28, 42, and 54 h after infection. Titrations were done in U2OS cells.
STING and IFI16 are eliminated in cells expressing UL46.
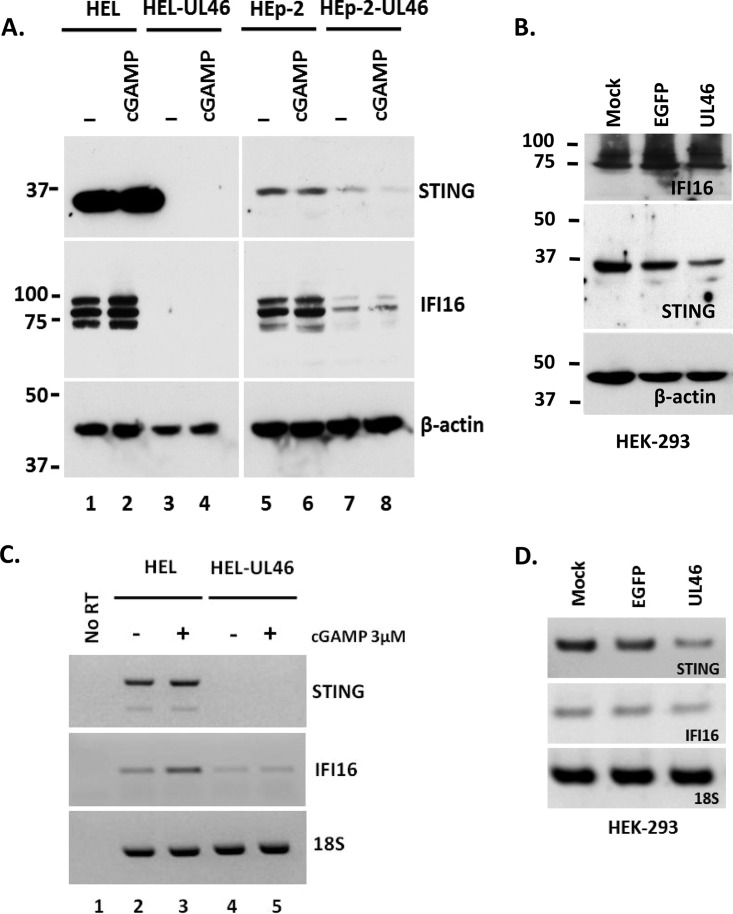
We next sought to determine how the HSV-1 UL46 protein suppresses the action of STING. A series of three experiments was performed. In the first experiment, the expression of STING was monitored in HEL and HEp-2 cells and their derivatives expressing UL46 that were exposed to 2′,3′-cGAMP or left untreated. STING was detected in both HEL and HEp-2 cells (Fig. 4A, lanes 1 and 5). An increase in the amounts of STING protein was observed in the HEL cells after treatment with 2′,3′-cGAMP (Fig. 4A, lane 2). Strikingly, in HEL-UL46 cells, the STING protein was undetectable, suggesting that STING is eliminated in the presence of the UL46 protein (Fig. 4A, lane 3). Treatment with 2′,3′-cGAMP did not restore the expression of STING in the HEL-UL46 cells (Fig. 4A, lane 4). Similarly, in the HEp-2-UL46 cells, a substantial decrease in the amounts of STING protein was detected compared to those in the HEp-2 cells, and treatment with 2′,3′-cGAMP did not reverse these results (Fig. 4A, compare lanes 7 and 8 to lanes 5 and 6). The differences in the elimination of STING between the HEp-2 and HEL cell lines could be because in the cancer cell line HEp-2, the STING protein does not function as a restriction factor for HSV-1, as was previously reported (10). We conclude that STING is eliminated in UL46-expressing cells in the absence of other viral functions.
FIG 4.
Elimination of STING and IFI16 in cells expressing UL46. (A) HEL, HEL-UL46, HEp-2, and HEp-UL46 cells were either treated with 2′,3′-cGAMP (3 μM) or left untreated. The cells were harvested at 8 h after the addition of 2′,3′-cGAMP, and protein analysis was done with antibodies against STING, IFI16, and β-actin. (B) HEK-293 cells were either mock transfected or transfected with the pcDNA 3.1 Myc-UL46 or with the pEGFP-N3 plasmid. The cells were harvested at 72 h after transfection, and immunoblotting was done with the STING, IFI16, and β-actin antibodies. (C and D) Total RNA was extracted from replicate cultures of cells described for panels A and B, and the STING and IFI16 transcripts were semiquantified by PCR. 18S rRNA served as a loading control.
In the next experiment, we monitored the levels of the interferon gamma-inducible protein 16 (IFI16) in the HEL-UL46 and HEp-2-UL46 cell lines, since a correlation in the accumulation of STING and of IFI16 was previously reported (10). Three isoforms of IFI16 were detected in HEL and HEp-2 cells. Interestingly, all forms of IFI16 were eliminated in the presence of UL46 (Fig. 4A, compare lanes 3 and 4 to lanes 1 and 2 and lanes 7 and 8 to lanes 5 and 6). We conclude that UL46 triggers the elimination of IFI16 in tandem with the elimination of STING. In an alternative approach, HEK-293 cells were transfected with a UL46-expressing plasmid or a control plasmid encoding enhanced green fluorescent protein (EGFP) or left untreated. At 72 h posttransfection, the cells were harvested and lysed, and equal amounts of proteins were analyzed for the accumulation of STING and IFI16 proteins. As shown in Fig. 4B, a reduction in the amounts of the STING protein was observed in the presence of UL46 compared to that in the control cells expressing EGFP or in the nontransfected cells. No substantial reduction in the amounts of IFI16 was observed. One possible explanation for the lack of elimination of IFI16 in the HEK-293 cells is that the amounts of STING protein at 72 h posttransfection with the UL46-expressing plasmid are sufficient to sustain the accumulation of IFI16. Another explanation is that the link between STING and IFI16 is missing in the HEK-293 cells. Finally, IFI16 might have defects in HEK-293 cells. Notably, the mobility of IFI16 in HEK-293 cells was different from that in HEL or HEp-2 cells (compare Fig. 4B to Fig. 4A). Differences in the mobility and defects in the activity of IFI16 in HEK-293 cells have been previously reported (28). We conclude that transient expression of UL46 in HEK-293 cells is sufficient to cause a reduction in the levels of STING protein.
In the third experiment, we investigated the mechanism of STING and IFI16 elimination in HEL-UL46 cells. The transcripts of STING and IFI16 were derived from HEL or HEL-UL46 cells that were treated with 2′,3′-cGAMP or left untreated. Semiquantification of these transcripts was done by PCR analysis. The transcripts of STING derived from the HEL cells were present in two forms that correspond to splicing variants of STING (Fig. 4C). In HEL cells treated with 2′,3′-cGAMP a small increase in the amounts of the STING transcripts was observed, which is consistent with the increase in the levels of the STING protein (Fig. 4C). The transcripts of STING were undetectable in the HEL-UL46 cell line (Fig. 4C), which correlated well with the loss of protein (Fig. 4A). Similarly, the levels of the IFI16 transcripts declined in the UL46-expressing HEL cells, which was consistent with the elimination of the protein (Fig. 4C). Similar results were obtained with the HEp-2-UL46 cell line (data not shown). Semiquantification of the STING and IFI16 transcripts derived from HEK-293 cells transfected with the UL46-expressing plasmid demonstrated a moderate reduction in the amounts of the STING transcripts that was reflected in the levels of the protein (Fig. 4D). The set of primers that was used to semiquantify the STING transcripts derived from the HEK-293 cells cannot distinguish between the two isoforms of STING, which is why only one STING mRNA product is depicted in Fig. 4D. We conclude that expression of UL46, in the absence of other viral proteins, results in elimination of the STING and IFI16 proteins and a reduction in the amounts of their transcripts.
A ΔUL46 virus failed to block innate immunity triggered by 2′,3′-cGAMP.
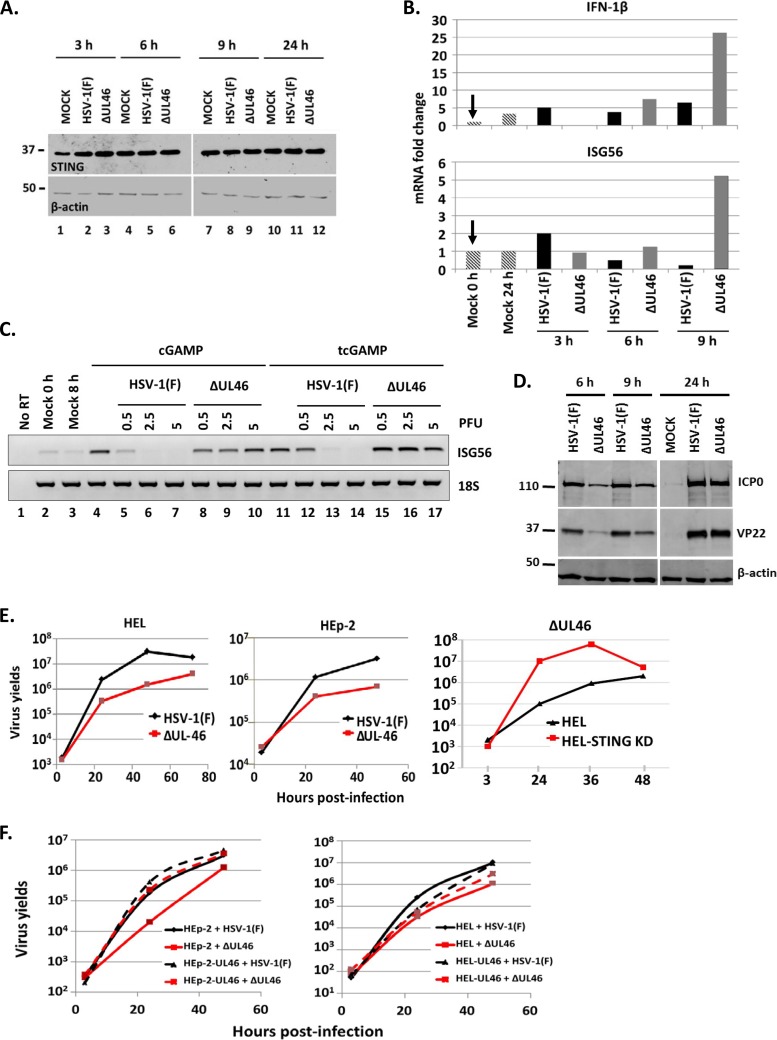
HEL cells were infected with either the ΔUL46 virus or HSV-1(F) (0.1 PFU/cell), a limited-passage isolate. The accumulation of STING was monitored over a period of 24 h after infection. The results shown in Fig. 5A demonstrated that the STING protein remained stable throughout the infection with the wild-type virus or the ΔUL46 virus.
FIG 5.
The ΔUL46 virus failed to block innate immunity triggered by the ligand of STING, 2′,3′-cGAMP. (A) HEL cells were infected with HSV-1(F) or the ΔUL46 virus (0.1 PFU/cell). The cells were harvested at 3, 6, 9, and 24 h after infection, and equal amounts of proteins were analyzed with antibodies against STING and β-actin. (B) HEL cells were infected with HSV-1(F) or the ΔUL46 virus (0.1 PFU/cell). The cells were harvested at 3, 6, and 9 h after infection, and total RNA was extracted and used for quantification of the IFN-β and ISG56 transcripts by real-time PCR. The experiment was repeated three independent times. 18S rRNA served as a loading control. (C) HEL cells were either mock infected (lanes 2 to 4 and 11) or exposed to HSV-1(F) (lanes 5 to 7 and 12 to 14) or to the ΔUL46 virus (lanes 8 to 10 and 15 to 17) (0.5, 2.5, or 5 PFU/cell). Two hours after infection, the cells were treated with 2′,3′-cGAMP (3 μM) that was either added to the cultures (cGAMP) or transfected to the cells (tcGAMP). The cells were harvested at 8 h after infection, and total RNA was extracted and used for semiquantification of the ISG56 transcripts by PCR. 18S rRNA was used as a control. (D) HEL cells were either mock infected or exposed to HSV-1(F) or to the ΔUL46 virus (5 PFU/cell). The cells were harvested at 6, 9, or 24 h after infection, and equal amounts of proteins were analyzed by immunoblot analysis using antibodies against ICP0, VP22, and β-actin. (E) HEL, HEp-2, or STING-depleted HEL cells were infected with either HSV-1(F) or the ΔUL46 virus (0.01 PFU/cell). The cells were harvested at 3, 24, 48, or 72 h after infection, and titrations were done in Vero cells. (F) HEL or HEp-2 cells or their derivatives expressing UL46 were infected with either HSV-1(F) or the ΔUL46 virus (0.01 PFU/cell). The cells were harvested at 3, 24, and 48 h after infection, and titrations were done in Vero cells.
Next, we tested whether the ΔUL46 virus infection activates innate immunity. HEL cells were exposed to HSV-1(F) or ΔUL46 virus (0.1 PFU/cell) or remained untreated. The cells were harvested at 3, 6, and 9 h after infection, total RNA was extracted, and quantification of the ΙFN-β and ISG56 transcripts was done by real-time PCR analysis. As shown in Fig. 5B, none of the viruses activated IFN-1β or ISG56 gene transcription up to 6 h after infection. Induction of ISG56 and IFN-1β gene transcription was recorded only at 9 h after infection with the ΔUL46 virus. The results were reproducible in two more independent experiments. We conclude that the ΔUL46 virus activates innate immunity gene expression.
We also investigated whether the ΔUL46 virus could block innate immunity triggered by the ligand of STING, 2′,3′-cGAMP. HEL cells were exposed to either the wild-type virus or the ΔUL46 virus (0.5, 2.5 or 5 PFU/cell) 2 h prior to treatment with 2′,3′-cGAMP (3 μM), which was either added to the medium of the cultures or transfected into the cells. The cells were harvested at 8 h after infection, total RNA was extracted, and the ISG56 transcripts were semiquantified by PCR analysis. As shown in Fig. 5C, treatment with 2′,3′-cGAMP induced ISG56 gene transcription (lanes 4 and 11) which was completely blocked by the wild-type virus at 2.5 PFU/cell (compare lanes 5 to 7 to lane 4 and lanes 12 to 14 to lane 11). In contrast, the ΔUL46 virus failed to block ISG56 gene transcription even at the highest multiplicity of infection (5 PFU/cell) (compare lanes 8 to 10 to lane 4 and lanes 15 to 17 to lane 11). We conclude that UL46 is required for the blockage of innate immunity initiated by the ligand of STING, 2′,3′-cGAMP.
Expression of viral genes by the ΔUL46 virus was compared to that of the wild-type virus. HEL cells were exposed to both viruses (5 PFU/cell), the cells were harvested at 6, 9, and 24 h after infection, and accumulation of viral proteins was analyzed by immunoblot analysis. As shown in Fig. 5D, the virus lacking the VP11/12 gene that encodes UL46 had a delay in expression of all classes of viral genes at early time points after infection, but later, no difference between the two viruses was detected. Similar results were obtained at 0.5 and 2.5 PFU/cell (data not shown). The growth properties of the ΔUL46 virus were examined in HEL cells, in their STING-depleted derivatives, and in the HEp-2 cell line. Replicate cultures of these cells were infected with either the ΔUL46 virus or HSV-1(F) (0.01 PFU/cell). The cells were harvested at 3, 24, 48, or 72 h postinfection, and titrations were done in Vero cells. At a low multiplicity of infection, the ΔUL46 virus yields were at least 10-fold lower in HEL and HEp-2 cells than for HSV-1(F) (Fig. 5E). The ΔUL46 virus yields were restored in the HEL STING knockdown cells. The results were reproducible in two more independent experiments. We conclude that HSV-1 UL46 is required for suppression of antiviral responses mediated by the DNA sensor STING.
In addition, we compared the growth properties of HSV-1(F) and the ΔUL46 virus in the HEL and HEp-2 cell lines and their derivatives expressing UL46. Replicate cultures of these cells were infected with either the ΔUL46 virus or the wild-type virus at 0.01 PFU/cell. The cells were harvested at 3, 24, and 48 h postinfection, and titrations were done in Vero cells. Consistent with the data above, the ΔUL46 virus displayed growth defects in the HEp-2 and HEL cells, which were completely (HEp-2) or partially (HEL) rescued in both cell lines expressing UL46. These data suggest that ectopic expression of UL46 restores the defects of the ΔUL46 virus (Fig. 5F, compare the red line for ΔUL46 virus in HEL cells or HEp-2 cells to the dashed red line for ΔUL46 virus in the UL46-expressing cell lines). The wild-type virus displayed higher yields in the HEp-2 cell line expressing the UL46 protein than in the parental HEp-2 cell line [Fig. 5F, compare the black line for HSV-1(F) in HEp-2 cells to the dashed black line for HSV-1(F) virus in the HEp-2-UL46-expressing cell line]. No substantial differences in the growth properties of HSV-1(F) were noticed between HEL and the HEL-UL46 cells (Fig. 5F). These data suggest that the inhibition of innate immune responses by UL46 benefits the ΔUL46 virus and also the wild-type virus, which is consistent with previous data (10).
STING and TBK1 bind on separate domains of the UL46 protein.
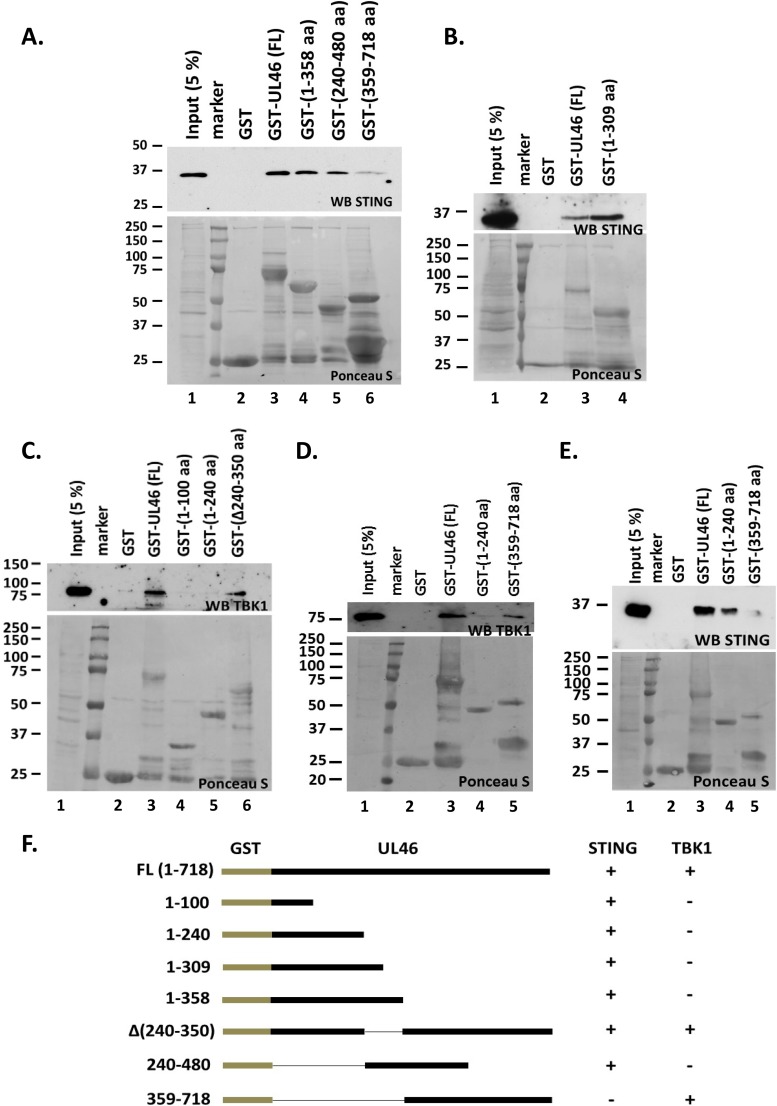
In the first series of experiments, we mapped the binding domain of STING to UL46. We developed three C-terminally truncated forms of UL46 fused to GST, one containing the first 240 amino acids (aa), the second containing the first 309 aa, and the third containing the first 358 aa of UL46. In addition, we developed an N-terminally truncated form of UL46 containing aa 359 to 718 and an internal fragment containing aa 240 to 480 of UL46 fused to GST. Equal amounts of lysates derived from HEL cells were reacted with either the full-length purified GST-UL46 (FL) or the purified GST fusion fragments of UL46. Purified GST protein alone served as a negative control. Full-length UL46 (Fig. 6A and B, lanes 3) and the amino-terminal fragment of UL46 (Fig. 6A and B, lanes 4) pulled down STING, but the carboxy-terminal fragment (aa 359 to 718) of UL46 pulled down only a trace amount of STING (Fig. 6A, lane 6). The internal fragment of UL46 (aa 240 to 480) showed weaker binding than did the N-terminal fragment (Fig. 6A, compare lane 5 to lane 4). One possibility is that STING has two binding sites on UL46, one within the first 100 aa and the second between aa 240 and 480.
FIG 6.
STING and TBK1 associate with UL46 via separate domains. (A, B, and E) Bacterially purified GST, GST-UL46 (full length [FL]), and N- or C-terminally truncated forms of UL46 fused to GST were incubated with equal amounts of lysates derived from HEL cells. The electrophoretically separated protein complexes bound to the beads were probed with antibody to STING. Also shown is STING protein present in 5% of the input of HEL cell lysates used for pulldown. Ponceau S staining of the purified proteins and their quantities used in the pulldown assay are depicted. (C and D) Procedures were done as described for panels A, B, and E except that immunoblotting was done with the TBK1 antibody. (F) Diagram summarizing the interactions between UL46, STING, and TBK1.
Next we examined whether TBK1, a binding partner of STING, is also pulled down by UL46. The approach was similar to that described above. We found that full-length UL46 (Fig. 6C and D, lanes 3), but not the N-terminal fragments of UL46 that interact with STING (compare Fig. 6C and D to Fig. 6A), pulled down TBK1. An internal deletion mutant of UL46 lacking aa 240 to 350 also pulled down TBK1 (Fig. 6C, lane 6). Intriguingly, the C-terminal fragment of UL46 (aa 359 to 718), which did not pull down STING (Fig. 6E, lane 5, and Fig. 6A, lane 6), pulled down TBK1 (Fig. 6D, lane 5). The reduced signal of STING that was detected in the pulldown reaction using the C-terminal fragment of UL46 (Fig. 6A, lane 6, and Fig. 6E, lane 5) could reflect an indirect association of STING through its binding partner, TBK1. A summary of the interactions between UL46 with STING and TBK1 is depicted in Fig. 6F. We conclude that both STING and TBK1 bind UL46 and that different domains of UL46 mediate the interactions with STING and TBK1.
DISCUSSION
To successfully colonize humans, HSV must overcome strong antiviral responses. The DNA sensor STING is an innate immune component that is activated upon HSV infection and restricts virus replication and dissemination (2–4, 29). STING is a transmembrane protein in the endoplasmic reticulum. STING senses foreign DNA or noncanonical cyclic dinucleotides and functions as an adaptor for the activation of IRF3 and NF-κΒ, which activates type I interferon and proinflammatory responses (2–4, 29). In addition to STING, the DNA sensor IFI16 has received attention because it localizes both in the nucleus and in the cytoplasm and is considered to have better chances of sensing the viral DNA after its release from the capsid into the nucleus (6, 9). IFI16 interacts with STING and under certain conditions can augment its activity (6). Although IFI16 can influence the innate immunity against HSV, it is eliminated during the early steps of HSV infection. On the other hand, HSV appears to stabilize STING (10). Moreover, STING was found in extracellular vesicles (EVs) released from infected cells (11). These data suggested that STING may be utilized by the virus. An attractive hypothesis is that HSV augments the packaging of STING in EVs and delivery to uninfected cells to control its dissemination in the human body (10, 11). Another implication of these data is that viral genes are involved in modifying the functions of STING.
Elucidation of the mechanisms by which HSV genes and their products evade the STING and IFI16 DNA-sensing pathways is ongoing. A few studies have linked the elimination of IFI16 to the immediate early protein of the virus ICP0, but the E3 ligase of ICP0 is neither necessary nor sufficient for the elimination of IFI16 (8, 30, 31). Recently, the immediate early protein of the virus ICP27 was linked to the suppression of the TBK1-STING pathway in human macrophages (32).
We report here that the tegument protein UL46 of HSV-1 associates with both STING and TBK1 through separate domains and that it blocks this DNA-sensing pathway. A ΔUL46 virus failed to block innate immunity after treatment with the ligand of STING, 2′,3′-cGAMP. Moreover, the ΔUL46 virus activated innate immunity gene expression later after infection. The ΔUL46 virus growth was compromised, especially at a low multiplicity of infection, but it was fully restored in STING knockdown cells. Viral gene expression was delayed at early hours after infection with the ΔUL46 virus compared to the wild-type virus.
We also found that in cells expressing the UL46 protein alone, the STING and IFI16 proteins were eliminated and the levels of their transcripts were reduced. The consequence of elimination of the STING and IFI16 proteins in the UL46-expressing cells was impaired innate immunity following exposure of cells to ligands of STING, such as 2′,3′-cGAMP or infection with the ΔICP0 mutant virus. Therefore, in UL46-expressing cells the growth of the ΔICP0 mutant virus was rescued. In addition, the ΔUL46 virus and the wild-type virus yielded higher titers in the UL46-expressing cells due to the lack of the activity of STING. However, during the course of HSV-1 infection STING is stable, as has been reported before (10). Moreover, in wild-type virus-infected cells, STING is stabilized, and a fraction of STING is packaged in EVs and delivered to uninfected cells (11). These data suggest that the tandem elimination of STING and IFI16 proteins in UL46-expressing cells is most likely an interrelated event that is augmented by UL46. These data also support our previous hypothesis that STING and IFI16 share common regulators or partners (10). One possible hypothesis that could explain this loss of STING and IFI16 proteins is that UL46 disrupts the functions of STING. As the functions of STING are required for its own expression, we assume that in the stable cell lines expressing UL46, prolonged inhibition of STING will result in the loss of the protein (33, 34). IFI16 is maybe indirectly regulated by genes affected by STING, in the presence of UL46. In infected cells, the effect of UL46 on STING is most likely transient. The tegument UL46 may interfere with the functions of STING early after infection, but later UL46 probably acquires modifications and changes subcellular compartments as it becomes part of the tegument (35). Therefore, during infection UL46 does not interfere with the accumulation of STING. Taken together, these data imply that multiple viral proteins regulate the functions and accumulation of STING during infection.
Our proposed model is based on three observations. First, STING is stable in HSV-1-infected cells (10). Second, STING is rendered innocuous during HSV infection. And third, STING is transported in EVs out of the infected cells (11). Thus, in light of the fact that UL46 interacts with STING and could trigger its elimination, in the absence of other viral proteins, the implication is that other viral proteins are involved in the stabilization of STING during HSV infection either directly or indirectly by altering the properties of UL46. Hence, one interpretation of these data is that UL46 interferes with functions of STING. In the absence of other viral proteins, STING is eliminated. In the context of the infection, STING is functionally disabled early after infection, perhaps through the actions of the abundant UL46 protein that is present in the tegument of the incoming virus. Following viral DNA synthesis, UL46 relocalizes to positions of viral assembly and egress and most likely does not associate anymore with STING. A fraction of STING is packaged in EVs to be released out of the infected cells. Despite the interaction of UL46 with STING, we found that STING but not UL46 is excreted in EVs (data not shown). One reason that HSV could support this complex series of events is to translocate STING by EVs to uninfected cells to control its dissemination. Why UL46 interacts with both STING and TBK1 and whether through UL46 the virus alters properties of TBK1 is a subject for investigation.
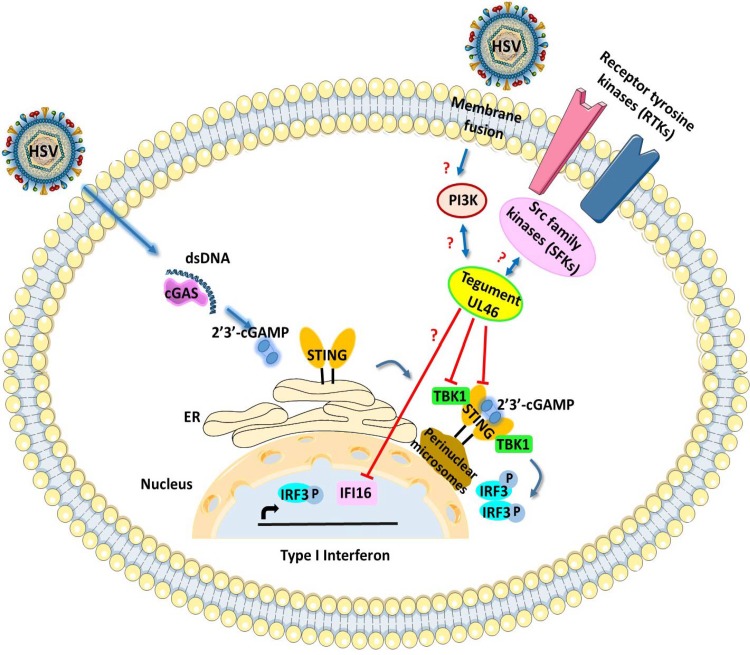
Overall, our studies identified a novel function for one of the least-studied proteins of HSV, the UL46 tegument protein. This function involves the inactivation of the STING DNA-sensing pathway that plays major role in mounting antiviral responses against HSV. A model summarizing the interplay of UL46 with STING and TBK1 based on the data discussed above is illustrated in Fig. 7. Based on this model, UL46 interacts with both STING and TBK1 via separate domains and possibly disrupts their functions. IFI16 is affected by UL46 in a less clear mechanism. Members of the Src family kinases also interact with UL46 (21–23).
FIG 7.
Model summarizing the immunomodulatory functions of HSV-1 UL46. HSV-1 UL46 interacts with both STING and TBK1 via separate domains. Most likely these interactions abrogate the functions of STING and of TBK1. UL46 also influences the accumulation of IFI16. Other partners of UL46 include members of the Src family kinases (21–23).
In principle, other HSV proteins such as ICP0, ICP27, and γ1 34.5 can interfere with downstream effectors of the STING DNA-sensing pathway (32, 36–38). UL46 is present only in alphaherpesviruses; thus, other members of the family involve different gene products to block the STING DNA-sensing pathway (39). STING is also a target for most DNA and RNA viruses, as signals from several pattern recognition receptors (PRRs) and adaptors merge on STING (39). The multifaceted strategy of HSV to compromise the STING DNA-sensing pathway highlights that STING is a key restriction factor for HSV.
MATERIALS AND METHODS
Cells and viruses.
The culture conditions for HEp-2 (human epithelial; ATCC), HEL (telomerase transformed human embryonic lung fibroblasts; kindly provided by Thomas Shenk, Princeton University), Vero (green monkey kidney epithelial; ATCC), U2OS (human osteosarcoma; ATCC), and HEK-293 cells (ATCC) have been reported elsewhere (10). HSV-1(F), a limited-passage isolate, is the prototype strain used in this laboratory (40). The properties of R7910, a ΔICP0 mutant virus, have been described before (41). The ΔUL46 virus has been described elsewhere (15). Titration of HSV-1(F) and of the ΔUL46 virus was done in Vero cells. Titration of ΔICP0 virus was done in U2OS cells.
Development of stable cell lines expressing UL46 using lentiviral vectors.
The procedures for development of stable cell lines expressing UL46 were as before (10). Briefly, The UL46 open reading frame (ORF) was PCR amplified from the HSV-1 genome, digested with EcoRV, and inserted into the EcoRV site of a pcDNA 3.1 Myc plasmid in frame with the Myc tag. The Myc-tagged UL46 ORF was cloned into the BamHI/SalI-blunt-ended sites of the PLKO.1 cytomegalovirus (CMV) EGFP plasmid (Addgene) after removal of the EGFP sequence. For the development of lentiviral vectors, HEK-293 cells seeded in a 25-cm2 flask were cotransfected with 8 μg of a plasmid carrying the Myc-UL46 ORF, 8 μg of Gag-Pol-expressing plasmid, and 1 μg of a plasmid encoding vesicular stomatitis virus glycoprotein (VSV-G) with Turbofect, according to the manufacturer's instructions. At 48 h after transfection, the supernatant from the cultures was collected, filtered through 0.45-μm-pore-size filters, and used to infect HEp-2 or HEL cells. Puromycin selection was initiated 24 h after exposure to lentiviruses and continued until only resistant clones survived.
Immunoblot analysis.
The procedures for immunoblotting have been described elsewhere (42). Mouse monoclonal antibodies to STING (R&D systems), IFI16 (Abcam), β-actin (Sigma), and the Myc tag (Santa Cruz) were used at a 1:1,000 dilution. Rabbit polyclonal antibodies against VP22 and ICP0 were kindly provided by B. Roizman (University of Chicago). The rabbit polyclonal antibody against TBK1 (Cell Signaling Technology) was used in a 1:1,000 dilution. Protein bands for β-actin, ICP0, and VP22 that generate a strong signal were visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (Fisher Scientific). ECL Western blotting detection reagents (Life Technologies) were used for the less abundant proteins. Treatment with 2′,3′-cGAMP (Sigma) was done as indicated in the legends to the figures.
Immunofluorescence.
The procedures for immunofluorescence have been described elsewhere (42). c-Myc mouse monoclonal antibody (Santa Cruz) and STING rabbit polyclonal antibody (Cell Signaling Technology) were used in a dilution of 1:800. Images were captured with a Zeiss confocal microscope.
Purification of the GST-UL46 fusion proteins and pulldown assays.
Full-length UL46 was PCR amplified from the HSV-1(F) genome, digested with EcoRV, and inserted into the SmaI site of pGEX-4T2, in frame with glutathione S-transferase (GST). Similarly, all the truncated forms of UL46 were PCR amplified from the full-length ORF and inserted into the pGEX-4T2 vector, in frame with GST. The purification of all the GST fusion proteins was done as previously described (43). For the GST pulldown assays, 2 × 106 HEp-2 or HEL cells were lysed in HEPES–1% Triton X-100 buffer consisting of 50 mM HEPES (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease cocktail inhibitor (Sigma) supplemented with 1% Triton X-100. The lysates were reacted overnight with equal amounts of GST fusion proteins immobilized on glutathione beads and rinsed three times with the HEPES–1% Triton X-100 buffer, and the bound protein complexes were dissolved in loading buffer composed of 4% SDS, 100 mM Tris-Cl (pH 6.8), 20% glycerol, and 0.2% bromophenol blue, supplemented with β-mercaptoethanol, and boiled for 5 min. After centrifugation to remove the beads, the eluted proteins were subjected to electrophoretic analysis.
Total RNA extraction and semiquantitative or real-time PCR analysis.
The procedures for total RNA extraction and PCR analysis have been described elsewhere (43, 44). Briefly, semiquantitative PCR analysis was performed using GoTaqG2 Hot Start polymerase (Promega) and an equal volume (1 μl) of cDNA that was generated from 1 μg of total RNA per sample. For the semiquantitative PCR analysis, we analyzed the products of the reactions between cycles 18 and 28. We found that at cycle 25, the amplifications were in exponential range and the quantities of the products were adequate to be appreciated by ethidium bromide analysis in 2% agarose gels. Standard curves were performed to optimize the conditions for each primer set. The annealing temperature was set up 4°C lower than the lowest melting temperature (Tm) between a primer set. Real-time PCR analyses were performed using SYBR green reagent (Invitrogen) or TaqMan (Applied Biosystems) according to the manufacturer's recommendations. The 18S rRNA primers (Universal Primers; Ambion) and the eukaryotic 18S rRNA endogenous control (FAM/MGB probe) (Thermo Fisher) were used for normalization. The primers for ICP22, gI, and TK1 have been reported elsewhere (43, 44). Predesigned probes (FAM/MGB) for ISG15, IFN-β, and IL-1β transcripts were obtained through Thermo Fisher. The primers for ISG56, IL-6, STING, and IFI16 are listed in Table 1.
TABLE 1.
Primers used for semiquantitative or real-time PCR analysisa
| Primer | Sequence |
|---|---|
| ISG56 (f) | 5′ GGA AAA AAA GCC CAC ATT TGA GGT 3′ |
| ISG56 (r) | 5′ CTTTTG AAA TTC CTG AAA CCG ACC A 3′ |
| IL-6 (f) | 5′ AGT ACC CCC AGG AGA AGA TTC CAA AG 3′ |
| IL-6 (r) | 5′ TTGTTTTCT GCC AGT GCCTCTTTG C 3′ |
| STING (qPCR) (f) | 5′ TTC GAA CTT ACA ATC AGC ATT ACA A 3′ |
| STING (qPCR) (r) | 5′ CTC ATA GAT GCT GTT GCT GTA AAC C 3′ |
| IFI16 (f) | 5′ TCA TCA ACA GAG CAA AGG AAA 3′ |
| IFI16 (r) | 5′ GAC ATT GTC CTG TCC CCA CT 3′ |
| STING (f) (two isoforms) | 5′ CCA TTG GAC TGT GGG GTG CCT GAT AAC C 3′ |
| STING (r) (two isoforms) | 5′ GAG GTC TTC AAG CTG CCC ACA GTA ACC T 3′ |
f, forward; r, reverse; qPCR, quantitative PCR.
ACKNOWLEDGMENTS
We thank Bernard Roizman (University of Chicago) for kindly providing the R7910 virus and the ICP0 and VP22 antibodies. We thank Edward Stephens (University of Kansas Medical Center) and graduate student Hope Waisner (University of Kansas Medical Center) for editing the manuscript.
M. Kalamvoki is funded through University of Kansas Medical Center startup funds and COBRE grant P20GM113117.
REFERENCES
- 1.Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed). 2013. Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrady CD, Zheng M, Fitzgerald KA, Liu C, Carr DJ. 2012. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol 5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orzalli MH, Knipe DM. 2014. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol 68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalamvoki M, Du T, Roizman B. 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci U S A 111:E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol 14:640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, McKnight JL. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol 67:1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato K, Daikoku T, Goshima F, Kume H, Yamaki K, Nishiyama Y. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch Virol 145:2149–2162. doi: 10.1007/s007050070045. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Sirko DA, McKnight JL. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J Virol 65:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brignati MJ, Loomis JS, Wills JW, Courtney RJ. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J Virol 77:4888–4898. doi: 10.1128/JVI.77.8.4888-4898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata T, Goshima F, Daikoku T, Inagaki-Ohara K, Takakuwa H, Kato K, Nishiyama Y. 2000. Mitochondrial distribution and function in herpes simplex virus-infected cells. J Gen Virol 81:401–406. doi: 10.1099/0022-1317-81-2-401. [DOI] [PubMed] [Google Scholar]
- 18.Nozawa N, Yamauchi Y, Ohtsuka K, Kawaguchi Y, Nishiyama Y. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp Cell Res 299:486–497. doi: 10.1016/j.yexcr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Willard M. 2002. Rapid directional translocations in virus replication. J Virol 76:5220–5232. doi: 10.1128/JVI.76.10.5220-5232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy MA, Bucks MA, O'Regan KJ, Courtney RJ. 2008. The HSV-1 tegument protein pUL46 associates with cellular membranes and viral capsids. Virology 376:279–289. doi: 10.1016/j.virol.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Wagner MJ, Smiley JR. 2009. Herpes simplex virus requires VP11/12 to induce phosphorylation of the activation loop tyrosine (Y394) of the Src family kinase Lck in T lymphocytes. J Virol 83:12452–12461. doi: 10.1128/JVI.01364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner MJ, Smiley JR. 2011. Herpes simplex virus requires VP11/12 to activate Src family kinase-phosphoinositide 3-kinase-Akt signaling. J Virol 85:2803–2812. doi: 10.1128/JVI.01877-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strunk U, Saffran HA, Wu FW, Smiley JR. 2013. Role of herpes simplex virus VP11/12 tyrosine-based motifs in binding and activation of the Src family kinase Lck and recruitment of p85, Grb2, and Shc. J Virol 87:11276–11286. doi: 10.1128/JVI.01702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benetti L, Roizman B. 2006. Protein kinase B/Akt is present in activated form throughout the entire replicative cycle of deltaU(S)3 mutant virus but only at early times after infection with wild-type herpes simplex virus 1. J Virol 80:3341–3348. doi: 10.1128/JVI.80.7.3341-3348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev 24:2627–2639. doi: 10.1101/gad.1978310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuluunbaatar U, Mohr I. 2011. A herpesvirus kinase that masquerades as Akt: you don't have to look like Akt, to act like it. Cell Cycle 10:2064–2068. doi: 10.4161/cc.10.13.16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman KL, Sarnow P. 2010. Herpes simplex virus is Akt-ing in translational control. Genes Dev 24:2583–2586. doi: 10.1101/gad.2004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110:E4492–E4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuchet-Lourenço D, Anderson G, Sloan E, Orr A, Everett RD. 2013. The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J Virol 87:13422–13432. doi: 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knipe DM. 2015. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology 479-480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR. 2016. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G. 2015. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep 16:202–212. doi: 10.15252/embr.201439366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 35.Eaton HE, Saffran HA, Wu FW, Quach K, Smiley JR. 2014. Herpes simplex virus protein kinases US3 and UL13 modulate VP11/12 phosphorylation, virion packaging, and phosphatidylinositol 3-kinase/Akt signaling activity. J Virol 88:7379–7388. doi: 10.1128/JVI.00712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. 2004. The herpes simplex virus ICP0 RING finger domain inhibits. J Virol 78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paladino P, Collins SE, Mossman KL. 2010. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS One 5:e10428. doi: 10.1371/journal.pone.0010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verpooten D, Ma Y, Hou S, Yan Z, He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J Biol Chem 284:1097–1105. doi: 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z, Damania B. 2016. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 41.Kawaguchi Y, Van SC, Roizman B. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalamvoki M, Roizman B. 2010. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A 107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalamvoki M, Roizman B. 2010. Interwoven roles of cyclin D3 and cdk4 recruited by ICP0 and ICP4 in the expression of herpes simplex virus genes. J Virol 84:9709–9717. doi: 10.1128/JVI.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deschamps T, Kalamvoki M. 13 April 2017. Impaired STING pathway in the human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]