Abstract
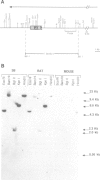
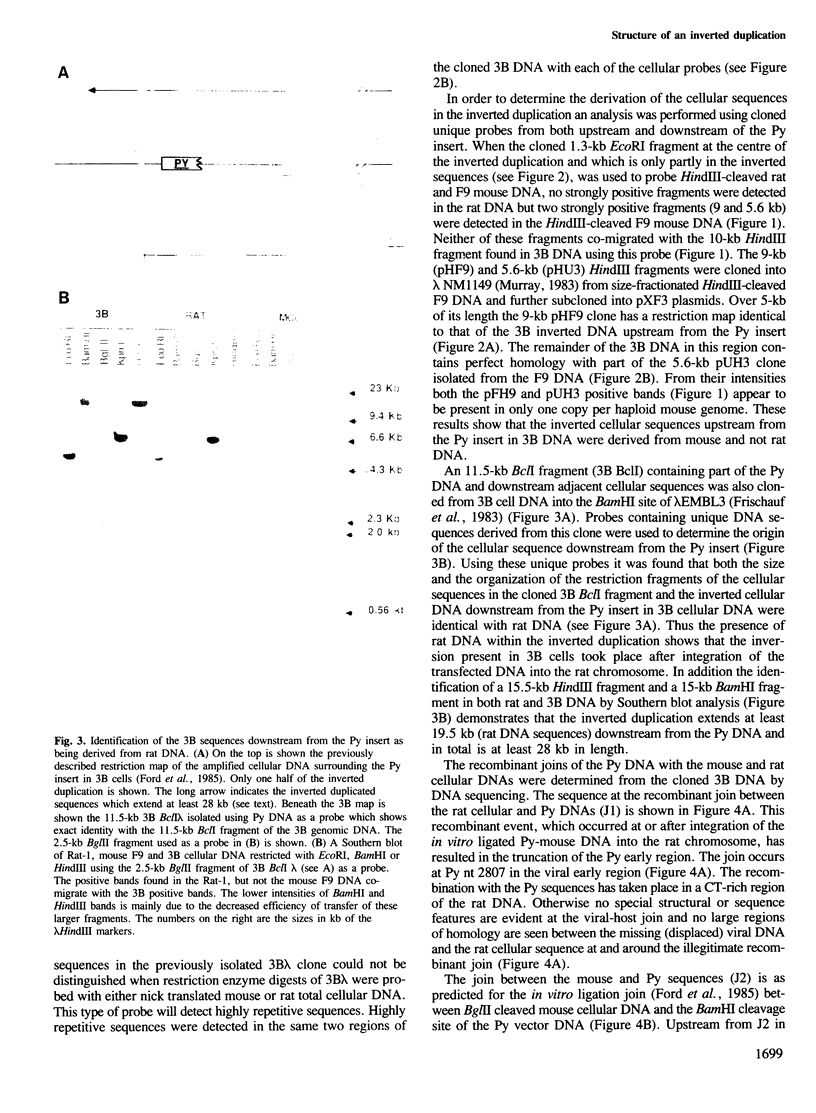
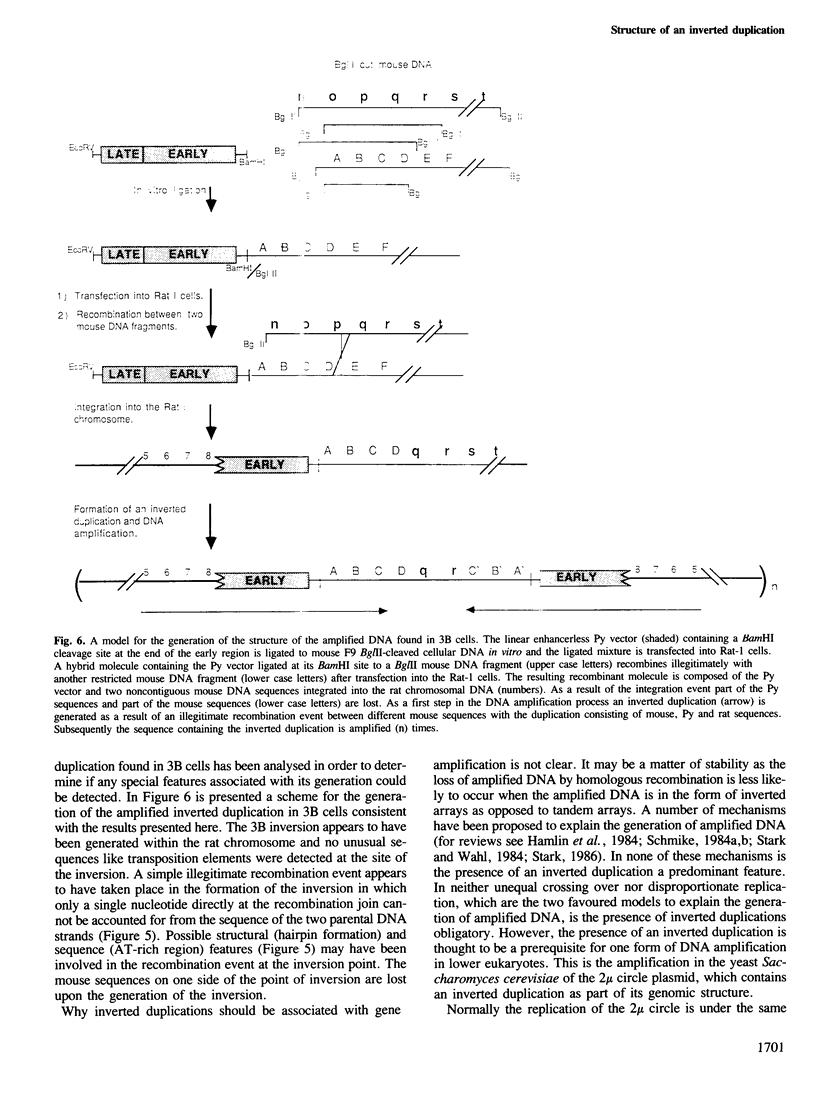
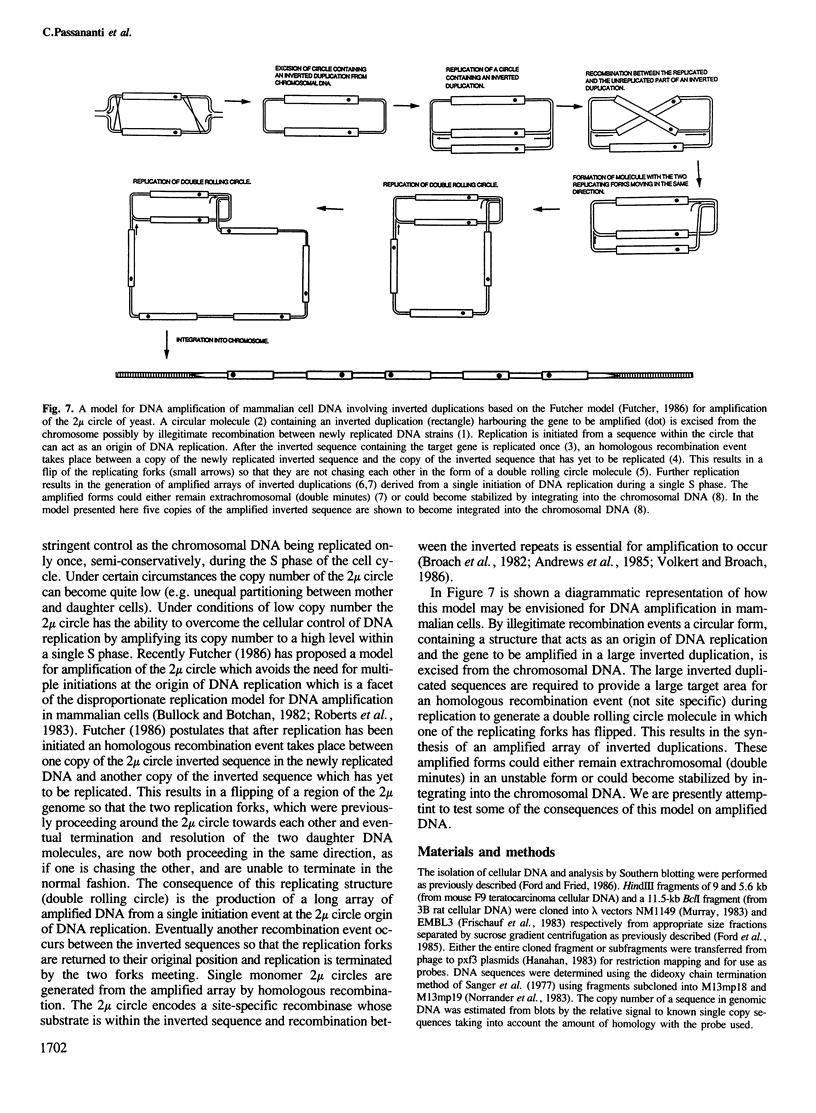
Inverted duplications have been observed to be a common feature of gene amplification in mammalian cells and appear to be generated as a primary event in the amplification process (Ford et al., 1985; Ford and Fried, 1986). The structural features of the amplified inverted duplication, containing the polyoma virus oncogene middle T-antigen, were analysed in transformed 3B rat cells. No unusual sequences such as transposition elements were detected at the site of the inversion. The inversion was generated by a simple illegitimate recombination event in which only a single nucleotide directly at the point of the inversion cannot be accounted for from the sequence of the two parental strands. Possible structural (hairpin formation) and sequence (rich AT) features may have been involved in the illegitimate recombination event at the inversion join. In the cellular DNA near one of its joins with polyoma virus DNA an unusual sequence of 198 bp composed of 99 consecutive purine-pyrimidine pairs has been detected. A model for the generation of amplified DNA containing inverted duplications is proposed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Ford M., Davies B., Griffiths M., Wilson J., Fried M. Isolation of a gene enhancer within an amplified inverted duplication after "expression selection". Proc Natl Acad Sci U S A. 1985 May;82(10):3370–3374. doi: 10.1073/pnas.82.10.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Futcher A. B. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986 Mar 21;119(2):197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Krystal M., Arnheim N. A member of a new repeated sequence family which is conserved throughout eucaryotic evolution is found between the human delta and beta globin genes. Nucleic Acids Res. 1981 Nov 25;9(22):5931–5947. doi: 10.1093/nar/9.22.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Paridaens R. J., Leclercq G., Piccart M. J., Kiss R., Mattheiem W. H., Heuson J. C. Comments on the treatment of breast cancer. Cancer Surv. 1986;5(3):447–461. [PubMed] [Google Scholar]
- Perucho M., Hanahan D., Wigler M. Genetic and physical linkage of exogenous sequences in transformed cells. Cell. 1980 Nov;22(1 Pt 1):309–317. doi: 10.1016/0092-8674(80)90178-6. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Roberts J. M., Buck L. B., Axel R. A structure for amplified DNA. Cell. 1983 May;33(1):53–63. doi: 10.1016/0092-8674(83)90334-3. [DOI] [PubMed] [Google Scholar]
- Saito I., Stark G. R. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured animal cells. Cell. 1984 Jul;37(3):705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification, drug resistance, and cancer. Cancer Res. 1984 May;44(5):1735–1742. [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]