Abstract
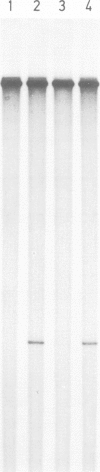
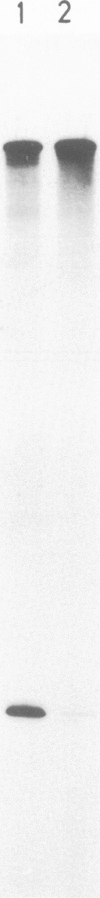
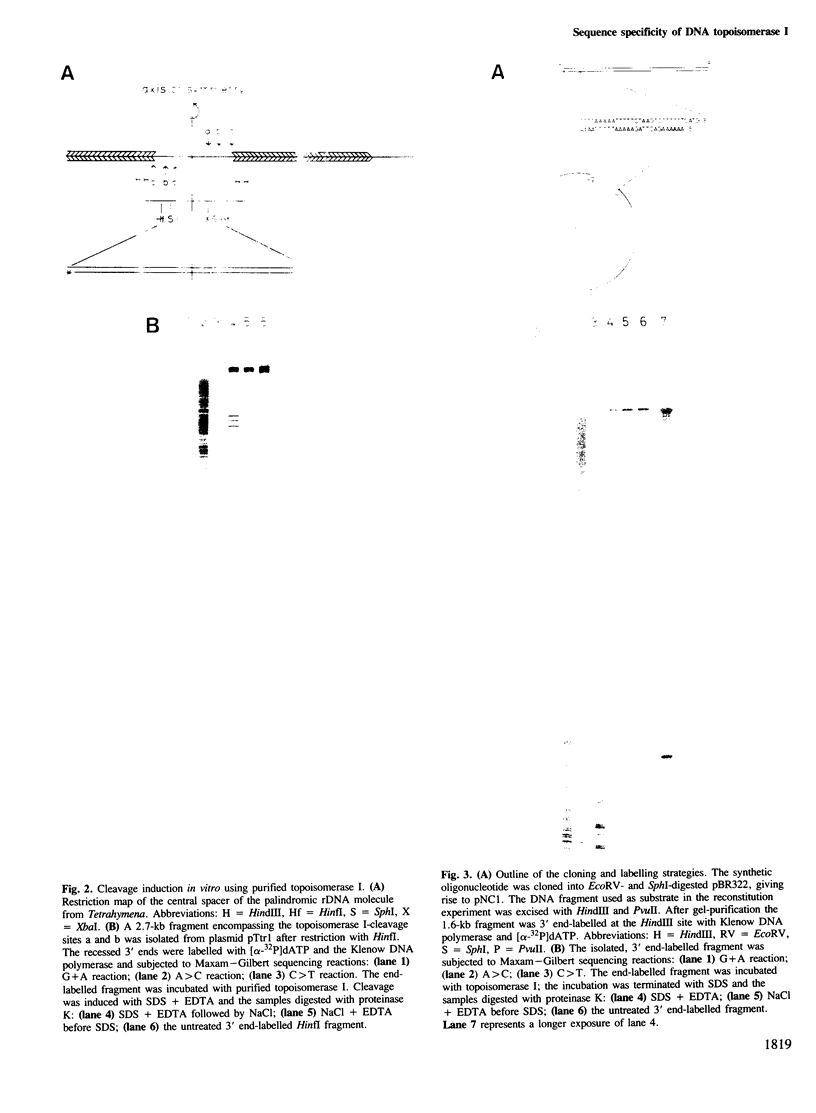
Previously, we have demonstrated that in Tetrahymena DNA topoisomerase I has a strong preference in situ for a hexadecameric sequence motif AAGACTTAGAAGAAAAAATTT present in the non-transcribed spacers of r-chromatin. Here we characterize more extensively the interaction of purified topoisomerase I with specific hexadecameric sequences in cloned DNA. Treatment of topoisomerase I-DNA complexes with strong protein denaturants results in single strand breaks and covalent linkage of DNA to the 3' end of the broken strand. By mapping the position of the resulting nicks, we have analysed the sequence-specific interaction of topoisomerase I with the DNA. The experiments demonstrate that: the enzyme cleaves specifically between the sixth and seventh bases in the hexadecameric sequence; a single base substitution in the recognition sequence may reduce the cleavage extent by 95%; the sequence specific cleavage is stimulated 8-fold by divalent cations; 30% of the DNA molecules are cleaved at the hexadecameric sequence while no other cleavages can be detected in the 1.6-kb fragment investigated; the sequence specific cleavage is increased 2- to 3-fold in the presence of the antitumor drug camptothecin; at high concentrations of topoisomerase I, the cleavage pattern is altered by camptothecin; the equilibrium dissociation constant for interaction of topoisomerase I and the hexadecameric sequence can be estimated as approximately 10(-10) M.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson H. T., Penman S. Selective interruption of high molecular weight RNA synthesis in HeLa cells by camptothecin. Nat New Biol. 1972 May 31;237(74):144–146. doi: 10.1038/newbio237144a0. [DOI] [PubMed] [Google Scholar]
- Andersen A. H., Gocke E., Bonven B. J., Nielsen O. F., Westergaard O. Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res. 1985 Mar 11;13(5):1543–1557. doi: 10.1093/nar/13.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Challoner P. B., Amin A. A., Pearlman R. E., Blackburn E. H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985 Apr 11;13(7):2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J Biol Chem. 1981 May 25;256(10):4805–4809. [PubMed] [Google Scholar]
- Christiansen K., Bonven B. J., Westergaard O. Mapping of sequence-specific chromatin proteins by a novel method: topoisomerase I on Tetrahymena ribosomal chromatin. J Mol Biol. 1987 Feb 5;193(3):517–525. doi: 10.1016/0022-2836(87)90264-6. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke E., Leer J. C., Nielsen O. F., Westergaard O. Transcriptional properties of nucleoli isolated from Tetrahymena. Nucleic Acids Res. 1978 Nov;5(11):3993–4006. doi: 10.1093/nar/5.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Cozzarelli N. R. The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res. 1982 Nov 11;10(21):6833–6847. doi: 10.1093/nar/10.21.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Ishii K., Hasegawa T., Fujisawa K., Andoh T. Rapid purification and characterization of DNA topoisomerase I from cultured mouse mammary carcinoma FM3A cells. J Biol Chem. 1983 Oct 25;258(20):12728–12732. [PubMed] [Google Scholar]
- Kessel D. Effects of camptothecin on RNA synthesis in leukemia L1210 cells. Biochim Biophys Acta. 1971 Aug 26;246(2):225–232. doi: 10.1016/0005-2787(71)90131-6. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Ross W., Rowe T., Glisson B., Yalowich J., Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984 Dec;44(12 Pt 1):5857–5860. [PubMed] [Google Scholar]
- Takeda Y., Folkmanis A., Echols H. Cro regulatory protein specified by bacteriophage lambda. Structure, DNA-binding, and repression of RNA synthesis. J Biol Chem. 1977 Sep 10;252(17):6177–6183. [PubMed] [Google Scholar]
- Tewey K. M., Chen G. L., Nelson E. M., Liu L. F. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Jul 25;259(14):9182–9187. [PubMed] [Google Scholar]
- Trask D. K., Muller M. T. Biochemical characterization of topoisomerase I purified from avian erythrocytes. Nucleic Acids Res. 1983 May 11;11(9):2779–2800. doi: 10.1093/nar/11.9.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]