Abstract
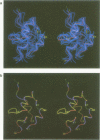
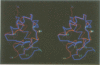
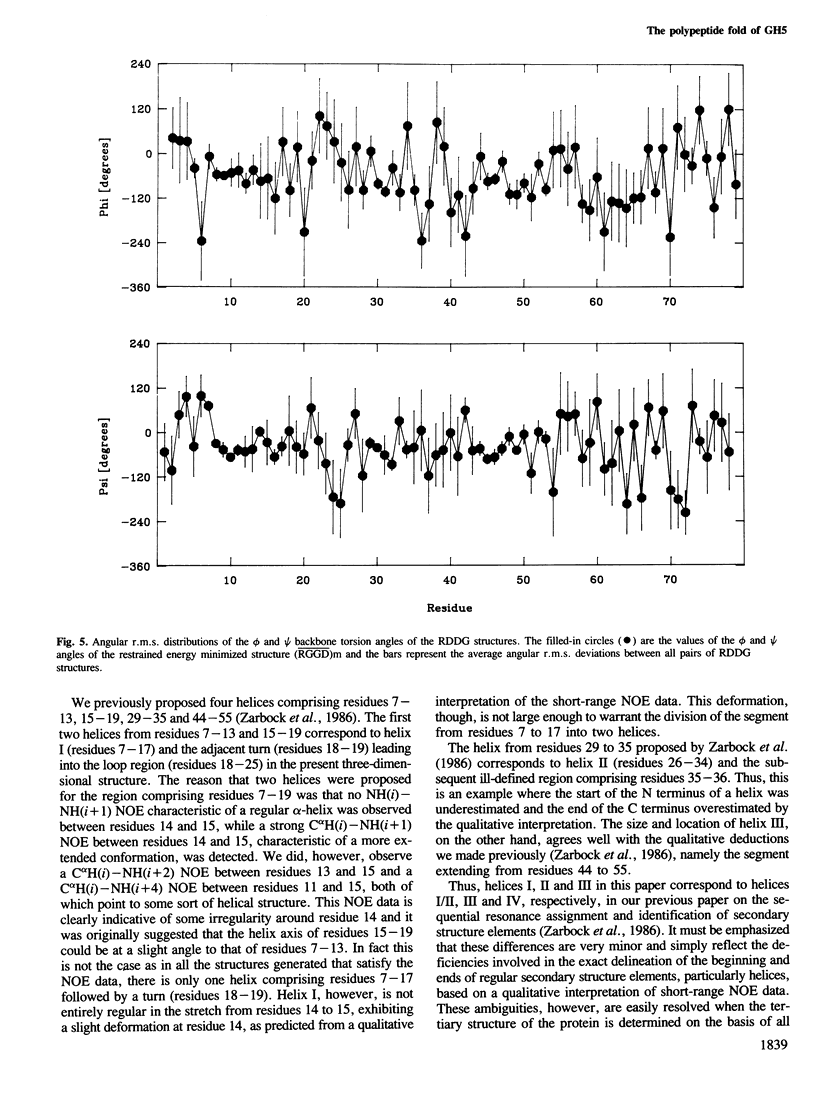
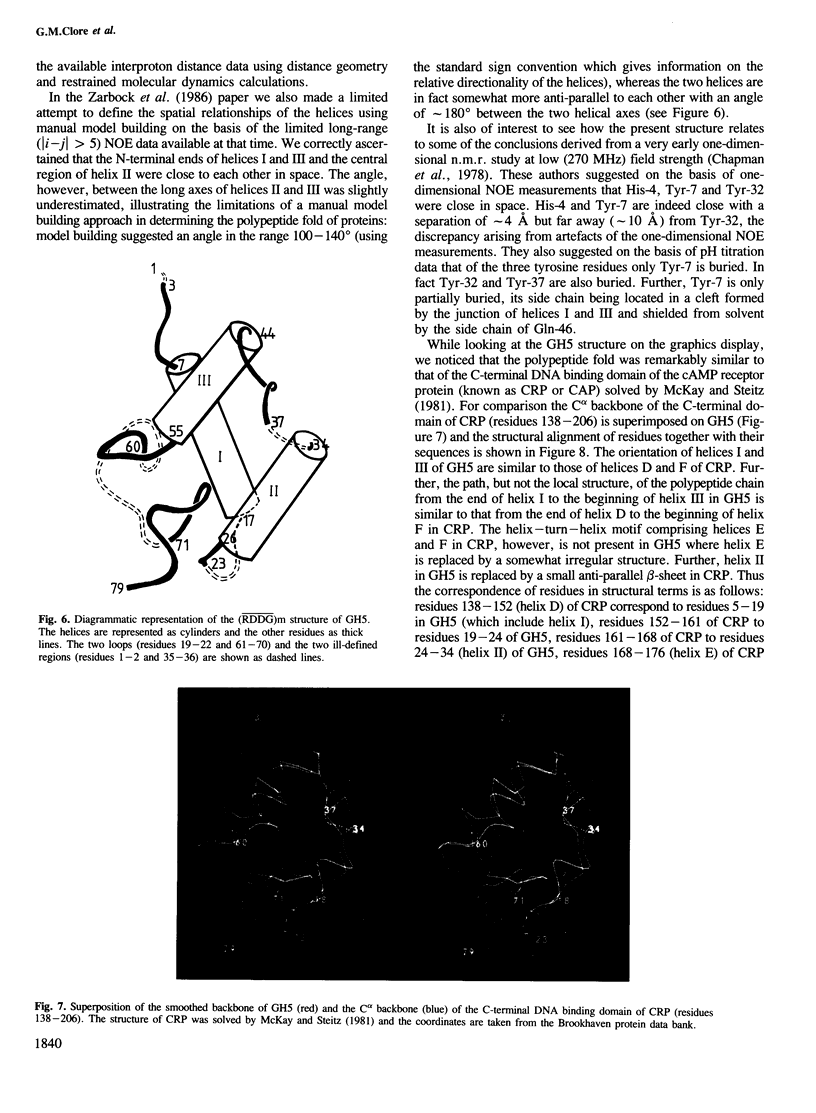
The polypeptide fold of the 79-residue globular domain of chicken histone H5 (GH5) in solution has been determined by the combined use of distance geometry and restrained molecular dynamics calculations. The structure determination is based on 307 approximate interproton distance restraints derived from n.m.r. measurements. The structure is composed of a core made up of residues 3-18, 23-34, 37-60 and 71-79, and two loops comprising residues 19-22 and 61-70. The structure of the core is well defined with an average backbone atomic r.m.s. difference of 2.3 +/- 0.3 A between the final eight converged restrained dynamics structures and the mean structure obtained by averaging their coordinates best fitted to the core residues. The two loops are also well defined locally but their orientation with respect to the core could not be determined as no long range ([i-j[ greater than 5) proton-proton contacts could be observed between the loop and core residues in the two-dimensional nuclear Overhauser enhancement spectra. The structure of the core is dominated by three helices and has a similar fold to the C-terminal DNA binding domain of the cAMP receptor protein.
Full text
PDF
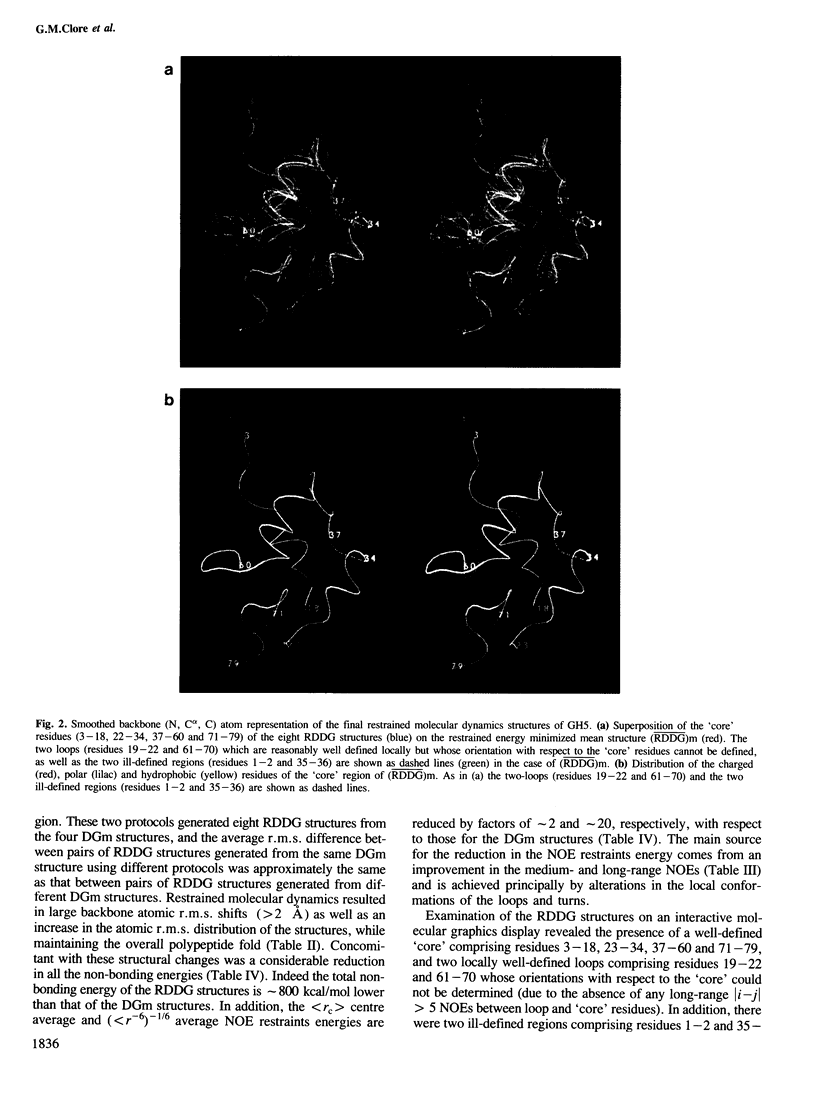
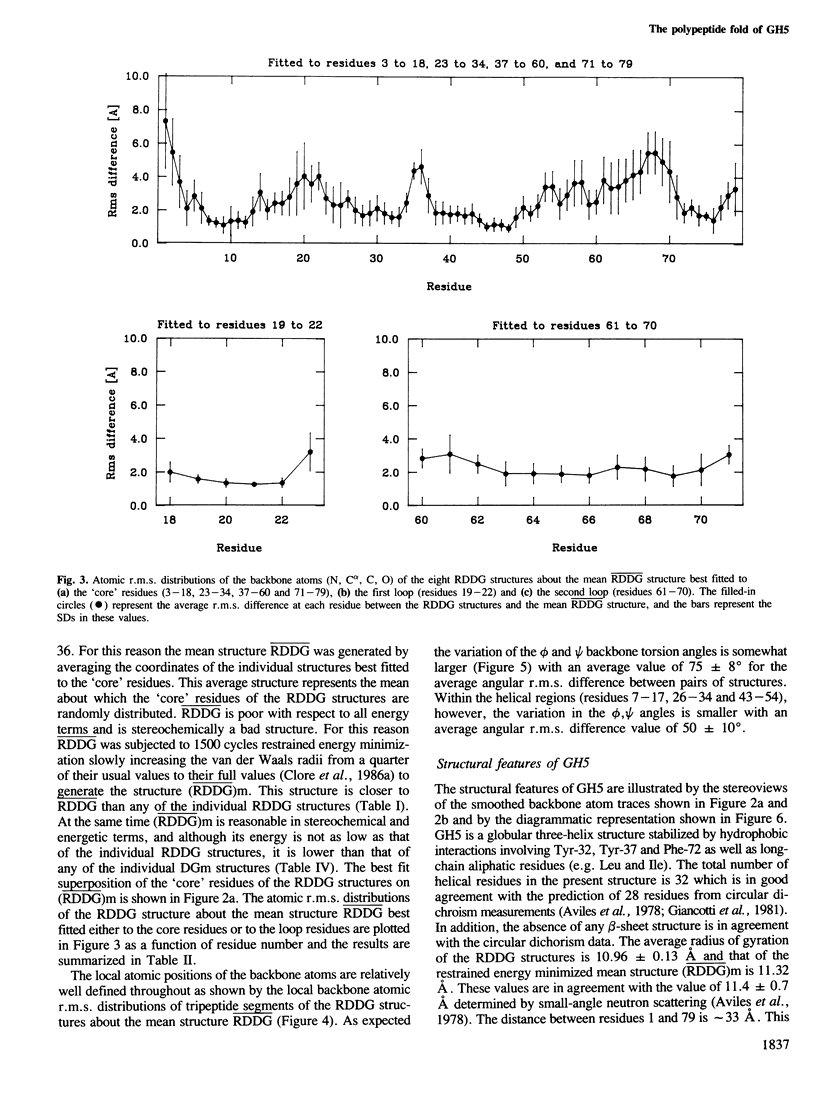
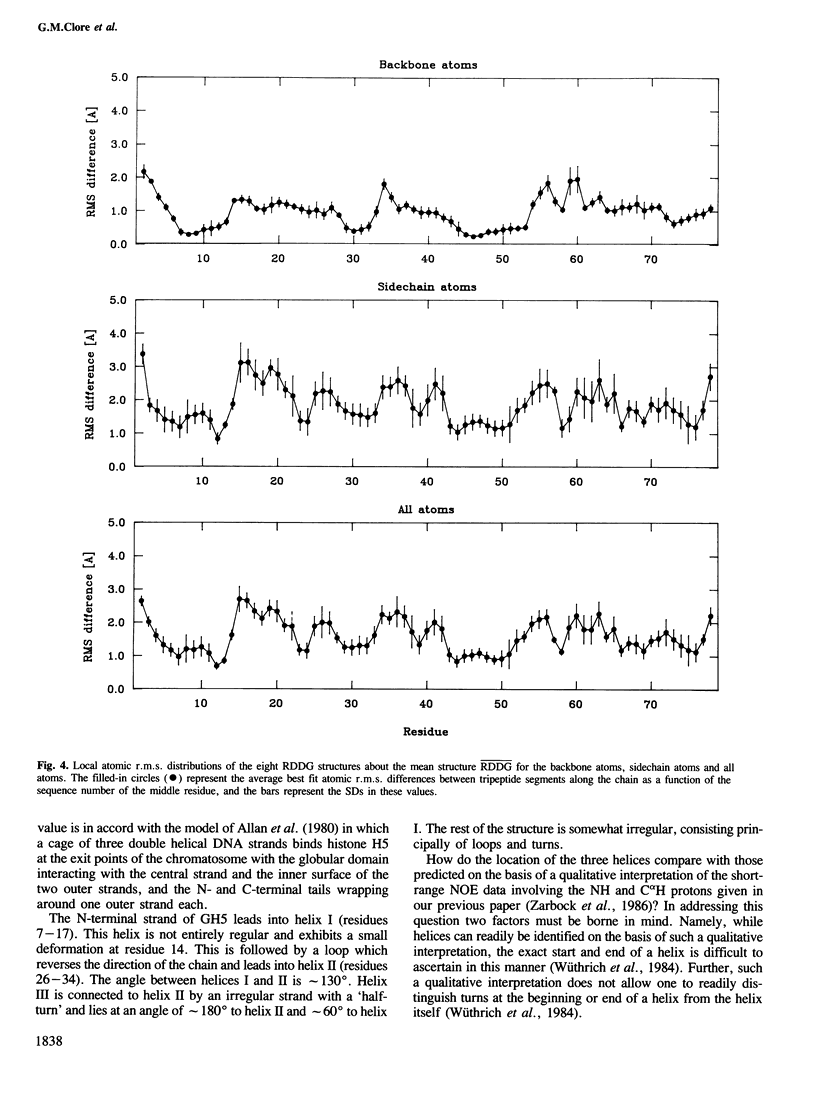


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Aviles F. J., Chapman G. E., Kneale G. G., Crane-Robinson C., Bradbury E. M. The conformation of histone H5. Isolation and characterisation of the globular segment. Eur J Biochem. 1978 Aug 1;88(2):363–371. doi: 10.1111/j.1432-1033.1978.tb12457.x. [DOI] [PubMed] [Google Scholar]
- Briand G., Kmiecik D., Sautiere P., Wouters D., Borie-Loy O., Biserte G., Mazen A., Champagne M. Chicken erythrocyte histone H5. IV. Sequence of the carboxy-termined half of the molecule (96 residues) and complete sequence. FEBS Lett. 1980 Apr 7;112(2):147–151. doi: 10.1016/0014-5793(80)80167-0. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Clore G. M., Gronenborn A. M., Karplus M. Three-dimensional structure of proteins determined by molecular dynamics with interproton distance restraints: application to crambin. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3801–3805. doi: 10.1073/pnas.83.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G. E., Aviles F. J., Crane-Robinson C., Bradbury E. M. A nuclear-magnetic-resonance study of the globular structure of the H5 histone. Eur J Biochem. 1978 Oct;90(2):287–296. doi: 10.1111/j.1432-1033.1978.tb12602.x. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Brünger A. T., Karplus M., Gronenborn A. M. Application of molecular dynamics with interproton distance restraints to three-dimensional protein structure determination. A model study of crambin. J Mol Biol. 1986 Oct 5;191(3):523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Solution conformation of a heptadecapeptide comprising the DNA binding helix F of the cyclic AMP receptor protein of Escherichia coli. Combined use of 1H nuclear magnetic resonance and restrained molecular dynamics. J Mol Biol. 1985 Nov 20;186(2):435–455. doi: 10.1016/0022-2836(85)90116-0. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Nilges M., Sukumaran D. K., Brünger A. T., Karplus M., Gronenborn A. M. The three-dimensional structure of alpha1-purothionin in solution: combined use of nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1986 Oct;5(10):2729–2735. doi: 10.1002/j.1460-2075.1986.tb04557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Sukumaran D. K., Nilges M., Zarbock J., Gronenborn A. M. The conformations of hirudin in solution: a study using nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1987 Feb;6(2):529–537. doi: 10.1002/j.1460-2075.1987.tb04785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P., Gicquel-Sanzey B. Cloning and sequence of the crp gene of Escherichia coli K 12. Nucleic Acids Res. 1982 Feb 25;10(4):1363–1378. doi: 10.1093/nar/10.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Molecular basis of DNA sequence recognition by the catabolite gene activator protein: detailed inferences from three mutations that alter DNA sequence specificity. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7274–7278. doi: 10.1073/pnas.81.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Gent M. E., Gronenborn A. M., Davies R. W., Clore G. M. Probing the sequence-specific interaction of the cyclic AMP receptor protein with DNA by site-directed mutagenesis. Biochem J. 1987 Mar 15;242(3):645–653. doi: 10.1042/bj2420645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti V., Russo E., Cosimi S., Cary P. D., Crane-Robinson C. Secondary and tertiary structural differences between histone H1 molecules from calf thymus and sea-urchin (Sphaerechinus granularis) sperm. Biochem J. 1981 Sep 1;197(3):655–660. doi: 10.1042/bj1970655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman P. G., Chapman G. E., Moss T., Bradbury E. M. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 1977 Jul 1;77(1):45–51. doi: 10.1111/j.1432-1033.1977.tb11639.x. [DOI] [PubMed] [Google Scholar]
- Havel T. F., Wüthrich K. An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol. 1985 Mar 20;182(2):281–294. doi: 10.1016/0022-2836(85)90346-8. [DOI] [PubMed] [Google Scholar]
- Kaptein R., Zuiderweg E. R., Scheek R. M., Boelens R., van Gunsteren W. F. A protein structure from nuclear magnetic resonance data. lac repressor headpiece. J Mol Biol. 1985 Mar 5;182(1):179–182. doi: 10.1016/0022-2836(85)90036-1. [DOI] [PubMed] [Google Scholar]
- Levitt M. Protein folding by restrained energy minimization and molecular dynamics. J Mol Biol. 1983 Nov 5;170(3):723–764. doi: 10.1016/s0022-2836(83)80129-6. [DOI] [PubMed] [Google Scholar]
- Losa R., Thoma F., Koller T. Involvement of the globular domain of histone H1 in the higher order structures of chromatin. J Mol Biol. 1984 Jun 5;175(4):529–551. doi: 10.1016/0022-2836(84)90183-9. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Neuhaus D., Wagner G., Vasák M., Kägi J. H., Wüthrich K. Systematic application of high-resolution, phase-sensitive two-dimensional 1H-NMR techniques for the identification of the amino-acid-proton spin systems in proteins. Rabbit metallothionein-2. Eur J Biochem. 1985 Sep 2;151(2):257–273. doi: 10.1111/j.1432-1033.1985.tb09096.x. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Structure refinement of oligonucleotides by molecular dynamics with nuclear Overhauser effect interproton distance restraints: application to 5' d(C-G-T-A-C-G)2. J Mol Biol. 1986 Apr 5;188(3):455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Wilson C. M. Selective radiolabelling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J. 1986 Dec 20;5(13):3531–3537. doi: 10.1002/j.1460-2075.1986.tb04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M. P., Havel T. F., Wüthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985 Mar 20;182(2):295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter M., Braun W. Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton-proton distances. J Mol Biol. 1984 Dec 15;180(3):715–740. doi: 10.1016/0022-2836(84)90034-2. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter M., Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol. 1983 Oct 5;169(4):949–961. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]
- Yaguchi M., Roy C., Dove M., Seligy V. Amino acid sequence homologies between H1 and H5 histones. Biochem Biophys Res Commun. 1977 May 9;76(1):100–106. doi: 10.1016/0006-291x(77)91673-4. [DOI] [PubMed] [Google Scholar]
- Yaguchi M., Roy C., Seligy V. L. Complete amino acid sequence of goose erythrocyte H5 histone and the homology between H1 and H5 histones. Biochem Biophys Res Commun. 1979 Oct 29;90(4):1400–1406. doi: 10.1016/0006-291x(79)91191-4. [DOI] [PubMed] [Google Scholar]
- Zarbock J., Clore G. M., Gronenborn A. M. Nuclear magnetic resonance study of the globular domain of chicken histone H5: resonance assignment and secondary structure. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7628–7632. doi: 10.1073/pnas.83.20.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]