Abstract
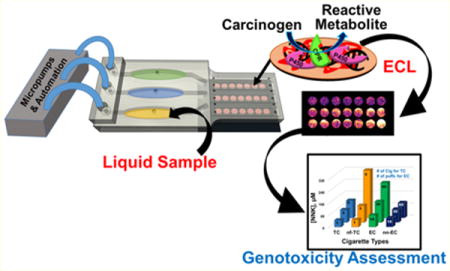
A novel, automated, low cost, three-dimensional (3-D) printed microfluidic array was developed to detect DNA damage from metabolites of chemicals in environmental samples. The electrochemiluminescent (ECL) detection platform incorporates layer-by-layer (LbL) assembled films of microsomal enzymes, DNA and an ECL-emitting ruthenium metallopolymer in ∼10 nm deep microwells. Liquid samples are introduced into the array, metabolized by the human enzymes, products react with DNA if possible, and DNA damage is detected by ECL with a camera. Measurements of relative DNA damage by the array assess the genotoxic potential of the samples. The array analyzes three samples simultaneously in 5 min. Measurement of cigarette and e-cigarette smoke extracts and polluted water samples was used to establish proof of concept. Potentially genotoxic reactions from e- cigarette vapor similar to smoke from conventional cigarettes were demonstrated. Untreated wastewater showed a high genotoxic potential compared to negligible values for treated wastewater from a pollution control treatment plant. Reactivity of chemicals known to produce high rates of metabolite-related DNA damage were measured, and array results for environmental samples were expressed in terms of equivalent responses from these standards to assess severity of possible DNA damage. Genotoxic assessment of wastewater samples during processing also highlighted future on-site monitoring applications.
Keywords: 3-D printing, genotoxicity, automation, e-cigarettes, environmental samples, electrochemiluminescence
Graphical abstract
Genotoxicity refers to the ability of chemicals or their metabolites to interact with genetic material. Reactions with DNA include covalent adduct formation, oxidation, strand breaks, and noncovalent intercalations. When damage to DNA is not repaired, subsequent mutations occur that may lead to cancer.1 For environmental chemicals and drugs, a battery of tests is typically used to predictively assess potential genotoxicity and other toxicities. Genotoxicity tests such as Comet, Ames, micronucleus, and mouse lymphoma assays are very useful for toxicity predictions, but are limited by ease of use and metabolic generality.2 We previously developed microfluidic toxicity screening arrays with the ability to uncover multiple chemical pathways of genotoxicity by measuring DNA damage.2 These high throughput electrochemiluminescent (ECL) and electrochemical assays include DNA and human metabolic enzymes that convert chemicals to metabolites, which are most often the DNA-reactive species in genotoxic pathways.2
Automated, disposable devices that rapidly detect genotoxic chemistry in environmental samples can serve to evaluate the potential influence of toxic chemicals and on-site risks to human health.2,3 We recently reported a paper microfluidic device that estimated the genotoxic potential of water samples.4 In this paper, 3-D printing is used to develop a more sophisticated and sensitive general automated array to assess genotoxic potential of environmental samples; 3-D printing was used to afford cheap, fast design and optimization. It has been used to print fluidic devices that detect pathogenic bacteria, biomedical markers, food allergens, and heavy metals, as well as in nanoparticle and chemical synthesis.5,6 3-D printed flow cells were designed for microdroplet generation, electrochemical sensing, and microfluidic devices.7 We used desktop 3-D printers to develop devices for flow injection amperometry, flow cells to measure ECL,8 and microfluidic immunoarrays to detect cancer biomarker proteins.9
The current device is designed for rapid assays of liquid samples. We focus here on two types of samples for reasons outlined below. The first is cigarette and e-cigarette smoke, since smoking is a major contributor to heart disease and cancer.10 The second is contaminated water, a major public health concern.
Smoking causes more deaths than human immunodeficiency virus, illegal drug use, alcohol, motor vehicle accidents, and fire related deaths combined.11 Electronic (e-)cigarettes are battery powered devices that vaporize nicotine, and were designed as an alternative to tobacco cigarettes. Additives in recent e-cigarettes make the vapor less harsh and allow more rapid delivery of nicotine to the brain, fostering use and increasing chances of addiction. Between 2011 and 2015 e-cigarette use increased from 1.5 to 16% among United States high school students and from 0.6 to 5.3% among middle school children.12 In addition, e-cigarette vapor contains toxic metals such as cadmium, lead, and nickel at levels of 0.022–0.057 ng in 15 puffs of the aerosol.13
Currently one-third of available fresh water is used for agriculture, industrial and domestic purpose.14 Chemical pollution of fresh water lakes and rivers is endemic in populated areas.15 Toxic pollutants found in water bodies include nitrogenous and phosphorus species, organic chemicals, metals, and biologically generated compounds.16 Coexistence of these chemicals in mixtures has been suggested as an origin of elevated genotoxic effects.14,17
In this paper, we report the first disposable, fully automated 3-D printed array designed to assess genotoxic potential of liquid environmental samples. This device can analyze vapor extract samples from cigarettes and water samples in 5 min for less than $1.00 (Figure 1). The arrays assess potential genotoxicity based on DNA reactivity of metabolites generated by enzymes on the array. To our knowledge, this is the first low cost 3-D printed microfluidic array capable of evaluating the metabolite-dependent genotoxic potential of environmental samples. Results suggest that e-cigarettes can have similar or enhanced genotoxic potential compared to tobacco cigarettes, depending on use patterns. The genotoxic potential of untreated wastewater was high, but was decreased to very low levels by reclamation in a sewage treatment facility.18
Figure 1.
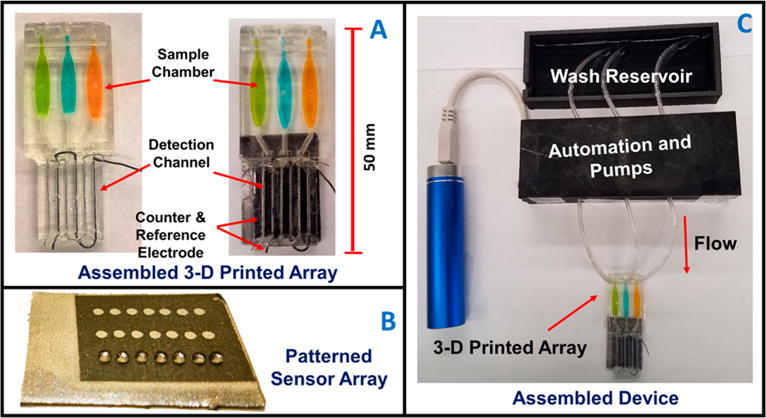
Automated genotoxicity screening array: (A) 3-D printed devices without (left) and with (right) microwell chip and counter electrode wires inserted showing the sample chambers containing dye solutions. (B) Microwell-patterned pyrolytic graphite detection array showing the first row holding 1 μL water droplets retained by the hydrophobic microwell boundaries. Each row is fed by a separate sample line. The working array features films of DNA, metabolic enzymes, and RuPVP in each microwell. (C) Assembled array system showing box enclosing electronic microprocessors and micropumps driven by a rechargeable battery and connected to the 3-D printed array below with a wash reservoir (top) containing pH 7.4 buffer.
MATERIALS AND METHODS
Safety note
Benzo[a]pyrene (B[a]P), 4-[methyl(nitroso)amino]-1-(3-pyridinyl)-1-butanone (NNK), N′-nitroso-2-(3-pyridyl)pyrolidine (NNN), 2-acetylaminofluorene (2-AAF), 2-naphthylamine (2-NA), aflatoxin-B1 (AFB1) and their metabolites are potential carcinogens. Handling these chemicals involved protections including gloves, safety glasses, and working in a hood.
Chemicals and Materials
B[a]P (MW 252.31), NNK (MW 207.23), NNN (MW 177.20), 2-AAF (MW 223.28), 2-NA (MW 143.19), AFB1 (MW 312.28), poly(diallyldimethylammonium chloride) (PDDA, avg MW = 100 000–200 000), poly(acrylic acid) (PAA, avg MW = 1800), calf thymus DNA (Type I), and other chemicals were from Sigma-Aldrich. Pooled male human liver microsomes were from BD Gentest. [Ru(bpy)2(PVP)10]2+ {RuPVP; (bpy = 2,2-bipyridyl; PVP = poly(4-vinylpyridine)} was synthesized and characterized as described previously.19 Pyrolytic graphite (PG) sheets are from Panasonic PGS-P13689-ND 70 μm thick.
3-D Printed Microfluidic Arrays
Microfluidic arrays were printed from clear acrylate resin using a Formlabs Form1+ stereolithographic 3-D printer. Design files are available on our Web site.20 Briefly, CAD files incorporating the design were converted to printer instruction files for input to the printer (details in the Supporting Information (SI)). After printing, devices were rinsed internally and externally successively with isopropanol and water, then spray coating with clear acrylic spray (Krylon).
Arrays were printed with three sample chambers that feed three detection channels designed so that sample solutions flow directly across shallow 1 mm wide, 10 nm deep microwells on the detection chip to facilitate reactions with enzyme/DNA films in the wells (Figures 1A,B and S1). Detection chips were made from conductive pyrolytic graphite (PG) sheets cut to desired sizes. PG sheets were patterned with microwells using our print and peel technology21 to accommodate tiny volumes of reagents during layer-by-layer (LbL) film assembly (Figure 1B).22 A microwell template featuring 21 spots of diameter 1 mm in 3 rows with 7 spots per flow channel was inkjet printed onto glossy paper (Avery 5263) and heat pressed for 45 s at 275 °C to transfer onto these PG sheets (SI Figure S2, SEM). Patterned PG sheets were attached onto the 3-D printed array using double sided adhesive.
Array were 3-D printed in less than 1 h using 6 mL of resin at fabrication cost $0.80. They have dimensions 50 mm (length (L)) × 22 mm (width (W)) × 5 mm (height (H)). Sample chamber dimensions are 17 mm (L) × 5 mm (W) × 2.5 mm (H) and maximum sample or reagent volume 170 ± 5 μL. Sample volume was 150 μL, and detection channels are 23 mm (L) × 3 mm (W) × 0.65 mm (H) with volume 45 ± 5 μL, and are provided with holes and grooves to accommodate stainless steel wire counter (0.4 mm diameter) and Ag/AgCl wire reference (0.6 mm diameter) electrodes to complete the ECL electrochemical detection cell (Figure 1A).
Automation
Automation was achieved by interfacing three Mp6 micropumps (Bartels) with an “ATtiny85” microprocessor chip via Bartels OEM microcontrollers (Figures S1 and S3). Micropump control features printed-circuit board (PCB)-linked microcontrollers independently connected to the ATtiny85 chip. The inexpensive 8-bit ATtiny microcontroller chip runs Arduino programs for pump control at low power consumption (Figure S3),23 and provides ON/OFF commands to control voltage input to micropumps from a rechargeable lithium ion battery. Micropumps were adjusted to flow rate 120 ± 3 μL/min.
Layer by Layer Film Assembly
Sequential layers of ECL metallopolymer RuPVP, human liver microsome (HLM) enzyme source, and DNA were grown in microwells on the PG chip by layer-by-layer (LbL) alternate electrostatic assembly,22 depositing appropriate solutions sequentially and incubating 20 min for polyion layers and 30 min for enzyme and DNA layers at 4 °C with water washing between adsorption steps.24 Film architecture optimized for best signal/noise was PDDA/PAA/(RuPVP/DNA)2/RuPVP/enzyme/DNA.
Detection of Genotoxic Reactions
The assay protocol involves two steps. First, the natural cyt P450 catalytic cycle is activated by applying −0.65 V vs Ag/AgCl while flowing oxygenated solutions of test samples in 10 mM phosphate buffer pH 7.4 + 1% DMSO for 45 s. This generates metabolites from the test compounds that can react with DNA.25 Second, after washing the array with buffer, 1.25 V vs Ag/AgCl is applied for 180 s to generate ECL proportional to damage of coreactant DNA. This oxidizes RuIIPVP to RuIIIPVP, and RuIIIPVP reacts with guanine in a complex pathway to form excited state *RuIIPVP that emits ECL light at 610 nm which is captured with a CCD camera in a dark box.2 ECL generation involves oxidation of guanine by RuIIIPVP in catalytic pathway resulting in a guanine radical that further reacts with RuIIIPVP to form *RuIIPVP that emits light.19 Covalent metabolite-nucleobase adducts disrupt the DNA double helix and can also lead to abasic sites and strand breaks, all resulting in more accessible guanines that generate more ECL. Electrodes are disposable and are discarded after each use.
Prior to assays, sample chambers (Figure 1A) were prefilled with 150 μL of test solution through sample injection ports, which are then sealed. Micropumps are connected to inlets with common feed at the back from a buffer reservoir (Figure 1C). Voltage input was by a three-electrode hand-held potentiostat. The entire device resides inside a dark box. Initially pumps are off, then the program initiates a 135 s pumping cycle for three steps, 10 s filling the detection channel, 45 s electrolysis, and 80 s washing. Samples were pumped into the 3 separate detection channels and 45 s electrolysis was done at −0.65 V vs Ag/AgCl once channels were full (while continuing flow) to activate the natural catalytic cycle of cyt P450s.,25 After a subsequent 80 s wash cycle, pumps turn off and visible ECL light is generated by applying 1.25 V vs Ag/AgCl for 180 s and capturing light with a CCD camera. Timing was optimized for the best ECL signal/noise.
Sample Analyses
Smoke (vapor) extracts from e-cigarettes and filtered and nonfiltered tobacco cigarettes were collected using an artificial inhalation device (see the SI, Figure S4). Tubing connecting a syringe and the cigarette or e-cigarette was attached via a pipet tip plugged with cotton so that smoke passes through it and chemicals are trapped. This cotton was subsequently extracted with 2 mL of DMSO. To keep experimental conditions representative and relevant for vaping usage by smokers we extracted 100 puffs and smoke from 5 tobacco cigarettes for comparison. Vaping anywhere from 75 to 175 puffs from e-cigarettes is equivalent on average to 5–6 tobacco cigarettes per day.26 Approximately 15–30 puffs from an e-cigarette is considered equivalent to smoke from one tobacco cigarette.13,28
Polyaromatic hydrocarbons and tobacco specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) are major carcinogens present in cigarette smoke.27 Most chemicals in tobacco cigarettes and e-cigarettes are similar to slightly lower concentrations reported for e-cigarettes.13,28 Usually, contents of e-cigarettes are loaded into a cartridge and used with a battery operated inhalation device that heats and converts a nicotine solution with additives into an aerosol.13 The contents of the e-cigarette liquid quoted by manufacturer content lists show tobacco derived nicotine, propylene glycol, vegetable glycerin, and natural and artificial flavoring agents.
Our second test targets featured untreated sewage water, partially, treated water and completely treated reclaimed water as collected from University of Connecticut water pollution control facility.18 Water samples were passed through a 0.2 μM syringe filters (Thermo Scientific F2504-6) prior to genotoxic evaluation to remove particulate matter. Genotoxic chemicals present in wastewater 2-acetylaminofluorene (2-AAF),29 2-naphthylamine (2-NA),30 and aflatoxin-B1 (AFB1)31 were used as reference standards.
RESULTS
Cigarettes and E-Cigarettes
Responses to tobacco smoke components B[a]P, NNK, and NNN were measured first. One channel in the microfluidic array was used for each specific compound (Figure 2A). Plots of % ECL increase over the blank (1% DMSO in 10 mM phosphate buffer, pH 7.4) vs concentration of standard (Figure 2B) serve as standard calibration curves and their slopes reflect relative rates of DNA damage.2 Dynamic range was from 3 to 150 μM for all standards. Increase in ECL intensity was observed with increase in test chemical concentration (Figure 2). ECL increases in earlier versions of related genotoxicity arrays were confirmed as directly related to amounts of specific metabolite-DNA adducts detected by LC-MS/MS.2,4,24 In the present array, spot-to-spot variability was ±6% (n = 21) and array-to-array variability was ±7% (n = 3) (Figure S5). Toluene, with poorly DNA-reactive metabolites,32 was used as a negative control with negligible ECL change (Figure 2).
Figure 2.
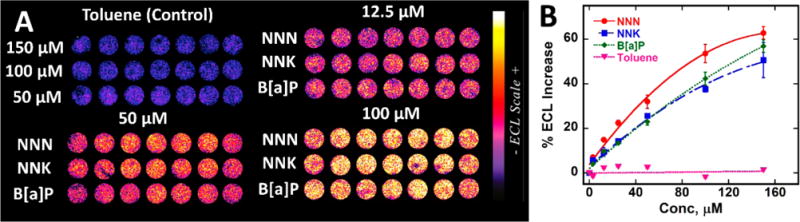
Array results for tobacco-related standards with DNA-reactive metabolites: (A) recolorized ECL data using arrays featuring RuPVP/enzyme/DNA microwells treated with oxygenated solutions of carcinogens B[a]P, NNK, and NNN and negative control toluene in 1% DMSO + 10 mM phosphate buffer pH 7.4 for 45 s at −0.65 V vs Ag/AgCl, with ECL captured by CCD camera after subsequently applying 1.25 V vs Ag/AgCl for 180 s. (B) Calibration plots of % ECL increase over 1% DMSO control vs concentration of standards. ECL intensity increases proportional to DNA damage that disorders ds-DNA and allows coreactant guanines in the DNA better access to RuIII sites of RuPVP.2
Previous studies of B[a]P, NNK and NNN confirmed that reactive metabolites of these pro-carcinogens react with DNA to form covalent DNA adducts (Scheme 1). B[a]P is a polyaromatic hydrocarbon present in coal tar, cigarette smoke, and grilled meat. Metabolic cyt P450s catalyze B[a]P oxidation to a 7,8-epoxide that is rapidly hydrolyzed to a diol by epoxide hydrolase. The diol is further oxidized by cyt P450 to an epoxide to form ±-antibenzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide (±-anti-BPDE, eq 1).33 Major metabolite ±-anti-BPDE is classified as a Group I carcinogen by the International Agency of Research on Cancer (IARC). It is a strong electrophile that reacts with DNA nucleobases of to form covalent adducts. The major covalent adduct is formed by ±-anti-BPDE reaction with the exocyclic amine of nucleobase guanine to form a stable covalent adduct (Scheme 1, eq 1). Similarly, the tobacco specific nitroso amines NNK and NNN undergo α-hydroxylation catalyzed by cyt P450s to form reactive metabolites that react with DNA to form adducts at the N7 position of guanine (Scheme 1, eqs 2 and 3).2,34,35
Scheme 1. Cytochrome P450 Mediated Bioactivation and DNA Reactivity of Standard Chemicals Used for Cigarette Studiesa.
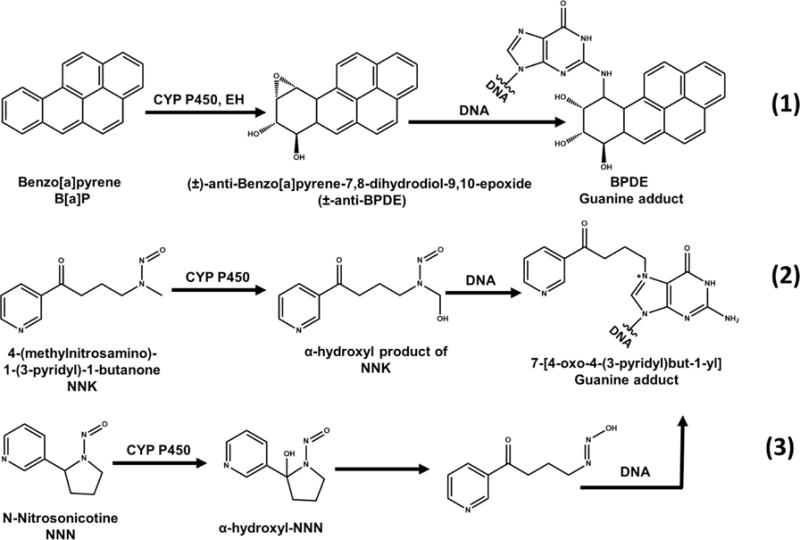
a(1) Benzo[a]pyrene (B[a]P), metabolized to benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide that intercalates and covalently binds predominantly with guanine base in DNA, Adapted from information in ref 33. (2) 4-(Methylnitrosoamino)-1-(3-pyridyl)-1-butanone (NNK) and (3) N-nitrosonicotine (NNN) form hydroxyl forms before binding to nucleobases within DNA. Adapted from information in ref 34.
Cigarette smoke and E-cigarette vapor trapped in the inhalation device (see Materials and Methods, and Figure S4, SI) was extracted into DMSO and then diluted 100-fold in pH 7.4 buffer prior to analysis. Vapor extract from 20 puffs of e-cigarettes was taken as equivalent to smoke from one tobacco cigarette.13,28 We found large increases in ECL intensity with increases in amount of extracted cigarette smoke and e-cigarette vapor (Figure 3A), suggesting increased amounts of DNA damage2,4 (Figure 3A,B). The most important finding is that % ECL values for equivalent numbers of puffs are slightly larger for e-cigarettes than for tobacco cigarettes, and much larger than for filtered tobacco cigarettes and non-nicotine e-cigarettes. These differences are significant at 95% confidence levels (t tests) and suggest that chemicals in the vapor of nicotine e-cigarettes metabolized in the array are at least as DNA-reactive as those in smoke of unfiltered tobacco cigarettes.
Figure 3.
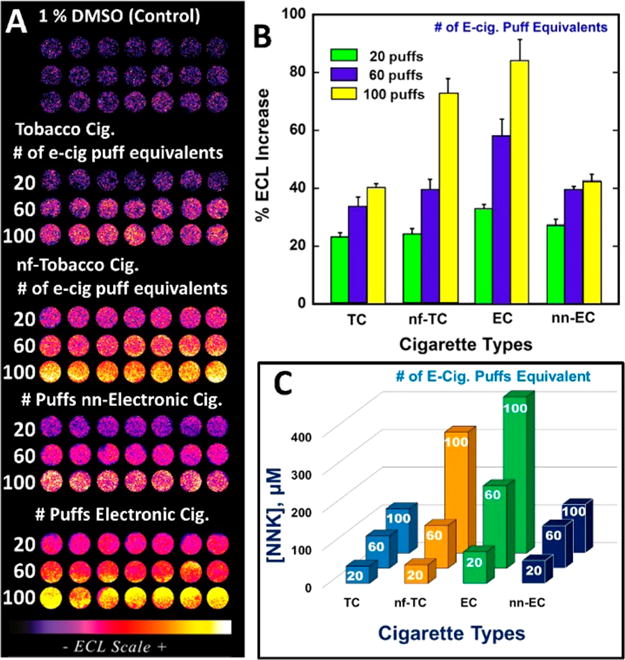
ECL array results comparing extracted vapor from e-cigarettes with extracted smoke from tobacco cigarettes using the conversion that 20 e-cigarette puffs equals smoke from one tobacco cigarette {Abbrev.: tobacco cigarettes (TC), e-cigarettes (EC), nonfiltered (nf) and non-nicotine (nn)}. (A) Recolorized ECL data from arrays. Each row represents microwells containing RuPVP/Enzyme/DNA layers treated with smoke extracted from 1, 3, and 5 TC and nf-TC (equivalent to 20, 60, and 100 puffs of e-cigarette) and 20, 60, and 100 puffs of EC and non-nicotine (nn)-EC in 1% DMSO containing buffer for 45 s under potential of −0.65 V vs Ag/AgCl. ECL captured while applying 1.25 V vs Ag/AgCl for 180 s. (B) % ECL increase over control (1% DMSO in buffer) vs cigarette samples. (C) NNK equivalents from %ECL for different cigarette samples.
The difference in %ECL increase between filtered and nonfiltered tobacco cigarettes for 20 puff e-cigarette equivalents was not statistically different at the 95% confidence interval. However, for 60 and 100 e-cigarette puff equivalents, %ECL was statistically larger for unfiltered cigarettes at the 95% confidence level (Figure 3B). DNA reactivity of the non-nicotine e-cigarettes and the filtered tobacco cigarettes denoted by array signals were comparable, and ∼1.8-fold smaller than that for unfiltered tobacco cigarettes.
To link these results to known DNA-reactive chemical metabolites, DNA-reactivity of the samples was expressed in terms of array responses of known tobacco component concentrations of B[a]P, NNK, and NNN using Figure 2B. This approach puts the results on a common footing related to major genotoxic components in tobacco smoke. Figure 3C shows the NNK-equivalent concentration in each of the cigarette types and suggest that nicotine e-cigarettes and unfiltered cigarettes are equivalent to quite significant levels of the potent tobacco carcinogen NNK. Equivalence of sample responses in terms of B[a]P and NNN are reported in Table S1 in the SI, and lead to qualitatively similar conclusions.
Water Samples
Known water pollutants 2-AAF, 2-NA, AFB1 were first tested as reference standards to assess genotoxic potential. Aflatoxins are metabolites of fungal organisms in polluted food and environmental samples and are associated with liver cancer.36 Aflatoxin B1 is one of the most potent aflatoxins, and requires cyt P450 bioactivation to become carcinogenic. It is oxidized by cyt P450s to the 8,9-epoxide, which reacts with the N7 position of guanine to form covalent adducts (Scheme 2, eq 4).37 Arylamines are commonly associated with bladder cancer.38 2-acetylaminofluorene (2-AAF) and 2-naphthylamine are converted by sequential reactions catalyzed by cyt P450s and acetyl transferase to arylnitrenium ions that react with nucleobases of DNA to form covalent adducts. A major adduct on the C8 position of guanine is shown in Scheme 2 (eqs 5, 6).35,39,40
Scheme 2. Cytochrome P450 Mediated Bioactivation and DNA Reactivity of Standard Chemicals Used for Water Samplesa.
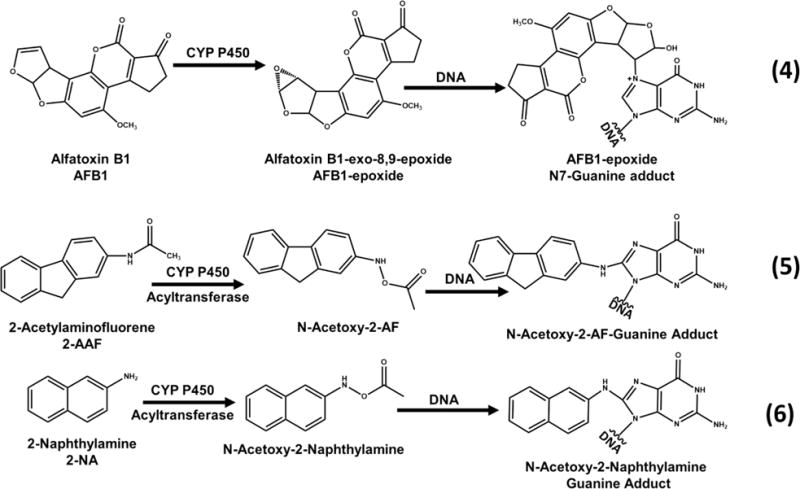
a(4) Aflatoxin B1 (AFB1), metabolically activated to its epoxide form that forms covalent adducts with DNA nucleobases. Adapted from information in ref 37. (5) 2-Acetylaminofluorene (2-AAF). Adapted from information in ref 38. (6) 2-naphthylamine (2-NA) form acetoxy forms upon bioactivation that form covalent adducts with DNAnucleobases. Adapted from information in ref 40.
Array results for these three compounds and toluene negative control are shown in Figure 4A. Plots of %ECL increase over blank (1% DMSO in pH 7.4 buffer) vs concentration of standard (Figure 4B) serve as calibration curves for water samples. Again, slopes reflect relative rates of DNA damage.2 Dynamic ranges were from 3 to 100 μM. Limits of detection as %ECL increase 3× the avgerage noise were ∼3 μM for these genotoxic compounds.
Figure 4.
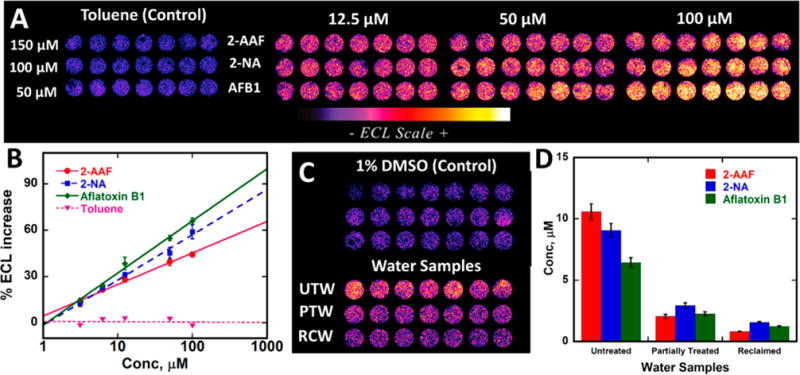
Array results for standards with known DNA-reactive metabolites: (A) Recolorized ECL data using arrays featuring RuPVP/enzyme/DNA microwells treated with oxygenated solutions of carcinogens (2-AAF, 2-NA, and aflatoxin B1 and negative control toluene in 1% DMSO + 10 mM phosphate buffer pH 7.4 for 45 s at −0.65 V vs Ag/AgCl, with ECL captured by CCD camera after subsequently applying 1.25 V vs Ag/AgCl for 180 s. (B) Calibration plots of %ECL increase over the blank vs concentration of standards. ECL intensity increases proportional to DNA damage. (C) ECL array results comparing ECL intensities obtained from untreated water (UTW), partially treated water (PTW), and fully treated reclaimed water (RCW) with respect to 1% DMSO controls. Recolorized ECL data from arrays with each row representing microwells containing RuPVP/enzyme/DNA layers treated with UTW, PTW, RCW, and 1% DMSO in buffer for 45 s at −0.65 V vs Ag/AgCL with ECL captured after subsequent application of 1.25 V vs Ag/AgCl for 180 s. (D) Bar graph showing chemical equivalents from %ECL response for different water samples.
Samples from the University of Connecticut water treatment facility were assayed on the array. Figure 4C shows a significant increase in ECL for untreated water samples compared to reclaimed and partially treated water samples. Results from Figure 4C are also expressed in terms of equivalent concentrations of the reference standards (Figure 4D) to provide comparisons of the relative genotoxicity potential of the samples. For example, the untreated wastewater is equivalent to about 10 μM of the parent chemical 2-AAF.
Compared to the untreated wastewater, the difference from the treated reclaimed water is significant at the 95% confidence level. Results suggest that more potentially genotoxic chemicals in the untreated wastewater are converted on the array to metabolites that are reactive with DNA compared to reclaimed or partially treated water samples. The larger %ECL for partially treated water compared to fully reclaimed water was also statistically significant at 95% confidence (Figure 4C, D). Genotoxic potential in terms of DNA reactivity of untreated water samples was ∼10-fold larger than that for fully reclaimed water and ∼2.4 fold larger than that for partially treated water.
DISCUSSION
The results above illustrate a novel, low cost, 3-D printed microfluidic array capable of assessing the genotoxic potential of environmental samples. The 3-D printed device with disposable microwell array containing enzyme/DNA/RuPVP films costs less than $1.00 to fabricate. Advantages of this device include multianalyte analysis, complete automation, on chip metabolite generation, and rapid detection of DNA damage (5 min). These disposable arrays are designed to “plugin” to a reusable automation platform featuring micro-controllers, micropumps, and battery that costs $150. The array here was equipped with 21 detection microwells, but it is possible to expand to larger size to accommodate multiple enzymes, multiple analytes, and higher sample throughput.
DNA reactivity related to metabolites from smoke or vapor extracts measured by the array clearly suggests comparable genotoxic potential of tobacco and nicotine e-cigarettes when assayed by the same protocol (Figure 3). Expression of the signals in terms of levels of known tobacco chemicals with metabolites having high rates of DNA damage, e.g., NNK (Figure 3D) provides a reference point to assess the severity of possible genotoxicity, without having to determine individual DNA adducts by expensive LC-MS/MS assays. E-cigarette vapor was reported to have low concentrations of chemicals with potential to cause DNA damage13 and could be assumed by some to be a safer alternative to tobacco cigarettes. However, our results suggest similar DNA damage from e-cigarette vapors and tobacco cigarette smoking.
Results also showed that genotoxic potential for non-nicotine e-cigarettes is about the same as that for filtered tobacco cigarettes, and 1.5–2-fold lower than that for e-cigarettes. DNA reactivity for 20 puffs of an e-cigarette was equivalent to about 83 μM NNK (1.6 μg/mL) (Figure 3C) compared to estimated levels of NNK in one tobacco cigarette of 46 μM (0.9 μg/mL). Unfiltered tobacco cigarettes gave DNA reactivity nearly 2 times greater than filtered tobacco cigarettes (Figure 3B and C). Even non-nicotine e-cigarettes showed significant DNA reactivity, similar to that of filtered tobacco cigarettes (Figure 3C).
The above results are consistent with recent reports using conventional assays that found significant DNA strand breaks, cytotoxic effects, and cell death caused by e-cigarette vapor with and without nicotine.41,42 Ease of use of e-cigarettes may also result in elevated use compared to tobacco cigarettes, which can result in escalated DNA damage. For example, DNA reactivity as NNK equivalents in vapor extract from two full e-cigarette cartridges was 1.1 mM, roughly equivalent to 0.9 mM for 20 tobacco cigarettes (SI Table S1).
The arrays also revealed genotoxic potential of water samples (Figure 4). ECL responses from the untreated wastewater were about 9 times larger than those for fully recovered water, suggesting significant presence of genotoxic chemicals. Successful analysis of samples during midtreatment suggests that the array can be used to monitor the success of intermediate stages of water treatment. Expression of these results in terms of water polluting chemicals that cause metabolite-related DNA damage again provide a rapid assessment of relative severity of the contamination (Figure 4D). Calibration range and LOD of 3 μM for standard water pollutants (Figure 4B) suggests applications in rapid identification of seriously polluted water. Array results for the untreated water samples are consistent with reports of genotoxic chemicals in domestic and industrial wastewater.43 ECL responses from fully reclaimed water did not show significant genotoxic potential when compared to controls, suggesting significant removal of genotoxic chemicals.
In summary, we described here a new, portable, low cost, automated, toxicity screening tool to detect metabolite-related genotoxicity chemistry from environmental samples. The 3-D printed array is fast and accurate in sensing effects of possible genotoxic chemicals. A unique feature is that test chemicals are converted to their metabolites so that metabolite reactivity toward genetic material can be measured rapidly and efficiently. This is a major attribute for assessment of possible genotoxic consequences of pollutant exposure from relevant samples.
Supplementary Material
Acknowledgments
The authors thank the National Institute of Environmental Health Sciences (NIEHS), NIH, Grant No. ES03154 for financial support. We thank Islam M. Mosa for SEM images.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.7b00118.
Sources of chemicals and reagents, 3-D print specifications, SEM images of PG sheets and microwells, schematic program upload and detailed instructions for PCB integration, artificial inhalation system for smoke/vapor extraction, reproducibility results, and table of genotoxic potential related standards (PDF)
ORCID
James F. Rusling: 0000-0002-6117-3306
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- 1.(a) Caldwell JC. DEHP: Genotoxicity and potential carcinogenic mechanisms–A review. Mutat Res, Rev Mutat Res. 2012;751:82–157. doi: 10.1016/j.mrrev.2012.03.001. [DOI] [PubMed] [Google Scholar]; (b) Chen RJ, Chang LW, Lin P, Wang YJ. Epigenetic effects and molecular mechanisms of tumorigenesis induced by cigarette smoke: an overview. J Oncol. 2011;2011:654931. doi: 10.1155/2011/654931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Hvastkovs EG, Schenkman JB, Rusling JF. Metabolic toxicity screening using electrochemiluminescence arrays coupled with enzyme-DNA biocolloid reactors and liquid chromatography-mass spectrometry. Annu Rev Anal Chem. 2012;5:79–105. doi: 10.1146/annurev.anchem.111808.073659. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hvastkovs EG, Rusling JF. State-of-the-Art Metabolic Toxicity Screening and Pathway Evaluation. Anal Chem. 2016;88:4584–4599. doi: 10.1021/acs.analchem.5b04772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Rogers K. Recent advances in biosensor techniques for environmental monitoring. Anal Chim Acta. 2006;568:222–231. doi: 10.1016/j.aca.2005.12.067. [DOI] [PubMed] [Google Scholar]; (b) Long F, Zhu A, Shi H. Recent advances in optical biosensors for environmental monitoring and early warning. Sensors. 2013;13:13928–13948. doi: 10.3390/s131013928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani V, Kadimisetty K, Malla S, Joshi AA, Rusling JF. Paper-based electrochemiluminescent screening for genotoxic activity in the environment. Environ Sci Technol. 2013;47:1937–1944. doi: 10.1021/es304426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM. Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Anal Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]; (b) Bishop GW, Satterwhite-Warden JE, Kadimisetty K, Rusling JF. 3D-printed bioanalytical devices. Nanotechnology. 2016;27:284002. doi: 10.1088/0957-4484/27/28/284002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Lee W, Kwon D, Choi W, Jung GY, Au AK, Folch A, Jeon S. 3D-printed microfluidic device for the detection of pathogenic bacteria using size-based separation in helical channel with trapezoid cross-section. Sci Rep. 2015;5:7717. doi: 10.1038/srep07717. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Coskun AF, Nagi R, Sadeghi K, Phillips S, Ozcan A. Albumin testing in urine using a smart-phone. Lab Chip. 2013;13:4231–4238. doi: 10.1039/c3lc50785h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wei Q, Luo W, Chiang S, Kappel T, Mejia C, Tseng D, Chan RYL, Yan E, Qi H, Shabbir F, et al. Imaging and sizing of single DNA molecules on a mobile phone. ACS Nano. 2014;8:12725–12733. doi: 10.1021/nn505821y. [DOI] [PubMed] [Google Scholar]; (d) Coskun AF, Wong J, Khodadadi D, Nagi R, Tey A, Ozcan A. A personalized food allergen testing platform on a cellphone. Lab Chip. 2013;13:636–640. doi: 10.1039/c2lc41152k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wei Q, Nagi R, Sadeghi K, Feng S, Yan E, Ki SJ, Caire R, Tseng D, Ozcan A. Detection and spatial mapping of mercury contamination in water samples using a smartphone. ACS Nano. 2014;8:1121–1129. doi: 10.1021/nn406571t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Kitson PJ, Rosnes MH, Sans V, Dragone V, Cronin L. Configurable 3D-Printed millifluidic and microfluidic ‘lab on a chip’reactionware devices. Lab Chip. 2012;12:3267–3271. doi: 10.1039/c2lc40761b. [DOI] [PubMed] [Google Scholar]; (g) Kitson PJ, Symes MD, Dragone V, Cronin L. Combining 3D printing and liquid handling to produce user-friendly reactionware for chemical synthesis and purification. Chemical Science. 2013;4:3099–3103. [Google Scholar]; (h) Dragone V, Sans V, Rosnes MH, Kitson PJ, Cronin L. 3D-printed devices for continuous-flow organic chemistry. Beilstein J Org Chem. 2013;9:951–959. doi: 10.3762/bjoc.9.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Bhargava KC, Thompson B, Malmstadt N. Discrete elements for 3D microfluidics. Proc Natl Acad Sci USA. 2014;111:15013–15018. doi: 10.1073/pnas.1414764111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Snowden ME, King PH, Covington JA, Macpherson JV, Unwin PR. Fabrication of versatile channel flow cells for quantitative electroanalysis using prototyping. Anal Chem. 2010;82:3124–3131. doi: 10.1021/ac100345v. [DOI] [PubMed] [Google Scholar]; (c) Erkal JL, Selimovic A, Gross BC, Lockwood SY, Walton EL, McNamara S, Martin RS, Spence DM. 3D printed microfluidic devices with integrated versatile and reusable electrodes. Lab Chip. 2014;14:2023–2032. doi: 10.1039/c4lc00171k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Saggiomo V, Velders AH. Simple 3D printed scaffold-removal method for the fabrication of intricate microfluidic devices. Advanced Science. 2015;2:1500125. doi: 10.1002/advs.201500125. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lee KG, Park KJ, Seok S, Shin S, Park JY, Heo YS, Lee SJ, Lee TJ. 3D printed modules for integrated microfluidic devices. RSC Adv. 2014;4:32876–32880. [Google Scholar]
- 8.(a) Bishop GW, Satterwhite JE, Bhakta S, Kadimisetty K, Gillette KM, Chen E, Rusling JF. 3D-printed fluidic devices for nanoparticle preparation and flow-injection amperometry using integrated prussian blue nanoparticle-modified electrodes. Anal Chem. 2015;87:5437–5443. doi: 10.1021/acs.analchem.5b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bishop GW, Satterwhite-Warden JE, Bist I, Chen E, Rusling JF. Electrochemiluminescence at bare and DNA-coated graphite electrodes in 3D-printed fluidic devices. ACS Sens. 2016;1:197–202. doi: 10.1021/acssensors.5b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadimisetty K, Mosa IM, Malla S, Satterwhite-Warden JE, Kuhns TM, Faria RC, Lee NH, Rusling JF. 3D-printed supercapacitor-powered electrochemiluminescent protein immunoarray. Biosens Bioelectron. 2016;77:188–193. doi: 10.1016/j.bios.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thun MJ, Carter BD, Feskanich D, Freedman ND, Prentice R, Lopez AD, Hartge P, Gapstur SM. 50-year trends in smoking-related mortality in the United States. N Engl J Med. 2013;368:351–364. doi: 10.1056/NEJMsa1211127. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Alexandrov LB, Ju YS, Haase K, Van LooP, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. Mutational signatures associated with tobacco smoking in human cancer. bioRxiv. 2016:051417. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 12.(a) Singh T, Arrazola RA, Corey CG, et al. Tobacco use among middle and high school students–United States, 2011–2015. MMWR Morbidity and mortality weekly report. 2016;65:361–367. doi: 10.15585/mmwr.mm6514a1. [DOI] [PubMed] [Google Scholar]; (b) McMillen RC, Gottlieb MA, Shaefer RM, Winickoff JP, Klein JD. Trends in Electronic Cigarette Use Among U.S. Adults: Use is Increasing in Both Smokers and Nonsmokers. Nicotine Tob Res. 2015;17:1195–1202. doi: 10.1093/ntr/ntu213. [DOI] [PubMed] [Google Scholar]
- 13.(a) Grana R, Benowitz N, Glantz SA. E-cigarettes: a scientific review. Circulation. 2014;129:1972–1986. doi: 10.1161/CIRCULATIONAHA.114.007667. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Trtchounian A, Williams M, Talbot P. Conventional and electronic cigarettes (e-cigarettes) have different smoking characteristics. Nicotine Tob Res. 2010;12:905–912. doi: 10.1093/ntr/ntq114. [DOI] [PubMed] [Google Scholar]
- 14.Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, von Gunten U, Wehrli B. The challenge of micropollutants in aquatic systems. Science. 2006;313:1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- 15.Schwarzenbach RP, Egli T, Hofstetter TB, von Gunten U, Wehrli B. Global water pollution and human health. Annual Review of Environment and Resources. 2010;35:109–136. [Google Scholar]
- 16.(a) Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]; (b) Filippelli GM. The global phosphorus cycle: past, present, and future. Elements. 2008;4:89–95. [Google Scholar]; (c) Jorgenson AK. Political-economic Integration, Industrial Pollution and Human Health: A Panel Study of Less-Developed Countries, 1980–2000. International Sociology. 2009;24:115–143. [Google Scholar]; (d) Watson SB. Aquatic taste and odor: a primary signal of drinking-water integrity. J Toxicol Environ Health, Part A. 2004;67:1779–1795. doi: 10.1080/15287390490492377. [DOI] [PubMed] [Google Scholar]
- 17.Ohe T, Watanabe T, Wakabayashi K. Mutagens in surface waters: a review. Mutat Res, Rev Mutat Res. 2004;567:109–149. doi: 10.1016/j.mrrev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.University of Connecticut (UCONN) water treatment facility. http://ecohusky.uconn.edu/water/rwf.html; last accessed 13 Feb 2017.
- 19.Dennany L, Forster RJ, Rusling JF. Simultaneous direct electrochemiluminescence and catalytic voltammetry detection of DNA in ultrathin films. J Am Chem Soc. 2003;125:5213–5218. doi: 10.1021/ja0296529. [DOI] [PubMed] [Google Scholar]
- 20.http://web2.uconn.edu/rusling/3D_printing.html last accessed 13 Feb 2017.
- 21.Tang CK, Vaze A, Rusling JF. Fabrication of immunosensor microwell arrays from gold compact discs for detection of cancer biomarker proteins. Lab Chip. 2012;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusling JF, Wasalathanthri DP, Schenkman JB. Thin multicomponent films for functional enzyme devices and bioreactor particles. Soft Matter. 2014;10:8145–8156. doi: 10.1039/c4sm01679c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.http://www.atmel.com/devices/attiny85.aspx; last accessed 13 Feb 2017
- 24.Krishnan S, Hvastkovs EG, Bajrami B, Choudhary D, Schenkman JB, Rusling JF. Synergistic metabolic toxicity screening using microsome/DNA electrochemiluminescent arrays and nanoreactors. Anal Chem. 2008;80:5279–5285. doi: 10.1021/ac800763r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Krishnan S, Wasalathanthri D, Zhao L, Schenkman JB, Rusling JF. Efficient bioelectronic actuation of the natural catalytic pathway of human metabolic cytochrome P450s. J Am Chem Soc. 2011;133:1459–1465. doi: 10.1021/ja108637s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Screening reactive metabolites bioactivated by multiple enzyme pathways using a multiplexed microfluidic system. Analyst. 2013;138:171–178. doi: 10.1039/c2an35993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Etter J, Bullen C. A longitudinal study of electronic cigarette users. Addict Behav. 2014;39:491–494. doi: 10.1016/j.addbeh.2013.10.028. [DOI] [PubMed] [Google Scholar]; (b) Etter J, Bullen C. Electronic cigarette: users profile, utilization, satisfaction and perceived efficacy. Addiction. 2011;106:2017–2028. doi: 10.1111/j.1360-0443.2011.03505.x. [DOI] [PubMed] [Google Scholar]; (c) Etter J. Electronic cigarettes: a survey of users. BMC Public Health. 2010;10:231. doi: 10.1186/1471-2458-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shihadeh A, Saleh R. Polycyclic aromatic hydrocarbons, carbon monoxide,“tar”, and nicotine in the mainstream smoke aerosol of the narghile water pipe. Food Chem Toxicol. 2005;43:655–661. doi: 10.1016/j.fct.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 28.(a) Grana RA, Ling PM. Smoking revolution”: a content analysis of electronic cigarette retail websites. Am J Prev Med. 2014;46:395–403. doi: 10.1016/j.amepre.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goniewicz ML, Kuma T, Gawron M, Knysak J, Kosmider L. Nicotine levels in electronic cigarettes. Nicotine Tob Res. 2013;15:158–166. doi: 10.1093/ntr/nts103. [DOI] [PubMed] [Google Scholar]
- 29.López-Barea J, Pueyo C. Mutagen content and metabolic activation of promutagens by molluscs as biomarkers of marine pollution. Mutat Res, Fundam Mol Mech Mutagen. 1998;399:3–15. doi: 10.1016/s0027-5107(97)00262-5. [DOI] [PubMed] [Google Scholar]
- 30.Le Curieux F, Marzin D, Erb F. Comparison of three short-term assays: results on seven chemicals: Potential contribution to the control of water genotoxicity. Mutat Res, Genet Toxicol Test. 1993;319:223–236. doi: 10.1016/0165-1218(93)90082-o. [DOI] [PubMed] [Google Scholar]
- 31.Žegura B, Heath E, Černoša A, Filipič M. Combination of in vitro bioassays for the determination of cytotoxic and genotoxic potential of wastewater, surface water and drinking water samples. Chemosphere. 2009;75:1453–1460. doi: 10.1016/j.chemosphere.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 32.Pan S, Zhao L, Schenkman JB, Rusling JF. Evaluation of electrochemiluminescent metabolic toxicity screening arrays using a multiple compound set. Anal Chem. 2011;83:2754–2760. doi: 10.1021/ac200050n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(a) Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]; (b) Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Cogliano V. Carcinogenicity of polycyclic aromatic hydrocarbons. Lancet Oncol. 2005;6:931. doi: 10.1016/s1470-2045(05)70458-7. [DOI] [PubMed] [Google Scholar]
- 34.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 35.Wasalathanthri DP, Li D, Song D, Zheng Z, Choudhary D, Jansson I, Lu X, Schenkman JB, Rusling JF. Elucidating organ-specific metabolic toxicity chemistry from electrochemiluminescent enzyme/DNA arrays and bioreactor bead-LC-MS/MS. Chemical Science. 2015;6:2457–2468. doi: 10.1039/c4sc03401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warth B, Sulyok M, Krska R. LC-MS/MS-based multi-biomarker approaches for the assessment of human exposure to mycotoxins. Anal Bioanal Chem. 2013;405:5687–5695. doi: 10.1007/s00216-013-7011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW. Methods of DNA adduct determination and their application to testing compounds for genotoxicity. Environ Mol Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 38.Yu MY, Skipper PL, Tannenbaum SR, Chan KK, Ross RK. Arylamine exposures and bladder cancer risk. Mutat Res, Fundam Mol Mech Mutagen. 2002;506:21–28. doi: 10.1016/s0027-5107(02)00148-3. [DOI] [PubMed] [Google Scholar]
- 39.Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Screening reactive metabolites bioactivated by multiple enzyme pathways using a multiplexed microfluidic system. Analyst. 2013;138:171–178. doi: 10.1039/c2an35993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arlt VM, Schmeiser HH, Osborne MR, Kawanishi M, Kanno T, Yagi T, Phillips DH, Takamura-Enya T. Identification of three major DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in rat lung at the C8 and N2 position of guanine and at the N6 position of adenine. Int J Cancer. 2006;118:2139–2146. doi: 10.1002/ijc.21622. [DOI] [PubMed] [Google Scholar]
- 41.Yu V, Rahimy M, Korrapati A, Xuan Y, Zou AE, Krishnan AR, Tsui T, Aguilera JA, Advani S, Alexander LEC, et al. Electronic cigarettes induce DNA strand breaks and cell death independently of nicotine in cell lines. Oral Oncol. 2016;52:58–65. doi: 10.1016/j.oraloncology.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(a) Wei B, Blount BC, Xia B, Wang L. Assessing exposure to tobacco-specific carcinogen NNK using its urinary metabolite NNAL measured in US population: 2011–2012. J Exposure Sci Environ Epidemiol. 2016;26:249–256. doi: 10.1038/jes.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhong Y, Carmella SG, Upadhyaya P, Hochalter JB, Rauch D, Oliver A, Jensen J, Hatsukami D, Wang J, Zimmerman C, Hecht SS. Immediate consequences of cigarette smoking: rapid formation of polycyclic aromatic hydrocarbon diol epoxides. Chem Res Toxicol. 2011;24:246–252. doi: 10.1021/tx100345x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.(a) White PA, Rasmussen JB. The genotoxic hazards of domestic wastes in surface waters. Mutat Res, Rev Mutat Res. 1998;410:223–236. doi: 10.1016/s1383-5742(98)00002-7. [DOI] [PubMed] [Google Scholar]; (b) Thewes MR, Endres Junior D, Droste A. Genotoxicity biomonitoring of sewage in two municipal wastewater treatment plants using the Tradescantia pallida var. purpurea bioassay. Genet Mol Biol. 2011;34:689–693. doi: 10.1590/S1415-47572011005000055. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Žegura B, Heath E, Černoša A, Filipič M. Combination of in vitro bioassays for the determination of cytotoxic and genotoxic potential of wastewater, surface water and drinking water samples. Chemosphere. 2009;75:1453–1460. doi: 10.1016/j.chemosphere.2009.02.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


