SUMMARY
The human gastrointestinal tract is populated by a diverse, highly mutualistic microbial flora, which is known as the microbiome. Disruptions to the microbiome have been shown to be associated with severe pathologies of the host including metabolic disease, cancer and inflammatory bowel disease. Mood and behavior are also susceptible to alterations in the gut microbiota. A particularly striking example of the symbiotic effects of the microbiome is the immune system, whose cells depend critically on a diverse array of microbial metabolites for normal development and behavior. This includes metabolites that are produced by bacteria from dietary components; metabolites that are produced by the host and biochemically modified by gut bacteria; and metabolites that are synthesized de novo by gut microbes. In this review, we highlight the role of the intestinal microbiome in human metabolic and inflammatory diseases and focus in particular on the molecular mechanisms that govern the gut-immune axis.
Keywords: Microbiome, commensals, atherosclerosis, metabolic syndrome, inflammatory bowel disease, short-chain fatty acids, indole, polysaccharide A, polyamine, obesity
A Brief History of Microbiome Research
Scientific investigation of the non-pathogenic bacteria that populate the mammalian gastrointestinal tract, collectively referred to as the gut microbiome1, has a long history. Well over a century ago, studies demonstrated that dietary changes can alter the type of metabolites produced by bacteria living in the healthy intestine (Hirschler, 1886), and that the composition of the intestinal microbiome itself depends on the composition of the consumed diet (Cannon, 1921; Herter and Kendall, 1910). The seminal notion that some non-pathogenic gut bacteria are more beneficial to human health than others was proposed as early as 1908, when Élie Metchnikoff suggested that a targeted modification of the microbiome may prolong life and guard against senility (Metchnikoff, 1908). His recommendation to regularly consume milk artificially soured by the addition of the “Bulgarian bacillus” (Lactobacillus delbrueckii subsp. bulgaricus), or even pure cultures of that microbe, to stave off the frailties of old age spawned what may very well have been the first “probiotics” craze (despite a distinct lack of corroborating evidence) (Bested et al., 2013; Underhill et al., 2016); but scientific interest in the microbiome was soon overshadowed by the advent of powerful antibiotics and their revolutionary success in the treatment of bacterial infections. Over the past two decades, however, microbiome research has experienced a remarkable renaissance, aided by technological advances that enabled more rigorous experimental designs and the identification of previously unknown signaling pathways.
Recent estimates suggest that the body of an average human male is colonized by around 3.8·1013 individual microbes, the vast majority of which are prokaryotes and reside in the colon. This slightly exceeds the number of human cells that constitute the host, which are estimated to number 3.0·1013 to 3.7·1013 cells for males. For human females, bacteria are thought to outnumber host cells by a factor of two, with 4.4·1013 bacteria to 2.1·1013 host cells (Bianconi et al., 2013; Sender et al., 2016a; 2016b). The genetic diversity of the microbiome is even more staggering. Metagenomic sequencing of fecal samples has identified 3.3·106 non-redundant microbial genes from up to 1,150 different species, outnumbering human protein-coding genes by ca. 150-fold. Each individual human is estimated to host at least 160 different species (Qin et al., 2010). Interestingly, this extensive diversity at the species level is in stark contrast to the limited diversity at the phylum level: the gut microbiome is dominated by members of only two phyla, the Firmicutes (no true outer membrane, mostly Gram-positive) and the Bacteroidetes (outer membrane, Gram-negative). Members of the phylum Proteobacteria are also represented frequently but far less prominently (Eckburg et al., 2005; Hold et al., 2002; Human Microbiome Project Consortium, 2012). Even though the bacteria of the microbiome are still frequently described as commensal, i.e. neither harmful nor beneficial, their relationship with the human host is often highly mutualistic: at the most basic level, the host provides the bacteria with easy access to food and shelter, while the bacteria aid the host in digesting complex food and synthesize essential metabolites, such as Vitamins B and K (Cummings and Macfarlane, 1997). In fact, it has been estimated that 10% of metabolites found in mammalian blood are derived from the gut microbiota (Wikoff et al., 2009). In a very real sense, humans and their microbiome thus form a composite organism, a so-called holobiont. Over the past two decades, it has become clear that this relationship is far more complex than originally thought. Perturbation of the microbiome, referred to as dysbiosis, can affect a remarkable array of different host systems, particularly metabolic and immune processes. This phenomenon blurs the lines between mutualists, commensals and pathogens, as bacteria that are harmless or even beneficial in the context of a healthy microbiome can promote chronic pathologies, such as atherosclerosis and obesity, when their surrounding ecosystem is altered. Conversely, the absence of a seemingly insignificant “commensal” can have deleterious effects on the development of immune cells. In the following sections, we discuss some of the most exciting discoveries connecting the microbiome to human metabolic diseases and describe how bacterial metabolites shape the immune system.
The Role of the Microbiome in Metabolic Diseases
Obesity
Obesity is one of the main risk factors that constitute the Metabolic Syndrome, which predisposes patients to cardiovascular disease and type 2 diabetes (Lam and LeRoith, 2015). Unexpectedly, the gut microbiome can be an active driver of obesity, either in combination with the interrelated factors of diet and genetic predisposition, or by itself (Cox and Blaser, 2013; Shapiro et al., 2017). The composition of the microbiome differs between obese and lean individuals in humans and in animal models: obesity is associated with a relative increase in Firmicutes at the expense of Bacteroidetes (Ley et al., 2005; 2006). Crucially, the transfer of the microbiota from obese mice to germ-free animals results in a significantly larger weight increase than an equivalent transfer from lean mice, clearly demonstrating that body weight is in part controlled by the microbiome (Turnbaugh et al., 2008; 2006). This observation also holds true when the microbiota from obese or lean humans is transferred to germ-free mice (Ridaura et al., 2013; Tremaroli et al., 2015). These findings have prompted proposals to develop fecal matter transplants (FMTs) as a novel therapeutic approach to treat obesity and other disorders associated with the metabolic syndrome (Marotz and Zarrinpar, 2016). A technique that dates back to the 4th century CE, FMTs have most recently been used as experimental treatments for recurrent infection with Clostridium difficile, with promising results (Rao and Safdar, 2016; van Nood et al., 2013; F. Zhang et al., 2012). Bacterial strains transferred in FMTs have been shown to persist in the recipient colon for at least three months, raising the possibility of long-term health benefits (S. S. Li et al., 2016). Interestingly, a small study of FMTs from lean, healthy donors to obese recipients with type 2 diabetes found no reduction in weight but a significant increase in insulin sensitivity in the recipient population six weeks after transplantation (Vrieze et al., 2012). Large-scale, long-term clinical studies will be necessary to determine whether FMTs can ameliorate obesity and/or other disorders associated with the Metabolic Syndrome.
A comprehensive model for the influence of the microbiome on the host’s body weight has been developed (Sonnenburg and Bäckhed, 2016): the microbiota extracts energy from dietary components that are not digested by the host and converts that energy to nutrients the host can absorb, thereby altering the overall energy balance of the host. Bacterial species able to most efficiently utilize the components of a given diet gain a growth advantage and displace other species, explaining the rapid changes in the composition of the microbiome upon dietary modifications described a century ago and studied in great detail more recently (Cannon, 1921; Cotillard et al., 2013; David et al., 2014; Herter and Kendall, 1910; Koeth et al., 2013; Walker et al., 2011). An obesity-inducing, Western-style diet, which is typically low in fiber and high in caloric content from sucrose and unsaturated fat, thus selects for a less diverse microbiota that efficiently extracts energy from these nutrients (mostly members of the Firmicutes). Conversely, a plant-based diet promotes the enrichment of microbial species that efficiently ferment fiber (Bacteroidetes) (De Filippo et al., 2010; Wu et al., 2011). When a germ-free mouse receives the microbiota of an obese individual, the ability to effectively utilize high-caloric nutrients is transferred along with it, resulting in obesity of the recipient.
Consistent with this model of microbial selection based on biochemical capacity, the representation of biochemical pathways in the collective genome of the gut microbiome is remarkably similar between healthy individuals, whereas the composition of the microbiome on a species level varies significantly. Obese individuals share a functionally similar gut microbiome between them, with reduced but still considerable species diversity (Human Microbiome Project Consortium, 2012; Kurokawa et al., 2007; Qin et al., 2010; Turnbaugh et al., 2009). While studies have mapped in impressive detail the shifts in the genetic repertoire and the species composition of the microbiome that are associated with obesity and other disorders, surprisingly little is known about which species drive the functional changes. A recent study by Manor and Borenstein described a bioinformatic framework, termed FishTaco (for functional shifts’ taxonomic contributors), to integrate taxonomic and functional information and determine contributions of individual species to functional shifts within the microbiome (Manor and Borenstein, 2017). This area of research is likely to yield further fascinating insights in the near future and enhance our understanding of the microbiome significantly. Another interesting question to be explored is which endogenous host factors affect the composition of the microbiome, in addition to the established environmental factors. Host genetic variation has been shown to be associated with variation in the microbiome composition, but genome-wide approaches to identify individual genes and their role in shaping the microbiome are only beginning to emerge (Blekhman et al., 2015; Spor et al., 2011).
In addition to changes in energy metabolism, obesity has also been linked to a deterioration of the intestinal mucosa and resultant chronic, systemic inflammation, analogous to type 2 diabetes (see below) (Boutagy et al., 2016). This is consistent with the notion that the intestinal microbiome is an important regulator of mucosal barrier function, which will be discussed in further detail in subsequent sections. The gut bacterium Akkermansia muciniphila is an excellent example of such a regulator. Abundance of A. muciniphila is decreased in obese (and diabetic) mice, while reintroduction of the bacterium by oral gavage improves epithelial barrier function and reduces diet-induced obesity (Everard et al., 2013). Recently, Plovier et al. reported that much of this effect could be replicated by administration of a membrane protein purified from A. muciniphila, suggesting the appealing possibility of a novel therapeutic application (Plovier et al., 2017).
Type 2 Diabetes
In addition to obesity, the gut microbiome has also been implicated directly in the development of the two primary complications of Metabolic Syndrome, type 2 diabetes (T2D) and cardiovascular disease (Fändriks, 2016; Jonsson and Bäckhed, 2017; Khan et al., 2014). T2D is associated with inflammation of the insulin-producing β cells of the pancreatic islets and the white adipose tissue itself, which results in decreased insulin production and increased insulin resistance, respectively (comprehensively reviewed by (Donath et al., 2013)). The microbiome of patients suffering from T2D, which is also markedly different from healthy individuals, is believed to promote these inflammatory events by two distinct mechanisms (Karlsson et al., 2013; Qin et al., 2012). First, the gut microbiota associated with T2D produces lower levels of short-chain fatty acids (SCFAs, discussed in detail below), which have been shown in vitro and in vivo to support the integrity of the intestinal mucosa in a variety of ways (Fukuda et al., 2011; Gaudier et al., 2004; Macia et al., 2015; Willemsen et al., 2003; Wrzosek et al., 2013). In the absence of adequate SCFA production, the permeability of the intestinal mucosa increases, allowing gut bacteria to cross and enter the bloodstream. Pathogen-associated molecular patterns (PAMPs) of invading bacteria, such as the cell-wall component lipopolysaccharide (LPS), bind to pattern recognition receptors (PRRs) expressed by immune and adipose cells and elicit pro-inflammatory cytokine production, resulting in a generalized state of chronic, low-level inflammation. Second, microbiota-derived LPS has been suggested to traverse the mucosal barrier of the intestine on chylomicrons, lipoprotein complexes that transport long-chain fatty acids from the gut lumen to the lymph and blood (Ghoshal et al., 2009; Khan et al., 2014; Sonnenburg and Bäckhed, 2016). Correspondingly, patients suffering from T2D have been reported to exhibit increased plasma levels of LPS (Creely et al., 2007). Interestingly, mice chronically exposed to low levels of LPS for four weeks gained weight and became insulin-resistant (Cani et al., 2007).
Atherosclerosis
The primary cause of cardiovascular disease is atherosclerosis, a process in which the microbiome has been implicated as well (Jonsson and Bäckhed, 2017; Sharon et al., 2014). Atherosclerotic plaques form as a result of excessive deposition of cholesterol, phospholipids and triglycerides associated with low-density lipoprotein (LDL) in the intima of arteries, where they are subject to oxidation and enzymatic modification (oxLDL). OxLDL has pro-inflammatory properties and elicits the recruitment of monocytes from the blood into the subendothelium, where they differentiate into macrophages and internalize large quantities of the lipids deposited in the intima until they eventually turn into dysfunctional foam cells laden with lipids. As foam cells, they secrete additional pro-inflammatory cytokines and thus attract more monocytes to the site of the incipient plaque, creating a positive feedback loop. Ultimately, the foam cells perish by apoptosis or necrosis, leaving behind a necrotic core in the plaque. This destabilizes the entire plaque, facilitating rupture and subsequent formation of an embolus that may result in myocardial infarction, stroke or other forms of cardiovascular disease (Brophy et al., 2017; Frostegård, 2013; Moore et al., 2013). Therefore, atherosclerosis is at least in part a disease of chronic inflammation, not unlike T2D. It is hence not surprising that the same systemic, pro-inflammatory events that promote T2D also promote atherosclerosis (see above). Beyond these general mechanisms, the gut microbiome affects atherogenesis more specifically through its degradation of phosphatidylcholine, which is abundant in eggs, cheese, seafood and red meat (Jonsson and Bäckhed, 2017; Sharon et al., 2014; Sonnenburg and Bäckhed, 2016). Bacteria of the gut microbiota process dietary phosphatidylcholine to yield trimethylamine (TMA), which is absorbed by the host and oxidized in the liver to trimethylamine N-oxide (TMAO). A seminal study by Hazen and colleagues discovered that plasma levels of TMAO and its precursors choline and betaine predicted the risk of cardiovascular disease in humans, thus identifying a novel connection between the microbiome and metabolic health. Consistent with this finding, supplementing food with choline or TMAO promoted atherosclerosis in mice. Elimination of the gut microbiota by treatment with antibiotics negated the choline-induced exacerbation of atherosclerosis, confirming the importance of the gut microbiome in this process (Z. Wang et al., 2011). These observations were further corroborated and extended by subsequent studies from the same group and others (X. S. Li et al., 2017; Mente et al., 2015; Tang et al., 2014; 2013; 2015; Trøseid et al., 2015; Z. Wang et al., 2014). Hazen and colleagues separately investigated l-carnitine, an amino acid found in red meat that is also converted to TMAO after processing by gut bacteria. They found that l-carnitine and its metabolite γ-butyrobetaine also promoted atherosclerosis in mice, and that elevated plasma levels of l-carnitine were associated with increased risk of cardiovascular disease in individuals with elevated TMAO plasma levels (Koeth et al., 2013; 2014). To identify specific members of the microbiome that produce TMA, Romano et al. recently screened 79 bacterial strains known to inhabit the human intestine and found nine (11.4%) that were able to produce TMA from dietary choline, including three different Clostridium species. Interestingly, the same strains did not produce TMA from l-carnitine under the same experimental conditions (Romano et al., 2015). The importance of the gut microbiota in atherosclerosis was further underscored by a study demonstrating that susceptibility to atherosclerosis in a mouse model can be transmitted via FMTs. Induction of atherosclerotic plaques was increased when Apoe−/− mice had received FMTs from a mouse strain that produced high levels of TMAO and was prone to atherosclerosis, compared to FMTs from a mouse strain that produced low levels of TMAO and was less susceptible to atherosclerosis (Gregory et al., 2015). The role of TMAO and its precursors in atherogenesis is not entirely straightforward, however. Many fish species contain large quantities of TMAO, yet fish-based diets have long been associated with reduced risk of cardiovascular disease. The cardioprotective effects of consuming fish are generally believed to be mediated by ω-3 fatty acids, which may outweigh any adverse effects of the simultaneously ingested TMAO (Maehre et al., 2015; Ussher et al., 2013). Dietary supplementation with l-carnitine has also been associated with a significant decrease in the risk of cardiovascular disease, in apparent contradiction to the reported association of elevated plasma l-carnitine with increased risk (DiNicolantonio et al., 2013; Ussher et al., 2013). More targeted, large-scale studies will be necessary to better understand the direct effects of dietary TMAO and l-carnitine on cardiovascular disease. Moreover, the molecular mechanism(s) underlying any effect of TMAO have remained elusive. A study by Collins et al. found that even excessive concentrations of TMAO did not affect uptake or efflux of cholesterol by macrophages in vitro, even though an up-regulation of the scavenger receptors CD36 and SR-A1 on macrophages from TMAO-fed mice had been described previously (Collins et al., 2016; Z. Wang et al., 2011). TMAO has been reported to inhibit reverse cholesterol transport, but details remain poorly understood (Koeth et al., 2013). Interestingly, a recent study found that TMAO induced the transcription of several genes encoding pro-inflammatory, atherosclerosis-promoting proteins in endothelial and smooth-muscle cells (but not peritoneal macrophages) through an unknown mechanism (Seldin et al., 2016). Beyond a direct effect on atherosclerosis, TMAO has also been reported to increase platelet responsiveness by enhancing intracellular Ca2+ release, thus increasing the likelihood of thrombotic events and thereby cardiovascular disease (W. Zhu et al., 2016). The precise mechanisms by which TMAO affects atherosclerosis remain unresolved and their continued study promises to offer fascinating insights into the relationship between microbiome and cardiovascular health.
Microbiome-Associated Metabolites that Shape the Immune System
One of the most surprising findings of recent microbiome research is the observation that the gut microbiota does not simply evade the immune system to persist in the intestinal tract. Instead, a complex interplay between the local microbiome, the intestinal epithelium and resident immune cells has begun to emerge, in which all participants actively foster gastrointestinal homeostasis. In this system, bacterially derived metabolites serve as important signals that continuously contribute to the proper function of the epithelial barrier and immune cells. Our understanding of the scope of these interactions is still in its infancy: most of the bacterial metabolome of the intestine remains unidentified, and many known metabolites have yet to be functionally characterized. For instance, of the 179 metabolites detected in the colonic lumen of mice by a mass-spectrometric study, 48 were not present in the supplied food. Of those 48 metabolites, 35 had altered concentrations in the colon of germ-free mice compared to controls, as did 88 metabolites that were also present in the food (Matsumoto et al., 2012). For the vast majority of these, it is entirely unknown if or how such changes in concentration could affect the host’s immune system.
While the true extent of microbially derived metabolites that shape the gut-immune axis remains to be elucidated, a comparatively small but diverse range of such molecules has been identified and keenly studied over the past two decades. These can broadly be grouped into three categories: (i) metabolites that are produced by bacteria directly from dietary components; (ii) metabolites that are produced by the host and biochemically modified by gut bacteria; and (iii) metabolites that are synthesized de novo by gut microbes (Table 1). In the following sections, we discuss the most striking examples from each of these groups.
Table 1.
Immunomodulatory metabolites that are produced by intestinal bacteria.
| Metabolite | Molecular Mechanism(s) of Action | Origin | Effects on Immune System | Key References |
|---|---|---|---|---|
| Metabolites that are produced by bacteria from dietary components | ||||
| Short-chain fatty acids (primarily acetate, propionate, butyrate) |
|
Fermentation of polysaccharides by colonic microbiota. Bacteroidetes: acetate and propionate. Firmicutes: butyrate. | Generally anti-inflammatory, protect against colitis. Promote integrity and function of the intestinal epithelium. Inhibit production of pro-inflammatory cytokines by innate immune cells. Promote function of microglia. Inhibit maturation of dendritic cells. Promote antibody production by B cells. Promote de novo differentiation and expansion of Tregs. | (Arpaia et al., 2013; Brown et al., 2003; Chang et al., 2014; Donohoe et al., 2012; Kalina et al., 2002; M. Kim et al., 2016; Le Poul et al., 2003; Macia et al., 2015; Maslowski et al., 2009; Millard et al., 2002; Nilsson et al., 2003; Singh et al., 2010; Smith et al., 2013; Thangaraju et al., 2009b; 2009a) |
| Indole derivatives | Activation of AhR and NR1I2. | Derived from dietary tryptophan by different intestinal bacteria. | AhR activation promotes maintenance of ILC3 cells, which strengthen integrity of intestinal mucosa by secreting IL-22. NR1I2 activation also enhances epithelial barrier function. | (Kiss et al., 2011; Lee et al., 2011; Y. Li et al., 2011; J. Qiu et al., 2012; Venkatesh et al., 2014; Zelante et al., 2013) |
| Polyamines (primarily putrescine, spermidine, spermine) | Unclear. Inhibit expression of pro-inflammatory cytokines in conjunction with AHSG. Also inhibit activation of NLRP6 inflammasome. | Derived from arginine by host and bacteria. | Enhance development and maintenance of intestinal mucosa and resident immune cells. Inhibit expression of pro-inflammatory cytokines by LPS-stimulated monocytes and macrophages. | (Dufour et al., 1988; Levy et al., 2015; Pérez-Cano et al., 2010; H. Wang et al., 1997; M. Zhang et al., 1999) |
| Metabolites that are produced by the host and biochemically modified by gut bacteria | ||||
| Secondary bile acids |
|
Derived from host-produced primary bile acids by intestinal microbiota. | Inhibit NF-κB-dependent transcription of pro-inflammatory genes in monocytes, macrophages, dendritic cells. Inhibit production of pro-inflammatory cytokine osteopontin by NKT cells. | (Cipriani et al., 2011; C. Guo et al., 2015; Kawamata et al., 2003; Mencarelli et al., 2009; Sayin et al., 2013; Vavassori et al., 2009; Y.-D. Wang et al., 2011) |
| Taurine | Enhancement of NLRP6 inflammasome activation. | Derived from host-produced primary bile salts by intestinal microbiota. | Enhances epithelial barrier function and maintenance by promoting epithelial production of IL-18. | (Levy et al., 2015) |
| Metabolites that are synthesized de novo by gut microbes | ||||
| ATP | Activation of P2X and P2Y receptors. | Actively secreted by subset of intestinal bacteria. | Limits numbers of Tfh cells in Peyer's patches, thus reducing secretion of bacteria-specific IgA by B cells across intestinal epithelium. Promotes differentiation of TH17 cells in intestinal mucosa. May promote epithelial barrier function by activating NLRP3 inflammasome and subsequent IL-18 secretion by macrophages. | (Atarashi et al., 2008; Kusu et al., 2013; Mariathasan et al., 2006; Nuttle and Dubyak, 1994; Perruzza et al., 2017; Sutterwala et al., 2006) |
| Polysaccharide A (PSA) | Activation of TLR2 on DCs and Tregs. Presentation of PSA fragments with MHC-II to CD4+ T cells. | Bacteroides fragilis (required for colonization). | Potent anti-inflammatory effects: induces secretion of IL-10 from CD4+ T cells, directly and indirectly. Skews TH1:TH2 ratio towards TH1 cells. | (Cobb et al., 2004; Dasgupta et al., 2014; Johnson et al., 2015; Mazmanian et al., 2008; Round et al., 2011) |
Metabolites that are produced by bacteria from dietary components
Short-chain fatty acids (SCFAs)
SCFAs are among the most-thoroughly investigated bacterial metabolites and comprise fatty acids with a backbone of one to six carbon atoms (Fig. 1). In general, SCFAs have anti-inflammatory properties in the intestinal mucosa (Ferreira et al., 2014). Several studies have demonstrated a protective role of SCFAs in mouse models of inflammatory bowel disease (IBD) (Furusawa et al., 2013; Maslowski et al., 2009; Smith et al., 2013). Consistent with these findings, IBD in humans is associated with a decrease of SCFA-producing microbial species in the gut, and dietary changes that promote production of SCFAs by the microbiome can ameliorate symptoms (Frank et al., 2007; Kanauchi et al., 2002; Machiels et al., 2014; Sokol et al., 2009). Direct application of the SCFA butyrate by enema has also been reported to alleviate IBD (Breuer et al., 1991; Harig et al., 1989).
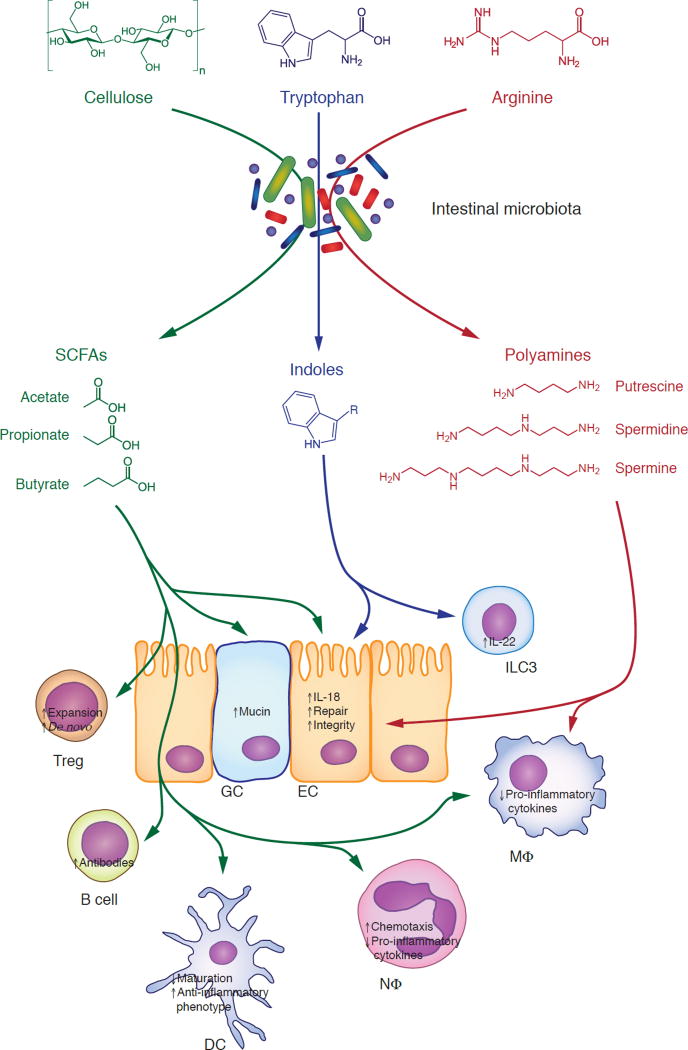
Fig. 1. Intestinal bacteria convert dietary nutrients to immunomodulatory metabolites.
Polysaccharides that cannot be digested by host enzymes, such as cellulose, are metabolized by the intestinal microbiota to short-chain fatty acids (SCFAs, green). These exert a plethora of anti-inflammatory effects, such as inducing the expansion and de novo differentiation of regulatory T cells; enhancing the barrier function of the intestinal mucosa through epithelial cells and goblet cells; facilitating production of antibodies by B cells; inhibiting the maturation of dendritic cells and promoting an anti-inflammatory phenotype; and reducing production of pro-inflammatory cytokines by innate immune cells. Dietary tryptophan is degraded to indole derivatives (blue). Indoles promote epithelial barrier function, e.g. by supporting the maintenance of type 3 innate lymphoid cells, which are the primary producers of IL-22. Dietary arginine is metabolized to polyamines (red), which promote the integrity of the intestinal epithelium and reduce the production of pro-inflammatory cytokines by macrophages. See text for details. DC, dendritic cell; EC, epithelial cell; GC, goblet cell; ILC3, type 3 innate lymphoid cell; MΦ, macrophage; NΦ, neutrophil; Treg, regulatory T cell.
Origin and distribution of short-chain fatty acids
SCFAs are produced in the large intestine by bacterial fermentation of plant-derived polysaccharides, primarily cellulose, that cannot be digested by host enzymes. Acetate (C2), propionate (C3) and butyrate (C4) are by far the most abundant of the intestinal SCFAs (Cummings et al., 1987; Morrison and Preston, 2016). Members of the Bacteroidetes provide most of the acetate and propionate whereas Firmicutes, which tend to utilize polysaccharides less efficiently, are believed to be the primary producers of butyrate (Flint et al., 2015; Kaoutari et al., 2013; Levy et al., 2016; Louis et al., 2010). While a comprehensive species inventory of SCFA producers remains elusive, studies investigating individual species have uncovered interesting microbial cross-feeding dynamics (recently reviewed by (Flint et al., 2015) and (Ríos-Covián et al., 2016)). For instance, one of the main producers of acetate, Bacteroides thetaiotaomicron, utilizes polysaccharides and produces acetate, while one of the main producers of butyrate, Faecalibacterium prausnitzii, can utilize acetate as a substrate (Duncan et al., 2004a). An in vivo study by Wrzosek et al. suggested that F. prausnitzii could indeed utilize acetate produced by B. thetaiotaomicron under physiological conditions, as gut bicolonization of rats with both bacterial species resulted in increased butyrate and decreased acetate concentrations in the cecum, compared to animals monocolonized with B. thetaiotaomicron (Wrzosek et al., 2013). Similarly, lactate, which is produced by many members of the human gut microbiome, has been shown to serve as substrate for bacterial production of propionate (e.g., Coprococcus catus) and butyrate (e.g., Eubacterium hallii and Anaerostipes caccae) (Duncan et al., 2004b; Reichardt et al., 2014).
Available SCFAs are readily taken up by the host but reach radically different levels of systemic distribution. Butyrate serves as a major energy source for colonic enterocytes and is largely metabolized within the epithelial mucosa (Clausen and Mortensen, 1995; Donohoe et al., 2012). Remaining butyrate is degraded in the liver (Bloemen et al., 2009; van der Beek et al., 2015). Propionate and acetate travel largely intact across the epithelium to the liver, where a substantial portion of propionate is degraded while acetate is released into systemic circulation (Bloemen et al., 2009; Morrison and Preston, 2016). As a result, butyrate is restricted to the enteric system, while bacterially derived acetate is disseminated throughout the circulation. Propionate exhibits an intermediate distribution.
Molecular mechanisms of SCFA signaling
SCFAs act as both extracellular and intracellular signaling molecules. Outside of the cell, they function as agonists for several G-protein-coupled receptors (GPCRs), namely the free fatty acid receptors 2 (FFAR2, also known as GPR43) and 3 (FFAR3 or GPR41) (Brown et al., 2003; Le Poul et al., 2003; Nilsson et al., 2003). Butyrate, but not acetate or propionate, can also activate the hydroxycarboxylic acid receptor 2 (HCAR2 or GPR109A) (Thangaraju et al., 2009b). FFAR2, FFAR3 and HCAR2 are expressed by a variety of cell types, including intestinal epithelial cells and immune cells (comprehensively reviewed by (Koh et al., 2016)). Of note, HCAR2 is largely absent from lymphocytes (Wise et al., 2003). A fourth SCFA-responsive GPCR, the olfactory receptor 51E2 (OR51E2, also known as Olfr78 in mice), is expressed by some cells of the intestinal epithelium but has not been connected to the immune system (Fleischer et al., 2015; Pluznick et al., 2013). FFAR2, FFAR3 and HCAR2 all signal through heterotrimeric Gi/o proteins. FFAR2 is additionally capable of utilizing Gq proteins, but physiological coupling of FFAR2 and Gq has only been observed in enteroendocrine L cells (a subset of which, interestingly, expresses OR51E2) (Brown et al., 2003; Fleischer et al., 2015; Le Poul et al., 2003; Nilsson et al., 2003; Y. Shi et al., 2017; Tolhurst et al., 2012; Tunaru et al., 2003; Wise et al., 2003). SCFA stimulation of GPCRs activates the mitogen-activated protein kinases (MAPKs) extracellular-signal-regulated kinase (ERK) 1 and 2, and to a lesser extent c-Jun N-terminal kinase (JNK) and p38/MAPK (Seljeset and Siehler, 2012). In addition to acting as ligands on the cell surface, SCFAs can enter cells via active transport by the sodium-coupled monocarboxylate transporter 1 (SLC5A8) (Miyauchi et al., 2004; Thangaraju et al., 2008). Once inside the cell, butyrate broadly affects transcriptional regulation by directly inhibiting nuclear class I histone deacetylases (HDACs) and activating histone acetyltransferases (HATs) (Donohoe et al., 2012; Koh et al., 2016; Steliou et al., 2012). Specifically, butyrate has been reported to inhibit HDAC1 and HDAC3 (Thangaraju et al., 2009a). Propionate is also a functional HDAC inhibitor, albeit less potently so than butyrate. Finally, intracellular butyrate can act as an agonist for the peroxisome proliferator-activated receptor gamma (PPAR-γ) (Alex et al., 2013). A direct relevance of this interaction for the immune system has not been described yet, but exposure to bacterial strains present in the healthy gut microbiome has been shown to affect PPAR-γ activity (Are et al., 2008; Kelly et al., 2004). Given the anti-inflammatory effects described for PPAR-γ, future studies are likely to further explore this topic (Croasdell et al., 2015).
Effects of SCFAs on the immune system
SCFAs manifest their anti-inflammatory properties by influencing nearly every aspect of the intestinal immune system. In a mouse model of colitis, SCFAs were shown to mitigate disease severity by activating the NACHT, LRR and PYD domains-containing protein 3 (NLRP3) inflammasome through FFAR2 and HCAR2, inducing the secretion of the epithelial repair cytokine interleukin (IL-) 18 (Kalina et al., 2002; Macia et al., 2015; Singh et al., 2014). This is an excellent example of SCFAs promoting the integrity of the intestinal epithelium. Other mechanisms include increased production of mucin by goblet cells and modification of tight junctions, although some important details remain to be elucidated (Fukuda et al., 2011; Gaudier et al., 2004; Willemsen et al., 2003; Wrzosek et al., 2013).
Cells of the innate immune system are also broadly affected by SCFAs. FFAR2 is essential for neutrophil chemotaxis at the intestinal mucosa, and exposure to SCFAs reduces the production of pro-inflammatory cytokines from LPS-stimulated neutrophils. Consequently, Ffar2−/− mice are significantly more susceptible to colitis induced by dextrane sodium sulfate (DSS) (Le Poul et al., 2003; Maslowski et al., 2009; Sina et al., 2009; Venkatraman et al., 2003; Vinolo et al., 2011a; 2011b). Similarly, Chang et al. demonstrated that butyrate limits the secretion of pro-inflammatory cytokines from LPS-stimulated macrophages in the colonic lamina propria. This effect, however, is mediated by HDAC inhibition independently of GPCR activation. Interestingly, the inhibition seems to be restricted to so-called secondary response genes, which require an initial round of protein synthesis and chromatin remodeling prior to transcription after LPS stimulation (Chang et al., 2014; Ramirez-Carrozzi et al., 2009). This mechanism is quite different from the previously reported general hyporesponsiveness of intestinal macrophages that was based on starkly reduced expression of several innate response receptors (Smythies et al., 2005). There are also conflicting reports on whether or not butyrate directly inhibits activation of the transcription factor NF-κB (nuclear factor kappa-light-chain enhancer of activated B cells), likely due to varying experimental methodologies and cell types (Chang et al., 2014; Lührs et al., 2002; Segain et al., 2000). Further studies will undoubtedly clarify the physiological relevance of each of the observed phenomena.
Beyond intestinal macrophages, SCFAs have also been reported to be functionally important for microglia, a macrophage-like cell type of the central nervous system. Germ-free mice exhibited microglia dysfunction that could be rescued by treatment with SCFAs. Ffar2−/− mice had a similar defect, although no FFAR2 expression could be detected on microglia, leaving the connection between SCFAs and microglia unresolved (Erny et al., 2015). A tentative connection between expression of HCAR2 on microglia and Parkinson’s disease has also been proposed, inviting further investigation (Wakade et al., 2014).
Several studies have demonstrated that butyrate and propionate inhibit the maturation of dendritic cells (DCs), which form a central bridge between the innate and adaptive immune systems (Lu Liu et al., 2012; Millard et al., 2002; Singh et al., 2010; B. Wang et al., 2008). This appears to be primarily mediated by HDAC inhibition, as the effect is dependent on the SCFA transporter SLC5A8 (Singh et al., 2010). However, SCFAs have also been shown to affect DC maturation and function through HCAR2 and FFAR3, depending on the context (Singh et al., 2014; Trompette et al., 2014).
A role for SCFA in the regulation of B-cell function was reported nearly 20 years ago (Kudoh et al., 1998). In a recent study, Kim et al. shed further light on this observation and demonstrated that SCFAs support antibody responses by regulating gene expression through HDAC inhibition. Intriguingly, they further found that SCFAs in parallel modulate the energy metabolism of B cells to promote production of antibodies (M. Kim et al., 2016). This latter finding is of particular interest as it represents a direct link between microbial metabolites and cellular immunometabolism, an area of research that promises to garner much attention in the near future.
Perhaps the most potent anti-inflammatory property of SCFAs is their promotion of regulatory T cells (Tregs), which suppress the activity of effector T cells. This effect is mediated through several different mechanisms: acetate and propionate, but not butyrate, stimulate the expansion of pre-existing colonic Tregs (cTregs), whereas propionate and butyrate, but not acetate, increase the de novo differentiation of naive T cells into Tregs (Arpaia et al., 2013; Furusawa et al., 2013; Singh et al., 2014; Smith et al., 2013). Each of these functions operates independently, as numbers of cTregs in germ-free mice, which are starkly reduced in the absence of treatment, can be rescued by separate provision of acetate, propionate or butyrate (Arpaia et al., 2013; Smith et al., 2013). The specificity of each respective effect for a different subset of SCFAs reflects the underlying molecular mechanism. The expansion of pre-existing cTregs is mediated by activation of FFAR2, which is not triggered effectively by butyrate (Smith et al., 2013). Conversely, the de novo differentiation of cTregs relies on HDAC inhibition in both DCs and T cells, which acetate does not promote (Arpaia et al., 2013; Furusawa et al., 2013). Separately, butyrate has also been shown to enhance Treg generation by promoting an anti-inflammatory phenotype in macrophages and DCs through activation of HCAR2 (Singh et al., 2014). In addition to these mechanisms, Atarashi et al. showed that exposure of a colorectal cancer cell line to a mixture of SCFAs could elicit the production of transforming growth factor beta (TGF-β), which promotes the differentiation of naive T cells into Tregs (Atarashi et al., 2013; Fu et al., 2004). If this observation reflects a physiological mechanism in intestinal epithelial cells, it may allow SCFAs to further increase the colonic Treg population. The regulation of T-cell biology may be even more complex, however, as SCFAs have also been reported to promote generation of both effector and regulatory T cells, depending on the immunological context (Park et al., 2015).
In summary, microbiome-derived SCFAs are key regulators of virtually every aspect of the intestinal immune system. Their functional diversity results in staggering biological complexity that will likely elude comprehensive characterization and distillation into a simplistic model for years to come, even as a steady stream of fascinating new discoveries continues to elucidate new mechanisms and functions. One particularly interesting facet are the physiologically relevant concentrations of different SCFAs. In vivo studies use widely varying methods of administration and concentrations of SCFAs, including oral uptake via enriched starch or drinking water, enema, and intraperitoneal injection. Concentrations of SCFAs provided with drinking water ranged from 36 mM to 300 mM in the studies cited above. It remains unclear which method(s) most closely resemble the physiological state, but there is evidence that these differences affect experimental outcomes. For instance, Smith et al. reported an increase in cTregs after providing butyrate at a concentration of 150 mM with drinking water, while Arpaia et al. detected no significant changes at a concentration of 36 mM, likely because this lower quantity of butyrate was largely absorbed by the upper gastrointestinal tract before reaching the colon (Arpaia et al., 2013; Smith et al., 2013). Direct application of 50 mM butyrate by enema, however, did elicit an increase in cTregs, emphasizing the importance of administration route and SCFA concentration for in vivo experiments (Arpaia et al., 2013).
Indole derivatives
Beyond SCFAs, bacteria of the gut microbiome produce a plethora of other immunologically important metabolites from food components (Fig. 1). Dietary tryptophan, for instance, is processed to several different indole derivatives, which can act as ligands for the aryl hydrocarbon receptor (AhR) in host cells (Wikoff et al., 2009). The AhR is a ligand-activated transcription factor with a central role in the intestinal mucosa (Stockinger et al., 2014). Ahr−/− mice suffer from multiple immunological deficits, including reduced resistance to infection with the bacteria Listeria monocytogenes or Citrobacter rodentium, as well as the fungus Candida albicans; exacerbated DSS-induced colitis; and even altered susceptibility to experimental autoimmune encephalomyelitis (EAE) (Kiss et al., 2011; Lee et al., 2011; Y. Li et al., 2011; J. Qiu et al., 2012; Rothhammer et al., 2016; L. Z. Shi et al., 2007; Veldhoen et al., 2008; Zelante et al., 2013). Protection against these pathologies is mediated by expression of IL-22, a cytokine that supports the integrity of the intestinal mucosa by inducing the secretion of antimicrobial peptides from epithelial cells, production of mucins and proliferation of intestinal goblet cells. Under homeostatic conditions, IL-22 is primarily produced by a specialized subset of intraepithelial lymphocytes, the type 3 innate lymphoid cells (ILC3s) (Melo-Gonzalez and Hepworth, 2017). Nearly ten years ago, it was demonstrated that expression of IL-22 requires the AhR (Martin et al., 2009; Veldhoen et al., 2008). A few years later, four groups independently discovered that the AhR is in fact required for postnatal maintenance of IL-22-producing ILC3s, as well as the formation of isolated lymphoid follicles (Kiss et al., 2011; Lee et al., 2011; Y. Li et al., 2011; J. Qiu et al., 2012). In Ido1−/− mice, which cannot produce the endogenous AhR ligand l-kynurenine, bacteria that are able to metabolize dietary tryptophan and produce indole derivatives gain a selective advantage due to the increased availability of tryptophan. Indole derivatives produced by these bacteria can then compensate for the lack of endogenously produced AhR ligands, reflecting an interesting mutualistic balance between the needs of microbiome and host (Zelante et al., 2013). The pathophysiological relevance of microbiome-derived AhR ligands is further underscored by the observation that mice deficient for Card9, a gene that in humans is associated with IBD susceptibility, have an altered microbiota that produces lower amounts of indole derivatives, similar to the microbiota of IBD patients (Lamas et al., 2016).
In addition to AhR, the promiscuous nuclear receptor subfamily 1 group I member 2 (NR1I2, also known as pregnane X receptor, PXR) has been identified as a functional receptor for indole-3-propionic acid (IPA), which is produced in mice monocolonized with Clostridium sporogenes but absent in the gut of germ-free mice (Venkatesh et al., 2014; Wikoff et al., 2009). Nr1i2−/− mice exhibit impaired epithelial barrier function, and administration of synthetic NR1I2 agonists exerts protective effects in mouse models of colitis (Dou et al., 2013; Garg et al., 2016; Shah et al., 2007; Terc et al., 2014; Venkatesh et al., 2014). Interestingly, the combined knockout of Nr1i2 and the gene encoding the LPS sensor toll-like receptor 4 (Tlr4) can rescue the phenotype of Nr1i2−/− mice, possibly because it counteracts the accelerated cell death of inflammatory monocytes observed in NR1I2-deficient mice (Z. Qiu et al., 2016; Venkatesh et al., 2014).
Polyamines
Arginine is another amino acid that can be processed by the gut microbiota to produce immunomodulatory metabolites (Fig. 1). The highly reactive polyamines putrescine (a diamine, N2), spermidine (N3) and spermine (N4) all are arginine derivatives produced and secreted by intestinal bacteria (Michael, 2016). Polyamines are present in every living cell and fulfill important roles in gene expression and proliferation; therefore their endogenous production in eukaryotic cells is strictly regulated (Miller-Fleming et al., 2015). Oral administration of polyamines enhances the development and maintenance of the intestinal mucosa and resident immune cells (Buts et al., 1993; Dufour et al., 1988; Löser et al., 1999; Pérez-Cano et al., 2010). Conversely, blocking endogenous production of polyamines with a chemical inhibitor of ornithine decarboxylase negatively affects maintenance, repair and function of the intestinal epithelium, although it is not clear if or how this relates to dietary and microbially derived polyamine absorption (Chen et al., 2007; X. Guo et al., 2005; 2003; Lan Liu et al., 2009; Lux et al., 1980). Unexpectedly, and contrary to these observations, Levy et al. recently reported that exogenous polyamines can reduce the release of IL-18, a cytokine that promotes epithelial repair and barrier function. Spermine and the monoamine histamine both inhibit activation of the NLRP6 inflammasome, a protein complex expressed by epithelial cells that regulates IL-18 secretion. Intriguingly, taurine, another microbial metabolite that derives from host-produced bile acids (see below), counteracts the effects of spermine and histamine, indicating a finely tuned balance of opposing bacterial stimuli (Levy et al., 2015).
In addition to epithelial cells, polyamines can also exert direct influence on innate immune cells. Extraneous spermine is taken up by monocytes and macrophages and selectively inhibits the LPS-induced expression of pro-inflammatory cytokines (M. Zhang et al., 1999; 1997). Indeed, spermine administration exhibits protective anti-inflammatory effects in mouse models of both localized and systemic inflammation (Steege et al., 1999; M. Zhang et al., 1997; S. Zhu et al., 2009). The cellular protein alpha-2-HS-glycoprotein (AHSG, also known as Fetuin-A) has been reported to be a required co-factor for the anti-inflammatory activity of spermine, but the underlying molecular mechanism remains unclear (H. Wang et al., 1997).
Interestingly, the metabolism of dietary arginine by the microbiome may have effects on the immune system beyond the actual polyamine products. Arginine itself is an important modulator of the immunometabolism of macrophages and T cells and thus affects their effector functions (O'Neill et al., 2016). Germ-free mice retain significantly higher amounts of arginine in their intestines than mice colonized by gut microbes. This indicates that the microbiome metabolizes substantial quantities of dietary arginine and may thus regulate the availability of arginine to the immune system to a certain extent (Mardinoglu et al., 2015; Matsumoto et al., 2012). However, further studies are needed to determine whether this phenomenon has a measurable effect on immune function.
Even though polyamines have been known to affect the intestinal mucosa and various immune functions for decades, surprisingly little is understood of the extent and the mechanisms by which they function biologically. This is undoubtedly a result of their ubiquitous presence and broad functionality, which pose daunting obstacles to detailed investigation. However, as increasingly sophisticated techniques allow for ever finer dissection of metabolic pathways, further insights are likely to emerge. A particularly interesting question will be the extent to which dietary, microbial and host-derived polyamines each contribute to the holobiont’s biology.
Bile acids: metabolites produced by the host and biochemically modified by gut bacteria
The enterohepatic circulation of bile acids
Intestinal microbes not only metabolize dietary components but also substrates that are secreted by the host into the gastrointestinal lumen (Fig. 2). This includes bile acids, which are released into the duodenum from the gall bladder to facilitate the absorption of dietary lipids and lipophilic vitamins. Primary bile acids, such as cholic acid and chenodeoxycholic acid, are synthesized in the liver from cholesterol and conjugated to glycine or taurine prior to secretion. After passage through the small intestine, about 95% of bile acids are reabsorbed in the terminal ileum by active transport, mostly in their conjugated form, and recycled by the host. About half of the remaining bile acids are taken up in the colon by passive absorption, the rest is excreted with the feces (Martinez-Augustin and Sanchez de Medina, 2008). Primary bile acids are converted to secondary bile acids by the local microbiota, partly in the small intestine and predominantly in the colon. This involves multiple steps and typically begins with the initial deconjugation of glycine or taurine, followed by dehydroxylation (Ridlon et al., 2006). As discussed above, free taurine generated by deconjugation can enhance the activation of the NLRP6 inflammasome and thereby increase production of IL-18 by the intestinal epithelium, which supports epithelial barrier function and maintenance (Levy et al., 2015). Primary bile acids are more readily absorbed in the small intestine, whereas secondary bile acids are more hydrophobic and can thus be taken up more efficiently in the colon (Martinez-Augustin and Sanchez de Medina, 2008). Consequently, the gut microbiota plays an important role in regulating the extent to which bile acids can be recycled by the host. Indeed, the composition of bile acids varies dramatically between germ-free and control animals in feces, liver, heart, kidney and plasma (Duboc et al., 2013; Sayin et al., 2013; Swann et al., 2011).
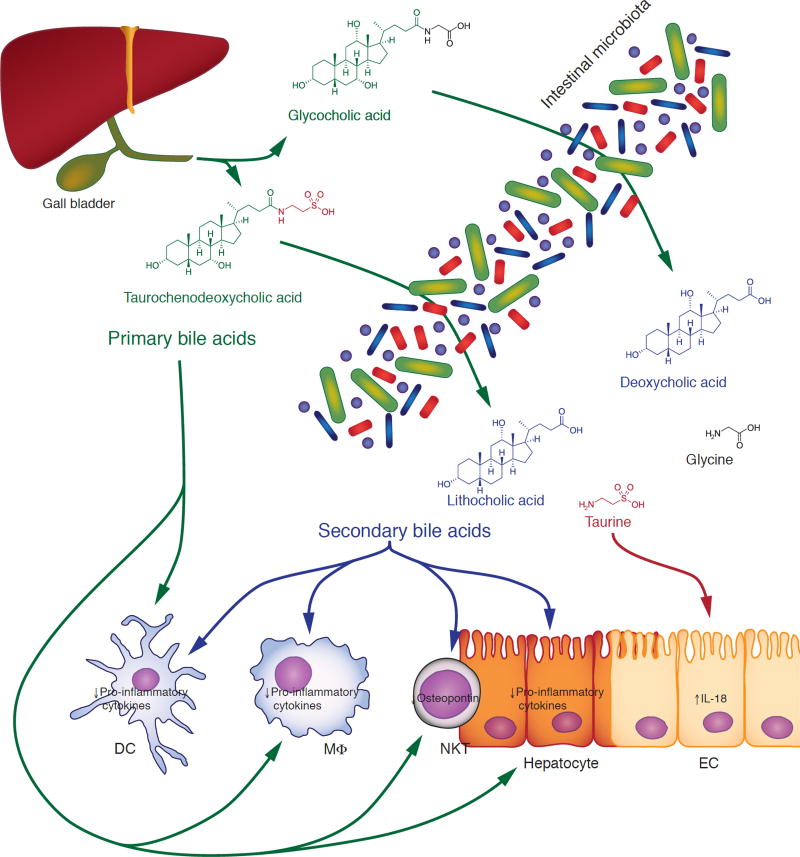
Fig. 2. Immunomodulatory function of primary and secondary bile acids.
Primary bile acids (green), such as cholic acid and chenodeoxycholic acid, are synthesized by the host in the liver and conjugated to glycine or taurine before secretion into the duodenum. In the intestinal tract, primary bile acids are converted by the microbiota to secondary bile acids (blue), such as deoxycholic acid and lithocholic acid, through a multi-step process that involves deconjugation and dehydroxylation. Primary and secondary bile acids can inhibit the secretion of pro-inflammatory cytokines by macrophages, dendritic cells and hepatocytes, as well as the production of osteopontin by NKT cells. During the deconjugation process, free taurine (red) is released into the intestinal lumen, where it can promote the integrity of the mucosal epithelium by increasing autocrine production of IL-18. See text for details. DC, dendritic cell; EC, epithelial cell; MΦ, macrophage.
Immunomodulatory effects of bile acids and their molecular mechanisms
Beyond their role in digestion, bile acids also function as signaling molecules for several tissues. This includes cells of the immune system: bile acids reduce the expression of pro-inflammatory cytokines from monocytes, macrophages, DCs and the hepatic macrophages known as Kupffer cells (Brestoff and Artis, 2013; Martinot et al., 2017). These effects are primarily mediated by two different host receptors, the bile acid receptor (BAR; also known as nuclear receptor subfamily 1 group H member 4, NR1H4, or farnesoid X-activated receptor, FXR) and the G-protein-coupled bile acid receptor 1 (GPBAR1; also known as membrane-type receptor for bile acids, M-BAR, or TGR5). Both receptors can be activated by primary and secondary bile acids alike (Kawamata et al., 2003; Makishima et al., 1999; Maruyama et al., 2002; Parks et al., 1999; H. Wang et al., 1999). Bile acids have also been reported to function as agonists to additional receptors, but the physiological relevance of these pathways remains incompletely understood (Thomas et al., 2008).
BAR is a nuclear, ligand-activated transcription factor that forms heterodimers with the retinoic-acid receptor RXR. Activation of BAR in LPS-stimulated macrophages by synthetic agonists induces the transcription of BAR-dependent genes; but it also causes the sumoylation-dependent stabilization of the nuclear receptor corepressor (NCoR) protein on the promoter of NF-κB-dependent genes, thus inhibiting their transcription. This results in a significant decrease in the expression of pro-inflammatory cytokines (Pascual et al., 2005; Vavassori et al., 2009). Correspondingly, treatment with a synthetic agonist of BAR has protective effects in mouse models of colitis, whereas BAR-deficient (Nr1h4−/−) mice exhibit exacerbated pathology (Gadaleta et al., 2011; Vavassori et al., 2009). Nr1h4−/− mice also develop spontaneous hepatic inflammation and liver tumors (I. Kim et al., 2007; Yang et al., 2007). Interestingly, the inflammatory effects of BAR deficiency on the liver may not only be mediated by Kupffer cells, but also by hepatocytes and NKT cells (Mencarelli et al., 2009; Y.-D. Wang et al., 2008).
GPBAR1, as the name suggests, is a Gs-protein-coupled receptor. Binding by bile acids triggers an increase in intracellular cAMP levels and the activation of ERK (Kawamata et al., 2003; Maruyama et al., 2002). Similar to BAR, activation of GPBAR1 on macrophages and Kupffer cells by synthetic agonists inhibits the expression of LPS-induced pro-inflammatory cytokines by interfering with NF-κB-dependent transcription. This has been linked to direct inhibition of NF-κB activation, but is also consistent with a model of transcriptional competition between NF-κB and the cAMP-activated transcription factor cAMP response element binding protein (CREB) (C. Guo et al., 2015; Kawamata et al., 2003; Keitel et al., 2008; Pols et al., 2011; Y.-D. Wang et al., 2011; Wen et al., 2010). Gpbar1−/− mice, like their Nr1h4−/− counterparts, develop more severe disease than wildtype controls in several animal models of inflammatory disorder, including colitis, hepatitis and gastritis (Cipriani et al., 2011; C. Guo et al., 2015; Y.-D. Wang et al., 2011).
Bile acids and IBD
Due to this significant role in the regulation of the immune system, dysregulation of bile acids as a result of alterations in the gut microbiome have been proposed as mediators of IBD. Indeed, changes in the composition of the bile acid pool, an increase in colonic expression of GPBAR1 and a decrease in ileal activity of BAR have all been detected in patients suffering from IBD (Cipriani et al., 2011; Duboc et al., 2013; Lenz et al., 1976; Nijmeijer et al., 2011; Rutgeerts et al., 1979). However, no conclusive evidence for a causative role of bile-acid imbalances in the pathogenesis of IBD has been uncovered so far. A study evaluating genetic variations in NR1H4 found no association between single-nucleotide polymorphisms in this gene and IBD risk (Nijmeijer et al., 2011). The efficiency of bile-acid recycling depends on their biotransformation by the microbiome; however, the de novo synthesis of bile acids is subject to a strict feedback mechanism, which may limit the effect of increased excretion of bile acids in the absence of a fully functional microbiota (Martinot et al., 2017). Furthermore, while secondary bile acids are exclusively produced by gut bacteria, both primary and secondary bile acids are able to activate BAR and GPBAR1, and it is not clear what functional effects a shift in the balance of primary and secondary bile acids might have. Studies investigating the role of bile acids in the regulation of the immune response typically utilize synthetic bile-acid receptor agonists, which do not allow for a distinction between the effect of primary and secondary bile acids. What, if any, physiological consequences to the microbial processing of bile acids may have in the context of chronic inflammatory disorders therefore remains an open question.
Metabolites that are synthesized de novo by gut microbes
ATP
Gut microbes do not only modify dietary components and host metabolites, they also secrete signaling molecules after de novo synthesis (Fig. 3). For instance some, but not all, intestinal bacteria actively secrete ATP, and germ-free mice have lower ATP concentrations in their gut than controls (Atarashi et al., 2008; Hironaka et al., 2013; Iwase et al., 2010).
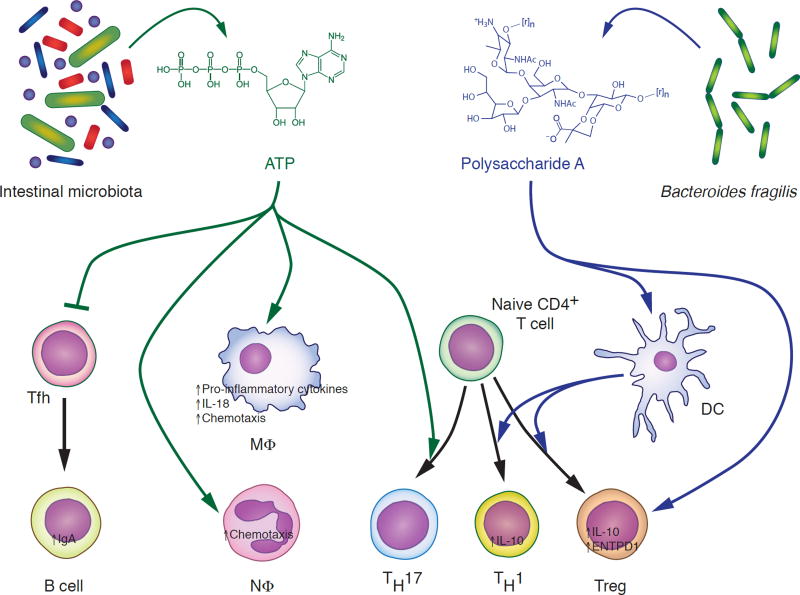
Fig. 3. The effects of ATP and Polysaccharide A on the immune system.
Extracellular ATP (green) secreted by bacteria of the intestinal tract has pro- and anti-inflammatory effects. While it promotes secretion of pro-inflammatory cytokines, chemotaxis and differentiation of naive T cells into TH17 cells, ATP also inhibits release of IgA into the intestinal lumen by reducing the number of B-cell-activating T follicular helper cells. Polysaccharide A (PSA, blue) is produced by the intestinal bacterium Bacteroides fragilis. It promotes production of the anti-inflammatory cytokine IL-10 by T cells directly and indirectly through dendritic cells. See text for details. DC, dendritic cell; MΦ, macrophage; NΦ, neutrophil; Tfh, T follicular helper cell; Treg, regulatory T cell. Chemical structure of PSA subunit based on (Mazmanian and Kasper, 2006). [r]n indicates where PSA subunit repeats branch off.
Effects of ATP on the immune system
Unlike the other microbiota-derived compounds discussed so far, extracellular ATP primarily exhibits pro-inflammatory properties. Within the host, ATP is released passively from necrotic cells and actively from stressed or stimulated cells, e.g. during an inflammatory response. As such, it serves as an endogenous danger-associated molecular pattern (DAMP) that can drive chemotaxis of and cytokine release by immune cells (interestingly, low concentrations of ATP tend to have the opposite effect) (Faas et al., 2017). Germ-free mice show a severe reduction of TH17 lymphocytes in the lamina propria of the colonic epithelium, which can be rescued by the direct administration of ATP. Specifically, ATP elicits the production of cytokines that favor TH17 differentiation by CD70high CD11clow antigen-presenting cells (APCs) in the lamina propria (Atarashi et al., 2008). Analogously, mice deficient for the epithelial ATP-degrading enzyme ectonucleoside triphosphate diphosphohydrolase 7 (ENTPD7) exhibit higher levels of luminal ATP and an increase in TH17 cells in the lamina propria of the small-intestinal epithelium (Kusu et al., 2013). It remains unclear which bacterial species constitute the main producers of microbially derived ATP under physiological conditions. Interestingly, segmented filamentous bacteria (SFB), which have been shown to induce TH17 cells in the lamina propria of the murine small intestine, do not produce high levels of ATP (Ivanov et al., 2009). Instead, presentation of SFB-derived antigens by intestinal APCs was shown to be sufficient for TH17 cell development (Goto et al., 2014; Panea et al., 2015). The capability of other TH17-inducing gut bacteria to produce ATP has not been determined yet, but a common property appears to be their ability to adhere to the intestinal epithelium (Atarashi et al., 2015; Tan et al., 2016). Further studies will be needed to elucidate whether microbially derived ATP and epithelial-adhesive bacterial strains are functionally overlapping, additive, synergistic or redundant.
Molecular functions of extracellular ATP
On a biochemical level, extracellular ATP is the sole agonist for receptors of the P2X family, which are ligand-activated ion channels expressed by a variety of tissues including innate and adaptive immune cells (Faas et al., 2017). The P2X purinoceptor 7 isoform (P2X7) appears to have a particularly important role in the ATP-mediated induction of inflammation. Binding of ATP to P2X7 results in the influx of Na+ and Ca2+ ions with simultaneous K+ efflux. Importantly, this has been shown to elicit activation of the NLRP3 inflammasome in LPS-stimulated macrophages, which causes secretion of IL-18 (but also of the pro-inflammatory cytokine IL-1β) and may thus promote maintenance and function of the epithelial barrier (Mariathasan et al., 2006; Sutterwala et al., 2014; 2006; Yaron et al., 2015). Long-term activation of P2X7, however, results in formation of a large membrane pore and cell death (Nuttle and Dubyak, 1994; Smart et al., 2003). Specifically in the gut mucosa, P2X7 has been shown to be an important regulator of IgA secretion into the intestinal lumen. T follicular helper (Tfh) cells in the Peyer’s patches foster the development of B cells that produce immunoglobulins with specificity for intestinal bacteria, which can be secreted across the epithelium as IgA. Mice deficient for P2X7 (P2rx7−/−) have elevated Tfh cell numbers in the intestinal mucosa, presumably because they are resistant to ATP-induced cell death. This results in an increase in high-affinity IgA specific for intestinal bacteria secreted into the intestinal lumen and thereby a depletion of the gut microbiota. The subsequent reduction in B1-cell stimulation is believed to cause increased susceptibility of P2rx7−/− mice to polymicrobial sepsis (Proietti et al., 2014). Importantly, Perruzza et al. recently demonstrated that microbially derived ATP can in fact modulate the host immune response through this pathway. Monocolonization with a Shigella flexneri mutant unable to secrete ATP was associated with a significant increase in Tfh and germinal-center B cells, as well as elevated levels of bacteria-specific IgA in the intestine, compared to colonization with the ATP-secreting wild-type S. flexneri (Perruzza et al., 2017). Thus, microbially derived ATP appears to protect the intestinal microbiota from excessive antibody production under physiological conditions, which allows for mutually beneficial colonization of the host.
ATP and intestinal inflammation
These protective effects notwithstanding, intestinal ATP has also been implicated in the development of IBD, likely due to the generally pro-inflammatory properties of ATP. For instance, intraperitoneal injection of ATP exacerbated experimental colitis in mice (Atarashi et al., 2008). Another study investigated the role of ENTPD1 (also known as CD39), a membrane-bound extracellular hydrolase expressed on the surface of immune and endothelial cells. ENTPD1 converts ATP to AMP, which is then quickly metabolized to the anti-inflammatory adenosine by 5’-nucleotidase (also known as CD73) (Allard et al., 2017). Entpd1−/− mice, which cannot remove extracellular ATP through this mechanism, where highly susceptible to experimental colitis (Friedman et al., 2009). Conversely, P2rx7−/− mice were protected against experimental induction of colitis, as were rats treated prophylactically with a chemical P2X7 antagonist (Marques et al., 2014; Neves et al., 2014). In humans, polymorphisms of ENTPD1 were shown to be associated with IBD, and expression of P2X7 was increased in the intestinal epithelium and lamina propria of IBD patients (Friedman et al., 2009; Neves et al., 2014). If a direct link between microbially derived ATP and IBD is established by further studies, this may in fact open the possibility of novel therapeutic avenues.
Polysaccharide A (PSA)
Another important microbial immune modulator is the zwitterionic PSA, produced by the common gut bacterium Bacteroides fragilis (Pantosti et al., 1991; Sharma et al., 2017) (Fig. 3). PSA induces secretion of the potent anti-inflammatory cytokine IL-10 by CD4+ T cells and promotes the differentiation of naive CD4+ T cells into TH1 cells over TH2 cells. The production of IL-10 is primarily mediated by DCs, which internalize PSA and present fragments to CD4+ T cells in conjunction with the major histocompatibility complex class II (MHC-II), thus stimulating activation and clonal expansion of T cells. Importantly, PSA also activates toll-like receptor (TLR) 2 expressed on the surface of DCs, which triggers the release of cytokines that promote IL-10 production by T cells (Cobb et al., 2004; Dasgupta et al., 2014; Johnson et al., 2015; Mazmanian et al., 2005; 2008; Telesford et al., 2015; Q. Wang et al., 2006). This process is supplemented by direct binding of PSA to TLR2 expressed on FoxP3+ Tregs, further enhancing IL-10 production (Round et al., 2011). The PSA-induced production of IL-10 inhibits the activity of mucosal effector T cells, in particular of TH17 cells, and thereby protects mice in experimental models of colitis (Dasgupta et al., 2014; Mazmanian et al., 2008). The presence of PSA is a prerequisite for effective colonization of the colonic mucosa by its producer B. fragilis, which further underscores the mutualistic nature of the interactions between PSA and the immune system (Round et al., 2011).
Interestingly, PSA exposure increases expression of ENTPD1 on FoxP3+ Tregs, thus suggesting a potential intersection between PSA and (microbially derived) extracellular ATP in the regulation of the immune system (Telesford et al., 2015). This observation is particularly intriguing as an increase in ENTPD1 expression on peripheral-blood Tregs has been associated with remission of IBD (Gibson et al., 2015). Furthermore, the up-regulation of ENTPD1 has been implicated in the protective effect of PSA in experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis (Ochoa-Reparaz et al., 2010; Y. Wang et al., 2014). This confirms that the immunomodulatory effects of PSA are not restricted to the intestinal compartment but have systemic consequences.
Beyond cells of the immune system, Jiang et al. recently reported that fetal enterocytes exposed to PSA produce lower levels of the IL-1β-induced, pro-inflammatory cytokine IL-8, which may play a role in the development of necrotizing enterocolitis in prematurely born infants (Jiang et al., 2017).
Concluding remarks
More than a century of scientific research has revealed the staggering complexity and breadth of the interactions between microbiome and host. Indeed, the gut microbiota appears to influence nearly every aspect of host physiology, including energy balance, cardiovascular health, chronic inflammation, development of adaptive immune cells and many more systems that fall outside of the scope of this review (Rooks and Garrett, 2016; Sharon et al., 2014). A comprehensive model for the inner workings of the holobiont, however, remains elusive. To build such a model, three avenues of research will have to be explored in concert:
What are the molecular underpinnings of the established observations? Even within the confines of known host-microbiome interactions, many details remain poorly understood, as discussed for several examples above. A particularly interesting question is bioavailability. When, where and how do microbially derived metabolites cross the epithelial barrier? How tightly is this process regulated (Sharon et al., 2014)?
What else is out there? Until recently, the extensive diversity of the gut microbiome and its myriad biochemical processes have defied comprehensive analysis. Novel high-throughput techniques are beginning to unravel the complexities of the intestinal compartment – and are illuminating how many unknown variables are left to investigate. For instance, most of the known bacterially derived metabolites in the intestinal tract remain functionally uncharacterized, and it is unclear how many other, hitherto unidentified metabolites are yet to be discovered. The situation is similar with regard to bacterial species. In a recent study, Geva-Zatorsky et al. monocolonized germ-free mice separately with 53 different bacterial species resident in the human intestine (less than five percent of the estimated total!) and found that each one of them had some sort of immunomodulatory effect on the host (Geva-Zatorsky et al., 2017).
How does it all fit together? Virtually all functional studies of the microbiome have been limited to single bacterial species or metabolites. This, of course, simply reflects current technical limitations, as the gut is not an easily accessible organ system. However, the effect of individual bacterial species or metabolites is merely the first layer of complexity in host-microbiome interactions. Microbially derived metabolites signal not only to the host but also to other gut bacteria, and multiple interacting communities may influence the host in an additive, synergistic, redundant or even subtractive fashion (Adair and Douglas, 2016). Considering that up to 1,150 different bacterial species are believed to reside in the human intestinal tract, and at least 160 of them are present in any given individual, the possible variations of this communication network appear astronomical. Another dimension is added by the gut virome, which is currently emerging as an exciting new chapter of microbiome research (Pfeiffer and Virgin, 2016). Computational analysis of high-throughput data sets is one approach to address this complexity, but another is to make the gut itself, or something reasonably similar, more accessible to experimental manipulation. Yissachar et al. recently reported the development of a highly functional, microfluidics-based intestinal organ culture system. They found that previous observations made in vivo could be robustly reproduced, while being able to extract compelling new insights due to the tightly controlled nature of their system (Yissachar et al., 2017). Novel systems like this will undoubtedly be crucial in the elucidation of more complex connections between host and microbiome, and ultimately allow the development of new therapeutic approaches to treat ailments ranging from IBD to depression.
Acknowledgments
We apologize to the many investigators whose important contributions to this expansive field could not be mentioned here. We wish to thank Drs. Nicholas Arpaia, Suneeta Krishnareddy and Ivaylo Ivanov (all Columbia University) for helpful discussions and critical reading of this manuscript. Some of the chemical structures used in the figures were derived from images released into the public domain via Wikimedia. We thank the designers for their generosity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Usage note: We use the term “microbiome” in its broader sense, meaning the entirety of a microbial community colonizing a given habitat. A more narrow definition refers exclusively to the genetic information encoded by that community.
AUTHOR CONTRIBUTIONS
TSP and SG conducted the literature review, devised and wrote the manuscript.
References
- Adair KL, Douglas AE. Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 2016;35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, Koppen A, Mudde K, Haenen D, Al-Lahham S, Roelofsen H, Houtman R, van der Burg B, Mandrup S, Bonvin AMJJ, Kalkhoven E, Müller M, Hooiveld GJ, Kersten S. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Molecular and Cellular Biology. 2013;33:1303–1316. doi: 10.1128/MCB.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B, Longhi MS, Robson SC, Stagg J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017;276:121–144. doi: 10.1111/imr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson J-A, Pettersson S, Arulampalam V. Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL-10 levels in colonic epithelial cells. Proceedings of the National Academy of Sciences. 2008;105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S-I, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nuñez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. CELL. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Bested AC, Logan AC, Selhub EM. Intestinal microbiota, probiotics and mental health: from Metchnikoff to modern advances: Part I - autointoxication revisited. Gut Pathog. 2013;5:5. doi: 10.1186/1757-4749-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013;40:463–471. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, Clark AG. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemen JG, Venema K, van de Poll MC, Olde Damink SW, Buurman WA, Dejong CH. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr. 2009;28:657–661. doi: 10.1016/j.clnu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Boutagy NE, McMillan RP, Frisard MI, Hulver MW. Metabolic endotoxemia with obesity: Is it real and is it relevant? Biochimie. 2016;124:11–20. doi: 10.1016/j.biochi.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer RI, Buto SK, Christ ML, Bean J, Vernia P, Paoluzi P, Di Paolo MC, Caprilli R. Rectal irrigation with short-chain fatty acids for distal ulcerative colitis. Preliminary report. Dig. Dis. Sci. 1991;36:185–187. doi: 10.1007/BF01300754. [DOI] [PubMed] [Google Scholar]
- Brophy ML, Dong Y, Wu H, Rahman HNA, Song K, Chen H. Eating the Dead to Keep Atherosclerosis at Bay. Front Cardiovasc Med. 2017;4:2. doi: 10.3389/fcvm.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Buts JP, De Keyser N, Kolanowski J, Sokal E, Van Hoof F. Maturation of villus and crypt cell functions in rat small intestine. Role of dietary polyamines. Dig. Dis. Sci. 1993;38:1091–1098. doi: 10.1007/BF01295726. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti J-F, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cannon PR. The effects of diet on the intestinal flora. J. Infect. Dis. 1921;29:369–385. [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rao JN, Zou T, Liu L, Marasa BS, Xiao L, Zeng X, Turner DJ, Wang J-Y. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G568–76. doi: 10.1152/ajpgi.00201.2007. [DOI] [PubMed] [Google Scholar]
- Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen MR, Mortensen PB. Kinetic studies on colonocyte metabolism of short chain fatty acids and glucose in ulcerative colitis. Gut. 1995;37:684–689. doi: 10.1136/gut.37.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BA, Wang Q, Tzianabos AO, Kasper DL. Polysaccharide processing and presentation by the MHCII pathway. CELL. 2004;117:677–687. doi: 10.016/j.cell.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins HL, Drazul-Schrader D, Sulpizio AC, Koster PD, Williamson Y, Adelman SJ, Owen K, Sanli T, Bellamine A. L-Carnitine intake and high trimethylamine N-oxide plasma levels correlate with low aortic lesions in ApoE(−/−) transgenic mice expressing CETP. Atherosclerosis. 2016;244:29–37. doi: 10.1016/j.atherosclerosis.2015.10.108. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto J-M, Renault P, ANR MicroObes consortium. Doré J, Zucker J-D, Clément K, Ehrlich SD. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Cox LM, Blaser MJ. Pathways in microbe-induced obesity. Cell Metab. 2013;17:883–894. doi: 10.1016/j.cmet.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creely SJ, McTernan PG, Kusminski CM, Fisher FM, Da Silva NF, Khanolkar M, Evans M, Harte AL, Kumar S. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 2007;292:E740–7. doi: 10.1152/ajpendo.00302.2006. [DOI] [PubMed] [Google Scholar]
- Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. PPARγ and the Innate Immune System Mediate the Resolution of Inflammation. PPAR Res. 2015;2015:549691. doi: 10.1155/2015/549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr. 1997;21:357–365. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker H-C, Kasper DL. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host and Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNicolantonio JJ, Lavie CJ, Fares H, Menezes AR, O'Keefe JH. L-carnitine in the secondary prevention of cardiovascular disease: systematic review and meta-analysis. Mayo Clin. Proc. 2013;88:544–551. doi: 10.1016/j.mayocp.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Donath MY, Dalmas É, Sauter NS, Böni-Schnetzler M. Inflammation in obesity and diabetes: islet dysfunction and therapeutic opportunity. Cell Metab. 2013;17:860–872. doi: 10.1016/j.cmet.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Molecular Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou W, Zhang J, Zhang E, Sun A, Ding L, Chou G, Wang Z, Mani S. Chrysin ameliorates chemically induced colitis in the mouse through modulation of a PXR/NF-κB signaling pathway. The Journal of pharmacology and experimental therapeutics. 2013;345:473–482. doi: 10.1124/jpet.112.201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H, Rajca S, Rainteau D, Benarous D, Maubert M-A, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, Bridonneau C, Dumetz F, Grill J-P, Masliah J, Beaugerie L, Cosnes J, Chazouillères O, Poupon R, Wolf C, Mallet J-M, Langella P, Trugnan G, Sokol H, Seksik P. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- Dufour C, Dandrifosse G, Forget P, Vermesse F, Romain N, Lepoint P. Spermine and spermidine induce intestinal maturation in the rat. Gastroenterology. 1988;95:112–116. doi: 10.1016/0016-5085(88)90298-3. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 2004a;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004b;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermöhlen O, Chun E, Garrett WS, McCoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, Prinz M. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas MM, Sáez T, de Vos P. Extracellular ATP and adenosine: The Yin and Yang in immune responses? Mol. Aspects Med. 2017 doi: 10.1016/j.mam.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Fändriks L. Roles of the gut in the metabolic syndrome: an overview. J Intern Med. 2016 doi: 10.1111/joim.12584. [DOI] [PubMed] [Google Scholar]
- Ferreira CM, Vieira AT, Vinolo MAR, Oliveira FA, Curi R, Martins FDS. The central role of the gut microbiota in chronic inflammatory diseases. J Immunol Res. 2014;2014:689492. doi: 10.1155/2014/689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J, Bumbalo R, Bautze V, Strotmann J, Breer H. Expression of odorant receptor Olfr78 in enteroendocrine cells of the colon. Cell Tissue Res. 2015;361:697–710. doi: 10.1007/s00441-015-2165-0. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Presented at the The Proceedings of the Nutrition Society. 2015:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DJ, Künzli BM, A-Rahim YI, Sevigny J, Berberat PO, Enjyoji K, Csizmadia E, Friess H, Robson SC. From the Cover: CD39 deletion exacerbates experimental murine colitis and human polymorphisms increase susceptibility to inflammatory bowel disease. Proceedings of the National Academy of Sciences. 2009;106:16788–16793. doi: 10.1073/pnas.0902869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostegård J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013;11:117. doi: 10.1186/1741-7015-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Zhang N, Yopp AC, Chen D, Mao M, Chen D, Zhang H, Ding Y, Bromberg JS. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 − precursors. Am. J. Transplant. 2004;4:1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen ECL, Renooij W, Murzilli S, Klomp LWJ, Siersema PD, Schipper MEI, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SWC. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- Garg A, Zhao A, Erickson SL, Mukherjee S, Lau AJ, Alston L, Chang TKH, Mani S, Hirota SA. Pregnane X Receptor Activation Attenuates Inflammation-Associated Intestinal Epithelial Barrier Dysfunction by Inhibiting Cytokine-Induced Myosin Light-Chain Kinase Expression and c-Jun N-Terminal Kinase 1/2 Activation. The Journal of pharmacology and experimental therapeutics. 2016;359:91–101. doi: 10.1124/jpet.116.234096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier E, Jarry A, Blottière HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G1168–74. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, Kasper DL. Mining the Human Gut Microbiota for Immunomodulatory Organisms. CELL. 2017;168:928–943. e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J. Lipid Res. 2009;50:90–97. doi: 10.1194/jlr.M800156-JLR200. [DOI] [PubMed] [Google Scholar]
- Gibson DJ, Elliott L, McDermott E, Tosetto M, Keegan D, Byrne K, Martin ST, Rispens T, Cullen G, Mulcahy HE, Cheifetz AS, Moss AC, Robson SC, Doherty GA, Ryan EJ. Heightened Expression of CD39 by Regulatory T Lymphocytes Is Associated with Therapeutic Remission in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015;21:2806–2814. doi: 10.1097/MIB.0000000000000566. [DOI] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL. Transmission of atherosclerosis susceptibility with gut microbial transplantation. Journal of Biological Chemistry. 2015;290:5647–5660. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Qi H, Yu Y, Zhang Q, Su J, Yu D, Huang W, Chen W-D, Wang Y-D. The G-Protein-Coupled Bile Acid Receptor Gpbar1 (TGR5) Inhibits Gastric Inflammation Through Antagonizing NF-κB Signaling Pathway. Front Pharmacol. 2015;6:287. doi: 10.3389/fphar.2015.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Rao JN, Liu L, Zou T, Keledjian KM, Boneva D, Marasa BS, Wang J-Y. Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G1159–69. doi: 10.1152/ajpgi.00407.2004. [DOI] [PubMed] [Google Scholar]
- Guo X, Rao JN, Liu L, Zou T-T, Turner DJ, Bass BL, Wang J-Y. Regulation of adherens junctions and epithelial paracellular permeability: a novel function for polyamines. Am. J. Physiol., Cell Physiol. 2003;285:C1174–87. doi: 10.1152/ajpcell.00015.2003. [DOI] [PubMed] [Google Scholar]
- Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- Herter CA, Kendall AI. The influence of dietary alterations on the types of intestinal flora. The Journal of Biological Chemistry. 1910;7:203–239. [Google Scholar]
- Hironaka I, Iwase T, Sugimoto S, Okuda K-I, Tajima A, Yanaga K, Mizunoe Y. Glucose triggers ATP secretion from bacteria in a growth-phase-dependent manner. Appl. Environ. Microbiol. 2013;79:2328–2335. doi: 10.1128/AEM.03871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschler A. Ueber den Einfluss der Kohlehydrate und einiger anderer Körper der Fettsäurereihe auf die Eiweissfäulniss. Zeitschrift für Physiologische Chemie. 1886;10:306–317. [Google Scholar]
- Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 2002;39:33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. CELL. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Shinji H, Tajima A, Sato F, Tamura T, Iwamoto T, Yoneda M, Mizunoe Y. Isolation and identification of ATP-secreting bacteria from mice and humans. J. Clin. Microbiol. 2010;48:1949–1951. doi: 10.1128/JCM.01941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Meng D, Weng M, Zhu W, Wu W, Kasper D, Walker WA. The symbiotic bacterial surface factor polysaccharide A on Bacteroides fragilis inhibits IL-1β-induced inflammation in human fetal enterocytes via toll receptors 2 and 4. PLoS ONE. 2017;12:e0172738. doi: 10.1371/journal.pone.0172738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Jones MB, Cobb BA. Polysaccharide A from the capsule of Bacteroides fragilis induces clonal CD4+ T cell expansion. Journal of Biological Chemistry. 2015;290:5007–5014. doi: 10.1074/jbc.M114.621771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson AL, Bäckhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14:79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- Kalina U, Koyama N, Hosoda T, Nuernberger H, Sato K, Hoelzer D, Herweck F, Manigold T, Singer MV, Rossol S, Böcker U. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur. J. Immunol. 2002;32:2635–2643. doi: 10.1002/1521-4141(200209)32:9<2635::AID-IMMU2635>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kanauchi O, Suga T, Tochihara M, Hibi T, Naganuma M, Homma T, Asakura H, Nakano H, Takahama K, Fujiyama Y, Andoh A, Shimoyama T, Hida N, Haruma K, Koga H, Mitsuyama K, Sata M, Fukuda M, Kojima A, Bamba T. Treatment of ulcerative colitis by feeding with germinated barley foodstuff: first report of a multicenter open control trial. J. Gastroenterol. 2002;37(Suppl 14):67–72. doi: 10.1007/BF03326417. [DOI] [PubMed] [Google Scholar]
- Kaoutari El A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Micro. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Keitel V, Donner M, Winandy S, Kubitz R, Häussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochemical and Biophysical Research Communications. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AGP, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab. 2014;20:753–760. doi: 10.1016/j.cmet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host and Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC, Org E, Wu Y, Li L, Smith JD, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20:799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. CELL. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Kudoh K, Shimizu J, Wada M, Takita T, Kanke Y, Innami S. Effect of indigestible saccharides on B lymphocyte response of intestinal mucosa and cecal fermentation in rats. J. Nutr. Sci. Vitaminol. 1998;44:103–112. doi: 10.3177/jnsv.44.103. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusu T, Kayama H, Kinoshita M, Jeon SG, Ueda Y, Goto Y, Okumura R, Saiga H, Kurakawa T, Ikeda K, Maeda Y, Nishimura J-I, Arima Y, Atarashi K, Honda K, Murakami M, Kunisawa J, Kiyono H, Okumura M, Yamamoto M, Takeda K. Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J. Immunol. 2013;190:774–783. doi: 10.4049/jimmunol.1103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam DW, LeRoith D. Metabolic Syndrome. In: De Groot LJ, Chrousos G, Dungan K, et al., editors. Endotext. MDText.com, Inc.; South Dartmouth (MA): 2015. [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay J-M, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul E, Loison C, Struyf S, Springael J-Y, Lannoy V, Decobecq M-E, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz K, Jensen KB, Jarnum S. Bile acid metabolism and plasma protein turnover in Crohn's disease. Scand. J. Gastroenterol. 1976;11:721–727. [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes & Development. 2016;30:1589–1597. doi: 10.1101/gad.284091.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, Pevsner-Fischer M, Shapiro H, Christ A, Harmelin A, Halpern Z, Latz E, Flavell RA, Amit I, Segal E, Elinav E. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. CELL. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. SciU.SA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, Huerta-Cepas J, Nieuwdorp M, Salojärvi J, Voigt AY, Zeller G, Sunagawa S, de Vos WM, Bork P. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- Li XS, Obeid S, Klingenberg R, Gencer B, Mach F, Räber L, Windecker S, Rodondi N, Nanchen D, Muller O, Miranda MX, Matter CM, Wu Y, Li L, Wang Z, Alamri HS, Gogonea V, Chung Y-M, Tang WHW, Hazen SL, Lüscher TF. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur. Heart J. 2017 doi: 10.1093/eurheartj/ehw582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. CELL. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Liu Lan, Guo X, Rao JN, Zou T, Xiao L, Yu T, Timmons JA, Turner DJ, Wang J-Y. Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am. J. Physiol., Cell Physiol. 2009;296:C801–10. doi: 10.1152/ajpcell.00620.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Lu, Li L, Min J, Wang J, Wu H, Zeng Y, Chen S, Chu Z. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cellular Immunology. 2012;277:66–73. doi: 10.1016/j.cellimm.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Löser C, Eisel A, Harms D, Fölsch UR. Dietary polyamines are essential luminal growth factors for small intestinal and colonic mucosal growth and development. Gut. 1999;44:12–16. doi: 10.1136/gut.44.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux GD, Marton LJ, Baylin SB. Ornithine decarboxylase is important in intestinal mucosal maturation and recovery from injury in rats. Science. 1980;210:195–198. doi: 10.1126/science.6774420. [DOI] [PubMed] [Google Scholar]
- Lührs H, Gerke T, Müller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 2002;37:458–466. doi: 10.1080/003655202317316105. [DOI] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6(6734) doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- Maehre HK, Jensen I-J, Elvevoll EO, Eilertsen K-E. ω-3 Fatty Acids and Cardiovascular Diseases: Effects, Mechanisms and Dietary Relevance. Int J Mol Sci. 2015;16:22636–22661. doi: 10.3390/ijms160922636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Manor O, Borenstein E. Systematic Characterization and Analysis of the Taxonomic Drivers of Functional Shifts in the Human Microbiome. Cell Host and Microbe. 2017;21:254–267. doi: 10.1016/j.chom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A, Shoaie S, Bergentall M, Ghaffari P, Zhang C, Larsson E, Bäckhed F, Nielsen J. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol. Syst. Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Marotz CA, Zarrinpar A. Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. Yale J Biol Med. 2016;89:383–388. [PMC free article] [PubMed] [Google Scholar]
- Marques CC, Castelo-Branco MT, Pacheco RG, Buongusto F, do Rosário A, Schanaider A, Coutinho-Silva R, de Souza HSP. Prophylactic systemic P2X7 receptor blockade prevents experimental colitis. Biochim. Biophys. Acta. 2014;1842:65–78. doi: 10.1016/j.bbadis.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J. Gastroenterol. 2008;14:5630–5640. doi: 10.3748/wjg.14.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinot E, Sèdes L, Baptissart M, Lobaccaro J-M, Caira F, Beaudoin C, Volle DH. Bile acids and their receptors. Mol. Aspects Med. 2017 doi: 10.1016/j.mam.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochemical and Biophysical Research Communications. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. CELL. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Melo-Gonzalez F, Hepworth MR. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology. 2017;150:265–275. doi: 10.1111/imm.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli A, Renga B, Migliorati M, Cipriani S, Distrutti E, Santucci L, Fiorucci S. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J. Immunol. 2009;183:6657–6666. doi: 10.4049/jimmunol.0901347. [DOI] [PubMed] [Google Scholar]
- Mente A, Chalcraft K, Ak H, Davis AD, Lonn E, Miller R, Potter MA, Yusuf S, Anand SS, McQueen MJ. The Relationship Between Trimethylamine-N-Oxide and Prevalent Cardiovascular Disease in a Multiethnic Population Living in Canada. Can J Cardiol. 2015;31:1189–1194. doi: 10.1016/j.cjca.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. The Prolongation of Life: Optimistic Studies. G. P. Putnam's Sons; 1908. [Google Scholar]
- Michael AJ. Biosynthesis of polyamines and polyamine-containing molecules. Biochem J. 2016;473:2315–2329. doi: 10.1042/BCJ20160185. [DOI] [PubMed] [Google Scholar]
- Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin Exp Immunol. 2002;130:245–255. doi: 10.1046/j.0009-9104.2002.01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015;427:3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- Miyauchi S, Gopal E, Fei Y-J, Ganapathy V. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na(+)-coupled transporter for short-chain fatty acids. J. Biol. Chem. 2004;279:13293–13296. doi: 10.1074/jbc.C400059200. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves AR, Castelo-Branco MTL, Figliuolo VR, Bernardazzi C, Buongusto F, Yoshimoto A, Nanini HF, Coutinho CMLM, Carneiro AJV, Coutinho-Silva R, de Souza HSP. Overexpression of ATP-activated P2X7 receptors in the intestinal mucosa is implicated in the pathogenesis of Crohn's disease. Inflamm. Bowel Dis. 2014;20:444–457. doi: 10.1097/01.MIB.0000441201.10454.06. [DOI] [PubMed] [Google Scholar]
- Nijmeijer RM, Gadaleta RM, van Mil SWC, van Bodegraven AA, Crusius JBA, Dijkstra G, Hommes DW, de Jong DJ, Stokkers PCF, Verspaget HW, Weersma RK, van der Woude CJ, Stapelbroek JM, Schipper MEI, Wijmenga C, van Erpecum KJ, Oldenburg B Dutch Initiative on Crohn, Colitis (ICC) Farnesoid X receptor (FXR) activation and FXR genetic variation in inflammatory bowel disease. PLoS ONE. 2011;6:e23745. doi: 10.1371/journal.pone.0023745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson NE, Kotarsky K, Owman C, Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochemical and Biophysical Research Communications. 2003;303:1047–1052. doi: 10.1016/s0006-291x(03)00488-1. [DOI] [PubMed] [Google Scholar]
- Nuttle LC, Dubyak GR. Differential activation of cation channels and non-selective pores by macrophage P2z purinergic receptors expressed in Xenopus oocytes. J. Biol. Chem. 1994;269:13988–13996. [PubMed] [Google Scholar]
- O'Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscsó B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infection and Immunity. 1991;59:2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Shortchain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruzza L, Gargari G, Proietti M, Fosso B, D'Erchia AM, Faliti CE, Rezzonico-Jost T, Scribano D, Mauri L, Colombo D, Pellegrini G, Moregola A, Mooser C, Pesole G, Nicoletti M, Norata GD, Geuking MB, McCoy KD, Guglielmetti S, Grassi F. T Follicular Helper Cells Promote a Beneficial Gut Ecosystem for Host Metabolic Homeostasis by Sensing Microbiota-Derived Extracellular ATP. Cell Rep. 2017;18:2566–2575. doi: 10.1016/j.celrep.2017.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cano FJ, González-Castro A, Castellote C, Franch A, Castell M. Influence of breast milk polyamines on suckling rat immune system maturation. Dev. Comp. Immunol. 2010;34:210–218. doi: 10.1016/j.dci.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JK, Virgin HW. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 2016;351 doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen J-P, Belzer C, de Vos WM, Cani PD. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan L-X, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proceedings of the National Academy of Sciences. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TWH, Nomura M, Harach T, Sasso, Lo G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, Auwerx J, Schoonjans K. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti M, Cornacchione V, Rezzonico-Jost T, Romagnani A, Faliti CE, Perruzza L, Rigoni R, Radaelli E, Caprioli F, Preziuso S, Brannetti B, Thelen M, McCoy KD, Slack E, Traggiai E, Grassi F. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer's patches to promote host-microbiota mutualism. Immunity. 2014;41:789–801. doi: 10.1016/j.immuni.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen Z-ME, Fish K, Fu Y-X, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Cervantes JL, Cicek BB, Mukherjee S, Venkatesh M, Maher LA, Salazar JC, Mani S, Khanna KM. Pregnane X Receptor Regulates Pathogen-Induced Inflammation and Host Defense against an Intracellular Bacterial Infection through Toll-like Receptor 4. Sci Rep. 2016;6:31936. doi: 10.1038/srep31936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. CELL. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K, Safdar N. Fecal microbiota transplantation for the treatment of Clostridium difficile infection. J Hosp Med. 2016;11:56–61. doi: 10.1002/jhm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6:e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao C-C, Patel B, Yan R, Blain M, Alvarez JI, Kébir H, Anandasabapathy N, Izquierdo G, Jung S, Obholzer N, Pochet N, Clish CB, Prinz M, Prat A, Antel J, Quintana FJ. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgeerts P, Ghoos Y, Vantrappen G. Bile acid studies in patients with Crohn's colitis. Gut. 1979;20:1072–1077. doi: 10.1136/gut.20.12.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Segain JP, Raingeard de la Blétière D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottière HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seljeset S, Siehler S. Receptor-specific regulation of ERK1/2 activation by members of the “free fatty acid receptor” family. J. Recept. Signal Transduct. Res. 2012;32:196–201. doi: 10.3109/10799893.2012.692118. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. Plos Biol. 2016a;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. CELL. 2016b;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1114–22. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- Shapiro H, Suez J, Elinav E. Personalized microbiome-based approaches to metabolic syndrome management and prevention. J Diabetes. 2017;9:226–236. doi: 10.1111/1753-0407.12501. [DOI] [PubMed] [Google Scholar]
- Sharma S, Erickson KM, Troutman JM. Complete Tetrasaccharide Repeat Unit Biosynthesis of the Immunomodulatory Bacteroides fragilis Capsular Polysaccharide A. ACS Chem. Biol. 2017;12:92–101. doi: 10.1021/acschembio.6b00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20:719–730. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Faith NG, Nakayama Y, Suresh M, Steinberg H, Czuprynski CJ. The aryl hydrocarbon receptor is required for optimal resistance to Listeria monocytogenes infection in mice. J. Immunol. 2007;179:6952–6962. doi: 10.4049/jimmunol.179.10.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lai X, Ye L, Chen K, Cao Z, Gong W, Jin L, Wang C, Liu M, Liao Y, Wang JM, Zhou N. Activated niacin receptor HCA2 inhibits chemoattractant-mediated macrophage migration via Gβγ/PKC/ERK1/2 pathway and heterologous receptor desensitization. Sci Rep. 2017;7:42279. doi: 10.1038/srep42279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, Franke A, Ott S, Häsler R, Nikolaus S, Fölsch UR, Rose-John S, Jiang H-P, Li J, Schreiber S, Rosenstiel P. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. 2009;183:7514–7522. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, Ganapathy V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. Journal of Biological Chemistry. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart ML, Gu B, Panchal RG, Wiley J, Cromer B, Williams DA, Petrou S. P2X7 receptor cell surface expression and cytolytic pore formation are regulated by a distal C-terminal region. J. Biol. Chem. 2003;278:8853–8860. doi: 10.1074/jbc.M211094200. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Micro. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Steege ter JC, Forget PP, Buurman WA. Oral spermine administration inhibits nitric oxide-mediated intestinal damage and levels of systemic inflammatory mediators in a mouse endotoxin model. Shock. 1999;11:115–119. doi: 10.1097/00024382-199902000-00008. [DOI] [PubMed] [Google Scholar]
- Steliou K, Boosalis MS, Perrine SP, Sangerman J, Faller DV. Butyrate histone deacetylase inhibitors. Biores Open Access. 2012;1:192–198. doi: 10.1089/biores.2012.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann. N. Y. Acad. Sci. 2014;1319:82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galán JE, Askenase PW, Flavell RA. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proceedings of the National Academy of Sciences. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu H-JJ, Kasper DL, Benoist C, Mathis D. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proceedings of the National Academy of Sciences. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Fan Y, Levison B, Hazen JE, Donahue LM, Wu Y, Hazen SL. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J. Am. Coll. Cardiol. 2014;64:1908–1914. doi: 10.1016/j.jacc.2014.02.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Shrestha K, Borowski AG, Wu Y, Troughton RW, Klein AL, Hazen SL. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. J. Card. Fail. 2015;21:91–96. doi: 10.1016/j.cardfail.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, Begum-Haque S, Kasper DL, Kasper LH. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes. 2015;6:234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terc J, Hansen A, Alston L, Hirota SA. Pregnane X receptor agonists enhance intestinal epithelial wound healing and repair of the intestinal barrier following the induction of experimental colitis. Eur J Pharm Sci. 2014;55:12–19. doi: 10.1016/j.ejps.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Thangaraju M, Carswell KN, Prasad PD, Ganapathy V. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J. 2009a;417:379–389. doi: 10.1042/BJ20081132. [DOI] [PubMed] [Google Scholar]
- Thangaraju M, Cresci G, Itagaki S, Mellinger J, Browning DD, Berger FG, Prasad PD, Ganapathy V. Sodium-coupled transport of the short chain fatty acid butyrate by SLC5A8 and its relevance to colon cancer. J. Gastrointest. Surg. 2008;12:1773–81. doi: 10.1007/s11605-008-0573-0. discussion 1781–2. [DOI] [PubMed] [Google Scholar]
- Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, Prasad PD, Ganapathy V. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009b;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature reviews. Drug discovery. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, Fändriks L, le Roux CW, Nielsen J, Bäckhed F. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Trøseid M, Ueland T, Hov JR, Svardal A, Gregersen I, Dahl CP, Aakhus S, Gude E, Bjørndal B, Halvorsen B, Karlsen TH, Aukrust P, Gullestad L, Berge RK, Yndestad A. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med. 2015;277:717–726. doi: 10.1111/joim.12328. [DOI] [PubMed] [Google Scholar]
- Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host and Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Gordon S, Imhof BA, Nuñez G, Bousso P. Élie Metchnikoff (1845–1916): celebrating 100 years of cellular immunology and beyond. Nat Rev Immunol. 2016;16:651–656. doi: 10.1038/nri.2016.89. [DOI] [PubMed] [Google Scholar]
- Ussher JR, Lopaschuk GD, Arduini A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis. 2013;231:456–461. doi: 10.1016/j.atherosclerosis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- van der Beek CM, Bloemen JG, van den Broek MA, Lenaerts K, Venema K, Buurman WA, Dejong CH. Hepatic Uptake of Rectally Administered Butyrate Prevents an Increase in Systemic Butyrate Concentrations in Humans. J. Nutr. 2015;145:2019–2024. doi: 10.3945/jn.115.211193. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, Speelman P, Dijkgraaf MGW, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J-C, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman A, Ramakrishna BS, Shaji RV, Kumar NSN, Pulimood A, Patra S. Amelioration of dextran sulfate colitis by butyrate: role of heat shock protein 70 and NF-kappaB. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G177–84. doi: 10.1152/ajpgi.00307.2002. [DOI] [PubMed] [Google Scholar]
- Vinolo MAR, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly-Y M, Stephens L, Hawkins PT, Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS ONE. 2011a;6:e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo MAR, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 2011b;22:849–855. doi: 10.1016/j.jnutbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Vrieze A, van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JET, Bloks VW, Groen AK, Heilig HGHJ, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JBL, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–6. e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- Wakade C, Chong R, Bradley E, Thomas B, Morgan J. Upregulation of GPR109A in Parkinson's disease. PLoS ONE. 2014;9:e109818. doi: 10.1371/journal.pone.0109818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Morinobu A, Horiuchi M, Liu J, Kumagai S. Butyrate inhibits functional differentiation of human monocyte-derived dendritic cells. Cellular Immunology. 2008;253:54–58. doi: 10.1016/j.cellimm.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang M, Soda K, Sama A, Tracey KJ. Fetuin protects the fetus from TNF. Lancet. 1997;350:861–862. doi: 10.1016/S0140-6736(05)62030-2. [DOI] [PubMed] [Google Scholar]
- Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Begum-Haque S, Telesford KM, Ochoa-Reparaz J, Christy M, Kasper EJ, Kasper DL, Robson SC, Kasper LH. A commensal bacterial product elicits and modulates migratory capacity of CD39(+) CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes. 2014;5:552–561. doi: 10.4161/gmic.29797. [DOI] [PubMed] [Google Scholar]
- Wang Y-D, Chen W-D, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-D, Chen W-D, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tang WHW, Buffa JA, Fu X, Britt EB, Koeth RA, Levison BS, Fan Y, Wu Y, Hazen SL. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 2014;35:904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J. Immunol. 2010;185:6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52:1442–1447. doi: 10.1136/gut.52.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A, Foord SM, Fraser NJ, Barnes AA, Elshourbagy N, Eilert M, Ignar DM, Murdock PR, Steplewski K, Green A, Brown AJ, Dowell SJ, Szekeres PG, Hassall DG, Marshall FH, Wilson S, Pike NB. Molecular identification of high and low affinity receptors for nicotinic acid. J. Biol. Chem. 2003;278:9869–9874. doi: 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- Wrzosek L, Miquel S, Noordine M-L, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC biology. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- Yaron JR, Gangaraju S, Rao MY, Kong X, Zhang L, Su F, Tian Y, Glenn HL, Meldrum DR. K(+) regulates Ca(2+) to drive inflammasome signaling: dynamic visualization of ion flux in live cells. Cell death & disease. 2015;6:e1954. doi: 10.1038/cddis.2015.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yissachar N, Zhou Y, Ung L, Lai NY, Mohan JF, Ehrlicher A, Weitz DA, Kasper DL, Chiu IM, Mathis D, Benoist C. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. CELL. 2017 doi: 10.1016/j.cell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am. J. Gastroenterol. 2012;107:1755–6. doi: 10.1038/ajg.2012.251. 1755–author reply. [DOI] [PubMed] [Google Scholar]
- Zhang M, Borovikova LV, Wang H, Metz C, Tracey KJ. Spermine inhibition of monocyte activation and inflammation. Mol. Med. 1999;5:595–605. [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Caragine T, Wang H, Cohen PS, Botchkina G, Soda K, Bianchi M, Ulrich P, Cerami A, Sherry B, Tracey KJ. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J. Exp. Med. 1997;185:1759–1768. doi: 10.1084/jem.185.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Ashok M, Li J, Li W, Yang H, Wang P, Tracey KJ, Sama AE, Wang H. Spermine protects mice against lethal sepsis partly by attenuating surrogate inflammatory markers. Mol. Med. 2009;15:275–282. doi: 10.2119/molmed.2009.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. CELL. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]