Abstract
Head and neck squamous carcinoma (HNSCC) is the sixth most prevalent cancer and one of the most aggressive malignancies worldwide. Despite continuous efforts to identify molecular markers for early detection, and to develop efficient treatments, the overall survival and prognosis of HNSCC patients remain poor. Accumulated scientific evidences suggest that epigenetic alterations, including DNA methylation, histone covalent modifications, chromatin remodeling and non-coding RNAs, are frequently involved in oral carcinogenesis, tumor progression, and resistance to therapy. Epigenetic alterations occur in an unsystematic manner or as part of the aberrant transcriptional machinery, which promotes selective advantage to the tumor cells. Epigenetic modifications also contribute to cellular plasticity during tumor progression and to the formation of cancer stem cells (CSCs), a small subset of tumor cells with self-renewal ability. CSCs are involved in the development of intrinsic or acquired therapy resistance, and tumor recurrences or relapse. Therefore, the understanding and characterization of epigenetic modifications associated with head and neck carcinogenesis, and the prospective identification of epigenetic markers associated with CSCs, hold the promise for novel therapeutic strategies to fight tumors. In this review, we focus on the current knowledge on epigenetic modifications observed in HNSCC and emerging Epi-drugs capable of sensitizing HNSCC to therapy.
Keywords: epigenetics, HNSCC (Head and Neck Squamous Cell Carcinoma), DNA methylation, histone modifications, microRNA, cancer stem cell, chemoresistance, acetylation, histone H3
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous group of tumors characterized by lesions in the oral cavity, larynx, pharynx (including nasopharynx, oropharynx, and hypopharynx), salivary glands, and thyroid [1,2,3]. HNSCC is the sixth most prevalent cancer worldwide, with approximately 780,000 new cases diagnosed each year and close to 350,000 deaths annually [4,5]. Despite several efforts to identify biomarkers for early detection and develop new treatments, the overall survival rate and prognosis remain poor [2,6,7]. The risk factors and carcinogens that participate in the development of HNSCC are well recognized [3,8]. Local recurrence and metastasis are limiting factors for the success of the treatment [9,10].
Over the years, cancer research efforts have focused on the genetic basis of tumor development and progression, identifying mutations and characterizing pathways that activate oncogenes and inactivate tumor suppressor genes. More recently, the research has pointed to epigenetic alterations as critic changes involved in the initiation and progression of human cancers [11]. Epigenetic alterations occur in an unsystematic way or as part of aberrant transcriptional machinery, promoting selective advantage to the tumor through the silencing of tumor suppressor genes or dysfunction in DNA repair genes [12,13]. Epigenetic modifications also contribute to the cellular plasticity during tumor progression, and the formation of tumor-initiating cells or cancer stem cells (CSCs) [14,15,16,17,18].
In this review, we discuss the current literature associated with the impact of epigenetic modifications on the progression and therapy of HNSCC, and examine aberrant DNA methylation, histone covalent modifications, chromatin remodeling, and non-coding-RNAs in HNSCC. We provide insights into recent studies on the epigenetics of CSCs, and novel treatment modalities using epigenetic drugs (Epi-drugs) alone, or in combination with conventional therapies.
2. Epigenetic Mechanisms in Cancer
The meaning of epigenetic was first introduced by Conrad Waddington in 1942 to define stable changes in the cell phenotype without genetic alterations [16,19]. Currently, epigenetic refers to stable and heritable changes in gene expression without changes in DNA sequence [20]. Epigenetic changes are fundamental mechanisms for carcinogenesis, and can serve as possible methods for early detection, treatment, and prognostic assessment for the cancer patients [21]. Epigenetic modifications include DNA methylation, histone covalent modifications, chromatin remodeling, and the effect of non-coding RNAs and polycomb proteins in gene expression [22,23]. Next, we will discuss the basis of epigenetics modifications.
2.1. DNA Methylation
The methylation of cytosine in CpG dinucleotides at the 5-carbon (5-methylcytosine) is a covalent modification of the DNA, and was the first and most explored chromatin modification [24,25,26]. CpG dinucleotides are highly accumulated in the 5′ promoter region of genes referred as CpG islands, which are unmethylated in transcriptionally active genes. Once CpG islands become methylated, transcriptional repression occurs [27].
A family of enzymes called DNA methyltransferases (DNMTs) controls methylation by catalyzing the transfer of the methyl group from S-adenosylmethionine to the cytosine. Three DNMTs (i.e., DNMT1, DNMT3a, and DNMTb) are fundamental to the methylation in mammals. DNMT1, called maintenance enzyme, is involved in restoring the parental DNA methylation profile after DNA replication, and exhibits a preference for hemimethylated DNA, ensuring the methylation status to the future cell generations [28,29,30]. DNA methylation in CpG dinucleotides, previously unmethylated or de novo DNA methylation, is carried out by DNMT3a and DNMT3b [12,26,30,31,32]. Moreover, DNA methylation works in association with chromatin modifications to repress gene expression [25]. DNA methylation provides a platform for many methyl-binding proteins, including MBD1, MBD2, MBD3, and MeCP2. These proteins operate by recruiting histone-modifying enzymes to coordinate chromatin dynamics [26,33].
Loss of DNA methylation was the first epigenetic alteration characterized in benign and malignant cancer cells [12,24,30]. Global DNA hypomethylation in cancer targets various genomic sequences, including repetitive elements, transposons, CpG dinucleotides in introns, and gene deserts, increasing genomic instability and activating proto-oncogenes [34,35]. In contrast, hypermethylation of the CpG islands on gene promoters contributes to the carcinogenesis through the silencing of tumor suppressor genes. For example, hypermethylation of E-cadherin, pRB, p53, and CDKN2A (p16INK4a/p14ARF), are common findings observed in cancer cell lines and primary tumors that result in gene silencing [36]. In fact, the use of next generation sequencing platforms has shown outstanding rates of abnormal CpG promoter methylation (5% to 10%) in various cancer types [25,26].
2.2. Covalent Histone Modification
The basic unit of the chromatin is the nucleosome, which consists of ~147 bp of DNA wrapped around a histone octamer containing two copies of four histone proteins (i.e., H2A, H2B, H3, and H4). Chromatin-modifying enzymes dynamically execute post-translational modifications (PTMs) of histones and DNA in a tightly regulated mechanism [26,37]. Histone PTMs are also an important mechanism that regulates chromatin structure and function [38]. Alterations in the patterns of histone PTMs are present in cancer at particular genes and global levels [37,39]. Histone tail projections from the octamer at the nucleosome undergo several post-translational covalent modifications involving the addition of chemical groups, such as methyl, acetyl, and phosphate. Less frequent alterations include ubiquitination, sumoylation, and ADP-ribosylation. These changes occur on the histone proteins at amino acid residues lysine, arginine, and serine [35,40].
Acetylation of lysine is the most important histone modification associated with transcription, chromatin architecture, and DNA repair. The addition of the acetyl group neutralizes the positive charge of the histone, weakening the electrostatic interaction between histones and the negatively charged DNA, promoting relaxation of the chromatin conformation favoring gene transcription [41,42,43]. Besides modification of histone charge, histone acetylation may also regulate intracellular pH. It is interesting that many tumors display low cellular pH and reduced levels of acetylated histones. Furthermore, the presence of low pH in tumors is also associated with poor prognosis for cancer patients [44]. In addition, histone acetylation has a role in recruitment of the general transcription machinery. In eukaryotes, general transcription is mediated by RNA polymerase II after the assembly of the preinitiation complex by the Transcription factor II D (TFIID). TFIID recognizes and selectively binds to sites with multiply acetylated histone H4 at the promoter [45]. Remarkably, the function of TFIID itself is regulated by TAFII250, which also has an acetyltransferase activity [46].
The addition of acetyl groups to lysine at the histone tails is catalyzed by enzymes called histone acetyltransferases (HATs), while histone deacetylases (HDACs) are responsible for removing acetyl groups. Although HATs are commonly driving gene expression, combined activation of HAT and HDAC is required for proper regulation of transcription [37,39]. HATs are classified into three major groups with nuclear location: (I) MOZ/YBF2/SAS2/TIP60, which belong to the MYST family; (II) GCN5 N-acetyltransferase or GNAT family; and (III) CBP/p300 family [47,48]. Different HATs work as both oncogenes and tumor suppressors, suggesting that a balanced acetylation is critical for cellular homeostasis [49]. HATs, such as the p300/(CREB binding protein) associated factor (PCAF), p300, and the CREB-binding Protein (CBP), target multiple non-histone proteins for acetylation, resulting in a gain of function for proteins such as p53 and BCL-6 [37,48]. For example, the HAT, Tip60, is involved in tumorigenesis through the modulation of ATM and the DNA damage response pathway, as well as the transcriptional activation of p53 and Myc. Down-regulation of Tip60 leads to genomic instability and impairment of the apoptotic signaling cascade, thereby helping the establishment of malignant transformation [50,51].
There are 18 human HDACs that are grouped into four classes according to the sequence homology to yeast. Class I (i.e., Rpd3-like enzymes) is comprised of HDAC1, 2, 3 and 8; and Class II (i.e., Hda1-like enzymes) is subdivided into IIa and IIb subclasses. HDAC4, 5, 7 and 9 belong to class IIa, and HDAC6 and 10 are in class IIb. Class III (i.e., Sir-like enzymes) consists of seven members called Sirtuins (SIRTs) which are NAD-dependent deacetylases. Class IV contains only HDAC11, which shares sequence homology with both HDACs class I and II. HDACs can regulate tumorigenesis through different mechanisms, such as downregulation of tumor suppressor genes and activation of oncogenic cell-signaling pathways [52]. HDACs possess low substrate specificity, and each enzyme can deacetylate multiple divergent histone sites. Although mutations in HDACs are not frequent, overexpression of HDACs is common in cancer [37,39,40]. HDACs, as well as HATs, act directly on non-histone proteins that are involved in tumor migration and metastasis. For example, HDAC2 deacetylates p53 and Cyclin-dependent kinase inhibitors 1B, 1C and 2A, which deregulates the apoptotic machinery, and disrupts the cell cycle [37,53,54].
Histone methylation in lysine, arginine, and histidine residues, does not alter the overall charge of the molecule, but notably, activates or represses gene transcription. The well-characterized sites of methylation occur on lysine residues, which can be mono-, di-, or tri-methylated. While histone methylases (HMT) add methyl groups, histone demethylases counteract this action by removing methyl groups from the histone tails [55]. Additionally, active genes typically display histone methylation markers such as H3K4me3, H3K36me3, and H3K79me3, while transcriptionally silenced genes frequently present repressive markers such as H3K27me3, H3K9me2, and H3K9me3 [56]. Often observed in several types of cancers, a member of HMT family responsible for the transcriptional (MLL1) displays loss of function and alters the expression of critical genes associated with cellular differentiation [57].
2.3. Chromatin Remodeling and Associated Proteins
Histone covalent modifications and DNA methylation are associated with a higher organization of the chromatin structure. The general process of inducing alterations in chromatin organization is named chromatin remodeling, which is an important mechanism for gene transcription. It requires modifications on the nucleosome structure to allow the transcription machinery to gain access to promoter regions [35]. Chromatin remodeler enzymes act through multiple mechanisms to overcome the barrier created by a densely-packed nucleosome. Four families of chromatin remodelers utilize energy derived from ATP hydrolysis to modify nucleosome organization, granting access to chromatin and target genes. They are the SWI/SNF family (switching defective/sucrose non-fermenting), the ISWI family(imitation switch), the NuRD family (Mi-2/nucleosome remodeling and histone deacetylation), and the INO80 family (inositol requiring 80). Deregulation of chromatin remodelers constitutes a common event in cancer development, progression, and therapeutic resistance [42]. Cancer-associated modifications of chromatin remodelers such as SNF5 (part of the subunit of the SWI/SNF chromatin remodeling complex), Brahma-related gene-1 (BRG1), and MTA family members (Metastasis-associated gene), are frequently found mutated in malignancies, suggesting a tumor suppressor role [58,59,60]. Overexpression of NuRD family members is often associated with the invasive potential of multiple cancers. The extension of modifications on chromatin remodeler enzymes becomes more evident with cancer genome sequencing data, which identified that ~20% of human tumors contain mutations in at least one member of the SWI/SNF complex [58,59,60,61,62]. Furthermore, loss of function of SNF5, a member of SWI/SNF family, results in enhanced susceptibility to multiple early childhood cancers, mediated by elevated expression of the polycomb gene EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit), resulting in increased H3K27me3 and cell cycle progression [63,64,65]. Conversely, RSF1 (Remodeling and Spacing Factor 1) gain of function is observed in a variety of human cancers, and is directly associated with tumor aggressiveness, poor therapeutic response, reduced survival, and poor prognosis [66,67,68].
2.4. Non-Coding RNA
In recent years, there is increased knowledge about non-coding ribonucleic acid (ncRNA), which goes above and beyond the well-known transfer RNA (tRNA) and ribosomal RNA (rRNA). Notably, a significant portion of the eukaryotic genome is transcribed into RNAs without protein- or peptide-coding function [69,70]. Many ncRNAs have several regulatory functions of mammalian organisms, particularly gene regulation at the levels of transcription, RNA processing, and translation. ncRNAs exploit the power of base pairing to selectively bind and directly act on other nucleic acids, but also act on epigenetic regulators. The majority of ncRNAs are involved in epigenetic regulations, mediating changes in chromatin conformation by directly targeting promoter regions, and consequently activating or repressing transcription [71]. ncRNA can be classified in microRNAs (miRNAs), PIWI-associated small RNAs (piRNAs) and long non-coding RNAs (lncRNAs), small interfering RNAs (siRNAs), enhancer RNAs (eRNAs), promoter-associated RNAs (PARs), among other RNAs, which play essential roles in regulating stem cells, cellular homeostasis, and contribute to the natural history of many human diseases, including cancer [72,73].
MicroRNAs (miRNAs) constitute the largest class of ncRNAs that regulate diverse cellular functions, including apoptosis, metabolism, cell growth, and differentiation. Deregulation in miRNAs expression changes normal cellular functions and is associated with the development of human diseases, including cancer [74,75,76]. miRNAs are comprised of 18–24 nucleotides, which usually act as post-transcriptional regulators of gene expression that recognize and bind to complementary sequences of target genes. miRNAs also inhibit protein synthesis via degradation of mRNA transcripts or repression of the translational machinery [77,78]. Deregulation of miRNA expression is found in cancer initiation and progression of most human cancers [72,75]. miR-21, for example, targets the tumor suppressor genes PTEN (Phosphatase and Tensin Homolog) and PDCD4 (Programmed Cell Death 4) in several neoplasms [79,80]. The miRNAs can work as either tumor suppressors (e.g., miR-29b and miR-30) or display oncogenic characteristics (e.g., miR17-92 cluster) influencing cellular growth [74,81].
Interestingly, a subgroup of miRNAs also called epi-miRNA acts as target effectors of the epigenetic machinery, including DNMTs, HDACs, and polycomb genes, suggesting that miRNAs can indirectly regulate gene expression by interacting with epigenetic processes [35], for example, miR-29, that governs the expression of DNMT3a, 3b, and DNMT1 in lung cancer and acute myeloid leukemia, and miR-1, miR-140, and miR-449a, that regulate several HDACs in prostate cancer [71,74,82,83]. In contrast, several miRNAs are under epigenetic control in human cancers, due to its localization in CpG island regions, or epigenetic silencing mediated by promoter hypermethylation and histone modifications [84,85]. These facts expose the complexity of epigenetic changes in human carcinogenesis.
3. Epigenetic Modifications Associated with Tumor Progression and Drug Resistance in HNSCC
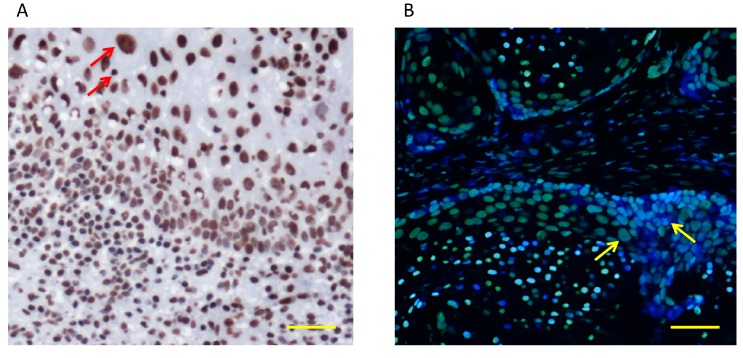
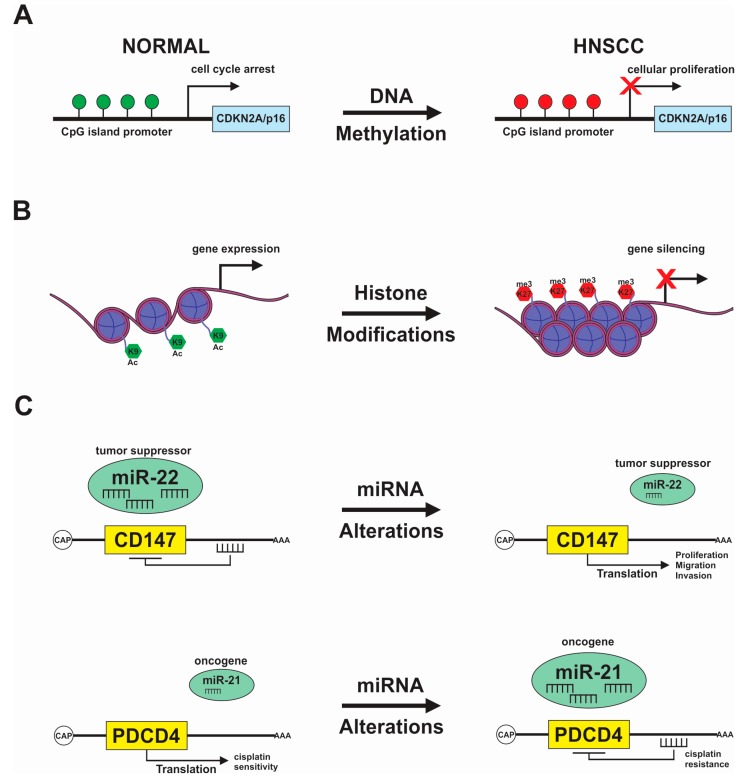
Despite recent advances in cellular and molecular biology techniques, the diagnosis of neoplasia is ultimately achieved by pathologists capable of identifying specific tissue characteristics and morphological features. Some of the microscopically-observed changes in tumor cells comprise alterations in the nuclear architecture and size (Figure 1). Indeed, changes in nuclear size, the presence of a condensed nuclear structure, prominent nucleoli, dense hyperchromatic chromatin, and a high nuclear-cytoplasmic ratio, are typical changes observed in HNSCC. These morphological changes suggest profound alterations in the chromatin structure and function of cancer cells [11]. Additionally, alterations in the nuclear morphology are predominantly associated with large-scale changes in gene expression mediated by the unbalance in global histone acetylation through redistribution of HDAC3 [86]. In HNSCC, reduction of nuclear size mediated by low levels of histone acetylation increased resistance to intercalating agents and reduced influx of DNA damage repair proteins to the nucleus [13]. Also, pharmacological inhibition of HDAC classes I and II decreased HNSCC proliferation and reduced the number of CSCs, a known subpopulation of cancer cells involved in tumor progression and the development of resistance to therapy [87]. The complexity of gene regulation by DNA methylation, histone modifications, chromatin remodeling and miRNAs are important mechanisms in tumor formation and progression; these mechanisms are currently active areas of research in head and neck cancer (Figure 2).
Figure 1.
Changes in nuclear morphology may be associated with epigenetic modifications and deregulation of gene expression. (A) Immunocytochemistry of a head and neck squamous carcinoma (HNSCC) tumor sample stained for acetyl-H3 (Lys9). Note differences in the nuclear size and high levels of histone acetylation (red arrows, dark brown); (B) Immunofluorescence of an HNSCC tumor sample stained for acetyl-H3 (Lys9). Note differences in the nuclear size and protein accumulation depicted by yellow arrows. Scale bars represent 50 μm. Experimental procedure is described in [88].
Figure 2.
Overview of epigenetic alterations involved in the HNSCC progression and resistance to therapy. (A) DNA hypermethylation of the tumor suppressor CDKN2A/p16INK4a is the most frequent epigenetic modification observed in HNSCC, leading to cellular proliferation; (B) Changes in the chromatin structure lead to histone deacetylation and methylation of lysine in the histone tails resulting in gene silencing; (C) MicroRNAs work as a tumor suppressor and oncogenes regulate genes involved in HNSCC tumorigenesis. miR-22 acts as a tumor suppressor, controlling the levels of CD147 and it is frequently downregulated in HNSCC, increasing processes like proliferation, migration, and invasion (upper image). miR-21 is accumulated in HNSCC, showing oncogenic characteristics by targeting PDCD4 gene increasing resistance to cisplatin treatment (lower image).
3.1. DNA Methylation Signature in HNSCC
Profiling DNA methylation constitutes a widely-applied tool to differentiate cancer from normal cells, as well as to identify subtypes of cancers, and to predict therapy outcome. Several studies have been carried out to explore the association between changes in DNA methylation and tumor progression of HNSCC. In fact, the DNA hypermethylation of tumor suppressor genes is an earlier event in carcinogenesis, and is responsible for uncontrolled cellular proliferation, which suggests that DNA hypermethylation is a potential marker for early diagnosis of HNSCC [89].
Over the years, DNA hypermethylation has been the most assessed epigenetic modification in HNSCC; however, the presence of global hypomethylation is a common finding during the tobacco-associated development of oral squamous cell carcinomas (OSCC) [90]. Methylation profiles analyzed in oral rinse revealed hypomethylation of LINE-1 (group member of the long interspersed nuclear elements family—LINEs) independent of the tumor grade, site, or risk factors, which suggests the potential use of LINE-1 as a biomarker for early detection of OSCC [77,91]. Hypomethylation of individual genes, such as the WISP1 gene (WNT-inducible-signaling pathway protein 1), was identified in OSCC associated with lymph node metastasis, suggesting its use as a potential diagnostic marker [92]. Survivin is a well-known protein related to tumor progression, and responsible for increased cellular proliferation and reduced apoptosis. Notably, promoter hypomethylation of BIRC5 (the gene that encodes surviving) is frequently found in OSCC, and leads to a more aggressive and invasive tumor phenotype [77,93]. DNA hypomethylation may also be involved in anticancer drug resistance, which results in accumulation of the ATP-binding cassette subfamily B member 1 (ABCB1), and cisplatin resistance in OSCC [94,95]. Furthermore, hypermethylation of CpG islands in the promoter region is associated with HNSCC formation and progression. Some examples of the frequently methylated genes in HNSCC are shown in Table 1 [96,97].
Table 1.
List of the frequently altered genes by DNA methylation in HNSCC. N/A: not applicable.
| Gene ID | HGNC ID | Function | Methylation Status | References |
|---|---|---|---|---|
| CDKN2A (p16; p14ARF) | HGNC:1787 | cell cycle arrest/apoptosis/senescence | hypermethylated | [5,77,89,98,99,100,101,102,103,104] |
| MGMT | HGNC:7059 | DNA damage repair | hypermethylated | [5,77,89,98,100,101,102,103,104] |
| DAPK | HGNC:2674 | apoptosis | hypermethylated | [5,89,98,100,101,102,103,104] |
| APC | HGNC:583 | cellular adhesion/migration | hypermethylated | [5,77,98,100,101,102] |
| RASSF1 | HGNC:9882 | cell cycle arrest/ cytoskeleton organization | hypermethylated | [5,77,89,101,103,104] |
| CDH1 | HGNC:1748 | cellular adhesion | hypermethylated | [77,100,102,103,104] |
| HOXA9 | HGNC:5109 | cell differentiation | hypermethylated | [91,98,105,106] |
| MLH1 | HGNC:7127 | DNA damage repair | hypermethylated | [5,77,101] |
| CDKN2B (p15) | HGNC:1788 | cell cycle arrest | hypermethylated | [5,77,100,101] |
| TIMP3 | HGNC:11822 | extracellular matrix degradation | hypermethylated | [89,98,103,104] |
| ATM | HGNC:795 | DNA damage repair | hypermethylated | [5,107] |
| MINT31 | N/A | chromatin remodeling | hypermethylated | [89,98,100] |
| CALCA | HGNC:1437 | cellular metabolism/ inflammatory response | hypermethylated | [99,103,106] |
| NPY | HGNC:7955 | cell proliferation | hypermethylated | [105,106] |
| HS3ST2 | HGNC:5195 | circadian rhythm | hypermethylated | [105,106] |
| ADGRE3 (EMR3) | HGNC:23647 | cellular surface receptor /inflammatory response | hypomethylated | [105] |
| PI3 | HGNC:8947 | inflammatory response | hypomethylated | [105] |
| AIM2 | HGNC:357 | apoptosis/ inflammatory response | hypomethylated | [105] |
| SPP1 | HGNC:11255 | cellular adhesion/ inflammatory response | hypomethylated | [105] |
Several studies show that p16INK4a (p14ARF) is one of the most hypermethylated genes in HNSCC. p16INK4a is a tumor suppressor gene encoded by the CDKN2A locus [5,77,98,99,100,101,102,105] The p16 function limits G1 cell cycle progression by inhibiting the cyclin-dependent kinases CDK4 and CDK6, and consequently compromises control of cellular proliferation, in addition to increased angiogenesis [103]. Several studies have suggested that p16INK4a hypermethylation could be used as a biomarker in the prediction of malignant transformation, due to the methylation-associated metastasis and poor survival in HNSCC [77]. Inactivation of p15INK4b (CDKN2B) associated with p16INK4a hypermethylation observed in precancerous oral tissues, suggests that methylation of these genes constitute early events in oral pathogenesis [99,104].
Hypermethylation of E-cadherin and DNA repair genes are also frequent events associated with tumor progression and invasion. Silencing of E-cadherin leads to the deregulation of essential cell functions, like adhesion, polarity, and morphogenesis, which results in increased motility and aggressive behavior of OSCC cells [5,99]. Hypermethylation of DNA repair genes such as MGMT (06-methylguanine-DNA methyltransferase), a DNA repair enzyme responsible for removing adducts from the DNA, results in reduced apoptosis and increased resistance of cancer cells to alkylating agents [5].
One of the most appealing areas of research in epigenetic modifications is the discovery and characterization of biomarkers. Epigenetic biomarkers have the potential to serve as markers for early stages of cancer to the identification of tumors with higher chances to develop resistance to therapy, or with great odds to produce local and distant metastasis. Discovery of biomarkers using samples of body fluids has been explored with the intent to improve screening accuracy and cost-effectiveness and decrease invasiveness. For head and neck cancer, saliva and oral rinse have been used to obtain DNA from the oral epithelium to analyze the DNA hypermethylation profile [101,108] Among potential biomarkers for oral carcinogenesis, HOXA9 (Homeobox A9) and NID2 (Nidogen 2) hold high promise due to their apparent sensitivity and specificity. Additional methylated genes, such as HS3ST2, NPY, EYA4, WT1, and the combination of E-cadherin, TMEFF2, and MGMT methylations, could also serve as potential biomarkers for early detection of HNSCC [78,99,107]. CHFR (i.e., Checkpoint with FHA and RING finger domains) hypermethylation is a late event in HNSCC, suggesting an association with tumor progression and potential use as a biomarker of late stage disease progression [99]. The identification of epigenetic biomarkers is paving the road to more attractive and reliable screenings to identify the development of HNSCC and better characterize subtypes of tumors to help implement personalized treatments [77].
Interestingly, few signaling pathways seem to be particularly vulnerable to DNA methylation in cancers. This is the case of the WNT molecular signaling, which is responsible for regulating the transcription of β-catenin, a key molecule involved in the carcinogenesis process, the proliferation of tumor cells, and the promotion of tumor survival. In oral cancer, several genes related to WNT signaling are found silenced by DNA methylation, including SFRP (Secreted frizzled-related protein), SOX17 (SRY-box 17), and WIF1 (WNT inhibitory factor 1) [100]. WNT signaling is a well-characterized pathway activated in embryogenesis and the maintenance of stem cells. Likewise, aberrant activation of WNT signaling is observed in cancers and leads to the accumulation of cancer stem cells, which is involved in the tumor progression, metastasis, and resistance to therapy [106,109].
3.2. Histone Acetylation and Chromatin Modifications in HNSCC
The dynamic mechanism of core histones is mediated by several posttranslational modifications that take place on histone tails as lysine acetylation. Histone acetylation is highly controlled by a balance between HATs and HDACs [39]. Given that histone modifications drive the chromatin organization and gene expression, it is no surprise that abnormal alterations in histone acetylation are associated with cancer development [52]. Chromatin is divided into two distinct conformations: heterochromatin, which is densely compacted and transcriptionally silenced, and euchromatin, which is less condensed and transcriptionally active. The structural status of the chromatin ultimately results in the transcriptional position of the gene [48].
The identification of a vast number of acetylation sites has been challenging because of the reversible nature and low abundance of this modification [110]; therefore, few studies described alterations in histone acetylation at specific lysine residues in HNSCC. Our group identified loss of histone H3K9ac as a marker for chemoresistance, associated with NFκB signaling [13] and accumulation of CSC [88,111]. We also showed that reduction of H3K9ac is associated with increased cell proliferation and activation of epithelial-mesenchyme transition (EMT) during oral carcinogenesis, suggesting the involvement of H3K9ac in tumor progression of HNSCC [87,112]. Furthermore, low levels of H3K4ac and high levels of H3K18ac are associated with OSCC progression and poor prognosis [113].
Changes in the expression of several HDACs were reported in multiple cancers and associated with the regulation of cell cycle-associated genes, cellular differentiation, apoptosis, angiogenesis, invasion, and migration [114,115,116]. For instance, HDAC6 is overexpressed in advanced stages of HNSCC, suggesting that its activity may be important in determining tumor aggressiveness in oral cancers [117]. HDAC6 can deacetylate α-tubulin increasing cell motility, a fundamental process in the development of tumor metastasis [77]. HDAC2 is overexpressed in OSCC that results in enhanced stability of the protein HIF-1α, which leads to increased invasion and migration of HNSCC [77,118]. HDAC7 is also overexpressed in head and neck cancer, and its accumulation activates c-MYC, promoting cellular proliferation [119]. Accumulation of HDAC8 induces proliferation by inhibiting activation of caspases and autophagy in OSCC [120]. HDAC9 has an oncogenic role in OSCC, increasing tumor growth by targeting pro-apoptotic genes, and its overexpression was correlated with reduced overall survival [114].
The SIRT family (Sirtuins) belongs to class III of NAD+-dependent histone deacetylases, which are implicated in the cell cycle, transcription regulation, and metabolism. SIRTs play opposite roles in cancer, acting as a tumor suppressor in some cancers, protecting against DNA damage and oxidative stress, and promoting cancer in others [121,122]. SIRT1, 2, 3, 5 and 7 are diminished in advanced stages of HNSCC and can be potential prognostic markers [123], while SIRT6 is accumulated in peripheral blood of patients with HNSCC [124].
Chromatin remodelers are important molecules responsible for controlling chromatin accessibility and nucleosome positioning in response to stimuli for DNA-driven biological processes. Disruption of chromatin remodelers is intimately implicated in cancer development, progression and therapeutic resistance, due to the impact on deregulated gene transcription machinery. Given these functions, a better insight into chromatin remodelers will provide the basis for the development of new anticancer therapeutic strategies [42]. ACTL6A is a subunit of SWI/SNF complex of chromatin remodeling. In HNSCC, ACTL6A is highly expressed, working as an oncogene changing chromatin organization of key genes, and is associated with loss of cellular differentiation and increasing of proliferation [125]. ING4 belong to ING family of chromatin remodelers, and has been associated with induction of apoptosis, cell cycle arrest, tumor growth, and angiogenesis [126,127,128]. ING4 is found either in the nucleus or the cytoplasmic in HNSCC. Increased levels of cytoplasmic ING4 are often involved in malignant progression, whereas nuclear ING4 may modulate the transactivation of target genes, promoting apoptosis and cell cycle arrest [129]. Another chromatin remodeler frequently deregulated in cancer is RSF-1, a member of ISWI family found up-regulated in OSCC, which displays increased tumor invasion, lymph node metastasis, and advanced tumor stages. RSF-1 also potentiates resistance to radiotherapy and chemotherapy through remodeling the chromatin of target genes [130].
Alterations in chromatin remodelers are not only important in the carcinogenesis process but also in the development of resistance to therapy. BRG1 and BRM are catalytic components of SWI/SNF, which mediate resistance to cisplatin in HNSCC. MTA1 overexpression induces cisplatin resistance in nasopharyngeal carcinoma by promoting cancer stem cell accumulation in a PI3K/AKT-dependent manner [131,132]. MTA1 is an important chromatin modifier that is upregulated in HNSCC. MTA1 regulates E-cadherin, and the WNT/β-catenin pathway activates EMT, favoring tumor invasion [133].
3.3. Non-Coding RNAs in HNSCC
MicroRNAs are a class of non-coding RNAs that is well investigated in HNSCC. Similar to proteins, miRNA dysfunction can be driven by alterations in miRNA expression resulting from gene mutations, epigenetic modifications, or deficiency in the miRNA processing. Genetic and epigenetic alterations in several miRNAs were correlated with cancer [74]. In OSCC, miR-26, -34b, -137, -193a and -203 are found downregulated via CpG hypermethylation [134]. Interestingly, miR-26a targets DNMT3B enzyme, stimulating cell proliferation and demonstrating the intimate association between different epigenetic mechanisms in oral carcinogenesis [135,136].
MicroRNAs can work as either tumor suppressor or oncogene to influence cellular growth [81]. For example, overexpression of miR-21 works as an oncogene when targeting PDCD4 gene, resulting in increased incidence of metastasis and enhanced invasive potential in OSCC [137]. miR-184 promotes the accumulation of c-MYC, inducing cell proliferation and overexpression of miR-24, that targets CDKN1B and decreases its levels promoting cell proliferation in oral cancer [138]. miR-211 is frequently overexpressed in cancer, and associated with tumor progression, nodal metastasis, vascular invasion, and poor prognosis of oral carcinoma [77].
In contrast, many miRNAs that play a role as tumor suppressors are often found downregulated in HNSCC. For example, miR-7 suppresses tumorigenesis by targeting the IGFR/AKT pathway and restricts cellular proliferation [139]. Similarly, downregulation of miR-22 results in the promotion of cell proliferation and increases tumor migration mediated by the overexpression of CD147, which suggests that miR-22 displays a tumor suppressor activity [140]. Other examples of reduced levels of miRNA leading to aggressive tumor behavior includes miR-195, which results in increased cell proliferation by targeting cyclin D and Bcl-2 [136], miR-222 and miR-34a, that results in increased MMP production in tongue squamous cell carcinoma [136,138], and miR-138, that results in enhanced migration and invasion through the accumulation of RhoC and ROCK2, and is also a potential therapeutic target for patients with oral cancer [139]. Moreover, miR-219 expression remarkably suppresses cell proliferation, colony formation, migration, and invasion by targeting PRKCI in HNSCC, suggesting that miRNAs play a significant role in tumor suppression [138].
Over 10% of all characterized miRNAs are associated with a chemoresistance phenotype by affecting central mechanisms involved in the development of cancer chemoresistance phenotypes, such as drug efflux, apoptotic dysfunction, cellular proliferation, and DNA damage repair [82,141,142,143,144,145,146,147]. miRNAs are also correlated with chemoresistance of HNSCC cells (Table 2). In HNSCC, increased levels of miR-21 are associated with low levels of PTEN and TPM1, as well as PDCD4. Accumulation of miR-21 has been widely observed in several cancers, and is related to its oncogenic function and role in resistance to cisplatin treatment [80,83]. miR-23a and -214 are also associated with chemoresistance. In oral cancer, both miRNAs are overexpressed, attenuating resistance to cisplatin by upregulating TOP2B [138].
Table 2.
Deregulation of miRNAs associated with chemoresistance in HNSCC.
| ID | Expression | Drug Resistance | Reference |
|---|---|---|---|
| miR-21 | increased | cisplatin | [148] |
| miR-634 | increased | paclitaxel | [149] |
| miR-181a | increased | cisplatin | [150] |
| miR-23a | increased | cisplatin | [151] |
| miR-10b | increased | cisplatin | [152] |
| miR-98 | increased | doxorubicin | [153] |
| miR-214 | increased | cisplatin | [140] |
| miR-200b | decreased | cisplatin | [154] |
| miR-15b | decreased | cisplatin | [154] |
4. Epigenetic Modifications of Cancer Stem Cells
The cancer stem cells (CSCs) model implicates that each tumor consists of a heterogeneous population of tumor cells and a small subpopulation of cancer cells endowed with stem cell properties and the ability to generate new tumors and metastasize. CSCs are undifferentiated, pluripotent, and have the ability of self-renewal, giving rise to other malignant daughter cells. Tumor cells are usually responsive to conventional chemotherapy, whereas CSCs are resistant to therapy, and will ultimately re-establish the tumor at the local or distant sites [35,148,155,156] Therefore, a better understanding of the regulatory mechanisms that control the population of CSCs is essential to the development of more efficient therapeutic strategies against HNSCC.
Despite the uncertain origin of CSCs, it is widely accepted that CSCs share key characteristics of embryonic stem cells, that include the capacity for self-renewal and the maintenance of an undifferentiated phenotype. CSCs can block differentiation by suppressing gene expression through epigenetic reprogramming, altering the profile of histone modifications and DNA methylation, similar to what is seen in embryonic stem cells [149]. Proteins from the polycomb group (PcG) are involved in the epigenetic regulation of gene expression, and play a fundamental role in the maintenance and differentiation of stem cells. EZH2 is a histone methyltransferase that works as a subunit of PcG. EZH2 catalyzes the trimethylation of lysine 27 on histone H3 (H3K27me3), regulating the expression of genes that determine the balance between cell differentiation and proliferation. Overexpression of EZH2 promotes self-renewal, prevents stem cells differentiation, and is associated with aggressiveness and poor prognosis in oral cancer [105,149].
BMI1 is a member of the polycomb group, which promotes chromatin silencing. BMI1 is required for the maintenance of adult stem cells, and is involved in carcinogenesis of different cancers, and may be a marker for cancer stem cells in HNSCC [150,151]. Wnt/β-catenin pathway has important functions in the self-renewal and differentiation programs of CSC. Besides promoter hypermethylation, deregulation of the Wnt/β-catenin pathway is also mediated by histone modifications [18,152]. Epigenetic mechanisms also upregulate several genes associated with Notch and Hedgehog signaling during CSC accumulation, self-renewal, and differentiation in carcinogenesis. Aberrant epigenetic modifications may deregulate a complex signaling process that modulates normal stem cells activities during tumor formation. These alterations ultimately contribute to CSC proliferation and maintenance, as well as tumor progression and invasion. Therefore, epigenetic regulation of key pathways may work as potential targets for therapy against CSCs [18].
CSCs tend to be more resistant to chemo and radiotherapy compared to non-CSC tumor cells, due to specific mechanisms like increased expression of drug efflux transporters [153,154,157,158], decreased expression of drug uptake transporters [159], increased drug detoxification [160], prolonged cell cycle arrest [161], increased DNA damage repair [162], activation of EMT [163,164], and epigenetic reprogramming [148]. Of note, our group and others have shown that treatment with cisplatin, the most common chemotherapeutic agent against head and neck cancer, promotes accumulation of CSCs, probably through mechanisms involving loss of global histone acetylation and epigenetic reprogramming [88,111,165].
Multidrug resistance refers to the ability of cancer cells to resist different anticancer drugs, which is one of the major clinical obstacles in cancer chemotherapy. Multidrug resistance can be caused by several mechanisms, including an increase of efflux transporters [166]. CSCs display high expression of drug efflux pumps, such as ATP-binding cassette (ABC) transporter family proteins, which are important for efflux of drugs across the plasma membrane [155,167]. Elevated levels of ABCG2 is a marker to identify CSCs in oral cancer [168]. Interestingly, ABCG2 is epigenetically regulated by miR-222 in tongue squamous cell carcinoma, and deregulation of the miR-222–ABCG2 contributes to cisplatin resistance, and cancer aggressiveness [169]. Also, ABCB1 and ABCG2 are expressed simultaneously in many cancer cells, including oral cancer [166]. They are found hypomethylated and associated with chemoresistance in solid and hematological malignances [124].
5. Epigenetic Drugs and HNSCC Therapy
One of the fundamental goals of cancer research is to translate new findings to a clinically relevant setting. A better understanding of the mechanisms associated with epigenetic modifications will result in the development of new epigenetic markers capable of early detection of tumors and maintenance of continuous surveillance. Also, the identification of distinct epigenetic profiles will corroborate to the identification of prognostic tools and a potential predictor of tumor response to therapy [104]. As we move towards personalized and precise medicine, it is important to remember that the genetic material is identical in every cell, while epigenetics is highly variable within different cells and tissues of an organism, and is also affected by aging and environmental factors [99].
Given the reversible nature of epigenetic modifications, it is clear that changes observed during disease progression become attractive targets for cancer therapy. An increasing number of epigenetic drugs are in development. Currently, they are classified into two groups: histone deacetylase inhibitors (HDACi), and DNA methyltransferase inhibitors (DNMTi). We will certainly see an increasing number of pharmacological classes as our knowledge on epigenetics evolves. Six US Food and Drug Administration (FDA)-approved epigenetic agents are applied in the clinic: two DNMTi, 5-azacytidine (i.e., Vidaza) and 5-aza-2′-deoxycytidine (i.e., Decitabine), and four HDACi, suberoylanilide hydroxamic acid (i.e., Vorinostat), F-228 (i.e., Romidepsin), LAQ-824 (i.e., Farydak), and LBH-589 (i.e., Panobinostat). Many others agents are in the pipeline [105,170]. Results from basic science and clinical research data are starting to pile up. Use of epigenetic inhibitors like the DNMTi, Zebularine, led to tumor growth inhibition, as evidenced by decreased of proliferation and cell cycle arrest in HNSCC cell lines [77]. The histone methylation inhibitor 3-deazaneplanocin (DZNep) reversed the aggressive characteristics of oral carcinoma cells through the dynamic regulation of epithelial plasticity and blocking EMT via the reprogramming of gene expression profile [171].
DNA hypermethylation is a common event in HNSCC that leads to gene transcription silencing of cell cycle controlling genes. Thus, DNA methyltransferases, the enzymes driving methylation dynamics, are important targets for cancer treatment [172,173]. The reversal of DNA hypermethylation by pharmaceutical agents like 5-azacytidine, a DNA methyltransferase inhibitor, constitutes an effective treatment of several cancers, including HNSCC. 5-Azacytidine, as single treatment, inhibited the proliferation of HNSCC tumor xenografts, however, it failed to reduce the severity of the lesions. In contrast, the association of 5-azacytidine with retinoic acid was efficient in reducing the effects of chemical carcinogenesis [174,175]. Alternatively, demethylation agents can also be administered as chemosensitizer agents against cancer. Administration of decitabine shows promising results in sensitizing HNSCC cell lines to cisplatin, reducing overall dose requirements for cisplatin-induced apoptosis. Combination treatment of cisplatin and decitabine significantly reduced HNSCC growth [176]. The DNMTi zebularine also works as an adjuvant drug to cisplatin and 5-fluoro-uracil, increasing apoptosis in HNSCC [77,177].
HDACi represents a growing class of anticancer agents that regulates gene expression through histone acetylation modulation. In several cancers, treatment with HDACi results in differentiation, cell cycle arrest, apoptosis, and decreasing of angiogenesis [26,178,179]. Through hyperacetylation of histone and non-histone targets, HDACi enables the reestablishment of cellular acetylation homeostasis, and restores normal expression and function of several proteins that may reverse cancer initiation, progression, and therapy resistance [52].
Although HDACi has been successfully used in the treatment of hematological malignancies, it shows limited clinical activity as a single agent in solid tumors. The most promising HDACi results were seen when it was combined with other therapeutic regimens, including its combination with radio and chemotherapy, which resulted in synergistic or additive effects. Indeed, HDACi is mostly effective against HNSCC when administrated with other therapeutic agents [178,180,181]. Treatment using combined Trichostatin A (TSA) and all-trans-retinoic acid (ATRA), synergistically increased the growth-inhibitory effect, and strongly induced transcriptional activation of target genes, which restored tumor sensitivity to retinoic acid in HNSCC cell lines [182]. Association of TSA and proteasome inhibitor PS-341 (Bortezomib) was also investigated in HNSCC. Although TSA alone did not induce apoptosis, it significantly enhanced PS-341-induced apoptosis through activation of caspase [1]. MS-275, a synthetic benzamide HDACi, also increased the cytotoxic effect of cisplatin in the treatment of OSCC [183].
Combined regimens seem to provide a better clinical outcome to HNSCC patients. An example of this strategy is the co-administration of HDACi and cisplatin in HNSCC, which displays synergistic effects in inducing greater cytotoxicity and apoptosis, compared to cisplatin alone [5,77,177,183,184]. Co-administration of HDACi and cisplatin also resulted in the activation of the miR-107 and miR-138 followed by the disruption of cellular migration and invasion of HNSCC [185]. Additionally, administration of TSA reversed chemoresistance to cisplatin in HNSCC cell lines by targeting NFκB nuclear accumulation [13].
In the search for novel therapeutic strategies capable of improving the efficacy of tumor treatment while limiting toxicity, a combination of radiotherapy with epigenetic drugs seems to be a valuable option. Preclinical radiosensitization using HDACi has been reported as a feasible strategy for solid tumors, including HNSCC. TSA, SAHA (suberoylanilide hydroxamic acid; Vorinostat), M344 (an analog of SAHA), and depsipeptide (FR90228), modulate cellular responses to ionizing radiation, and promote cell cycle arrest and apoptosis in HNSCC [186]. Additionally, demethylating agents, such as 5-aza-2′-deoxycytidine, target DNMTs, resulting in global demethylation, and are also radiosensitizing agents for HNSCC treatment [187,188].
As epigenetic mechanisms have important functions in modulating stem cell properties in cancer cells, targeting components of these epigenetic pathways would help to eradicate the population of CSCs and non-CSCs. Recent studies suggest that small doses of the DNMTi 5-aza-2′-deoxycytidine are sufficient to reduce the tumorigenicity by targeting CSCs [189,190]. We have recently found that changes in chromatin acetylation of HNSCC unexpectedly triggered the reduction of CSCs. Hyperacetylation of chromatin in HNSCC cells using TSA disrupted the ability of CSCs to generate and maintain tumor spheres. We also found that TSA reduces the enzymatic activity levels of ALDH, a well-stablished marker of CSCs [87]. HDAC inhibitors SAHA and TSA, also decreased cancer stem cell markers CD44 and ABCG2, and genes related with cellular stemness, as well as the EMT phenotype in oral carcinoma [191]. Likewise, low levels of SAHA also abolished the CSC population in salivary gland tumors [88,111].
6. Conclusions
Increasing knowledge on epigenetic modifications is contributing to the overall understanding of cancer biology. Epigenetic changes may be used as tools to diagnose, treat, and provide prognostic information for HNSCC patients. The clinical importance of research efforts to identify and validate novel epigenetic modifications associated with cancer chemoresistance cannot be underestimated. Discovery of new epigenetic biomarkers for individual cancer chemoresistance can open the possibility for the development of novel drugs (Epi-drugs), which can be used as an adjuvant therapy associated with conventional chemotherapeutic drugs, enhancing tumor sensitivity to traditional agents and ultimately increasing treatment effectiveness. The emerging understanding of the biological effects of HDACi over the population of CSCs is a breakthrough on epigenetic studies. The success of deploying HDACi as sensitizer agents before the administration of chemotherapeutic agents has underscored the importance of targeting CSCs using Epi-drugs for HNSCC and salivary tumors. Therefore, epigenetic modifications induced by Epi-drugs, aligned with the prospective identification of epigenetic biomarkers, are an exciting frontier in tumor biology waiting to be explored.
Acknowledgments
None of the authors has any competing interests in the manuscript. The funders had no role in study design, data collection, data analysis, decision to publish, or preparation of the paper. This study was funded by the University of Michigan School of Dentistry faculty grant.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kim J., Guan J., Chang I., Chen X., Han D., Wang C.Y. PS-341 and histone deacetylase inhibitor synergistically induce apoptosis in head and neck squamous cell carcinoma cells. Mol. Cancer Ther. 2010;9:1977–1984. doi: 10.1158/1535-7163.MCT-10-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papillon-Cavanagh S., Lu C., Gayden T., Mikael L.G., Bechet D., Karamboulas C., Ailles L., Karamchandani J., Marchione D.M., Garcia B.A., et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pullos A.N., Castilho R.M., Squarize C.H. HPV Infection of the Head and Neck Region and Its Stem Cells. J. Dent. Res. 2015;94:1532–1543. doi: 10.1177/0022034515605456. [DOI] [PubMed] [Google Scholar]
- 4.Argiris A., Karamouzis M.V., Raben D., Ferris R.L. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascolo M., Siano M., Ilardi G., Russo D., Merolla F., De Rosa G., Staibano S. Epigenetic disregulation in oral cancer. Int. J. Mol. Sci. 2012;13:2331–2353. doi: 10.3390/ijms13022331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisani P., Bray F., Parkin D.M. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int. J. Cancer. 2002;97:72–81. doi: 10.1002/ijc.1571. [DOI] [PubMed] [Google Scholar]
- 7.Molinolo A.A., Amornphimoltham P., Squarize C.H., Castilho R.M., Patel V., Gutkind J.S. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009;45:324–334. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.United States Public Health Service, Office of the Surgeon General . The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. Volume 2 U.S. Department of Health and Human Services, Public Health Service, Office of the Surgeon General; Rockville, MD, USA: 2014. [Google Scholar]
- 9.Le J.M., Squarize C.H., Castilho R.M. Histone modifications: Targeting head and neck cancer stem cells. World J. Stem Cells. 2014;6:511–525. doi: 10.4252/wjsc.v6.i5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang I., Wang C.Y. Inhibition of HDAC6 Protein Enhances Bortezomib-induced Apoptosis in Head and Neck Squamous Cell Carcinoma (HNSCC) by Reducing Autophagy. J. Biol. Chem. 2016;291:18199–18209. doi: 10.1074/jbc.M116.717793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg A.P., Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 13.Almeida L.O., Abrahao A.C., Rosselli-Murai L.K., Giudice F.S., Zagni C., Leopoldino A.M., Squarize C.H., Castilho R.M. NFkappaB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC) FEBS Open Bio. 2014;4:96–104. doi: 10.1016/j.fob.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Easwaran H., Tsai H.C., Baylin S.B. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 18.Toh T.B., Lim J.J., Chow E.K. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waddington C.H. The epigenotype. 1942. In. J. Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 20.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Ling S., Rettig E., Sobel R., Tan M., Fertig E.J., Considine M., El-Naggar A.K., Brait M., Fakhry C., et al. Epigenetic screening of salivary gland mucoepidermoid carcinoma identifies hypomethylation of CLIC3 as a common alteration. Oral Oncol. 2015;51:1120–1125. doi: 10.1016/j.oraloncology.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyn H., Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 23.Issa M.E., Takhsha F.S., Chirumamilla C.S., Perez-Novo C., Vanden Berghe W., Cuendet M. Epigenetic strategies to reverse drug resistance in heterogeneous multiple myeloma. Clin. Epigenet. 2017;9:17. doi: 10.1186/s13148-017-0319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 25.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson M.A., Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Langevin S.M., Eliot M., Butler R.A., Cheong A., Zhang X., McClean M.D., Koestler D.C., Kelsey K.T. CpG island methylation profile in non-invasive oral rinse samples is predictive of oral and pharyngeal carcinoma. Clin. Epigenet. 2015;7:125. doi: 10.1186/s13148-015-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 29.Chedin F., Lieber M.R., Hsieh C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulis M., Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 31.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Paredes M., Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 33.Klose R.J., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez J., Frigola J., Vendrell E., Risques R.A., Fraga M.F., Morales C., Moreno V., Esteller M., Capella G., Ribas M., et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 35.Hatziapostolou M., Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell. Mol. Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 37.Audia J.E., Campbell R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016;8:a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothbart S.B., Strahl B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi P., Allis C.D., Wang G.G. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 42.Kumar R., Li D.Q., Muller S., Knapp S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene. 2016;35:4423–4436. doi: 10.1038/onc.2015.513. [DOI] [PubMed] [Google Scholar]
- 43.Stricker S.H., Koferle A., Beck S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017;18:51–66. doi: 10.1038/nrg.2016.138. [DOI] [PubMed] [Google Scholar]
- 44.McBrian M.A., Behbahan I.S., Ferrari R., Su T., Huang T.W., Li K., Hong C.S., Christofk H.R., Vogelauer M., Seligson D.B., et al. Histone acetylation regulates intracellular pH. Mol. Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobson R.H., Ladurner A.G., King D.S., Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 46.Gegonne A., Weissman J.D., Singer D.S. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc. Natl. Acad. Sci. USA. 2001;98:12432–12437. doi: 10.1073/pnas.211444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G.G., Allis C.D., Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol. Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Ellis L., Atadja P.W., Johnstone R.W. Epigenetics in cancer: Targeting chromatin modifications. Mol. Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 49.Zhao D., Li F.L., Cheng Z.L., Lei Q.Y. Impact of acetylation on tumor metabolism. Mol. Cell. Oncol. 2014;1:e963452. doi: 10.4161/23723548.2014.963452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squatrito M., Gorrini C., Amati B. Tip60 in DNA damage response and growth control: Many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Sykes S.M., Mellert H.S., Holbert M.A., Li K., Marmorstein R., Lane W.S., McMahon S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y., Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung K.H., Noh J.H., Kim J.K., Eun J.W., Bae H.J., Xie H.J., Chang Y.G., Kim M.G., Park H., Lee J.Y., et al. HDAC2 overexpression confers oncogenic potential to human lung cancer cells by deregulating expression of apoptosis and cell cycle proteins. J. Cell. Biochem. 2012;113:2167–2177. doi: 10.1002/jcb.24090. [DOI] [PubMed] [Google Scholar]
- 54.Reichert N., Choukrallah M.A., Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell. Mol. Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Bedi U., Mishra V.K., Wasilewski D., Scheel C., Johnsen S.A. Epigenetic plasticity: A central regulator of epithelial-to-mesenchymal transition in cancer. Oncotarget. 2014;5:2016–2029. doi: 10.18632/oncotarget.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dorrance A.M., Liu S., Yuan W., Becknell B., Arnoczky K.J., Guimond M., Strout M.P., Feng L., Nakamura T., Yu L., et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J. Clin. Investig. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Versteege I., Sevenet N., Lange J., Rousseau-Merck M.F., Ambros P., Handgretinger R., Aurias A., Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 60.Wilson B.G., Roberts C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Perez A., Jene-Sanz A., Lopez-Bigas N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol. 2013;14:r106. doi: 10.1186/gb-2013-14-9-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun L., Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell. Mol. Life Sci. 2016;73:4493–4515. doi: 10.1007/s00018-016-2303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koschmann C., Nunez F.J., Mendez F., Brosnan-Cashman J.A., Meeker A.K., Lowenstein P.R., Castro M.G. Mutated Chromatin Regulatory Factors as Tumor Drivers in Cancer. Cancer Res. 2017;77:227–233. doi: 10.1158/0008-5472.CAN-16-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanton B.Z., Hodges C., Calarco J.P., Braun S.M., Ku W.L., Kadoch C., Zhao K., Crabtree G.R. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson B.G., Wang X., Shen X., McKenna E.S., Lemieux M.E., Cho Y.J., Koellhoffer E.C., Pomeroy S.L., Orkin S.H., Roberts C.W. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi J.H., Sheu J.J., Guan B., Jinawath N., Markowski P., Wang T.L., Shih Ie M. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69:1407–1415. doi: 10.1158/0008-5472.CAN-08-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tai H.C., Huang H.Y., Lee S.W., Lin C.Y., Sheu M.J., Chang S.L., Wu L.C., Shiue Y.L., Wu W.R., Lin C.M., et al. Associations of Rsf-1 overexpression with poor therapeutic response and worse survival in patients with nasopharyngeal carcinoma. J. Clin. Pathol. 2012;65:248–253. doi: 10.1136/jclinpath-2011-200413. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y.I., Ahn J.H., Lee K.T., Shih Ie M., Choi J.H. RSF1 is a positive regulator of NF-kappaB-induced gene expression required for ovarian cancer chemoresistance. Cancer Res. 2014;74:2258–2269. doi: 10.1158/0008-5472.CAN-13-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willingham A.T., Gingeras T.R. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 71.Costa F.F. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 72.Esteller M., Pandolfi P.P. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 2017;7:359–368. doi: 10.1158/2159-8290.CD-16-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D’Angelo B., Benedetti E., Cimini A., Giordano A. MicroRNAs: A Puzzling Tool in Cancer Diagnostics and Therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 75.Gangaraju V.K., Lin H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ivey K.N., Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 77.Gasche J.A., Goel A. Epigenetic mechanisms in oral carcinogenesis. Future Oncol. 2012;8:1407–1425. doi: 10.2217/fon.12.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ventura A., Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y., Zhu Y., Lv P., Li L. The role of miR-21 in proliferation and invasion capacity of human tongue squamous cell carcinoma in vitro. Int. J. Clin. Exp. Pathol. 2015;8:4555–4563. [PMC free article] [PubMed] [Google Scholar]
- 81.Esquela-Kerscher A., Slack F.J. Oncomirs—Micrornas with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 82.Ayers D., Vandesompele J. Influence of microRNAs and Long Non-Coding RNAs in Cancer Chemoresistance. Genes. 2017;8:95. doi: 10.3390/genes8030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorenchtein M., Poh C.F., Saini R., Garnis C. MicroRNAs in an oral cancer context—From basic biology to clinical utility. J. Dent. Res. 2012;91:440–446. doi: 10.1177/0022034511431261. [DOI] [PubMed] [Google Scholar]
- 84.Datta J., Kutay H., Nasser M.W., Nuovo G.J., Wang B., Majumder S., Liu C.G., Volinia S., Croce C.M., Schmittgen T.D., et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Lujambio A., Ropero S., Ballestar E., Fraga M.F., Cerrato C., Setien F., Casado S., Suarez-Gauthier A., Sanchez-Cespedes M., Git A., et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 86.Jain N., Iyer K.V., Kumar A., Shivashankar G.V. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. USA. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giudice F.S., Pinto D.S., Jr., Nor J.E., Squarize C.H., Castilho R.M. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE. 2013;8:e58672. doi: 10.1371/journal.pone.0058672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Almeida L.O., Guimaraes D.M., Martins M.D., Martins M.A.T., Warner K.A., Nor J.E., Castilho R.M., Squarize C.H. Unlocking the chromatin of adenoid cystic carcinomas using HDAC inhibitors sensitize cancer stem cells to cisplatin and induces tumor senescence. Stem Cell Res. 2017;21:94–105. doi: 10.1016/j.scr.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim Y., Sun C.X., Tran P., Punyadeera C. Salivary epigenetic biomarkers in head and neck squamous cell carcinomas. Biomark. Med. 2016;10:301–313. doi: 10.2217/bmm.16.2. [DOI] [PubMed] [Google Scholar]
- 90.Baba S., Yamada Y., Hatano Y., Miyazaki Y., Mori H., Shibata T., Hara A. Global DNA hypomethylation suppresses squamous carcinogenesis in the tongue and esophagus. Cancer Sci. 2009;100:1186–1191. doi: 10.1111/j.1349-7006.2009.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Subbalekha K., Pimkhaokham A., Pavasant P., Chindavijak S., Phokaew C., Shuangshoti S., Matangkasombut O., Mutirangura A. Detection of LINE-1s hypomethylation in oral rinses of oral squamous cell carcinoma patients. Oral Oncol. 2009;45:184–191. doi: 10.1016/j.oraloncology.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 92.Clausen M.J., Melchers L.J., Mastik M.F., Slagter-Menkema L., Groen H.J., van der Laan B.F., van Criekinge W., de Meyer T., Denil S., Wisman G.B., et al. Identification and validation of WISP1 as an epigenetic regulator of metastasis in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2016;55:45–59. doi: 10.1002/gcc.22310. [DOI] [PubMed] [Google Scholar]
- 93.Tanaka C., Uzawa K., Shibahara T., Yokoe H., Noma H., Tanzawa H. Expression of an inhibitor of apoptosis, survivin, in oral carcinogenesis. J. Dent. Res. 2003;82:607–611. doi: 10.1177/154405910308200807. [DOI] [PubMed] [Google Scholar]
- 94.Bebek G., Bennett K.L., Funchain P., Campbell R., Seth R., Scharpf J., Burkey B., Eng C. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum. Mol. Genet. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan C.H., Horng C.T., Lee C.F., Chiang N.N., Tsai F.J., Lu C.C., Chiang J.H., Hsu Y.M., Yang J.S., Chen F.A. Epigallocatechin gallate sensitizes cisplatin-resistant oral cancer CAR cell apoptosis and autophagy through stimulating AKT/STAT3 pathway and suppressing multidrug resistance 1 signaling. Environ. Toxicol. 2017;32:845–855. doi: 10.1002/tox.22284. [DOI] [PubMed] [Google Scholar]
- 96.Arantes L.M., de Carvalho A.C., Melendez M.E., Carvalho A.L., Goloni-Bertollo E.M. Methylation as a biomarker for head and neck cancer. Oral Oncol. 2014;50:587–592. doi: 10.1016/j.oraloncology.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 97.Jithesh P.V., Risk J.M., Schache A.G., Dhanda J., Lane B., Liloglou T., Shaw R.J. The epigenetic landscape of oral squamous cell carcinoma. Br. J. Cancer. 2013;108:370–379. doi: 10.1038/bjc.2012.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singh N.N., Peer A., Nair S., Chaturvedi R.K. Epigenetics: A possible answer to the undeciphered etiopathogenesis and behavior of oral lesions. J. Oral Maxillofac. Pathol. 2016;20:122–128. doi: 10.4103/0973-029X.180967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Worsham M.J., Stephen J.K., Chen K.M., Havard S., Shah V., Gardner G., Schweitzer V.G. Delineating an epigenetic continuum in head and neck cancer. Cancer Lett. 2014;342:178–184. doi: 10.1016/j.canlet.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Towle R., Truong D., Hogg K., Robinson W.P., Poh C.F., Garnis C. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013;49:1033–1042. doi: 10.1016/j.oraloncology.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 101.Carvalho A.L., Jeronimo C., Kim M.M., Henrique R., Zhang Z., Hoque M.O., Chang S., Brait M., Nayak C.S., Jiang W.W., et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin. Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 102.Righini C.A., de Fraipont F., Timsit J.F., Faure C., Brambilla E., Reyt E., Favrot M.C. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin. Cancer Res. 2007;13:1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 103.Coombes M.M., Briggs K.L., Bone J.R., Clayman G.L., El-Naggar A.K., Dent S.Y. Resetting the histone code at CDKN2A in HNSCC by inhibition of DNA methylation. Oncogene. 2003;22:8902–8911. doi: 10.1038/sj.onc.1207050. [DOI] [PubMed] [Google Scholar]
- 104.Glazer C.A., Chang S.S., Ha P.K., Califano J.A. Applying the molecular biology and epigenetics of head and neck cancer in everyday clinical practice. Oral Oncol. 2009;45:440–446. doi: 10.1016/j.oraloncology.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Lindsay C., Seikaly H., Biron V.L. Epigenetics of oropharyngeal squamous cell carcinoma: Opportunities for novel chemotherapeutic targets. J. Otolaryngol. Head Neck Surg. 2017;46:9. doi: 10.1186/s40463-017-0185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhan T., Rindtorff N., Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guerrero-Preston R., Herbstman J., Goldman L.R. Epigenomic biomonitors: Global DNA hypomethylation as a biodosimeter of life-long environmental exposures. Epigenomics. 2011;3:1–5. doi: 10.2217/epi.10.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Langevin S.M., Butler R.A., Eliot M., Pawlita M., Maccani J.Z., McClean M.D., Kelsey K.T. Novel DNA methylation targets in oral rinse samples predict survival of patients with oral squamous cell carcinoma. Oral Oncol. 2014;50:1072–1080. doi: 10.1016/j.oraloncology.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takebe N., Miele L., Harris P.J., Jeong W., Bando H., Kahn M., Yang S.X., Ivy S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Svinkina T., Gu H., Silva J.C., Mertins P., Qiao J., Fereshetian S., Jaffe J.D., Kuhn E., Udeshi N.D., Carr S.A. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Mol. Cell. Proteom. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guimaraes D.M., Almeida L.O., Martins M.D., Warner K.A., Silva A.R., Vargas P.A., Nunes F.D., Squarize C.H., Nor J.E., Castilho R.M. Sensitizing mucoepidermoid carcinomas to chemotherapy by targeted disruption of cancer stem cells. Oncotarget. 2016;7:42447–42460. doi: 10.18632/oncotarget.9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Webber L.P., Wagner V.P., Curra M., Vargas P.A., Meurer L., Carrard V.C., Squarize C.H., Castilho R.M., Martins M.D. Hypoacetylation of acetyl-histone H3 (H3K9ac) as marker of poor prognosis in oral cancer. Histopathology. 2017 doi: 10.1111/his.13218. [DOI] [PubMed] [Google Scholar]
- 113.Chen Y.W., Kao S.Y., Wang H.J., Yang M.H. Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer. 2013;119:4259–4267. doi: 10.1002/cncr.28356. [DOI] [PubMed] [Google Scholar]
- 114.Rastogi B., Raut S.K., Panda N.K., Rattan V., Radotra B.D., Khullar M. Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol. Cell. Biochem. 2016;415:183–196. doi: 10.1007/s11010-016-2690-5. [DOI] [PubMed] [Google Scholar]
- 115.Ropero S., Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Witt O., Deubzer H.E., Milde T., Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 117.Sakuma T., Uzawa K., Onda T., Shiiba M., Yokoe H., Shibahara T., Tanzawa H. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int. J. Oncol. 2006;29:117–124. doi: 10.3892/ijo.29.1.117. [DOI] [PubMed] [Google Scholar]
- 118.Chang H.H., Chiang C.P., Hung H.C., Lin C.Y., Deng Y.T., Kuo M.Y. Histone deacetylase 2 expression predicts poorer prognosis in oral cancer patients. Oral Oncol. 2009;45:610–614. doi: 10.1016/j.oraloncology.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 119.Ahn M.Y., Yoon J.H. Histone deacetylase 7 silencing induces apoptosis and autophagy in salivary mucoepidermoid carcinoma cells. J. Oral Pathol. Med. 2017;46:276–283. doi: 10.1111/jop.12560. [DOI] [PubMed] [Google Scholar]
- 120.Ahn M.Y., Yoon J.H. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol. Rep. 2017;37:540–546. doi: 10.3892/or.2016.5280. [DOI] [PubMed] [Google Scholar]
- 121.Haigis M.C., Guarente L.P. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 122.Noguchi A., Li X., Kubota A., Kikuchi K., Kameda Y., Zheng H., Miyagi Y., Aoki I., Takano Y. SIRT1 expression is associated with good prognosis for head and neck squamous cell carcinoma patients. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013;115:385–392. doi: 10.1016/j.oooo.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 123.Lai C.C., Lin P.M., Lin S.F., Hsu C.H., Lin H.C., Hu M.L., Hsu C.M., Yang M.Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34:1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- 124.Lv J.F., Hu L., Zhuo W., Zhang C.M., Zhou H.H., Fan L. Epigenetic alternations and cancer chemotherapy response. Cancer Chemother. Pharmacol. 2016;77:673–684. doi: 10.1007/s00280-015-2951-0. [DOI] [PubMed] [Google Scholar]
- 125.Saladi S.V., Ross K., Karaayvaz M., Tata P.R., Mou H., Rajagopal J., Ramaswamy S., Ellisen L.W. ACTL6A Is Co-Amplified with p63 in Squamous Cell Carcinoma to Drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell. 2017;31:35–49. doi: 10.1016/j.ccell.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hung T., Binda O., Champagne K.S., Kuo A.J., Johnson K., Chang H.Y., Simon M.D., Kutateladze T.G., Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol. Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ozer A., Wu L.C., Bruick R.K. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc. Natl. Acad. Sci. USA. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shiseki M., Nagashima M., Pedeux R.M., Kitahama-Shiseki M., Miura K., Okamura S., Onogi H., Higashimoto Y., Appella E., Yokota J., et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 129.Li X.H., Kikuchi K., Zheng Y., Noguchi A., Takahashi H., Nishida T., Masuda S., Yang X.H., Takano Y. Downregulation and translocation of nuclear ING4 is correlated with tumorigenesis and progression of head and neck squamous cell carcinoma. Oral Oncol. 2011;47:217–223. doi: 10.1016/j.oraloncology.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 130.Fang F.M., Li C.F., Huang H.Y., Lai M.T., Chen C.M., Chiu I.W., Wang T.L., Tsai F.J., Shih Ie M., Sheu J.J. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. Am. J. Pathol. 2011;178:2407–2415. doi: 10.1016/j.ajpath.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Feng X., Zhang Q., Xia S., Xia B., Zhang Y., Deng X., Su W., Huang J. MTA1 overexpression induces cisplatin resistance in nasopharyngeal carcinoma by promoting cancer stem cells properties. Mol. Cells. 2014;37:699–704. doi: 10.14348/molcells.2014.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kothandapani A., Gopalakrishnan K., Kahali B., Reisman D., Patrick S.M. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp. Cell Res. 2012;318:1973–1986. doi: 10.1016/j.yexcr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marzook H., Deivendran S., Kumar R., Pillai M.R. Role of MTA1 in head and neck cancers. Cancer Metastasis Rev. 2014;33:953–964. doi: 10.1007/s10555-014-9521-5. [DOI] [PubMed] [Google Scholar]
- 134.Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 135.Jia L.F., Huang Y.P., Zheng Y.F., Lyu M.Y., Wei S.B., Meng Z., Gan Y.H. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 136.Jia L.F., Wei S.B., Mitchelson K., Gao Y., Zheng Y.F., Meng Z., Gan Y.H., Yu G.Y. miR-34a inhibits migration and invasion of tongue squamous cell carcinoma via targeting MMP9 and MMP14. PLoS ONE. 2014;9:e108435. doi: 10.1371/journal.pone.0108435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reis P.P., Tomenson M., Cervigne N.K., Machado J., Jurisica I., Pintilie M., Sukhai M.A., Perez-Ordonez B., Grenman R., Gilbert R.W., et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol. Cancer. 2010;9:238. doi: 10.1186/1476-4598-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu X., Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J. Cell Mol. Med. 2016;20:10–16. doi: 10.1111/jcmm.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jiang L., Liu X., Chen Z., Jin Y., Heidbreder C.E., Kolokythas A., Wang A., Dai Y., Zhou X. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem. J. 2010;432:199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qiu K., Huang Z., Huang Z., He Z., You S. miR-22 regulates cell invasion, migration and proliferation in vitro through inhibiting CD147 expression in tongue squamous cell carcinoma. Arch. Oral Biol. 2016;66:92–97. doi: 10.1016/j.archoralbio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 141.Bourguignon L.Y., Wong G., Shiina M. Up-regulation of Histone Methyltransferase, DOT1L, by Matrix Hyaluronan Promotes MicroRNA-10 Expression Leading to Tumor Cell Invasion and Chemoresistance in Cancer Stem Cells from Head and Neck Squamous Cell Carcinoma. J. Biol. Chem. 2016;291:10571–10585. doi: 10.1074/jbc.M115.700021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu M., Wang J., Huang H., Hou J., Zhang B., Wang A. miR-181a-Twist1 pathway in the chemoresistance of tongue squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2013;441:364–370. doi: 10.1016/j.bbrc.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 143.Peng F., Zhang H., Du Y., Tan P. miR-23a promotes cisplatin chemoresistance and protects against cisplatin-induced apoptosis in tongue squamous cell carcinoma cells through Twist. Oncol. Rep. 2015;33:942–950. doi: 10.3892/or.2014.3664. [DOI] [PubMed] [Google Scholar]
- 144.Peng X., Cao P., He D., Han S., Zhou J., Tan G., Li W., Yu F., Yu J., Li Z., et al. MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel and inhibits cell growth both in vitro and in vivo. Int. J. Clin. Exp. Pathol. 2014;7:6784–6791. [PMC free article] [PubMed] [Google Scholar]
- 145.Yang G.D., Huang T.J., Peng L.X., Yang C.F., Liu R.Y., Huang H.B., Chu Q.Q., Yang H.J., Huang J.L., Zhu Z.Y., et al. Epstein-Barr Virus_Encoded LMP1 upregulates microRNA-21 to promote the resistance of nasopharyngeal carcinoma cells to cisplatin-induced Apoptosis by suppressing PDCD4 and Fas-L. PLoS ONE. 2013;8:e78355. doi: 10.1371/journal.pone.0078355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hebert C., Norris K., Scheper M.A., Nikitakis N., Sauk J.J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun L., Yao Y., Liu B., Lin Z., Lin L., Yang M., Zhang W., Chen W., Pan C., Liu Q., et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31:432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]
- 148.Naik P.P., Das D.N., Panda P.K., Mukhopadhyay S., Sinha N., Praharaj P.P., Agarwal R., Bhutia S.K. Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncol. 2016;62:122–135. doi: 10.1016/j.oraloncology.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 149.Momparler R.L., Cote S. Targeting of cancer stem cells by inhibitors of DNA and histone methylation. Expert Opin. Investig. Drugs. 2015;24:1031–1043. doi: 10.1517/13543784.2015.1051220. [DOI] [PubMed] [Google Scholar]
- 150.Shukla S., Meeran S.M. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim. Biophys. Acta. 2014;1840:3494–3502. doi: 10.1016/j.bbagen.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 151.Allegra E., Trapasso S., Pisani D., Puzzo L. The role of BMI1 as a biomarker of cancer stem cells in head and neck cancer: A review. Oncology. 2014;86:199–205. doi: 10.1159/000358598. [DOI] [PubMed] [Google Scholar]
- 152.Hoffmeyer K., Raggioli A., Rudloff S., Anton R., Hierholzer A., Del Valle I., Hein K., Vogt R., Kemler R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 153.An Y., Ongkeko W.M. ABCG2: The key to chemoresistance in cancer stem cells? Expert Opin. Drug Metab. Toxicol. 2009;5:1529–1542. doi: 10.1517/17425250903228834. [DOI] [PubMed] [Google Scholar]
- 154.Grimm M., Krimmel M., Polligkeit J., Alexander D., Munz A., Kluba S., Keutel C., Hoffmann J., Reinert S., Hoefert S. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer. 2012;48:3186–3197. doi: 10.1016/j.ejca.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 155.Colak S., Medema J.P. Cancer stem cells—Important players in tumor therapy resistance. FEBS J. 2014;281:4779–4791. doi: 10.1111/febs.13023. [DOI] [PubMed] [Google Scholar]
- 156.Paduch R. Theories of cancer origin. Eur. J. Cancer Prev. 2015;24:57–67. doi: 10.1097/CEJ.0000000000000024. [DOI] [PubMed] [Google Scholar]
- 157.Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wagner V.P., Martins M.A., Martins M.D., Warner K.A., Webber L.P., Squarize C.H., Nor J.E., Castilho R.M. Overcoming adaptive resistance in mucoepidermoid carcinoma through inhibition of the IKK-beta/IkappaBalpha/NFkappaB axis. Oncotarget. 2016;7:73032–73044. doi: 10.18632/oncotarget.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Matsumoto S., Tanaka T., Kurokawa H., Matsuno K., Hayashida Y., Takahashi T. Effect of copper and role of the copper transporters ATP7A and CTR1 in intracellular accumulation of cisplatin. Anticancer Res. 2007;27:2209–2216. [PubMed] [Google Scholar]
- 160.Singh A., Wu H., Zhang P., Happel C., Ma J., Biswal S. Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Mol. Cancer Ther. 2010;9:2365–2376. doi: 10.1158/1535-7163.MCT-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Harper L.J., Costea D.E., Gammon L., Fazil B., Biddle A., Mackenzie I.C. Normal and malignant epithelial cells with stem-like properties have an extended G2 cell cycle phase that is associated with apoptotic resistance. BMC Cancer. 2010;10:166. doi: 10.1186/1471-2407-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hsu D.S., Lan H.Y., Huang C.H., Tai S.K., Chang S.Y., Tsai T.L., Chang C.C., Tzeng C.H., Wu K.J., Kao J.Y., et al. Regulation of excision repair cross-complementation group 1 by Snail contributes to cisplatin resistance in head and neck cancer. Clin. Cancer Res. 2010;16:4561–4571. doi: 10.1158/1078-0432.CCR-10-0593. [DOI] [PubMed] [Google Scholar]
- 163.Harada K., Ferdous T., Ueyama Y. Establishment of 5-fluorouracil-resistant oral squamous cell carcinoma cell lines with epithelial to mesenchymal transition changes. Int. J. Oncol. 2014;44:1302–1308. doi: 10.3892/ijo.2014.2270. [DOI] [PubMed] [Google Scholar]
- 164.Liu S., Ye D., Guo W., Yu W., He Y., Hu J., Wang Y., Zhang L., Liao Y., Song H., et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6:6887–6901. doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nor C., Zhang Z., Warner K.A., Bernardi L., Visioli F., Helman J.I., Roesler R., Nor J.E. Cisplatin induces Bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia. 2014;16:137–146. doi: 10.1593/neo.131744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu T., Chen Z., To K.K., Fang X., Wang F., Cheng B., Fu L. Effect of abemaciclib (LY2835219) on enhancement of chemotherapeutic agents in ABCB1 and ABCG2 overexpressing cells in vitro and in vivo. Biochem. Pharmacol. 2017;124:29–42. doi: 10.1016/j.bcp.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 167.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 168.Qiao B., Johnson N.W., Chen X., Li R., Tao Q., Gao J. Disclosure of a stem cell phenotype in an oral squamous cell carcinoma cell line induced by BMP-4 via an epithelial-mesenchymal transition. Oncol. Rep. 2011;26:455–461. doi: 10.3892/or.2011.1299. [DOI] [PubMed] [Google Scholar]
- 169.Zhao L., Ren Y., Tang H., Wang W., He Q., Sun J., Zhou X., Wang A. Deregulation of the miR-222-ABCG2 regulatory module in tongue squamous cell carcinoma contributes to chemoresistance and enhanced migratory/invasive potential. Oncotarget. 2015;6:44538–44550. doi: 10.18632/oncotarget.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ceccacci E., Minucci S. Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br. J. Cancer. 2016;114:605–611. doi: 10.1038/bjc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hatta M., Naganuma K., Kato K., Yamazaki J. 3-Deazaneplanocin A suppresses aggressive phenotype-related gene expression in an oral squamous cell carcinoma cell line. Biochem. Biophys. Res. Commun. 2015;468:269–273. doi: 10.1016/j.bbrc.2015.10.115. [DOI] [PubMed] [Google Scholar]
- 172.Issa J.P. DNA methylation as a therapeutic target in cancer. Clin. Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 173.Issa J.P. Cancer prevention: Epigenetics steps up to the plate. Cancer Prev. Res. 2008;1:219–222. doi: 10.1158/1940-6207.CAPR-08-0029. [DOI] [PubMed] [Google Scholar]
- 174.McGregor F., Muntoni A., Fleming J., Brown J., Felix D.H., MacDonald D.G., Parkinson E.K., Harrison P.R. Molecular changes associated with oral dysplasia progression and acquisition of immortality: Potential for its reversal by 5-azacytidine. Cancer Res. 2002;62:4757–4766. [PubMed] [Google Scholar]
- 175.Tang X.H., Albert M., Scognamiglio T., Gudas L.J. A DNA methyltransferase inhibitor and all-trans retinoic acid reduce oral cavity carcinogenesis induced by the carcinogen 4-nitroquinoline 1-oxide. Cancer Prev. Res. 2009;2:1100–1110. doi: 10.1158/1940-6207.CAPR-09-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Viet C.T., Dang D., Achdjian S., Ye Y., Katz S.G., Schmidt B.L. Decitabine rescues cisplatin resistance in head and neck squamous cell carcinoma. PLoS ONE. 2014;9:e112880. doi: 10.1371/journal.pone.0112880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Suzuki M., Shinohara F., Nishimura K., Echigo S., Rikiishi H. Epigenetic regulation of chemosensitivity to 5-fluorouracil and cisplatin by zebularine in oral squamous cell carcinoma. Int. J. Oncol. 2007;31:1449–1456. doi: 10.3892/ijo.31.6.1449. [DOI] [PubMed] [Google Scholar]
- 178.Martins M.D., Castilho R.M. Histones: Controlling Tumor Signaling Circuitry. J. Carcinog. Mutagen. 2013;1(Suppl. S5):1–12. doi: 10.4172/2157-2518.S5-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Monneret C. Histone deacetylase inhibitors. Eur. J. Med. Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 180.Gan C.P., Hamid S., Hor S.Y., Zain R.B., Ismail S.M., Wan Mustafa W.M., Teo S.H., Saunders N., Cheong S.C. Valproic acid: Growth inhibition of head and neck cancer by induction of terminal differentiation and senescence. Head Neck. 2012;34:344–353. doi: 10.1002/hed.21734. [DOI] [PubMed] [Google Scholar]
- 181.Gillenwater A.M., Zhong M., Lotan R. Histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis through both mitochondrial and Fas (Cd95) signaling in head and neck squamous carcinoma cells. Mol. Cancer Ther. 2007;6:2967–2975. doi: 10.1158/1535-7163.MCT-04-0344. [DOI] [PubMed] [Google Scholar]
- 182.Whang Y.M., Choi E.J., Seo J.H., Kim J.S., Yoo Y.D., Kim Y.H. Hyperacetylation enhances the growth-inhibitory effect of all-trans retinoic acid by the restoration of retinoic acid receptor beta expression in head and neck squamous carcinoma (HNSCC) cells. Cancer Chemother. Pharmacol. 2005;56:543–555. doi: 10.1007/s00280-004-0970-3. [DOI] [PubMed] [Google Scholar]
- 183.Rikiishi H., Shinohara F., Sato T., Sato Y., Suzuki M., Echigo S. Chemosensitization of oral squamous cell carcinoma cells to cisplatin by histone deacetylase inhibitor, suberoylanilide hydroxamic acid. Int. J. Oncol. 2007;30:1181–1188. doi: 10.3892/ijo.30.5.1181. [DOI] [PubMed] [Google Scholar]
- 184.Sato T., Suzuki M., Sato Y., Echigo S., Rikiishi H. Sequence-dependent interaction between cisplatin and histone deacetylase inhibitors in human oral squamous cell carcinoma cells. Int. J. Oncol. 2006;28:1233–1241. doi: 10.3892/ijo.28.5.1233. [DOI] [PubMed] [Google Scholar]
- 185.Datta J., Islam M., Dutta S., Roy S., Pan Q., Teknos T.N. Suberoylanilide hydroxamic acid inhibits growth of head and neck cancer cell lines by reactivation of tumor suppressor microRNAs. Oral Oncol. 2016;56:32–39. doi: 10.1016/j.oraloncology.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang Y., Jung M., Dritschilo A., Jung M. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat. Res. 2004;161:667–674. doi: 10.1667/RR3192. [DOI] [PubMed] [Google Scholar]
- 187.Ferreira M.B., Lima J.P., Cohen E.E. Novel targeted therapies in head and neck cancer. Expert Opin. Investig. Drugs. 2012;21:281–295. doi: 10.1517/13543784.2012.651455. [DOI] [PubMed] [Google Scholar]
- 188.Smits K.M., Melotte V., Niessen H.E., Dubois L., Oberije C., Troost E.G., Starmans M.H., Boutros P.C., Vooijs M., van Engeland M., et al. Epigenetics in radiotherapy: Where are we heading? Radiother. Oncol. 2014;111:168–177. doi: 10.1016/j.radonc.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 189.Tian J., Lee S.O., Liang L., Luo J., Huang C.K., Li L., Niu Y., Chang C. Targeting the unique methylation pattern of androgen receptor (AR) promoter in prostate stem/progenitor cells with 5-aza-2′-deoxycytidine (5-AZA) leads to suppressed prostate tumorigenesis. J. Biol. Chem. 2012;287:39954–39966. doi: 10.1074/jbc.M112.395574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zagonel V., Lo Re G., Marotta G., Babare R., Sardeo G., Gattei V., De Angelis V., Monfardini S., Pinto A. 5-Aza-2′-deoxycytidine (Decitabine) induces trilineage response in unfavourable myelodysplastic syndromes. Leukemia. 1993;7(Suppl. S1):30–35. [PubMed] [Google Scholar]
- 191.Chikamatsu K., Ishii H., Murata T., Sakakura K., Shino M., Toyoda M., Takahashi K., Masuyama K. Alteration of cancer stem cell-like phenotype by histone deacetylase inhibitors in squamous cell carcinoma of the head and neck. Cancer Sci. 2013;104:1468–1475. doi: 10.1111/cas.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]