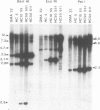
Abstract
We have identified, in the amplified domain of adenylate deaminase (AMPD) overproducing Chinese hamster fibroblasts, a 2.6 kb recombinogenic DNA region which is frequently involved in amplification-associated rearrangements. The nucleotide sequence reveals a mosaic organization of four Alu-equivalent repeats of the B1 and B2 families and eight long A + T-rich DNA segments. Part of this region is enriched with long imperfect palindromes. The center of one palindrome contains a putative topoisomerase I cleavage site and this site defines the position of a novel junction which was formed by illegitimate recombination with anther A + T-rich DNA sequence located far apart on the amplified DNA. These findings and their significance are discussed in the context of related data from other systems and in the light of current models for eukaryotic DNA recombination, replication and organization.
Full text
PDF
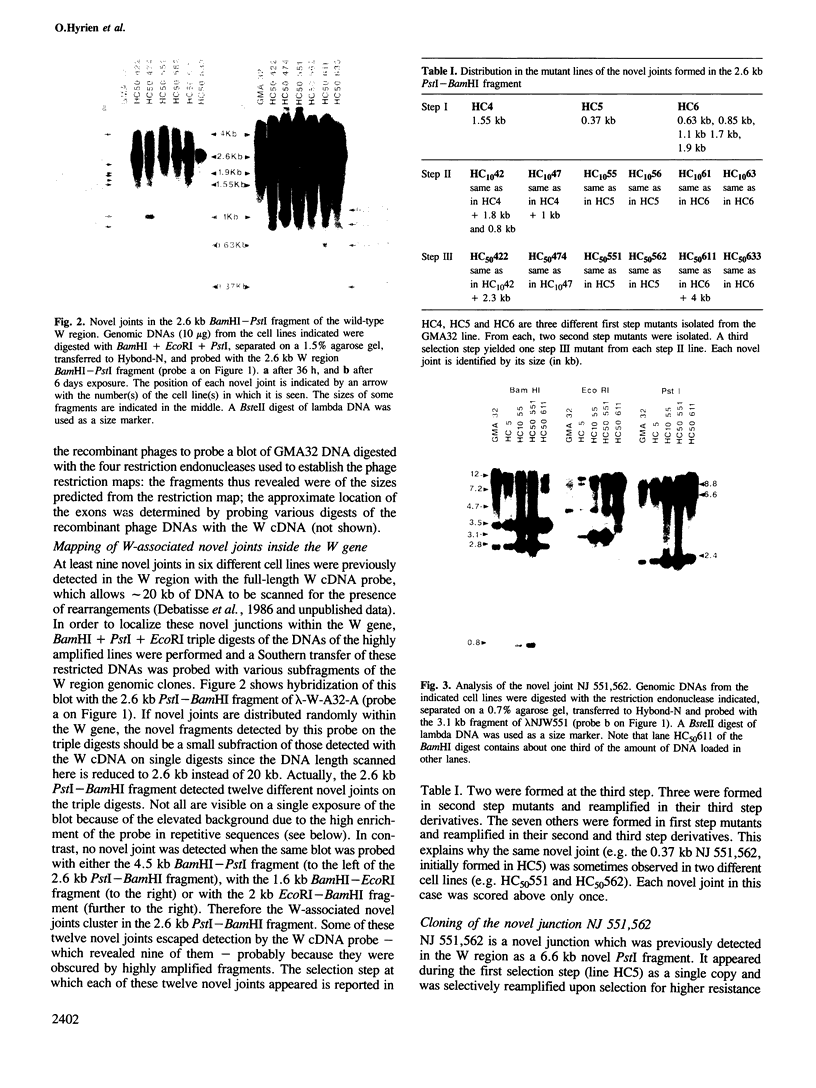
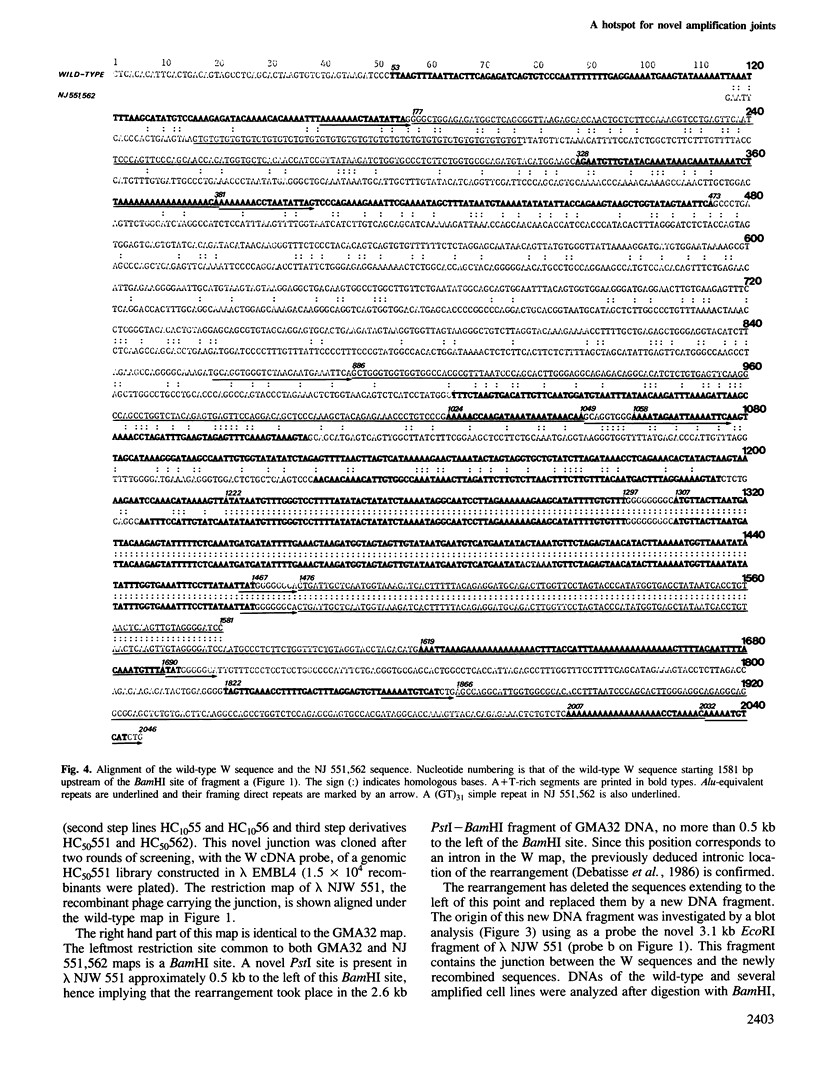
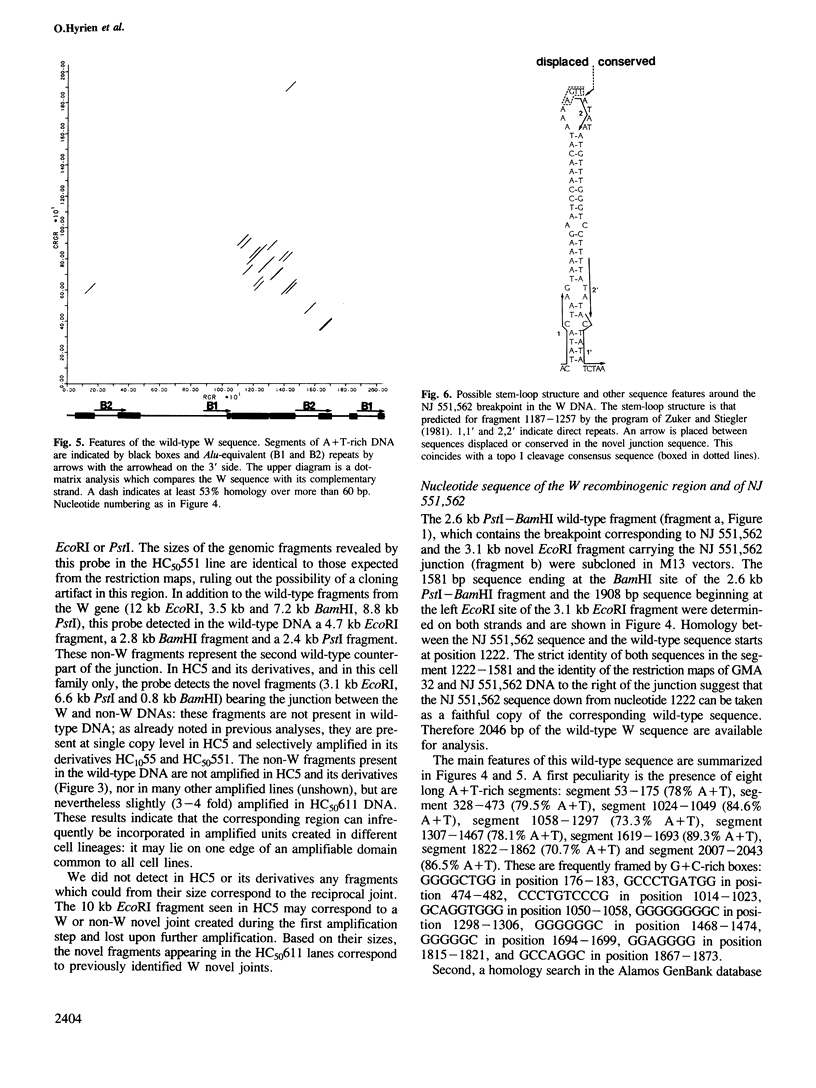




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anachkova B., Russev G., Altmann H. Identification of the short dispersed repetitive DNA sequences isolated from the zones of initiation of DNA synthesis in human cells as Alu-elements. Biochem Biophys Res Commun. 1985 Apr 16;128(1):101–106. doi: 10.1016/0006-291x(85)91650-x. [DOI] [PubMed] [Google Scholar]
- Ardeshir F., Giulotto E., Zieg J., Brison O., Liao W. S., Stark G. R. Structure of amplified DNA in different Syrian hamster cell lines resistant to N-(phosphonacetyl)-L-aspartate. Mol Cell Biol. 1983 Nov;3(11):2076–2088. doi: 10.1128/mcb.3.11.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh G. S., Seeburg P. H., Gelinas R. E. The human growth hormone gene family: structure and evolution of the chromosomal locus. Nucleic Acids Res. 1983 Jun 25;11(12):3939–3958. doi: 10.1093/nar/11.12.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Caizzi R., Bostock C. J. Gene amplification in methotrexate-resistant mouse cells. IV. Different DNA sequences are amplified in different resistant lines. Nucleic Acids Res. 1982 Nov 11;10(21):6597–6618. doi: 10.1093/nar/10.21.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta B., Robberson D. L., Barrera-Saldaña H. A., Lambrou T. P., Saunders G. F. Genome instability in a region of human DNA enriched in Alu repeat sequences. Nature. 1982 Mar 18;296(5854):219–225. doi: 10.1038/296219a0. [DOI] [PubMed] [Google Scholar]
- Claverie J. M. A common philosophy and FORTRAN 77 software package for implementing and searching sequence databases. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):397–407. doi: 10.1093/nar/12.1part1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Conformational constraints in nuclear DNA. J Cell Sci. 1976 Nov;22(2):287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Cowell J. K. Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Integration site preferences of the Alu family and similar repetitive DNA sequences. Nucleic Acids Res. 1985 Dec 20;13(24):8939–8954. doi: 10.1093/nar/13.24.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., Berry M., Buttin G. Stepwise isolation and properties of unstable Chinese hamster cell variants that overproduce adenylate deaminase. Mol Cell Biol. 1982 Nov;2(11):1346–1353. doi: 10.1128/mcb.2.11.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., Hyrien O., Petit-Koskas E., de Saint-Vincent B. R., Buttin G. Segregation and rearrangement of coamplified genes in different lineages of mutant cells that overproduce adenylate deaminase. Mol Cell Biol. 1986 May;6(5):1776–1781. doi: 10.1128/mcb.6.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., de Saint Vincent B. R., Buttin G. Expression of several amplified genes in an adenylate-deaminase overproducing variant of Chinese hamster fibroblasts. EMBO J. 1984 Dec 20;3(13):3123–3127. doi: 10.1002/j.1460-2075.1984.tb02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Wenink P. W., Poddighe J. Permanent attachment of replication origins to the nuclear matrix in BHK-cells. Nucleic Acids Res. 1986 Apr 25;14(8):3241–3249. doi: 10.1093/nar/14.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federspiel N. A., Beverley S. M., Schilling J. W., Schimke R. T. Novel DNA rearrangements are associated with dihydrofolate reductase gene amplification. J Biol Chem. 1984 Jul 25;259(14):9127–9140. [PubMed] [Google Scholar]
- Flanagan J. G., Lefranc M. P., Rabbitts T. H. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984 Mar;36(3):681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Furano A. V., Somerville C. C., Tsichlis P. N., D'Ambrosio E. Target sites for the transposition of rat long interspersed repeated DNA elements (LINEs) are not random. Nucleic Acids Res. 1986 May 12;14(9):3717–3727. doi: 10.1093/nar/14.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gilson E., Clément J. M., Brutlag D., Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984 Jun;3(6):1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulotto E., Saito I., Stark G. R. Structure of DNA formed in the first step of CAD gene amplification. EMBO J. 1986 Sep;5(9):2115–2121. doi: 10.1002/j.1460-2075.1986.tb04474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Hasson J. F., Mougneau E., Cuzin F., Yaniv M. Simian virus 40 illegitimate recombination occurs near short direct repeats. J Mol Biol. 1984 Jul 25;177(1):53–68. doi: 10.1016/0022-2836(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Jelinek W. R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6130–6134. doi: 10.1073/pnas.78.10.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Henthorn P. S., Mager D. L., Huisman T. H., Smithies O. A gene deletion ending within a complex array of repeated sequences 3' to the human beta-globin gene cluster. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs H. H., Lehrman M. A., Yamamoto T., Russell D. W. Polymorphism and evolution of Alu sequences in the human low density lipoprotein receptor gene. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7651–7655. doi: 10.1073/pnas.82.22.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Kato S., Anderson R. A., Camerini-Otero R. D. Foreign DNA introduced by calcium phosphate is integrated into repetitive DNA elements of the mouse L cell genome. Mol Cell Biol. 1986 May;6(5):1787–1795. doi: 10.1128/mcb.6.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Krawinkel U., Zoebelein G., Bothwell A. L. Palindromic sequences are associated with sites of DNA breakage during gene conversion. Nucleic Acids Res. 1986 May 12;14(9):3871–3882. doi: 10.1093/nar/14.9.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A., Marcaud L., Moreau J., Scherrer K. Conservation and variation in the large scale organisation of the globin gene domains of duck and chicken. Mol Gen Genet. 1986 May;203(2):193–201. doi: 10.1007/BF00333954. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman M. A., Goldstein J. L., Russell D. W., Brown M. S. Duplication of seven exons in LDL receptor gene caused by Alu-Alu recombination in a subject with familial hypercholesterolemia. Cell. 1987 Mar 13;48(5):827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Alu-Alu recombination deletes splice acceptor sites and produces secreted low density lipoprotein receptor in a subject with familial hypercholesterolemia. J Biol Chem. 1987 Mar 5;262(7):3354–3361. [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Exon-Alu recombination deletes 5 kilobases from the low density lipoprotein receptor gene, producing a null phenotype in familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3679–3683. doi: 10.1073/pnas.83.11.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman M. A., Schneider W. J., Südhof T. C., Brown M. S., Goldstein J. L., Russell D. W. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985 Jan 11;227(4683):140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D. L., Henthorn P. S., Smithies O. A Chinese G gamma + (A gamma delta beta)zero thalassemia deletion: comparison to other deletions in the human beta-globin gene cluster and sequence analysis of the breakpoints. Nucleic Acids Res. 1985 Sep 25;13(18):6559–6575. doi: 10.1093/nar/13.18.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M. P., Laemmli U. K. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979 Aug;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Montiel J. F., Norbury C. J., Tuite M. F., Dobson M. J., Mills J. S., Kingsman A. J., Kingsman S. M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Jan 25;12(2):1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau J., Marcaud L., Maschat F., Kejzlarova-Lepesant J., Lepesant J. A., Scherrer K. A + T-rich linkers define functional domains in eukaryotic DNA. Nature. 1982 Jan 21;295(5846):260–262. doi: 10.1038/295260a0. [DOI] [PubMed] [Google Scholar]
- Moreau J., Matyash-Smirniaguina L., Scherrer K. Systematic punctuation of eukaryotic DNA by A+T-rich sequences. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1341–1345. doi: 10.1073/pnas.78.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Meuth M. DNA amplification--deletion in a spontaneous mutation of the hamster aprt locus: structure and sequence of the novel joint. Nucleic Acids Res. 1986 Nov 11;14(21):8361–8371. doi: 10.1093/nar/14.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz H. R., Zehetner G., Frischauf A. M., Lehrach H. Rapid restriction mapping of DNA cloned in lambda phage vectors. Gene. 1984 Oct;30(1-3):195–200. doi: 10.1016/0378-1119(84)90120-3. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Kekelidze M. G., Lukanidin E. M., Scherrer K., Georgiev G. P. Replication origins are attached to the nuclear skeleton. Nucleic Acids Res. 1986 Oct 24;14(20):8189–8207. doi: 10.1093/nar/14.20.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G. E., Blanton H. M., Hager L. J., Zakian V. A. Isolation and characterization of sequences from mouse chromosomal DNA with ARS function in yeasts. Mol Cell Biol. 1983 Nov;3(11):1898–1908. doi: 10.1128/mcb.3.11.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Stark G. R. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured animal cells. Cell. 1984 Jul;37(3):705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Shih C. K., Linial M., Goodenow M. M., Hayward W. S. Nucleotide sequence 5' of the chicken c-myc coding region: localization of a noncoding exon that is absent from myc transcripts in most avian leukosis virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4697–4701. doi: 10.1073/pnas.81.15.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Broyles S. S., Pettijohn D. E. Perfect palindromic lac operator DNA sequence exists as a stable cruciform structure in supercoiled DNA in vitro but not in vivo. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1797–1801. doi: 10.1073/pnas.80.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Stark G. R. DNA amplification in drug resistant cells and in tumours. Cancer Surv. 1986;5(1):1–23. [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Recombination between poly[d(GT).d(CA)] sequences in simian virus 40-infected cultured cells. Mol Cell Biol. 1985 Jun;5(6):1247–1259. doi: 10.1128/mcb.5.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. A dominant influence of flanking sequences on a local structural transition in DNA. Cell. 1986 Dec 5;47(5):817–827. doi: 10.1016/0092-8674(86)90524-6. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Parker R. C., Varmus H. E., Bishop J. M. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1983 May;80(9):2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986 Aug 14;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Kaufmann G., Martin R. G. Mammalian DNA enriched for replication origins is enriched for snap-back sequences. J Mol Biol. 1984 Nov 15;179(4):577–586. doi: 10.1016/0022-2836(84)90156-6. [DOI] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Kaufmann G., Wang S. S., Lechner R. L., Karawya E., Hesse J., Martin R. G. Properties of some monkey DNA sequences obtained by a procedure that enriches for DNA replication origins. Mol Cell Biol. 1985 Jul;5(7):1621–1629. doi: 10.1128/mcb.5.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]