Abstract
The integration host factor of Escherichia coli (IHF) is a small, histone-like protein which participates in the integration of bacteriophage lambda into the E. coli chromosome and in a number of regulatory processes. Our recent footprinting analysis has shown that IHF binds specifically to the ends of the transposable element IS1, as well as to several sites within a short segment of the plasmid pBR322. We have extended our studies of the binding of the IHF molecule to these sites in vitro using a gel retardation assay. We report here that IHF bends the DNA upon binding, as judged from the strong cyclic dependence of the protein-induced mobility shift on the position of the binding site. Using cloned, synthetic ends of IS1 as substrates, we have found that some mutations within the conserved bases of the IHF consensus binding sequence abolish binding, and that alterations of the flanking sequences can greatly reduce IHF binding. The presence of multiple IHF sites on a single DNA fragment increases binding very little, indicating that IHF does not bind cooperatively in this complex. We discuss the possibility that DNA bending is related to the role IHF plays in forming and stabilizing nucleoprotein complexes, and suggest that bending at the IHF sites may be important to its diverse effects in the cell.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. A phage repressor-operator complex at 7 A resolution. Nature. 1985 Aug 15;316(6029):596–601. doi: 10.1038/316596a0. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- Better M., Lu C., Williams R. C., Echols H. Site-specific DNA condensation and pairing mediated by the int protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. The sequence of a region of bacteriophage phiX174 DNA coding for parts of genes A and B. J Mol Biol. 1977 Oct 15;116(1):1–28. doi: 10.1016/0022-2836(77)90115-2. [DOI] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 1983 Jan 11;11(1):141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. J., Carver D., Gellert M. Synergistic effect of himA and gyrB mutations: evidence that him functions control expression of ilv and xyl genes. J Bacteriol. 1984 Feb;157(2):484–489. doi: 10.1128/jb.157.2.484-489.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Burger A. C., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986 Jul;204(1):85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Gamas P., Caro L., Galas D., Chandler M. Expression of F transfer functions depends on the Escherichia coli integration host factor. Mol Gen Genet. 1987 May;207(2-3):302–305. doi: 10.1007/BF00331593. [DOI] [PubMed] [Google Scholar]
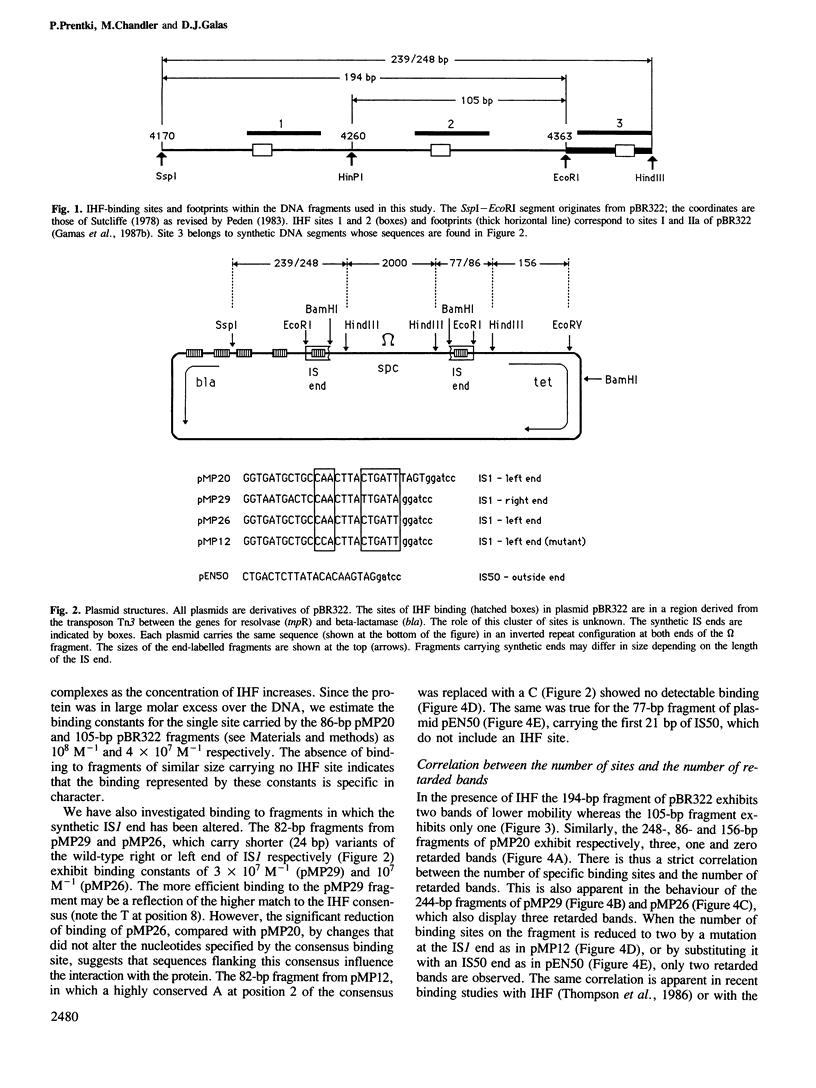
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van Heuvel M., Moolenaar G. F., van de Putte P. Regulation of Mu transposition. II. The escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu. Gene. 1984 Dec;32(3):419–426. doi: 10.1016/0378-1119(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Griffith J., Hochschild A., Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986 Aug 21;322(6081):750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- Gronenborn A. M., Nermut M. V., Eason P., Clore G. M. Visualization of cAMP receptor protein-induced DNA kinking by electron microscopy. J Mol Biol. 1984 Nov 15;179(4):751–757. doi: 10.1016/0022-2836(84)90166-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Grindley N. D. Analysis of gamma delta resolvase mutants in vitro: evidence for an interaction between serine-10 of resolvase and site I of res. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5429–5433. doi: 10.1073/pnas.83.15.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., Knight D. M., Das A., Miller H. I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982 Dec;31(3 Pt 2):565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978 Oct 25;253(20):7149–7157. [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lathe R., Kieny M. P., Skory S., Lecocq J. P. Linker tailing: unphosphorylated linker oligonucleotides for joining DNA termini. DNA. 1984;3(2):173–182. doi: 10.1089/dna.1984.3.173. [DOI] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Mahajna J., Oppenheim A. B., Rattray A., Gottesman M. Translation initiation of bacteriophage lambda gene cII requires integration host factor. J Bacteriol. 1986 Jan;165(1):167–174. doi: 10.1128/jb.165.1.167-174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Pollock T. J., Nash H. A. Knotting of DNA caused by a genetic rearrangement. Evidence for a nucleosome-like structure in site-specific recombination of bacteriophage lambda. J Mol Biol. 1983 Oct 15;170(1):1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Ross W., Shulman M., Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982 Apr 15;156(3):505–522. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
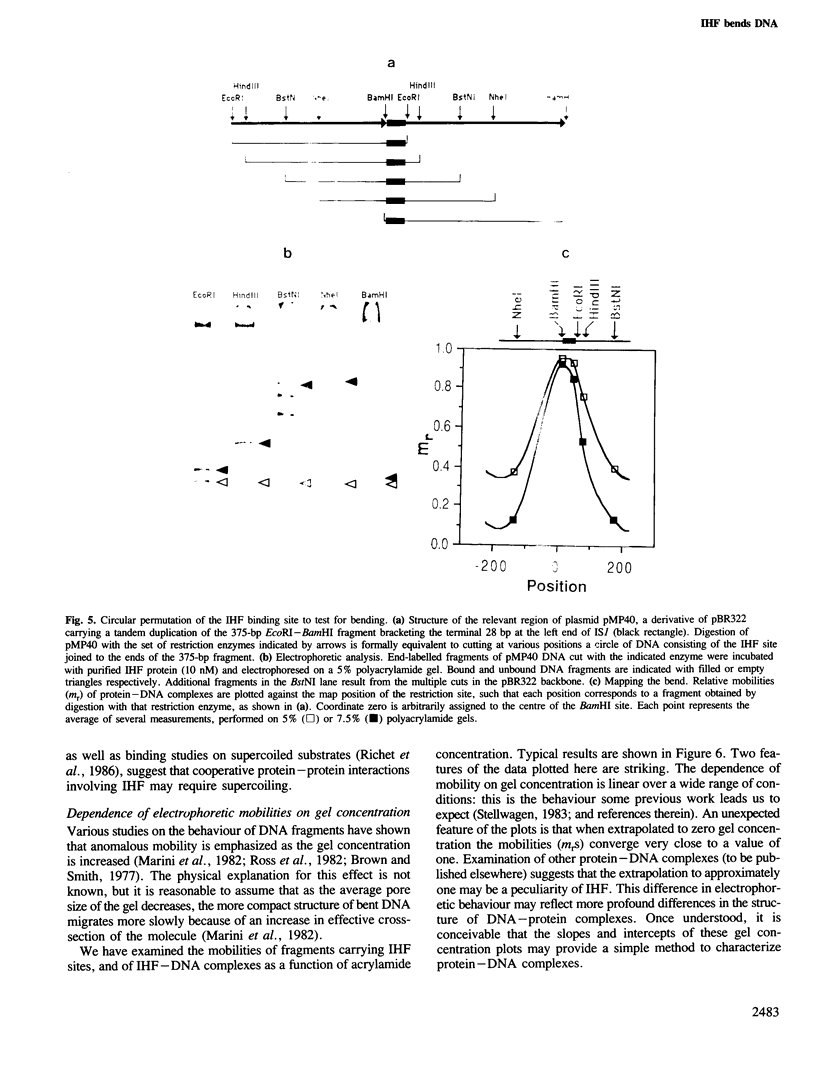
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R. B., Berg O. G., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981 Nov 24;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985 Dec 16;4(13A):3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D., Gamas P., Chandler M., Prentki P., Bass S., Galas D. Specificity of insertion of IS1. J Mol Biol. 1985 Oct 5;185(3):517–524. doi: 10.1016/0022-2836(85)90068-3. [DOI] [PubMed] [Google Scholar]