Abstract
The bacterial TATAAT −10 region sequence was the first promoter element to be identified, but how it functions is still not clear. Because the duplex element is melted during initiation, the effects of substitutions were studied in both single-and double-strand contexts. Band-shift results were particularly unexpected in the context of melted DNA. The effect of the lac UV5-melted −10 region on polymerase binding was found to include a large sequence nonspecific contribution. Instead the dominant role of single-stranded −10 region nucleotides was in directing the isomerization of the RNA polymerase to its heparin resistant form. This role becomes minimal when the melting is extended beyond the −10 region to encompass the transcription start site, as in the final open complex. The duplex binding results are in agreement with previous reports that showed positions −12T and −11A are of primary importance for promoter recognition. Thus the consensus −10 region sequences function in two ways, both before full promoter melting. They stabilize initial polymerase binding via duplex interactions and subsequently as single-stranded DNA they promote enzyme isomerization to the functional form.
The σ70-dependent bacterial promoters (reviewed in refs. 1–3) contain a series of moderately conserved promoter elements. Together these elements stabilize the holoenzyme at the promoter, direct the holoenzyme-dependent melting of the DNA, and allow for productive initiation of transcription. The first element to be discovered and the most conserved is the −10 region. On the nontemplate strand this has the sequence TATAAT, from −12 to −7, where the underlined nucleotides are ≈80% conserved and the others are ≈60% conserved (4). The element is recognized predominantly by region 2 of σ70. Numerous genetic and biochemical studies have demonstrated its importance. The specific roles of this region in promoter usage are not yet reliably established.
Determination of the biochemical roles of these nucleotides is complicated by several factors. First, most or all of the sequence typically becomes melted before transcription initiation. Thus the region may influence transcription either in the duplex or single-stranded form. Second, the preinitiation complex pathway includes several intermediates in which the conformational state of the DNA and the enzyme may vary (5–7). The population of these various states may depend on solution conditions and on the type of template used for binding or transcription studies. The final functional state is characterized by resistance to the single strand DNA analog heparin (8). In this state, the conformation of the DNA:enzyme complex is said to be fully isomerized (9). In addition, the −10 region acts in concert with other conserved promoter elements and with activators. There is a lesser reliance on the −10 sequences when other elements are well conserved or when activators are present (10). Thus the −10 region can contribute in a variety of forms and contexts and at diverse steps leading to transcription initiation.
Systematic studies of variant −10 regions have contributed to our understanding of its function. Competition binding studies demonstrated that the upstream half (TAT- - -) is dominant for duplex recognition by σ (11). Abortive initiation studies have shown the functional importance of the initial duplex TA- - - - sequence (12), but even the less well conserved positions can make important contributions (10). Binding competition studies by using diverse probes containing melted DNA are not in full agreement, with either the sequence - A - - AA - (1) or - A- - - T (13) being suggested as most important. Studies of bubble (11) and fork junction (14) templates have shown the importance of the −11A in the single-stranded state, which is a common conclusion in all studies. In this state, the nontemplate strand is dominant for binding (15). There have not been systematic studies comparing duplex and single strand effects. Nor have there been studies assessing the importance, if any, of these nucleotides on enzyme isomerization to the functional heparin resistant state (16).
Our goal in the current work is to bring these various types of information together and to extend them to include work that might be relevant to enzyme isomerization. Thus the experiments include comparative studies of binding in the duplex- and single-stranded forms and also studies of heparin-resistant complex formation. The approach is to use electrophoretic mobility-shift assay (EMSA) on DNA probes with conditions that attempt to mimic these conformational states. The effect of the presence of the −10 region and of every substitution at every position under each condition will be evaluated. The results are unexpected in that they reveal a critical role for nucleotide identity within single-stranded DNA in enzyme isomerization, as measured by formation of heparin-resistant complex formation. As expected, the region also plays an important role in sequence-specific duplex recognition. The role of single strand DNA sequence in holoenzyme binding appears to be minor compared to these effects.
Materials and Methods
The Escherichia coli RNA polymerase core enzyme was from Epicenter Technologies, (Madison, WI), σ70 was purified as described from pQE30-rpoD (11), and probes were prepared as described (14). In brief, the bottom strand was labeled with γ-[32P]ATP and annealed to all top strands used for an experiment. The 40-μl mixtures, containing 100 nmol of kinased DNA, 150 nmol top strand, 20 mM Hepes/pH 7.5, and 160 mM NaCl, were heated to 95°C and slowly cooled to room temperature. Annealing was monitored by electrophoresis. This method has the benefit of excluding all radioactive single strand DNA but at the expense of the presence of some unlabeled single-strand DNA. The annealed probes were diluted and used in Tris-EDTA buffer containing 80 mM NaCl.
EMSA were as described (17). In brief, 40 nM core was mixed with 100 nM σ70 in a 10-μl reaction mixture with 1 × buffer A [30 mM Tris⋅HCl, pH 7.9/100 mM KCl/3 mM MgCl2/0.1 mM EDTA/1 mM DTT /100 μg/ml BSA/6 ng/μl (dI-dC)/3.25% glycerol] and 1 nM annealed probe and then incubated for 20 min on ice. Some batches of core were less effective binders and these were preincubated at 37°C for 5 min, which enhanced efficiency. For heparin and cold duplex challenge experiments, 50 μg/ml heparin or DNA was added for an additional 5 min (17). Samples were run on 5% page with [45 mM Tris-borate/1 mM EDTA] (TBE) buffer, with the apparatus packed in ice. The internal temperature of the gel was not monitored, but all gels were run for ≈14 min at a constant 325 V. Binding at higher temperatures was generally less, and the effects of some mutations were somewhat exaggerated. The amount bound was determined by phosphorimager analysis of bound and free probes. All experiments were repeated at least three times.
Results
Duplex- and Single-Stranded Contributions to General Binding.
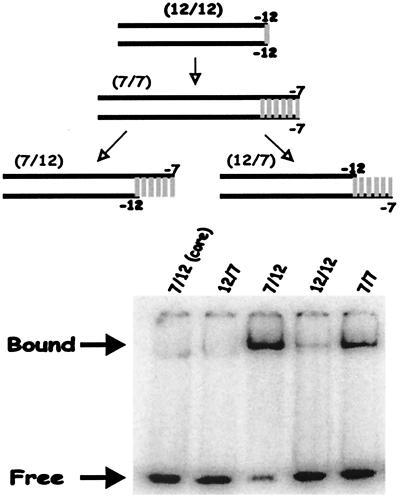
A collection of four probes is used to initially assess the importance of nucleotides within the −10 region for binding to holoenzyme (Fig. 1). The probes have in common the lac UV5-promoter sequence between positions −41 and −13. The lac UV5 −10 region from −12 to −7 matches the consensus but the other elements, −35, spacer and extended −10, do not. The four probes differ in how the −10 region is presented. The probes are denoted by pairs of numbers that identify the termini of the two oligonucleotides that were annealed to make the hybrid (Fig. 1). Probes 12/12 and 7/7 are duplexes, with only the latter containing the −10 region sequences from −11 to −7. Probes 7/12 and 12/7 are “fork” probes (14, 16) in which the −11 to −7 sequences are present in single-stranded form. The 7/12 probe contains the top, nontemplate single strand whereas the 12/7 probe contains the bottom, template strand.
Figure 1.
EMSA of lac UV5 (CAGGCTTTACACTTTATGCTTCCGGCTCGTATAAT) duplex and fork probes with bound and free probes indicated by arrows. The numbers refer to the position of the last base present in each strand with respect to the transcription start site. The probe names are in parenthesis. Vertical bars represent the −10 consensus region.
EMSA (band shift) experiments were done to assess how well each of these probes bound to holoenzyme. The experiments were done at low temperature to attempt to populate the unisomerized state of the enzyme and conform to our previous studies (17). The data show that only two probes generate a strong shifted band (Fig. 1). Fork probe 7/12, which contains the nontemplate −10 region in single strand form, is bound the most tightly. By contrast fork probe 12/7, which is identical except that the template single strand is present, bound at a very low level. This binding is barely above the background level seen when core polymerase is used with various probes (see lane “7/12 core” as an example). When the −10 region is present as a duplex in probe 7/7, binding is lower than on the 7/12 fork probe, as seen previously in different contexts (14, 17). Binding to the duplex 7/7 probe but not the fork probe 7/12 is abolished by addition of heparin (not shown and see below), suggesting that the band shift on the 7/7 probe predominantly reflects duplex recognition. When the −11 to −7 region is absent, as on the 12/12 probe, binding is barely detectable.
These data demonstrate that binding is highly sensitive to the presence of the −10 region and to how the −10 region sequences are presented. The region is required for recognition because the 12/12 probe is essentially unbound. Maximum binding occurs when the region is presented in the context of a fork probe in which the nontemplate strand is exposed in single-stranded form (probe 7/12).
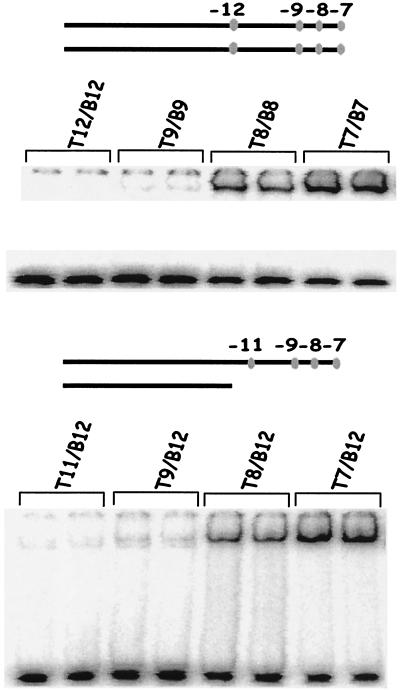
Fig. 2 shows how the binding is altered when the duplex and fork probes are truncated by progressive removal of −10 region nucleotides. The parent probes are the two with detectable binding, the duplex 7/7 (Upper) and the fork 7/12 (Lower). Each parent is truncated by removing bases or base pairs progressively from the downstream direction (the truncation points are shown as dots). The results show that there is a progressive loss of binding (right to left in each panel). In each case, the binding is lowered by removal of position −7 and nearly eliminated by additional removal of position −8. We infer that a contiguous −10 region is optimal for binding in both duplex and fork probe contexts.
Figure 2.
EMSA with either the duplex (Upper) or the template strand (Lower) cut back. The ovals indicate the terminus of the probe. The average binding is: 7/7, 40%; 8/8, 20%; 9/9 and 12/12, <2%, and for 1/1 probes (not shown), 50%. The average binding is: 7/12, 70%; 8/12, 20%; 9/12 and 11/12, <2%, and for 1/12 probes (not shown), 60%.
General Binding to Duplex Probes with All Substitutions in the −10 Region.
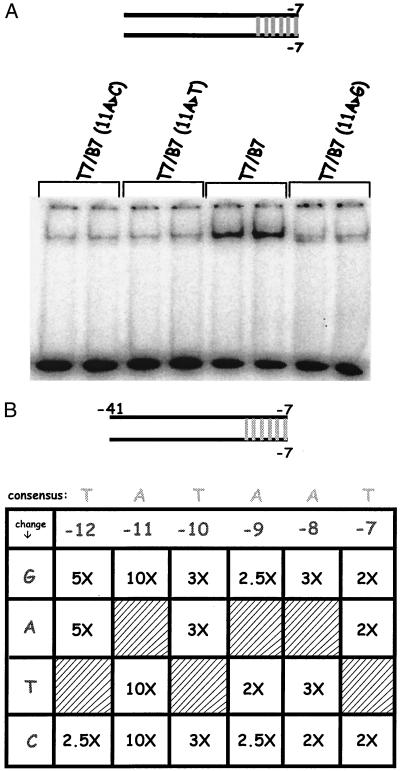
The next set of experiments investigated the effect of substitutions within the −10 region of duplex probes. The parent is the 7/7 probe containing the consensus sequence. Eighteen variants were constructed, encompassing all possible single substitutions within the 6-bp consensus. The EMSA experiments were repeated on this collection of duplex probes. An example, the strongest effect seen, is shown in Fig. 3A. In this case, any substitution for base pair −11 gives a significant reduction in binding, ≈10-fold. All other substitutions were analyzed similarly, and the results are compiled in Fig. 3B.
Figure 3.
EMSA on duplex probes with all substitutions in the −10 consensus. The parent is the 7/7 probe shown and each substitution is indicated. (A) Sample EMSA results when −11A is substituted. (B) The fold reduction caused by each substitution is shown. The consensus sequence is shown along the top and each change along the side.
Fig. 3B displays the fold reductions caused by each of the 18 substitutions. We note first that all substitutions have a detectable effect. Thus every consensus nucleotide makes some contribution to duplex recognition. The effects for base pairs −7, −8, −9, and −10 are very similar; essentially any individual substitution leads to a 2- to 3-fold reduction in the extent of binding. Substitutions at position −12 are somewhat more deleterious, giving 2.5- to 5-fold reductions. As noted above, −11 substitutions give 10-fold reductions. We infer that some positions are more important than others for duplex recognition but that all make a contribution. Some of these contributions in duplex form could be subject to small errors if there is some DNA melting at the low temperature of the experiment. The strong and selective reductions associated with −11 and −12 substitutions are consistent with previous reports (11, 12).
General Binding to Fork Probes with All Substitutions in the −10 Region.
Experiments of this type were repeated with probes based on the consensus parent fork probe 7/12. Each probe contains the −12 position base paired, and the remaining positions are presented as the nontemplate single strand. Eighteen probes were studied, representing all possible single substitutions. The results are presented in Fig. 4.
Figure 4.
EMSA on fork + tail probes with all substitutions in the −10 consensus. The fold reduction caused by each substitution is shown. The consensus sequence is shown along the top and each change along the side.
This collection of results is remarkable in the sense that most substitutions have little or no effect. Eleven of the 18 substitutions lead to no change in the extent to which the probe is bound by holoenzyme. Six of the remaining substitutions lead to only 2-fold reductions, with a single substitution, from A:T to G:C at −11, giving a 4-fold reduction. We infer that the sequence of this region is only modestly important for binding when the determinants are in single-stranded form.
The result is very surprising when contrasted with the data of Figs. 1 and 2. Recall that the Fig. 1 data showed that presenting the nontemplate strand in single-stranded form was very important for binding. That is, the 12/12 duplex did not bind detectably and adding the nontemplate single strand led to very tight binding. The results of Fig. 4 indicate that this increase in general binding seen upon addition of the nontemplate strand is largely sequence nonspecific. This proposal is consistent with the results by using truncated probes presented in Fig. 2. In that experiment the progressive removal of single strand nucleotides led to a progressive loss of binding. Taking the data at this point as a whole, we suggest the following: General duplex binding is very sequence dependent; the binding enhancement from exposing the nontemplate single strand is much less sequence dependent.
Binding in the Presence of Heparin: Evidence for a Role for Specific Nucleotides in Enzyme Isomerization.
Although nontemplate single strand sequence identity does not appear to play a major role in general binding, other functional roles are still possible. Chief among these would be contributing to the events that follow the initial binding of the holoenzyme. Evidence indicates that several isomerization events must follow the initial binding, leading to the formation of a functional open complex (5, 6). Recently, we proposed that the enzyme could isomerize independent of the state of the DNA (16). The proposal is based on results obtained in EMSA experiments done in the presence and absence of heparin.
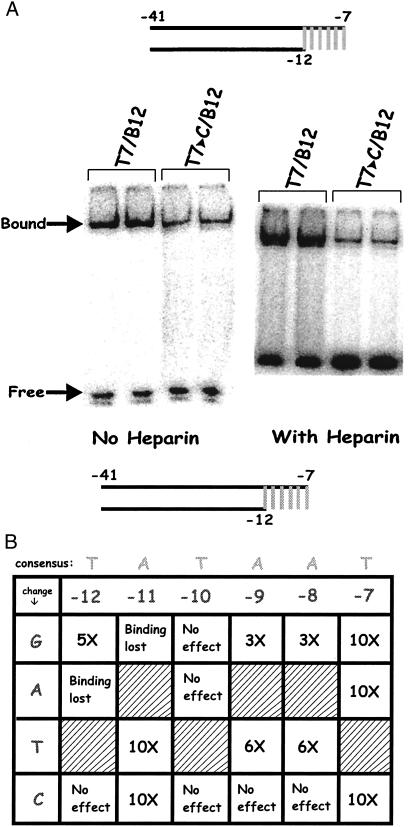
Heparin challenge has long been used to dissociate unstable closed complexes, while leaving open complexes largely intact. That is, the state of the enzyme in the open complex largely resists the effects of heparin. We wish to learn whether changes in the −10 region can influence the conversion to heparin-resistance. In the experiments using fork probes, the DNA is already melted so any conversion to heparin resistance must be due to changes in the enzyme rather than melting of the DNA. When complexes containing fork probe 7/12 and holoenzyme are challenged with heparin, a fraction remains intact, ≈1/3 under the low temperature conditions of our experiments.
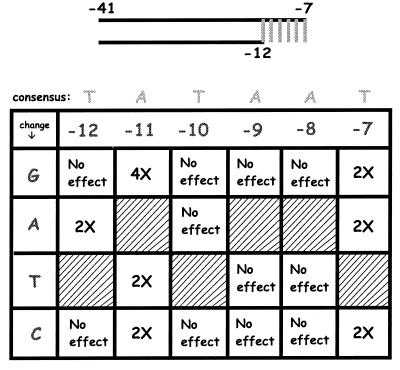
Fig. 5A compares binding to a fork probe with and without heparin when the −7T is substituted by C in the single-stranded tail. The control experiment without heparin (at the Left) confirms the 2-fold reduction caused by this change. When the same experiment is done using a heparin challenge, the effect of the substitution is now 10-fold (at the Right, overexposed to show low-level binding). We infer that changing the nucleotide identity at −7 in the single-stranded tail can drastically change the ability of the enzyme to resist the effects of heparin.
Figure 5.
EMSA on fork + tail probes in the presence of heparin. (A) EMSA comparison of probes with changes at −7 in the presence and absence of heparin (Right panel is overexposed to show low binding). (B) Data summary for all substitutions in the presence of heparin, as described in Fig. 4.
These heparin challenge experiments were repeated by using the entire set of 18 substitutions on the fork probe. That is, the experiments are identical to those of Fig. 4 except that heparin is used. The results are compiled in Fig. 5B. Note that the results of the two experiments differ remarkably. Whereas few sequence-specific effects were seen without heparin, the majority of substitutions have strong effects in the presence of heparin.
These larger changes include the effect of position −7 already noted and even stronger effects of certain substitutions at −12 and −11. Indeed, an A to T change at −11 and a T to A change at −12 nearly prevent the holoenzyme from binding in the presence of heparin. Substitutions at positions −8 and −9 can strongly reduce the binding in the presence of heparin. In both cases only the A to C change is without effect. Recall that in the absence of heparin no substitutions at these positions had a detectable effect on binding. Because the DNA on all these probes is already melted, the effects are clearly on the state of the holoenzyme and are not due to the melting of the DNA. We infer that specific nucleotide sequences within the nontemplate strand are critical for the conversion of the enzyme to a form that resists heparin challenge.
Selected experiments were repeated by using a duplex DNA challenge (50 μg/ml of cold UV5 T7/B7, which is a 10-fold excess over polymerase) to assess whether the effect was specific for heparin. Five of the six substitutions at the −12 and −11 nucleotides showed changes similar to that without heparin, except that the overall binding was slightly reduced (data not shown). The −11 A to G change showed an intermediate level between that with and without heparin. We infer that these mutations affect primarily the conversion to the heparin resistant form rather than lead to a general decrease in complex stability.
Lac UV5 and Wild Type: Mechanistic Sources of the Functional Difference.
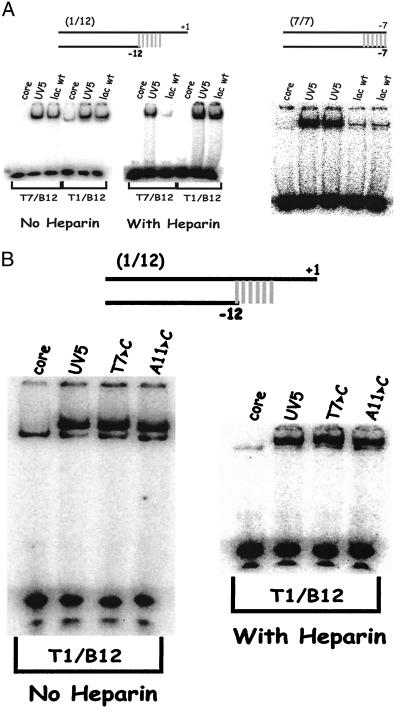
The parent for this work, the lac UV5 promoter, differs from lac wild type in that the latter contains nonconsensus nucleotides at positions −8 and −9 (TATGTT) (18). The consensus UV5 substitutions allow transcription to occur independent of activator and increase transcription from 5- to 50-fold (10, 19). The above experiments suggest that the UV5 changes should increase the binding to duplex DNA, have little effect on the binding to single-stranded fork DNA, and enhance isomerization to heparin resistance. We tested these predictions and the results are shown in Fig. 6A. The data are in accord with expectation: UV5 duplex binding is enhanced 10-fold compared to wild type (Right, overexposed to show low levels); UV5 fork binding is increased by 2-fold (T7/B12, Left); UV5 directs 10-fold higher isomerization to heparin resistance on this same fork template (T7/B12, Center).
Figure 6.
EMSA on lac wild type and UV5 by using probes that reach the start site. The UV5 mutation is a double change (−8T and −9G). (A Left) Comparative data for the longer 1/12 and shorter 7/12 probes. Lac wild-type binding is within a factor of two of the UV5 binding except on T7/B12 in the presence of heparin, which differs by a factor of 10. (Right) This panel is overexposed to show low binding; wild-type binding differs by a factor of 10 on duplex probe (7/7), but this difference is eliminated by addition of base pairs to + 1 (data not shown). (B) On the longer 1/12 probes, substitutions in important positions within the consensus have little effect.
Additional data suggest that once the isomerization process is complete, the UV5 mutation has little to do with stabilizing the final open complex. These data involve a series of 1/12 probes in which the nontemplate single strand is extended to include the transcription start site (Fig. 6A). Fig. 6A shows that lac UV5 and wild type have nearly identical behavior on such probes. There is less than a 2-fold difference on either binding or isomerization (Fig. 6A, T1/B12 lanes). Because the effect on isomerization is large when the strand is truncated at −7 (Fig. 6A Center T7/B12 lanes), it appears that when the melted DNA encompasses both the −10 region and the start site, the UV5 mutation is not very relevant. This effect also is seen when single substitutions are made in the more important positions −11 and −7 (Fig. 6B) within the UV5 promoter, suggesting that the primary effects of the −10 region at this promoter are on isomerization and duplex binding rather than on stability of the final functional complex.
Discussion
The bacterial −10 region sequence TATAAT was the first identified conserved core-promoter element (20). In this work, we have studied all possible substitutions within the −10 region by using EMSA assays on probes that attempt to mimic aspects of closed and open complexes. The results confirm the role of this region in duplex DNA binding and reveal a new and unexpected role of the sequence in single-stranded form.
The properties of the single-stranded −10 region were investigated in the context of a fork probe in which the upstream DNA remained in duplex form (14). This fork probe, with five of the six consensus nucleotides present as a consensus nontemplate single strand, bound significantly more holoenzyme than the analogous duplex. However, most substitutions in the consensus (11 of 18) had no significant effect on binding, and the remaining substitutions showed modest decreases in binding. Thus the binding affinity caused by the single strand was inferred to be largely sequence nonspecific. By contrast, when this same collection of complexes was challenged with heparin the effect of substitution was very great. Thirteen of the 18 led to clear reductions in binding, and in 7 of these cases, binding decreased by an order of magnitude or more. Thus the −10 region nucleotide sequence is critical for converting the holoenzyme into a form that can resist a heparin challenge. That is, the −10 region appears to play a significant sequence nonspecific role in stabilizing holoenzyme binding and a sequence-specific role in changing its conformation. As conversion to heparin resistance is often taken as the final event in complex isomerization, we suggest that the −10 region consensus nucleotides play a direct role in isomerizing RNA polymerase into its final heparin resistant form.
The data are in general agreement with previous studies, with some minor discrepancies. Previous studies on duplex DNA (11, 12) pointed to the importance of the TAT- - -, and the current study shows that the highly conserved TA is most important with all other positions making an accessory contribution. Two previous systematic studies (13, 21) on probes containing single-stranded DNA used competition to show significant effects of individual nucleotides on binding. The details of our study are in good agreement with one of these (13) and all three agree on the importance of the −11A nucleotide. Two aspects of the current study cause us to downplay these effects. First, we see a large sequence nonspecific contribution of this region in the single strand form. Second, and more important, the previous studies did not measure effects in the presence of heparin, which are by far the strongest sequence-dependent effects of single-stranded DNA in the current study. Overall, we believe that there are sequence-dependent effects of single-stranded DNA on binding but that these are significantly smaller than the effects of sequence on enzyme isomerizaton and also on duplex recognition. Because the enzyme is isomerized before initiation, processes that depend on melted DNA, such as the transition to elongation (22) and related regulatory events (23), could depend strongly on −10 region nucleotide identity.
Taken together these data suggest that the −10 region has its greatest effect on the closed complex, stabilizing the enzyme on duplex DNA and subsequently helping the bound enzyme to isomerize. The effects on single strand binding appear to be much less important. In the case of the lacUV5 mutant promoter, one can speculate that the substitution allows the following pathway to occur without activator. The −10 substitution allows the closed (unisomerized) enzyme to reside on closed DNA long enough for it to melt the −10 region. Melting then further stabilizes the enzyme nonspecifically. It also reveals the DNA substitutions in single-stranded form, which help to isomerize the enzyme as it extends the bubble to include the transcription start site. The mutation does not appear to significantly stabilize the complex once it is opened.
Caveats in interpreting these and other studies include the effects of promoter sequences outside the −10 region. A number of −10 region changes have effects only in some promoter contexts (unpublished and ref. 11). As an extreme example, it has long been known that certain stable RNA promoters completely fail to form heparin resistant preinitiation complexes (24, 25). These promoters are functionally competent and contain consensus −10 elements; yet they still require initiating nucleotides for the bound holoenzyme to attain heparin resistance. On the other hand, certain probes with optimal −35 and spacer elements can become bound even when much of the −10 region is truncated or mutated (14, 17). It should also be noted that mutations may reduce transcription rates by altering the properties of intermediates along the pathway to stable open complex formation (10). Moreover, the properties of the low temperature intermediates studied here may not fully conform to those in the physiological transcription pathway (6).
Natural promoters are tremendously diverse, and indeed there is no known promoter that fully matches the overall consensus. The purpose of this diversity is thought to be to suit each promoter to the demands of its function. So during initiation at natural promoters one could have very diverse pathways, which need not match that of lac UV5. As suggested previously (16), it is important to consider that both the enzyme and the DNA may exist predominantly in closed forms and need not always isomerize to open forms by the same pathway. Most promoters are subject to activation, which sometimes occurs at the overall “isomerization” step, and the suitability of a promoter toward activation can strongly depend on the core DNA sequence. The challenge is to understand how the core sequence sets up the potential pathways to open complex formation and to learn how the molecular contacts made by activators participate in enhancing these pathways to ensure appropriate transcription rates.
Acknowledgments
This work was supported by National Institutes of Health Grant GM35754.
Abbreviation
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Helmann J D, deHaseth P L. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 2.deHaseth P L, Zupancic M L, Record M T., Jr J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross C A, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 4.Lisser S, Margalit H. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roe J H, Burgess R R, Record M T., Jr J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 6.Buckle M, Pemberton I K, Jacquet M A, Buc H. J Mol Biol. 1999;285:955–964. doi: 10.1006/jmbi.1998.2391. [DOI] [PubMed] [Google Scholar]
- 7.Craig M L, Tsodikov O V, McQuade K L, Schlax P E, Jr, Capp M W, Saecker R M, Record M T., Jr J Mol Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 8.Zillig W, Zechel K, Rabussay D, Schachner M, Sethi V S, Palm P, Heil A, Seifert W. Cold Spring Harbor Symp Quant Biol. 1971;35:47–58. [Google Scholar]
- 9.McClure W R. Proc Natl Acad Sci USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiklejohn A L, Gralla J D. J Mol Biol. 1989;207:661–673. doi: 10.1016/0022-2836(89)90236-2. [DOI] [PubMed] [Google Scholar]
- 11.Dombroski A J. J Biol Chem. 1997;272:3487–3494. [PubMed] [Google Scholar]
- 12.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 13.Matlock D L, Heyduk T. Biochemistry. 2000;39:12274–12283. doi: 10.1021/bi001433h. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, Gralla J D. Proc Natl Acad Sci USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marr M T, Roberts J W. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Lew C M, Gralla J D. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenton M S, Lee S J, Gralla J D. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert W. RNA Polymerase. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. [Google Scholar]
- 19.Silverstone A E, Arditti R R, Magasanik B. Proc Natl Acad Sci USA. 1970;66:773–779. doi: 10.1073/pnas.66.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pribnow D. Proc Natl Acad Sci USA. 1975;72:784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu J, Helmann J D. Nucleic Acids Res. 1999;27:4541–4546. doi: 10.1093/nar/27.23.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpousis A J, Gralla J D. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 23.Ring B Z, Yarnell W S, Roberts J W. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 24.Gourse R L. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohlsen K L, Gralla J D. J Biol Chem. 1992;267:19813–19818. [PubMed] [Google Scholar]