Abstract
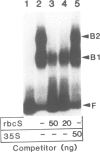
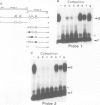
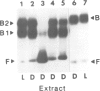
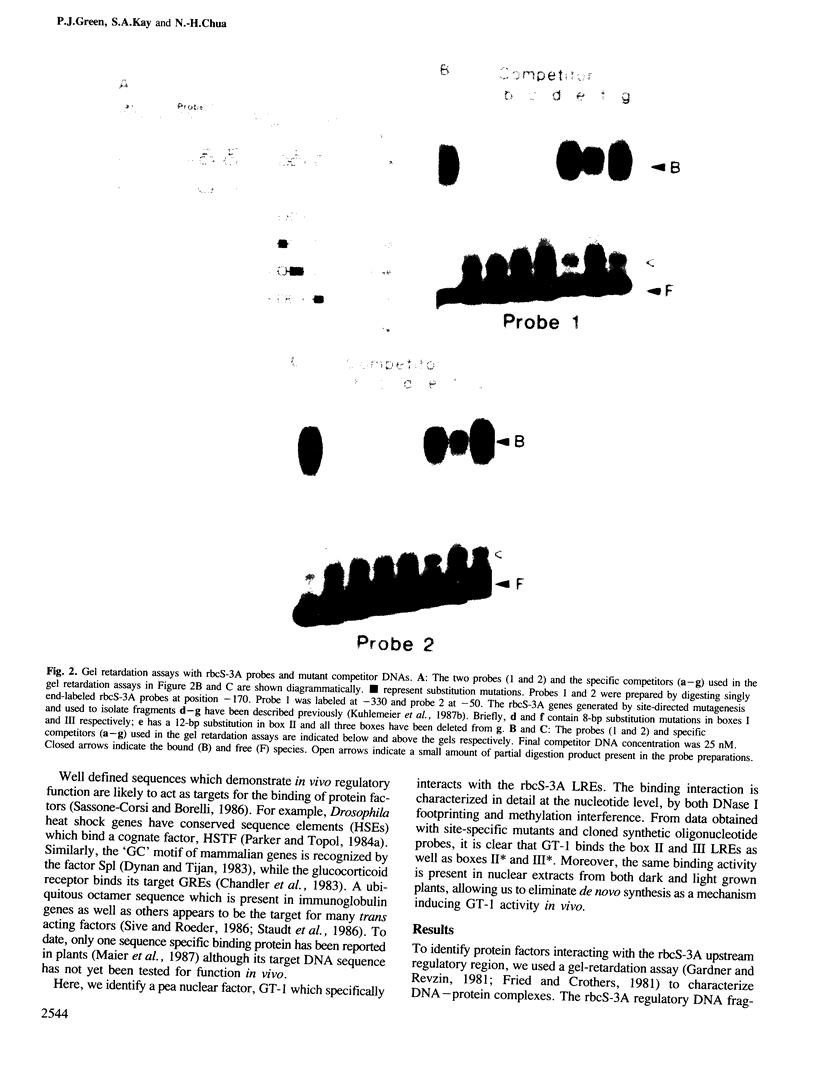
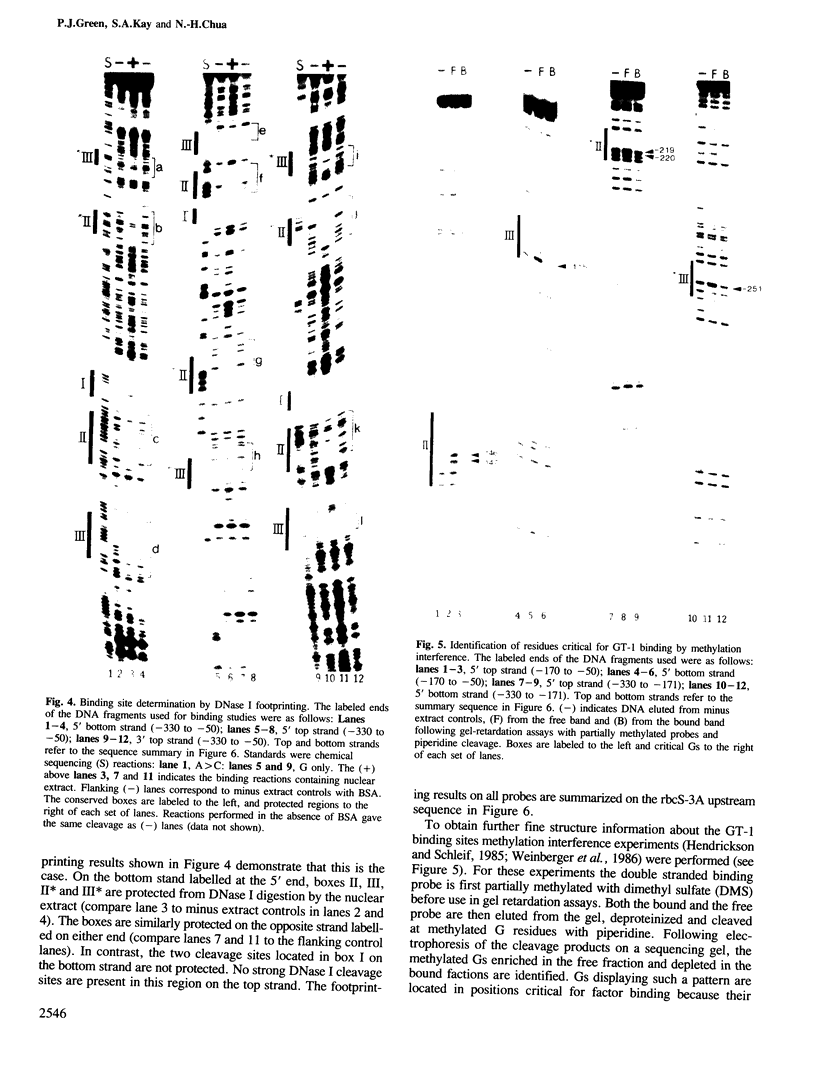
Pea nuclear extracts were used in gel retardation assays and DNase I footprinting experiments to identify a protein factor that specifically interacts with regulatory DNA sequences upstream of the pea rbcS-3A-gene. This factor, designated GT-1, binds to two short sequences (boxes II and III) in the -150 region that are known to function as light-responsive elements (LREs) in transgenic tobacco. Binding of GT-1 to homologous sequences further upstream (boxes II and III in the -220 region) indicates that these boxes comprise the redundant LRE that functions in vivo when boxes II and III are deleted. In both box II and box II, methylation interference experiments demonstrate that two adjacent G residues are critical for GT-binding. Single Gs present in boxes III and III are also important. Since GT-1 is present in nuclear extracts from leaves of light-grown and dark-adapted pea plants, its regulatory role does not depend on de novo synthesis. Thus if GT-1 binds differentially in vivo it must be postranslationally modified or sterically blocked from binding by another factor in response to light.
Full text
PDF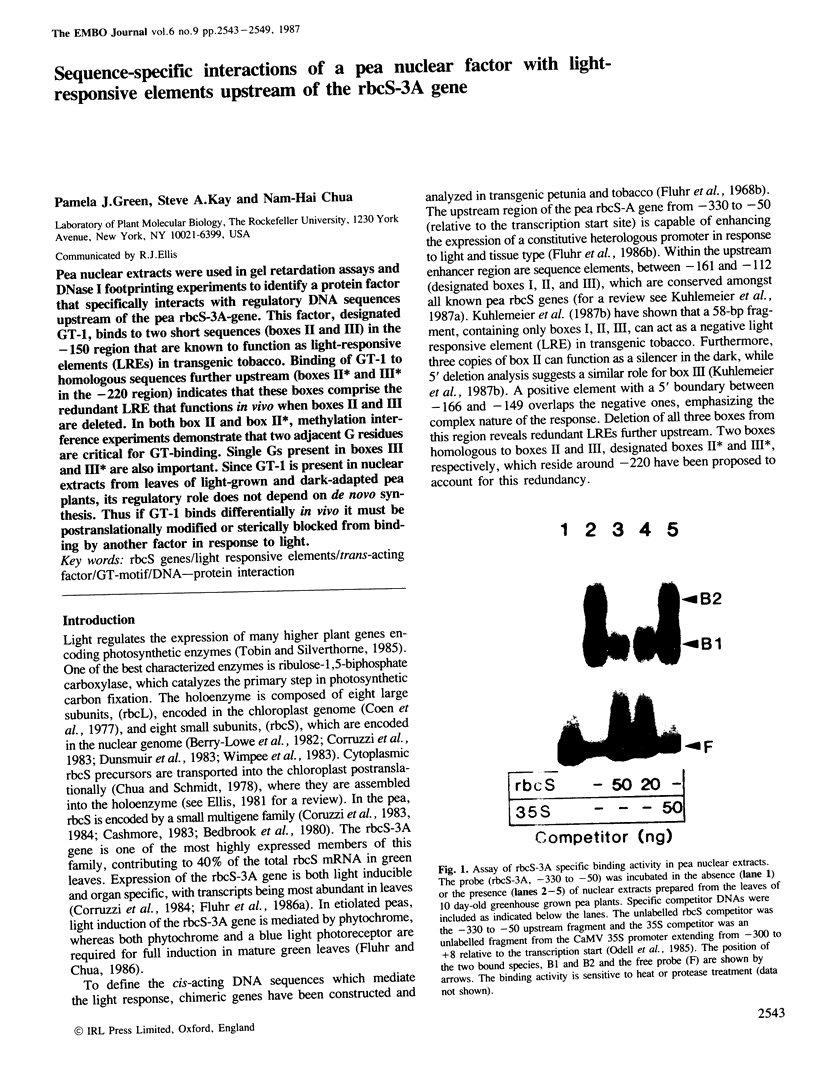




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Gloss B., Schmid W., Strähle U., Schütz G. In vivo protein-DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature. 1986 Dec 18;324(6098):686–688. doi: 10.1038/324686a0. [DOI] [PubMed] [Google Scholar]
- Berry-Lowe S. L., Mc Knight T. D., Shah D. M., Meagher R. B. The nucleotide sequence, expression, and evolution of one member of a multigene family encoding the small subunit of ribulose-1,5-bisphosphate carboxylase in soybean. J Mol Appl Genet. 1982;1(6):483–498. [PubMed] [Google Scholar]
- Chandler V. L., Maler B. A., Yamamoto K. R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983 Jun;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Chua N. H., Schmidt G. W. Post-translational transport into intact chloroplasts of a precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6110–6114. doi: 10.1073/pnas.75.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Bedbrook J. R., Bogorad L., Rich A. Maize chloroplast DNA fragment encoding the large subunit of ribulosebisphosphate carboxylase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5487–5491. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Broglie R., Edwards C., Chua N. H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J. 1984 Aug;3(8):1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J. Determination of fragment order through partial digests and multiple enzyme digests. Methods Enzymol. 1980;65(1):449–467. doi: 10.1016/s0076-6879(80)65055-1. [DOI] [PubMed] [Google Scholar]
- Dunsmuir P., Smith S., Bedbrook J. A number of different nuclear genes for the small subunit of RuBPCase are transcribed in petunia. Nucleic Acids Res. 1983 Jun 25;11(12):4177–4183. doi: 10.1093/nar/11.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Fluhr R., Chua N. H. Developmental regulation of two genes encoding ribulose-bisphosphate carboxylase small subunit in pea and transgenic petunia plants: Phytochrome response and blue-light induction. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2358–2362. doi: 10.1073/pnas.83.8.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R., Kuhlemeier C., Nagy F., Chua N. H. Organ-specific and light-induced expression of plant genes. Science. 1986 May 30;232(4754):1106–1112. doi: 10.1126/science.232.4754.1106. [DOI] [PubMed] [Google Scholar]
- Fluhr Robert, Moses Phyllis, Morelli Giorgio, Coruzzi Gloria, Chua Nam-Hai. Expression dynamics of the pea rbcS multigene family and organ distribution of the transcripts. EMBO J. 1986 Sep;5(9):2063–2071. doi: 10.1002/j.1460-2075.1986.tb04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T. F., Ellis R. J. Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J. 1982;1(12):1493–1498. doi: 10.1002/j.1460-2075.1982.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C., Fluhr R., Green P. J., Chua N. H. Sequences in the pea rbcS-3A gene have homology to constitutive mammalian enhancers but function as negative regulatory elements. Genes Dev. 1987 May;1(3):247–255. doi: 10.1101/gad.1.3.247. [DOI] [PubMed] [Google Scholar]
- Last T. A., Gantzer M. L., Tyler C. D. Ion-gated channel induced in planar bilayers by incorporation of (Na+,K+)-ATPase. J Biol Chem. 1983 Feb 25;258(4):2399–2404. [PubMed] [Google Scholar]
- Maier U. G., Brown J. W., Tologcyzki C., Feix G. Binding of a nuclear factor to a consensus sequence in the 5' flanking region of zein genes from maize. EMBO J. 1987 Jan;6(1):17–22. doi: 10.1002/j.1460-2075.1987.tb04712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell J. T., Nagy F., Chua N. H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. 1985 Feb 28-Mar 6Nature. 313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Prywes R., Roeder R. G. Inducible binding of a factor to the c-fos enhancer. Cell. 1986 Dec 5;47(5):777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- Seguin C., Hamer D. H. Regulation in vitro of metallothionein gene binding factors. Science. 1987 Mar 13;235(4794):1383–1387. doi: 10.1126/science.3103216. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986 Dec 26;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sive H. L., Roeder R. G. Interaction of a common factor with conserved promoter and enhancer sequences in histone H2B, immunoglobulin, and U2 small nuclear RNA (snRNA) genes. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6382–6386. doi: 10.1073/pnas.83.17.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weinberger J., Baltimore D., Sharp P. A. Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. 1986 Aug 28-Sep 3Nature. 322(6082):846–848. doi: 10.1038/322846a0. [DOI] [PubMed] [Google Scholar]
- Zinn K., Maniatis T. Detection of factors that interact with the human beta-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986 May 23;45(4):611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]