Summary
A central goal of genetics is to understand the links between genetic variation and disease. Intuitively, one might expect disease-causing variants to cluster into key pathways that drive disease etiology. But for complex traits, association signals tend to be spread across most of the genome–including near many genes without an obvious connection to disease. We propose that gene regulatory networks are sufficiently interconnected that all genes expressed in disease-relevant cells are liable to affect the functions of core disease-related genes and that most heritability can be explained by effects on genes outside core pathways. We refer to this hypothesis as an “omnigenic” model.
The longest-standing question in genetics is to understand how genetic variation contributes to phenotypic variation. In the early 1900s, there was fierce debate between the Mendelians—who were inspired by Mendel’s work on pea genetics and focused on discrete, monogenic phenotypes—and the biometricians, who were interested in the inheritance of continuous traits such as height. The biometricians believed that Mendelian genetics could not explain the continuous distribution of variation observed for many traits in humans and other species.
This debate was largely resolved in a seminal 1918 paper by RA Fisher, who showed that if many genes affect a trait, then the random sampling of alleles at each gene produces a continuous, normally-distributed phenotype in the population (Fisher, 1918). As the number of genes grows very large, the contribution of each gene becomes correspondingly smaller, leading in the limit to Fisher’s famous “infinitesimal model” (Barton et al., 2016).
Despite the success of the infinitesimal model in describing inheritance patterns, especially in plant and animal breeding, it was unclear throughout the 20th century how many genes would actually be important for driving complex traits. Indeed, human geneticists expected that even complex traits would be driven by a handful of moderate-effect loci–thus giving rise to large numbers of mapping studies that were, in retrospect, greatly underpowered. For example, an elegant 1999 analysis of allele-sharing in autistic siblings concluded from the lack of significant hits that there must be “a large number of loci (perhaps ≥15)”–this prediction was strikingly high at the time, but seems quaintly low now (Risch et al., 1999, Weiner et al., 2016).
Since around 2006, the advent of genome-wide association studies and, more recently exome-sequencing, has provided the first detailed understanding of the genetic basis of complex traits. One of the early surprises of the GWAS era was that for typical traits, even the most important loci in the genome have small effect sizes and that together, the significant hits only explain a modest fraction of the predicted genetic variance. This has been referred to as the mystery of the “missing heritability” (Manolio et al., 2009). The mystery has since been largely resolved by analyses showing that common single nucleotide polymorphisms (SNPs) with effect sizes well below genome-wide statistical significance account for most of the “missing heritability” of many traits (Yang et al., 2010, Shi et al., 2016). Rare variants with larger effect sizes also contribute genetic variance (Marouli et al., 2017), especially for diseases with major fitness consequences (Simons et al., 2014) such as autism and schizophrenia (De Rubeis et al., 2014, Fromer et al., 2014, Purcell et al., 2014).
A second surprise was that, in contrast to Mendelian diseases–which are largely caused by protein-coding changes (Botstein and Risch, 2003)–complex traits are mainly driven by noncoding variants that presumably affect gene regulation (Pickrell, 2014, Welter et al., 2014, Li et al., 2016). Indeed, many studies have shown that significant variants are highly enriched in regions of active chromatin such as promoters and enhancers in relevant cell types. For example, risk variants for autoimmune diseases show particular enrichment in active chromatin regions of immune cells (Maurano et al., 2012, Farh et al., 2015, Kundaje et al., 2015).
These observations are generally interpreted in a paradigm where complex disease is driven by an accumulation of weak effects on the key genes and regulatory pathways that drive disease risk (Furlong, 2013, Chakravarti and Turner, 2016). This model has motivated many studies that aim to dissect the functional impacts of individual disease-associated variants (Smemo et al., 2014, Sekar et al., 2016), or to aggregate hits to identify key disease pathways and processes (Califano et al., 2012, Jostins et al., 2012, Wood et al., 2014, Krumm et al., 2015). For several diseases, the leading hits have indeed helped to highlight specific molecular processes: for example uncovering the role of autophagy in Crohn’s disease (Jostins et al., 2012), and roles for adipocyte thermogenesis (Claussnitzer et al., 2015) and central nervous system genes in obesity (Locke et al., 2015).
But despite the success of these earlier studies, we argue that the enrichment of signal in relevant genes is surprisingly weak overall, suggesting that prevailing conceptual models for complex diseases are incomplete. We highlight some pertinent features of current data, and discuss what these may tell us about the genetic architecture of complex diseases.
Distribution of GWAS signals across the genome
Early practitioners of GWAS were dismayed to find that for most traits, the strongest genetic associations could explain only a small fraction of the genetic variance (Manolio et al., 2009). This was taken to imply that there must be many causal loci, each with small effect sizes (Goldstein et al., 2009). Subsequent analyses soon provided direct evidence for this in the case of schizophrenia (Purcell et al., 2009) and showed that, together, common variants can explain much of the expected heritability (Yang et al., 2010). While traits vary greatly in terms of both the importance of the largest-effect common variants and of higher-penetrance rare variants (Loh et al., 2015, Shi et al., 2016, Sullivan et al., 2017), it is now clear that polygenic effects are important across a wide variety of traits (Shi et al., 2016, Weiner et al., 2016).
One key question that has been under-studied to date is the extent to which causal variants are spread widely across the genome or clumped into disease-relevant pathways. However, it is known that the heritability contributed by each chromosome tends to be closely proportional to its physical length (Visscher et al., 2006, Shi et al., 2016), hinting that causal variants may be fairly uniformly distributed. And recent data show that causal variants can be surprisingly dispersed even at finer scales. A paper from Alkes Price and colleagues estimated that between 71–100% of 1MB windows in the genome contribute to heritability for schizophrenia (Loh et al., 2015).
Here we explore a second example, namely height, for which very large GWAS data sets are available (Figure 1). While height is often thought of as the quintessential polygenic trait, recent work shows that the genetic architecture of height is actually broadly similar to that of a wide variety of other quantitative traits and diseases ranging from diabetes or autoimmune diseases to BMI or cholesterol levels. Thus we use height to illustrate the extreme polygenicity typical of many complex traits (Shi et al., 2016, Chakravarti and Turner, 2016).
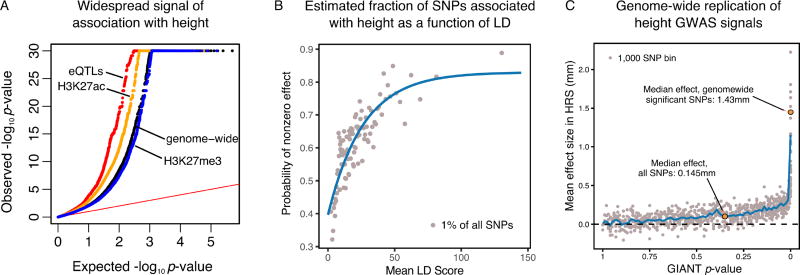
Figure 1.
Genome-wide signals of association with height. A. Genome-wide inflation of small p-values from the GWAS for height, with particular enrichment among expression quantitative trait loci and single nucleotide polymorphisms (SNPs) in active chromatin (H3K27ac). B. Estimated fraction of SNPs associated with non-zero effects on height (Stephens, 2016), as a function of Linkage Disequilibrium Score (i.e., the effective number of SNPs tagged by each SNP (Bulik-Sullivan et al., 2015b)). Overall we estimate that 62% of all SNPs are associated with a non-zero effect on height. The best-fit line estimates that 3.8% of SNPs have causal effects. C. Estimated mean effect size for SNPs, sorted by GIANT p-value with the direction (sign) of effect ascertained by GIANT. Replication effect sizes were estimated using data from the Health and Retirement Study (HRS). The points show averages of 1,000 consecutive SNPS in the p-value sorted list. The effect size on the median SNP in the genome is about 10% of that for genome-wide significant hits.
A height meta-analysis from the GIANT study reported 697 genome-wide significant loci which, together, explain 16% of the phenotypic variance (Wood et al., 2014). But a quantile-quantile plot comparing the distribution of p-values against the expected null distribution shows that the distribution of p-values is hugely shifted toward small p-values (Figure 1A), such that common variants together explain 86% of the expected heritability (Shi et al., 2016). The inflation is stronger in active chromatin and in regulatory quantitative trait loci (QTLs), consistent with the expected enrichment of signal in gene-regulatory regions.
We next used ashR to analyze the distribution of regression coefficients from the set of all SNPs (Stephens, 2016). ashR models the GWAS results as a mixture of SNPs that have a true effect size of exactly zero, with SNPs that have a true effect size that is not zero. Using this approach we estimated that–remarkably–62% of all common SNPs are associated with a non-zero effect on height (this includes both causal SNPs as well as nearby SNPs that are correlated through linkage disequilibrium; Figure 1B). Given that the typical extent of linkage disequilibrium (LD) is around 10kb–100kb (International HapMap Consortium, 2005), this implies that most 100kb windows in the genome include variants that affect height. Stratifying the ashR analysis by the LD Score for each SNP (Bulik-Sullivan et al., 2015b), we see a clear effect that SNPs with more LD partners are more likely to be associated with height. Under simplifying assumptions (see Supplementary Materials), the best-fit curve suggests that ~3.8% of 1000 Genomes SNPs have causal effects on height.
As validation, we used the regression estimate from each SNP in the height meta-analysis to predict its direction of effect on height (Figure 1C), and then examined the extent to which SNP effects are consistent in a smaller, independent data set from the Health and Retirement Study (Juster and Suzman, 1995). In brief, we computed the mean replication effect sizes of height-increasing alleles as determined by GIANT. Under the null hypothesis of no true signal, the replication effect sizes would be centered on zero; when there is true signal the observed mean effect sizes can be considered a lower bound on the true effect sizes due to occasional sign errors in GIANT.
Strikingly, we find clear enrichment of shared directional signal for most SNPs, even for SNPs with p-values as large as 0.5 (Figure 1C). Across all SNPs genome-wide, the median SNP is associated with an effect size of 0.14 mm, which is approximately one tenth the median effect size of genome-wide significant SNPs (1.43 mm). We also obtained similar results starting from a smaller family-based GWAS, confirming that the signals are not driven by confounding from population structure (Supplementary Materials). Putting the various lines of evidence together, we estimate that more than 100,000 SNPs exert independent causal effects on height, similar to an early estimate of 93,000 causal variants based on a different approach (Goldstein et al., 2009) (Supplementary Materials).
In summary, we conclude that there is an extremely large number of causal variants with tiny effect sizes on height and, moreover, that these are spread very widely across the genome, such that most 100kb windows contribute to variance in height. More generally, the heritability of complex traits and diseases is spread broadly across the genome (Loh et al., 2015, Shi et al., 2016), implying that a substantial fraction of all genes contribute to variation in disease risk. These observations seem inconsistent with the expectation that complex trait variants are primarily in specific biologically-relevant genes and pathways. To explore this further, we turn next to data on functional enrichment of signals.
Enrichment of genetic signals in transcriptionally active regions
As shown above for height, GWAS signals tend to be markedly enriched in predicted gene regulatory elements. In particular, many groups have shown that disease-associated SNPs are enriched in active chromatin, and particularly in chromatin that is active in cell types relevant to disease (Trynka et al., 2013, Farh et al., 2015, Finucane et al., 2015, Kundaje et al., 2015). Similarly, signals also aggregate near genes that are expressed in relevant cell types (Hu et al., 2011, Wood et al., 2014).
An intuitive interpretation is that the cell type-based regulatory maps point us toward cell type-specific regulatory elements that control specific functions of those cells and thereby drive disease biology. Indeed, the relevant papers often describe these analyses as highlighting “cell type-specific” aspects of regulation. But given that the heritability signal is so widespread, we wanted to understand whether the signal is specifically concentrated in chromatin that is active in just the relevant (or related) cell types, as opposed to chromatin that is broadly active.
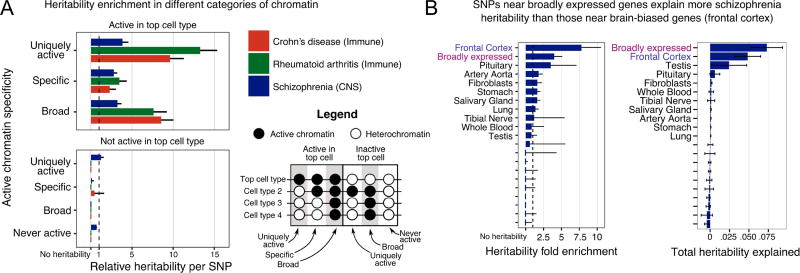
To explore this question, we used active chromatin data measured in ten broadly-defined cell-type groups (e.g., immune, central nervous system (CNS), cardiovascular, etc). A region was considered active in a cell type group if it was detected as active for any cell type in that group. We applied stratified LD score regression–a method that estimates how much different classes of SNPs contribute to heritability (Finucane et al., 2015). We focused on three well-powered GWAS studies that showed clear enrichment within a single cell-type group in a previous analysis: Crohn’s disease and rheumatoid arthritis (RA, immune), and schizophrenia (CNS) (Finucane et al., 2015).
While there are strong cell-type effects, these are largely independent of the breadth of chromatin activity. For example, we observed that SNPs in chromatin that is broadly active across most cell types make substantial contributions to heritability. On average, SNPs in broadly active elements contribute roughly as much to heritability as do SNPs in cell type-specific active chromatin (only for RA are these significantly different; Figure 2a). Meanwhile, SNPs in chromatin that is inactive, or active only in irrelevant cell types contribute little or no heritability, thus providing an important negative control.
Figure 2.
Heritability tends to be enriched in regions that are transcriptionally active in relevant tissues. (a) Contributions to heritability (relative to random SNPs) as a function of chromatin context. There is enrichment for signal among SNPs that are in chromatin active in the relevant tissue, regardless of the overall tissue breadth of activity. (b) Genes with brain-specific expression show the strongest enrichment of schizophrenia signal (left), but broadly expressed genes contribute more to total heritability due to their greater number (right).
For an alternative viewpoint, we also considered breadth of gene expression. We estimated the contribution of SNPs in or near exons for genes with different expression profiles. Based on GTEx data, we identified genes that are particularly highly expressed in particular tissue groups, as well as broadly expressed genes (GTEx Consortium, 2015). As shown for schizophrenia (Figure 2B), SNPs near genes that are expressed in the brain contribute substantially to heritability, while genes that are specifically expressed in other tissues contribute little or nothing. Perhaps intuitively, SNPs near genes expressed specifically in brain contribute more heritability per SNP than SNPs near genes with broad expression profiles. However, only a modest fraction of all brain-expressed genes are specifically up-regulated in brain. Hence broadly expressed genes actually contribute more heritability than do brain-specific genes.
In summary, genetic contribution to disease is heavily concentrated in regions that are transcribed or marked by active chromatin in relevant tissues, but there is little enrichment for cell type-specific regulatory elements versus broadly actively regions. As expected there appears to be little or no genetic contribution from regions that are inactive in these tissues. To investigate the question of GWAS specificity further, we next examined evidence for enrichment of associated genes in specific functional categories.
Weak enrichment of genetic signals by functional categories
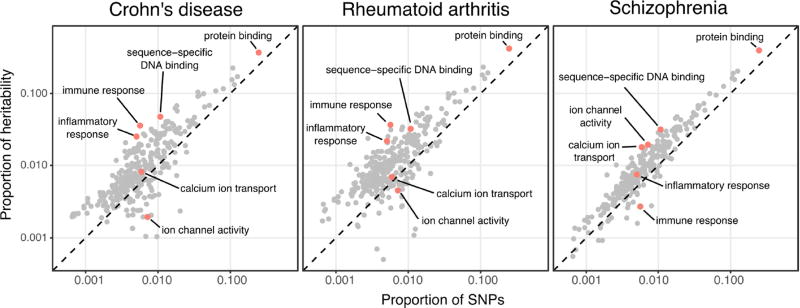
We considered the contributions of genes from different functional ontologies. As expected, we found that the genetic signals for the two autoimmune diseases (Crohn’s and RA), were most enriched in ontologies corresponding to “immune response” and “inflammatory response”, whereas schizophrenia heritability was most enriched in nervous system-related genes with ontologies such as “ion channel activity” and “calcium ion transport” (Figure 3). However these enrichments were relatively modest, and for all three diseases we observed a strong linear relationship between the sizes of the functional categories and the proportion of heritability that they contributed. Broad functional categories contribute more total trait heritability than do genes in apparently disease-relevant functional categories, and for all three diseases, the largest contributor to heritability was simply the largest category, namely protein-binding.
Figure 3.
Gene ontology enrichments for three diseases, with categories of particular interest labeled. The x-axis indicates the fraction of genes in each category; the y-axis shows the fraction of heritability assigned to each category relative to all other SNPs. Note that the diagonal indicates the genome-wide average across all SNPs; most GO categories lie above the line due to the general enrichment of signal in and around genes. Analysis by stratified LD score regression (Finucane et al., 2015).
Moreover, these results are markedly different from analysis of rare variants implicated in schizophrenia. Recent studies of rare variants have consistently found enrichment of synaptic genes, and other gene sets involved in neuronal functions within de novo, rare, and CNV polymorphism sets (Table 1). In contrast, analysis of the 108 genome-wide significant loci from GWAS found examples of hits in relevant genes, but no ontology categories that were significant overall (Ripke et al., 2014), consistent with the weak enrichment described above for the heritability analysis of the same data. Together, these results suggest that the types of genes detected in rare variant studies–which can detect highly deleterious variants with large effect sizes–play more direct roles in schizophrenia than do genes identified from GWAS based on common variants.
Table 1.
Summary of gene sets that show functional enrichment in recent large-scale papers on schizophrenia. Studies of rare and de novo variants, and CNVs—which tend to identify larger-effect variants—show clearer evidence of enrichment than seen in GWAS.
| Variant type | Gene Set/Ontology | Enrichment p-value | Reference |
|---|---|---|---|
| Rare | ARC | p = 1.6 × 10−3 | Purcell et al. (2014) |
| voltage-gated calcium channel | p = 1.9 × 10−3 | ||
|
| |||
| de novo | ARC | p = 4.8 × 10−4 | Fromer et al. (2014) |
| N-methyl-D-aspartate receptor (NMDAR) | p = 2.5 × 10−2 | ||
|
| |||
| CNV | ARC | p = 1.8 × 10−4 | PGC (2016) |
| Synaptic gene | p = 2.8 × 10−11 | ||
|
| |||
| GWAS | glutamatergic neurotransmission synaptic plasticity | not significant* | Ripke et al. (2014) |
The p-values are shown without multiple testing correction, but corrected p-values are < 0.05.
Consistent with studies of rare variants, Ripke et al. (2014) identified associated loci near several genes involved in glutamatergic neurotransmission and synaptic plasticity, but these categories did not show a statistically significant enrichment for GWAS hits.
ARC: activity-regulated cytoskeleton-associated scaffold protein.
An extended model for complex traits
In summary, for a variety of traits, the largest-effect variants are modestly enriched in specific genes or pathways that may play direct roles in disease. However, the SNPs that contribute the bulk of the heritability tend to be spread across the genome, and are not near genes with disease-specific functions. The clearest pattern is that the association signal is broadly enriched in regions that are transcriptionally active or involved in transcriptional regulation in disease-relevant cell types, but absent from regions that are transcriptionally inactive in those cell types. For typical traits, huge numbers of variants contribute to heritability, in striking consistency with Fisher’s century-old infinitesimal model.
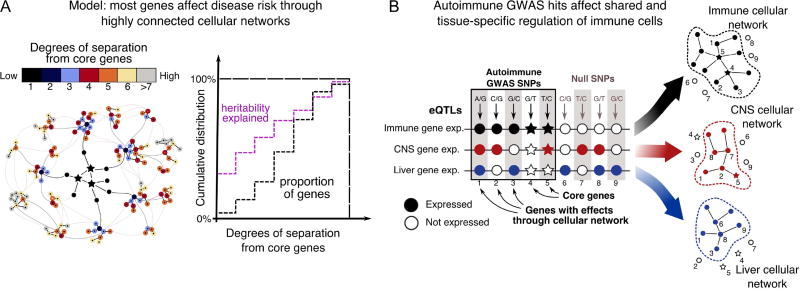
To make sense of these observations, we propose an “omnigenic” model of complex traits (Figure 4). First, we assume that most traits can be directly affected by a modest number of genes or gene pathways with specific roles in disease etiology, as well as their direct regulators (Chakravarti and Turner, 2016). We refer to these as “core genes”. Such genes will tend to have biologically interpretable roles in disease, such as the roles of IRX3 and IRX5 in controlling adipocyte differentiation, with consequent effects on obesity (Claussnitzer et al., 2015), or the role of the C4 genes on synaptic pruning in development, thereby affecting schizophrenia risk (Sekar et al., 2016). Furthermore, when core genes are damaged by loss-of-function or other particularly damaging mutations, we can anticipate that these will tend to have the strongest effects on disease risk (although the actual degree of increased risk conferred by the largest effect-size mutations varies greatly across traits (Krumm et al., 2015, Marouli et al., 2017)). In practice, the sorting of core genes from peripheral genes may be on a graduated scale as opposed to a binary classification.
Figure 4.
An omnigenic model of complex traits. (A) For any given disease phenotype, a limited number of genes have direct effects on disease risk. However by the small world property of networks, most expressed genes are only a few steps from the nearest core gene, and thus may have non-zero effects on disease. Since core genes only constitute a tiny fraction of all genes, most heritability comes from genes with indirect effects. (B) Diseases are generally associated with dysfunction of specific tissues; genetic variants are only relevant if they perturb gene expression (and hence network state) in those tissues. For traits that are mediated through multiple cell types or tissues, the overall effect size of any given SNP would be a weighted average of its effects in each cell type.
Second, we need to understand why core genes generally contribute just a small part of the total heritability, and how most genes expressed in relevant cell types could make non-zero contributions to heritability. To resolve this, we propose that cell regulatory networks are highly interconnected, to the extent that any expressed gene is likely to affect the regulation or function of core genes.
At this time, our understanding of cellular regulatory networks remains incomplete, but the relevant connections likely include all layers of interactions among cellular molecules, including transcriptional networks, post-translational modifications, protein-protein interactions, and intercellular signaling (Furlong, 2013). In particular cases, it has been possible to elucidate the most important wiring connections in gene regulatory networks that drive development or disease (Davidson, 2010, Chatterjee et al., 2016). However we still have very limited knowledge of how weaker effects such as expression QTLs percolate through the entire regulatory network. Nonetheless, research in network theory finds that most real-world networks tend to be highly interconnected–this is referred to as the “small world” property of networks (Watts and Strogatz, 1998, Strogatz, 2001). Specifically, many kinds of networks have structures consisting of distinct modules of connected nodes, but also frequent long-range connections. Under these conditions, any two nodes in the graph are usually connected by just a few steps.
If this is the case in cellular networks, then any gene that is expressed in a disease-relevant tissue is likely to be just a few steps from one or more core genes. Consequently, any variant that affects expression of a “peripheral” gene is likely to have non-zero effects on regulation of the core genes, and thereby incur a small effect on disease risk. Crucially, because the total set of expressed genes may outnumber core genes by 100:1 or more, the sum of small effects across peripheral genes can far exceed the genetic contribution of variants directly affecting the core genes themselves.
Our model posits that information flows from regulatory variants, e.g., by affecting chromatin activity, to cis-regulation of nearby genes and ultimately to affect the activity of other genes. Cis-eQTLs (cis-acting expression quantitative trait loci) may in turn affect mRNA or protein levels of other unlinked genes via the regulatory network (i.e., the variants would also be trans-acting eQTLs for genes elsewhere in the genome), but might also affect other functions such as post-translational modification or subcellular localization. At present, detection of trans-QTLs is challenging in current sample sizes (Westra et al., 2013, Jo et al., 2016), but it is estimated that ~70% of mRNA heritability is determined by trans-acting factors (Price et al., 2011). Moreover, many trans-QTLs may act through protein networks and thus not be detectable from RNA, though current data on trans-acting controls of proteins are very limited (Battle et al., 2015, Chick et al., 2016, Sun et al., 2017).
Lastly, many diseases are mediated through multiple cell types–for example different immune cell subsets for autoimmune disease, or even unrelated tissues such as brain and adipose tissue for obesity. Furthermore, although GWAS hits are highly enriched in active chromatin, only a modest fraction can currently be explained by known eQTLs (Chun et al., 2017). This gap may imply that many risk variants affect expression only in narrowly defined cell types or under precise conditions such as immune stimulation (Alasoo et al., 2017). When disease risk is mediated through multiple cell types, or highly specialized cell types, we anticipate that the cellular networks would vary across cell types (Price et al., 2011, Sonawane et al., 2017). The quantitative effect of any given variant would then be an average of its effect size in each cell type, weighted by cell type importance.
In summary, the omnigenic model of complex disease proposes that essentially any gene with regulatory variants in at least one tissue that contributes to disease pathogenesis is likely to have nontrivial effects on risk for that disease. Furthermore, the relative effect sizes are such that, since core genes are hugely outnumbered by peripheral genes, a large fraction of the total genetic contribution to disease comes from peripheral genes that do not play direct roles in disease.
Widespread pleiotropy
There has recently been considerable interest in identifying particular variants with pleiotropic effects on different traits (Cotsapas et al., 2011, Pickrell et al., 2016) as well as in identifying pairs of traits with correlated genetic effects (Bulik-Sullivan et al., 2015a). However, the observation that genetic signals are spread broadly across the genome implies that pleiotropy may be ubiquitous (Visscher and Yang, 2016).
Indeed the omnigenic model predicts that virtually any variant with regulatory effects in a given tissue is likely to have (weak) effects on all diseases that are modulated through that tissue. Many eQTLs are active in all tissues, and consequently these may have weak effects on most or even all traits.
We refer to this form of pleiotropy as “network pleiotropy”: i.e., the principle that a single variant may affect multiple traits because those traits are mediated through the same cell type(s) and hence regulated through the same network(s)–and not because the traits are directly causally related. Traits that share core genes, or whose genes are close in the network, will tend to have correlated effects. Conversely, traits that are mediated through the same tissue but have no overlap of core genes, may show little or no correlation in effects even though many causal variants are shared.
If network pleiotropy is widespread, this raises challenges for the interpretation of genetic correlations and for Mendelian Randomization studies (Bulik-Sullivan et al., 2015a, Davey Smith and Hemani, 2014). Mendelian Randomization generally assumes that pleiotropy between traits that are not causally related–also referred to as “Type I pleiotropy” (Wagner and Zhang, 2011)–is rare. It remains to be determined whether the effects of network pleiotropy would be strong enough to drive significant signals in practice, especially if the core genes are far apart in the network.
Evolutionary change of complex traits
The observation that many traits are affected by huge numbers of variants also has important implications for studies of evolutionary change. Within the evolutionary community, there has been great interest in identifying particular genetic variants that are responsible for adaptive changes, both within and between species (Vitti et al., 2013). While this work has produced a number of interesting examples, we argue that these are not likely to be representative of most evolutionary change. Instead, most adaptive changes may proceed by polygenic adaptation: i.e., species adapt by small allele frequency shifts of many causal variants across the genome (Pritchard et al., 2010). For example, if 105 variants affect height by 0.15mm each, then even a small shift in average allele frequencies could generate a large shift in average height: e.g., a 0.5% genome-wide increase in the frequency of “tall” alleles would generate a 15cm shift in average height. There is now a growing collection of examples of recent polygenic adaptation in humans, especially for morphometric traits including height, BMI and infant birth size (Turchin et al., 2012, Field et al., 2016).
We anticipate that many of the more dramatic phenotypic differences seen between species are also driven by an accumulation of tiny effects, and that larger-effect differences are likely to be exceptions to the rule. For example, there are ~40 million single nucleotide differences between humans and chimpanzees: if 1% of these affect chromatin function or other aspects of regulation then there could easily be half a million species-differences with small but nonzero effects on phenotypes (these need not all be adaptive), and these would likely dominate the contributions of a handful of large-effect loci.
Turning to the within-species level, one important open question is whether pleiotropic effects limit how many traits can be selected for at once. As described above, pleiotropy is likely ubiquitous in the genome. This may place constraints on the ability of selection to shift allele frequencies, as a change in the frequency of one variant must be balanced by changes at other sites. Does this effectively limit the number of independent polygenic traits that can be simultaneously selected? There has been previous consideration of the extent to which pleiotropy shapes variation and adaptation (Barton, 1990, Walsh and Blows, 2009), but we believe this area is ripe for further exploration in the light of modern data.
Future directions
Huge numbers of genes contribute to the heritability for complex diseases. This fact raises fundamental questions about how genetic variation perturbs genetic systems to produce phenotypes. We have proposed one possible model, and it will be important to test this, and perhaps others. There are deep challenges to fully understanding the impact of very small effects in organismal systems, so we believe there is great need to develop cell-based model systems that can recapitulate aspects of complex traits. Furthermore, we still have limited understanding of cellular networks, and it will be important to develop highly precise, high-throughput techniques for mapping networks in diverse cell types, especially at the protein level. We suggest the following key questions and tests of the omnigenic model:
For a variety of representative traits: how many distinct variants and how many genes contribute causal variation? What fraction of this variation is in non-core genes? Which traits are closer to (or further from) the omnigenic extreme?
Are there variants that affect expression in the cell types that drive a particular disease but have no effect on disease risk? While traits vary in terms of the importance of the largest-effect variants, the strongest form of the omnigenic model predicts that essentially all regulatory variants active in relevant cell types would contribute non-zero effects.
If most genetic variants act through cellular networks, then what mediates these connections? Transcriptional regulation, post-translational modification, protein-protein interaction, and intercellular signaling may all contribute. What is the nature and frequency of long-range interactions in cellular networks? How do network architectures vary across cell types and tissues?
As we get increasingly precise measurements of the percolation of genetic variation through cellular networks, can we infer the effects of peripheral genes from their relation to core genes?
Is the conceptual distinction between core genes and peripheral genes useful for understanding disease and, if so, how should core genes be defined? One possible formal definition is that, conditional on the genotype and expression levels of all core genes, then the genotypes and expression levels of peripheral genes no longer matter. Less formally, we might think of core genes as the genes that (if mutated or deleted) have the strongest effects, as seen for large-effect mutations in autism (Krumm et al., 2015). Or we might think of core genes simply as the genes with interpretable mechanistic links to disease. Alternatively, some diseases may not even have core genes–instead the global activity of all genes might help to set cellular system states that determine cellular function and disease risk (Preininger et al., 2013).
Our model also raises questions about the next generation of mapping studies. One goal of gene mapping is to identify core genes and pathways that drive disease. These provide mechanistic insights into disease biology and may suggest druggable targets. The biggest hits from GWAS have helped to pinpoint important core genes. After these have been found, the next most promising step is to hunt for lower frequency variants of larger effects, which likely contribute little to heritability, but may implicate additional core genes. Deep sequencing has not been uniformly successful for all traits (possibly due to insufficient sample sizes (Marouli et al., 2017)), but following the identification of the biggest association hits among common variants, large-scale sequencing is the most promising next step. In the short-term, exome sequencing is likely the most cost-effective approach, given current evidence that larger-effect variants are more likely to affect protein-coding sequences.
Nonetheless large-scale genotyping data will continue to be valuable for two reasons. First, very deep association data will be essential for developing personalized risk prediction. Second, these data will be essential for modeling the flow of regulatory information through cellular networks. For a complete understanding of disease genetics, we will want to know why increased expression of gene X increases risk for diseases Y and Z. For this we will need to understand cellular networks much better and to have estimates of disease risk in very large samples.
In summary, many complex traits are driven by enormously large numbers of variants of small effects, potentially implicating most regulatory variants that are active in disease-relevant tissues. To explain these observations, we propose that disease risk is largely driven by genes with no direct relevance to disease, and propagated through regulatory networks to a much smaller number of core genes with direct effects. If this model is correct, then it implies that detailed mapping of cell-specific regulatory networks will be an essential task for fully understanding human disease biology.
Supplementary Material
Acknowledgments
This work was supported by RO1 HG008140, the National Science Foundation graduate research fellowship program and the Howard Hughes Medical Institute. We thank many colleagues for helpful conversations or comments, including D. Golan, W. Greenleaf, A. Harpak, A. Marson, J. Pickrell, M. Przeworski, G. Sella and three anonymous reviewers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alasoo K, Rodrigues J, Mukhopadhyay S, Knights AJ, Mann AL, Kundu K, HIPSCI Consortium. Hale C, Dougan G, Gaffney DJ. Genetic effects on chromatin accessibility foreshadow gene expression changes in macrophage immune response. bioRxiv 102392. 2017 doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. Pleiotropic models of quantitative variation. Genetics. 1990;124(3):773–782. doi: 10.1093/genetics/124.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Etheridge AM, Veber A. The infinitesimal model. bioRxiv 039768 2016 [Google Scholar]
- Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. Impact of regulatory variation from RNA to protein. Science. 2015;347(6222):664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat, Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, Duncan L, Perry JR, Patterson N, Robinson EB, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015a;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015b;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet. 2012;44(8):841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Turner TN. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. BioEssays. 2016;38(6):578–586. doi: 10.1002/bies.201500203. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Kapoor A, Akiyama JA, Auer DR, Lee D, Gabriel S, Berrios C, Pennacchio LA, Chakravarti A. Enhancer Variants Synergistically Drive Dysfunction of a Gene Regulatory Network In Hirschsprung Disease. Cell. 2016;167(2):355–368. doi: 10.1016/j.cell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534(7608):500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Casparino A, Patsopoulos NA, Croteau-Chonka DC, Raby BA, De Jager PL, Sunyaev SR, Cotsapas C. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 2017 Feb;20 doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson PA, Hsu YH, Drucker DJ, Mellgren G, Hui CC, Hauner H, Kellis M. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abecasis GR, Barrett JC, Behrens T, Cho J, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8):e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468(7326):911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK-H, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Boyle EA, Telis N, Gao Z, Gaulton KJ, Golan D, Yengo L, Rocheleau G, Froguel P, McCarthy MI, et al. Detection of human adaptation during the past 2000 years. Science. 2016;354(6313):760–764. doi: 10.1126/science.aag0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat, Genet. 2015;47(11):1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52(02):399–433. [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong LI. Human diseases through the lens of network biology. Trends Genet. 2013;29(3):150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, et al. Common genetic variation and human traits. N. Engl. J. Med. 2009;360(17):1696. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet. 2011;89(4):496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo B, He Y, Strober BJ, Parsana P, Aguet F, Brown AA, Castel SE, Gamazon ER, Gewirtz A, Gliner G, et al. Distant regulatory effects of genetic variation in multiple human tissues. bioRxiv 074419 2016 [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juster FT, Suzman R. An overview of the Health and Retirement Study. J. Hum. Resour. 1995;30:S7–S56. [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He Z-X, et al. Excess of rare, inherited truncating mutations in autism. Nat. Gen. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352(6285):600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47(9):979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, de Candia TR, Lee SH, Wray NR, Kendler KS, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015a;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015b;47(3):284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542(7640):186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94(4):559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, S_gurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preininger M, Arafat D, Kim J, Nath AP, Idaghdour Y, Brigham KL, Gibson G. Blood-informative transcripts define nine common axes of peripheral blood gene expression. PLoS Genet. 2013;9(3):e1003362. doi: 10.1371/journal.pgen.1003362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Helgason A, Thorleifsson G, McCarroll SA, Kong A, Stefansson K. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7(2):e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20(4):R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, Ruderfer DM, McQuillin A, Morris DW, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JT, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, et al. A genomic screen of autism: evidence for a multilocus etiology. Am. J. Hum. Genet. 1999;65(2):493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MR, Hemani G, Medina-Gomez C, Mezzavilla M, Esko T, Shakhbazov K, Powell JE, Vinkhuyzen A, Berndt SI, Gustafsson S, et al. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 2015;47(11):1357–1362. doi: 10.1038/ng.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kichaev G, Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 2016;99:139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat, Genet. 2014;46(3):220. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, G_mez-Mar_n C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane AR, Platig J, Fagny M, Chen C-Y, Paulson JN, Lopes-Ramos CM, DeMeo DL, Quackenbush J, Glass K, Kuijjer ML. Understanding Tissue-specific Gene Regulation. bioRxiv 110601. 2017 doi: 10.1016/j.celrep.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. False discovery rates: a new deal. Biostatistics. 2016;18(2):275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz SH. Exploring complex networks. Nature. 2001;410(6825):268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik C, Andreassen OA, Borglum A, Breen G, Cichon S, Edenberg H, Faraone SV, Gelernter J, Mathews CA, Nievergelt CM, Smoller J, O’ Donovan M, TPGC Psychiatric Genomics: An Update and an Agenda. bioRxiv 115600. 2017 doi: 10.1176/appi.ajp.2017.17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Consequences Of Natural Perturbations In The Human Plasma Proteome. bioRxiv 134551 2017 [Google Scholar]
- The Psychiatric Genetics Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2016 Nov;21 doi: 10.1038/ng.3725. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45(2):124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin MC, Chiang CW, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN, of ANthropometric Traits (GIANT) Consortium GI, et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 2012;44(9):1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Medland SE, Ferreira MA, Morley KI, Zhu G, Cornes BK, Montgomery GW, Martin NG. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2(3):e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Yang J. A plethora of pleiotropy across complex traits. Nat. Genet. 2016;48:707–708. doi: 10.1038/ng.3604. [DOI] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu. Rev. Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 2011;12(3):204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- Walsh B, Blows MW. Abundant genetic variation + strong selection = multivariate genetic constraints: A geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 2009;40:41–59. [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein J, Okbay A, Bybjerg-Gauholm J, Werge T, Hougaard DM, Taylor J, Skuse D, Devlin B, Anney R, Sanders S, Bishop S, Bo Mortensen P, Borglum A, Davey Smith G, Daly MJ, Robinson EB. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. bioRxiv 089342. 2016 doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(D1):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45(10):1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.