Abstract
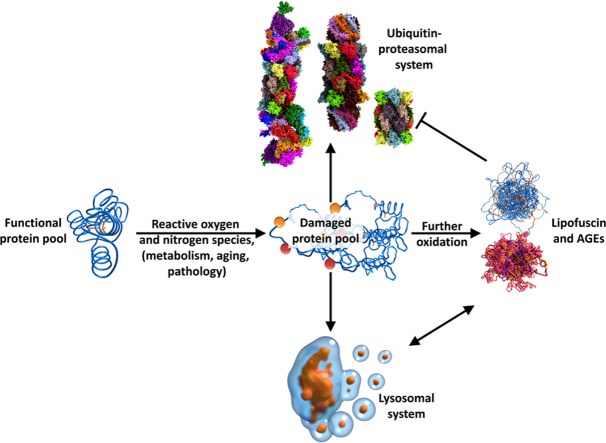
The production of reactive species is an inevitable by-product of metabolism and thus, life itself. Since reactive species are able to damage cellular structures, especially proteins, as the most abundant macromolecule of mammalian cells, systems are necessary which regulate and preserve a functional cellular protein pool, in a process termed “proteostasis”. Not only the mammalian protein pool is subject of a constant turnover, organelles are also degraded and rebuild. The most important systems for these removal processes are the “ubiquitin-proteasomal system” (UPS), the central proteolytic machinery of mammalian cells, mainly responsible for proteostasis, as well as the “autophagy-lysosomal system”, which mediates the turnover of organelles and large aggregates.
Many age-related pathologies and the aging process itself are accompanied by a dysregulation of UPS, autophagy and the cross-talk between both systems. This review will describe the sources and effects of oxidative stress, preservation of cellular protein- and organelle-homeostasis and the effects of aging on proteostasis in mammalian cells.
Keywords: Redox shift, Oxidative stress, Proteasome, Autophagy, Lysosome
Graphical abstract
1. Introduction
One of the main “primary” free radicals in mammalian cells is the superoxide radical anion (O2•−), resulting from electrons taken up by molecular oxygen. In a broad variety of secondary/further reactions, a large amount of different so-called “reactive oxygen species” (ROS) can be formed, either by chemical reactions or even catalyzed by cellular enzymes.
The term “ROS” summarizes “reactive oxygen species” (such as superoxide), “reactive nitrogen species” (RNS, including nitric oxide: •NO) and numerous other species with different properties; some of them are not radicals, some of them are highly reactive while others are less reactive. Besides superoxide, nitric oxide (•NO) and hydrogen peroxide (H2O2) are the main and most abundant primary reactive species formed in mammalian cells either as side products or even “intentionally” by enzymes.
In order to cope with oxidative stress or redox shifts, powerful antioxidative systems developed during evolution. Those systems include low molecular antioxidants that “compete” with ROS for cellular structures. This group contains mainly vitamins, glutathione, lipophilic antioxidants, uric acid (one of the major antioxidants in human blood-plasma [1]) and other small molecules, which mainly “disarm” ROS by a direct chemical reaction resulting in the formation of much less reactive or even inert products, that are no longer able to exert oxidative damage to cellular structures.
The mentioned glutathione (GSH) is the most important and abundant intracellular antioxidant. The ratio of GSH to its oxidized form glutathione disulfide (GSSG), and therefore the ratio of 2GSH/GSSG, can serve as an important indicator of the cellular redox state [2].
Furthermore, antioxidative enzymes which can catalyze the detoxification of certain ROS are expressed. Superoxide dismutases such as Cu,ZnSOD or its mitochondrial equivalent MnSOD convert superoxide (O2•‒) into hydrogen peroxide (H2O2), while H2O2 is converted into water and oxygen mediated by enzymes such as catalase or glutathione peroxidases. The glutathione disulfide produced by glutathione peroxidases during the reduction of peroxides is then restored again by glutathione reductase.
Other antioxidative proteins are able to bind redox-active transition metals to maintain them in an inactive form and are thus also part of the antioxidative defense network. For example, iron (as Fe2+) and copper (as Cu+) are both able to transfer an electron to H2O2, resulting in the formation of a hydroxide anion (‒OH) and the highly aggressive hydroxyl radical (•OH), which is virtually able to oxidize every cellular molecule. In a mammalian cell, the resulting Fe3+ or Cu2+ can be reduced by different electron donors like (amongst others) superoxide into their redox-active forms again, continuing this so-called “Fenton reaction”. Though, in the highly reducing environment of a living cell, also other electron donors like ascorbate (Fe3+ + Asc− → Fe2+ + Asc•−) are able to transfer an electron to Fe3+ in order to restore its redox-active state. And furthermore redox-cycling and ROS-generating reactions of both Fe2+/3+ with hydroperoxides and membrane phospholipids are possible [3]. In pathologic cases like Alzheimers disease, it was recently found that complexes of copper and amyloid-β peptides are able to produce hydroxyl radicals from H2O2 via Fenton [4].
Redox-active metals are thus strictly regulated and stored or transferred in a protein-bound manner, so that they are unable to catalyze the Fenton reaction.
Furthermore some enzymes are able to repair oxidatively damaged proteins. However, since only the two sulfur-containing amino acids methionine and cysteine can be reduced after moderate oxidative damage and under certain conditions (since only methionine sulfoxide or sulfenic acid at cysteine residues can be restored to methionine and cysteine, respectively), the cellular ability to completely restore proteins after oxidative damage is limited. In most cases, oxidatively damaged proteins are removed via proteolysis, mediated by either the ubiquitin-proteasomal or the lysosomal system. Furthermore, a large variety of lipases, RN-/DNases are available, that can terminally remove irreversibly oxidized cellular structures.
Cellular functionality significantly depends on permanent fine-tuning and turnover of the proteome, the entity of all cellular proteins. For this, two main cellular degradation systems have evolved the so-called ubiquitin-proteasomal system (UPS), which is responsible for degradation of both functional and dysfunctional proteins and the lysosomal system, that degrades whole organelles, large aggregates of proteins/macromolecules, as well as single proteins. The UPS is responsible for the proteolytic degradation of 80–90% of all cellular proteins, including many regulated, short-lived or misfolded/damaged ones [5].
The redox state of a mammalian cell mainly impacts both the proteome itself - in a range from reversible oxidative modification of a few redox-sensitive proteins up to severe irreversible oxidative damage of almost the whole protein pool. Also the process of aging affects the formation of reactive species and thus, the cellular redox state, as well as the ability of the cell to maintain its proteostasis.
2. Proteostasis maintenance systems
2.1. Counteracting protein oxidation by repair
Protein oxidation is divided into two general forms: reversible and irreversible modifications. Both occur under physiological conditions as well as under phases of oxidative stress, though, naturally, under oxidative stress the amount of irreversible (oxidative) protein modification increases.
Reversible oxidative modifications mainly affect the sulfur-containing amino acids methionine and cysteine, the only amino acids which can be restored by cellular antioxidative systems. Methionine – very susceptible to oxidation - can be modified in a one- (resulting in a sulfide radical cation) or two-electron (resulting in the formation of methionine sulfoxide) oxidation. Sulfide radical cations are highly instable and may form products that are irreversible posttranslational protein modifications and enter chemical pathways which may result in carbon-centered and/or peroxyl radicals – both representing starting points of further protein oxidation chains. One common product of oxidation is the methionine sulfoxide (MetSO), formed as a mixture of S- (often termed MetA) and R- (MetB) stereoisomers. Under stress, aging and pathologic/inflammatory processes, the amounts of MetSO are increased [6], [7], [8]. Thus, MetSO can be restored by cellular enzymes, in this special case the methionine sulfoxide reductases A (MSR-A) and B (MSR-B), catalyzing the reduction of the respective isomers to methionine in a thioredoxin (Th-(SH)2) consuming manner. The resulting Th-(S-S) is reduced by the enzyme thioredoxin-reductase using NADPH as redox-element. Decreased MSR-activities are associated with lowered stress-resistance as well as with reduction of maximal life span, while MSR-overexpression results in enhanced stress-resistance and extended life span [9].
The other main reversible posttranslational protein modification affects cysteine and the formation of disulfide bonds, leading to intra- or intermolecular cross-links. Oxidative modification of thiol switches is a widespread mechanism of redox signaling. In mammalian cells for example different peroxiredoxins (Prx) are available, that catalyze reduction of hydroperoxides such as R-OOH + Prxred → R-OH + Prxox + H2O, while R-OOH can also be H2O2[10]. Prxox is then reduced via thioredoxin to Prxred. Two main types of peroxiredoxins are known, the cytosolic ones (e.g. Prx2) [11] and the mitochondrial ones (e.g. Prx3) [12]. If Prx is further oxidized from sulfenic (Prx-S-OH) to sulfinic acid (Prx-SO-OH), the protein forms higher molecular weight aggregates and switches its function from an antioxidant to a molecular chaperone. In most cases, hyperoxidation from sulfenic to sulfinic acid is not reversible, only mitochondrial sulfiredoxins (Srx) [13] are able to reduce sulfinic acid in mitochondrial peroxiredoxins and restore the functional protein-SH-form (mainly of Prx3).
Interestingly, active-site cysteines are sometimes S-glutathionylated in order to prevent further irreversible (hyper)-oxidation. S-glutathionylation is a posttranslational modification of cysteine residues in proteins via addition of glutathione (GSH). The addition of GSH to a cysteine residue forms a so-called mixed protein-disulfide in a redox-dependent manner. The microenvironment of a cysteine residue in natively folded proteins determines via its pKs-value, how reactive the exposed SH-group is. If the cysteine is found as -S− its reactivity is enhanced compared to the protonated form and thus, cysteines are often used as “redox-switches” in redox-regulated proteins [14]. Oxidation, i.e. the formation of a sulfenic acid (-S-OH) or reaction with glutathione may change activity, specificity or localization of the respective protein. Such redox-switches are often used in redox-regulated kinases which induce signaling-cascades depending on the cellular redox state or are mediated by changes in ROS-concentrations. Those changes also shift the proportion of redox-sensitive cysteines from a more reduced to a more oxidized/modified state and affect the functionality of proteins/the cellular protein pool. About 200 different mammalian proteins are known which can be modified by thiol-disulfide exchange. Posttranslational S-glutathionylation mostly results in inhibitory effects as found in phosphofructokinase, carboanhydrase III, nuclear factor NF1, glyceraldehyde-3-phosphate dehydrogenase, protein tyrosine phosphatase 1B, the protein kinases Cα and A, creatine kinase, actin, protein phosphatase 2A, tyrosine hydroxylase, complex I of the mitochondrial respiratory chain, the transcription factor NF-κB and the IκB kinase (IKK) [15]. Other proteins are activated by S-glutathionylation including the microsomal S-glutathione transferase, the phosphatase of carbonic anhydrase III, HIV-1 protease, matrix metalloproteinases, HRAS GTPase, sarcoplasmic calcium ATPase, as well as complex II of the respiratory chain [16].
A further protein which is able to reduce disulfides to SH-groups is glutaredoxin (Grx), that plays a significant role in thiol-disulfide-exchange, regulates the activity of transcription factors and acts in apoptosis. During the reduction of disulfides, Grx is oxidized and later restored to its functional (reduced) form by glutathione or by thioredoxin reductase in a NADPH-dependent manner. In mammals four different isoforms are known: Grx1, -2, -3, and -5. Grx1 is found mainly in the cytosol, but also in the nucleus and mitochondrial intermembrane space. Grx2 was originally found in mitochondria, but also in cytosol and nucleoplasm of several tumor lines [15]. Whether Grx2 S-glutathionylizes a protein, or reverses which modification, depends on the cellular redox state (2GSH/GSSG-ratio). In the presence of a high 2GSH/GSSG-ratio, Grx restores SH-groups. If the 2GSH/GSSG-ratio is low, the substrate will be S-glutathionylized. Grx3 (a multidomain protein-complex) is also found in cytosol and nucleus, the monomeric Grx5 is only found in the mitochondria. First, it was suggested that Grx is responsible for reduction of disulfides and deglutathionylation of proteins, meanwhile, it is recognized that Grx isoforms are rather transfer proteins for iron-sulfur clusters (FeS) which use glutathione as ligand [15].
2.2. Counteracting protein oxidation by proteolysis
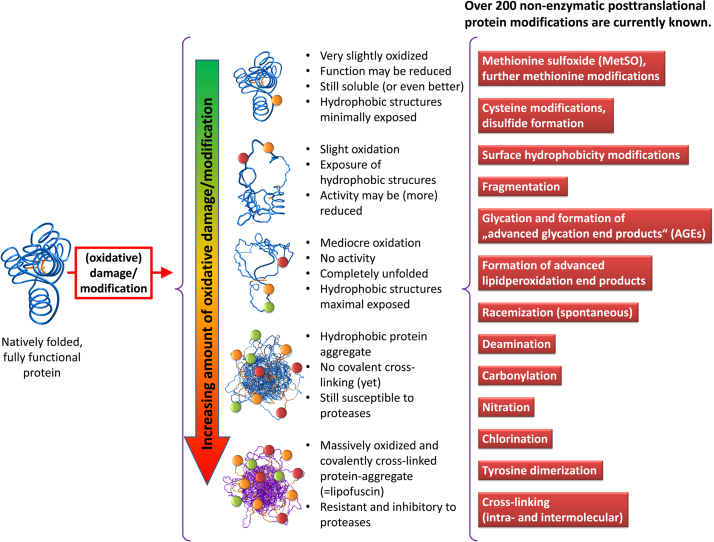
Despite the large variety of reversible oxidative modifications and reducing enzymes, most oxidative protein modifications are irreversible. Irreversibly oxidized proteins must be degraded and replaced by de novo synthesized ones in order to maintain functionality and proteostasis of a cell [17]. In the case a protein (globular and water-soluble) is oxidatively modified/damaged by ROS in an ongoing process, it undergoes a transition from slight functional decrease and increased solubility to a completely dysfunctional, unfolded and insoluble structure that may be even resistant to mammalian proteases due to covalent cross-linking, depending on the amount of oxidative modification. This transition is depicted in Fig. 1. The effect of a single reaction of ROS with a protein depends on the localization of the resulting modification. An enzyme may be completely inactivated if the active center (the amino acids essential for proper function) is modified. To reduce this probability, many proteins have amino acids on their surface that function as “ROS-scavengers” (mainly methionine-residues) and may prevent functional damage in a limited range [18], [19]. Oxidative damage to other amino acids besides of cysteine and methionine is not reversible and thus, irreversibly modified proteins need to be degraded. During evolution two main proteolytic systems evolved to fulfill this task: the “ubiquitin-proteasome system” (UPS) and the autophagy-lysosomal system [20].
Fig. 1.
Oxidation of a soluble protein. The degree of oxidative damage applied to a native protein is both time- and dose-dependent. Minimal amounts of damage may show only slight or no impact on protein function, solubility in this case may even increase, since additional charges are introduced into the protein. Further oxidation leads to a partial unfolding and exposure of hydrophobic residues that are normally buried inside soluble proteins, the overall solubility now decreases compared to the native form of the protein. Mediocre oxidation results in further/complete loss of activity and entire unfolding, hydrophobic structures are now fully exposed. Larger protein aggregates are formed by hydrophobic interactions of such unfolded proteins; formation of such aggregates is still reversible, since the single proteins are not covalently cross-linked. Further oxidation leads to a largely covalently cross-linked protein-aggregate; formation of those structures is irreversible, these products are highly resistant to mammalian proteases. The list on the right shows the most important of the over 200 currently known enzymatic and non-enzymatic posttranslational protein modifications.
2.2.1. The ubiquitin-proteasomal-system (UPS)
The UPS is one of the two most important proteolytic machineries of the mammalian cell. It has two main functions: Recognition and degradation of damaged (including oxidized), modified, dysfunctional proteins as well as the removal of fully functional and natively folded proteins which are no longer needed or undergo normal turnover. Removal of damaged proteins prevents an accumulation of dysfunctional proteins that tend to form aggregates which can be covalently cross-linked. Both functions preserve the cellular functionality and provide a constant fine-tuning of the (functional) proteome (proteostasis).
2.2.1.1. The 20S “core” proteasome
As explained above, (oxidative) damage to proteins is inevitable. This problem already occurred in the earliest known bacteria, the so-called archaea. Consequently, during evolution, proteolytic systems emerged, which are able to recognize and to remove dysfunctional proteins from a cell, in order to prevent intracellular accumulation. The most important protease, removing more than 90% of all oxidatively damaged proteins in eukaryotic cells, is the 20S proteasome [21].
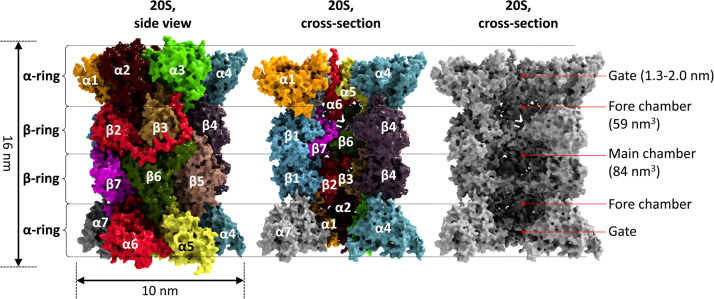
The eukaryotic 20S “core” proteasome, as shown in Fig. 2, is composed of four stacked rings, two alpha and two beta rings, each one containing seven different subunits, arranged in the sequence alpha-beta-beta-alpha, forming a cylindrical structure. The alpha rings are composed of different alpha subunits (alpha1 to alpha7), the beta rings of different beta subunits (beta1 to beta7). Thus, 20S is composed of 28 subunits, which altogether have a molecular weight of 700 kDa [17], [22]. While the outer alpha rings are responsible for substrate recognition and “gating” (regulation of substrate access into the inner proteolytic chamber of 20S), the inner beta rings provide the catalytic activity. The whole 20S complex has three inner chambers: two fore chambers (one between each alpha ring-beta ring-interface) and one main proteolytic chamber (between the two beta rings), where the active centers are localized. Whether there is a special function of the fore chambers is still not known.
Fig. 2.
Structure of the 20S proteasome. This image shows the mammalian (bovine) 20S proteasome as a reconstruction from X-ray crystallographic data with a resolution of 2.75 Å [197]. The left panel shows the structure of this large multicatalytic protease complex with color-coded individual subunits from a side-view including its dimensions. The center panel shows a cross section with several removed alpha and beta subunits and the right panel shows a greyscale image of that cross-section for a better visualization of the inner structure, subdivided into two fore chambers, separated by the gate from the environment and the central main chamber where the proteolytic centers are localized facing the inside of the proteasome.
Three of the seven beta subunits show proteolytic activities: beta1, beta2 and beta5, thus there are six proteolytic centers overall [23]. The ancient archaeal proteasome shows the same structure: four rings, each one containing seven subunits. In contrast to the mammalian one, in the archaeal proteasome the alpha rings are formed of seven identical alpha subunits and the beta rings contain also seven identical subunits which show all proteolytic activity (thus 14 catalytic centers per proteasome). The evolutionary more developed (mammalian) proteasome contains only six active centers per proteasome, but these exhibit different catalytic activities and specificities [24].
As mentioned above, proteolytic activities are localized on the beta subunits, facing the inside of the proteasome, rendering substrate degradation strictly regulated by substrate access. The substrate specificities of the single (mammalian) beta subunits are different [25]:
-
•
beta1 shows a peptidyl-glutamyl-peptide-hydrolyzing activity (a caspase-like one, which cleaves after acidic amino acids, also termed “post-glutamyl-peptide hydrolytic” activity),
-
•
beta2 has a trypsin-like activity and cleaves after basic amino acids, while
-
•
beta5 executes a chymotrypsin-like activity, cleaving after large hydrophobic amino acids.
Until now, the exact mechanism of 20S-substrate recognition remains still unclear. Though, it is strongly suggested that hydrophobic structures that are usually buried inside of natively folded and soluble proteins are recognized [26], [27]. After (oxidative) protein damage, those structures are exposed by unfolding, rendering (partially) unfolded proteins to ideal substrates for 20S proteasomal degradation [28], [29], working in an ATP-independent manner. A very similar mechanism of substrate recognition is found in heat shock proteins and other chaperones, that bind (partially) unfolded proteins in order to prevent their aggregation, enable refolding to their native form or assist in proteolytic degradation by handing over the bound substrates to proteases such as 20S or the lysosomal system [30], [31].
After binding to the exposed hydrophobic structures of a potential substrate protein, the conformation of the alpha rings that form a “gate”-like structure, blocking the entrance to the inner proteolytic chamber, changes. This conformation change widens the annulus (axial pore) of the gate from a diameter from about 0.9 nm to about 1.3–2.0 nm [32], enabling access for a linear and completely unfolded protein and even “hairpin”-like structures of proteins, providing also endopeptidase activity. The main products of 20S proteasomal degradation are oligopeptides with a length between 2 and 35 amino acids. Product peak values are found at 2–3, 8–10, and 20–30 amino acids, while the average length is 8–12 amino acids [33].
During evolution, several proteasomal regulators have evolved, which can bind to the 20S “core” proteasome, modulating its activity and substrate specificity. Those regulators are presented in detail in the next chapter.
2.2.1.2. Regulators of the 20S proteasome
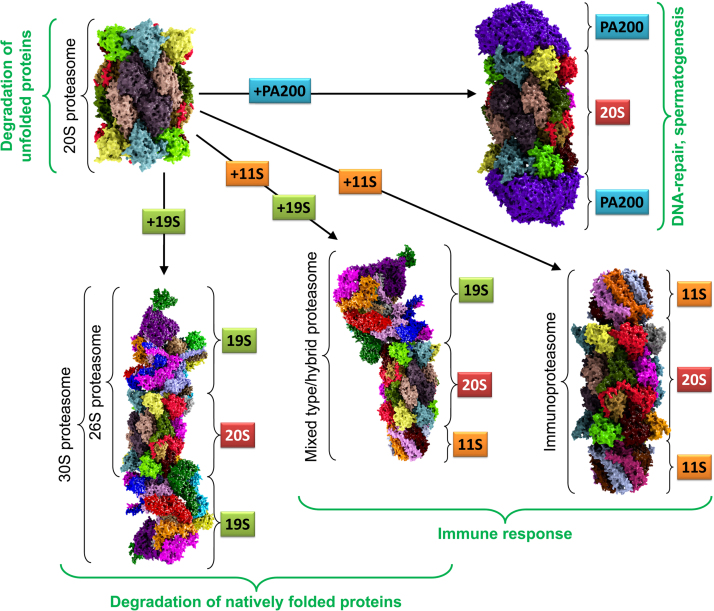
In this chapter a short overview of the most important regulators of the 20S proteasome is given. The single regulators and their combinations are depicted in Fig. 3.
Fig. 3.
The 20S proteasome and its regulators. This image shows the constitutive 20S “core” proteasome (top left) and its most important regulators, that can bind to one or both outer alpha-rings: the 26S (19S-20S) and the 30S proteasome (19S-20S-19S), respectively, often also referred to as the 26S proteasome (bottom left). The 19S regulator is formed of at least 19 different subunits (also color-coded); the so-called “mixed type” or “hybrid proteasome”, a 20S core bound to a 19S regulator and an 11S regulator at the same time (bottom center). The most stable exclusively cytosolic 11S regulator is a heteroheptameric structure (PA28(α3β4)), the exclusively nuclear form is a homoheptamer (PA28(γ7)); the single PA28 subunits are color-coded here. A 20S proteasome, that is bound to two 11S regulators (bottom right). 11S can bind to both the constitutive 20S proteasome as well as to the inducible immunoproteasome (i20S) and induce like all the other regulator proteins gate-opening by conformational changes, thus increasing the cores proteolytic activity. A 20S proteasome bound to two monomeric PA200 nuclear regulator proteins is also shown (top right).
2.2.1.2.1. 19S regulator and 26S proteasome
One of the most prominent proteasomal regulators is termed 19S. 19S is a large complex, composed of at least 19 different subunits, summarizing a molecular weight of about 1 MDa. Besides the known 19S subunits, other proteins exists which interact with the complex, without becoming an essential part of it. 19S can bind to an alpha ring of 20S, forming the so-called 26S proteasome (19S-20S, about 1.7 MDa). If two 19S regulators bind to the core proteasome (19S-20S-19S, about 2.7 MDa) the 30S proteasome is formed, even though this complex is also termed 26S in the literature [34], [35].
6 of the 19 subunits of 19S show ATPase activity (Rpt1-6), while the others do not (Rpn1-12 and Rpn15). The Rpt subunits form a hexameric ring that binds to the 20S proteasomal alpha ring, while the Rpn subunits form a lid-like structure that enables recognition and binding of substrates (via Rpn10 and Rpn13). 19S-binding activates the 20S proteasome in a process involving Rpt2, -3, and -5 [36].
In contrast to the free 20S proteasome, 26/30S is able to degrade natively folded and fully functional proteins in a both ATP-dependent and -consuming manner (in the absence of ATP/NAD(P)H and Ca2+ 20S detaches from the 19S regulator complex) [37], [38], [39], [40]. The energy provided by ATP is not necessary for proteolytic degradation of a substrate but for its unfolding.
For degradation of a fully functional and natively folded substrate, highly specific labeling is necessary – in this special case a short chain of ubiquitin (Ub)-molecules, termed poly-ubiquitin chain. The according process is called polyubiquitination. Ubiquitin, is a small ubiquitous found protein of 76 amino acids. The average Ub-concentration in different mammalian cell types is about 100 pmol/mg [41]. In HEK293 cells 60% of the whole Ub-pool are considered to be bound to other proteins (mono-ubiquitination), mainly histones, while about 11% of Ub are found in form of polyubiquitin chains [41].
Both polyubiquitination and deubiquitination of substrates are regulated by a very complex system still poorly understood. The substrate specificity necessary for polyubiquitination is provided by the so-called ubiquitin system, containing four different types of enzymes (E1–E4) [42].
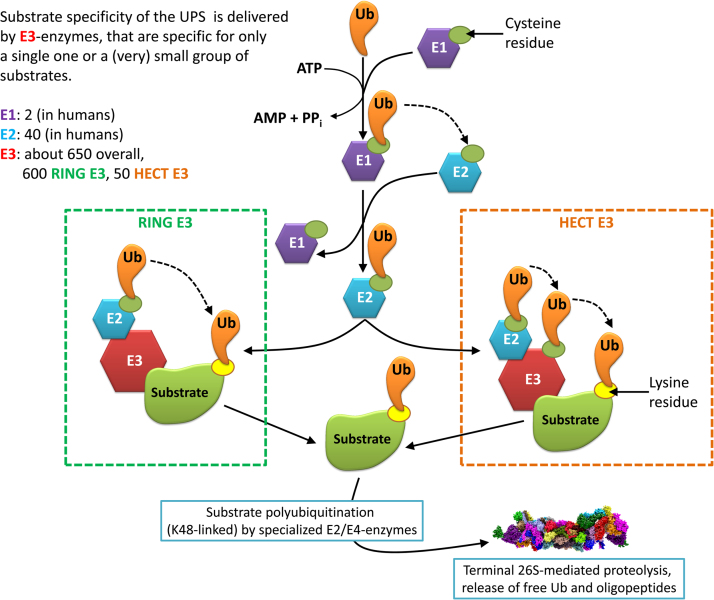
Ubiquitination (the whole process is depicted in Fig. 4) of a substrate starts with “ubiquitin activation” mediated by E1 enzymes: Until now, eight different mammalian E1 enzymes are known, two different ones are found in humans - for activation, one molecule of Ub is attached via formation of a thioester-bond (at the C-terminal glycine76 of Ub) to a cysteine residue in the active center of E1 [43]. The second step is the so-called “ubiquitin-conjugation”, mediated by E2 enzymes. Currently about 50–75 mammalian E2-forms are known [44], 40 of them are found in humans. From those 40, about 35 are dedicated to ubiquitin-conjugation. In the third step, Ub is transferred to substrate proteins in a highly specific manner by the E3 enzymes. About 650 E3 enzymes are currently known, 600 are of the so-called “RING” type (RING stands for “really interesting new gene”) [45], the remaining 50 are of the “HECT” (“homologous to the E6-AP carboxyl terminus”) type [46]. In both cases an intermediate complex is formed of Ub-loaded E2, E3 and the substrate protein. Ub is attached to the substrate by an amide bond, linking the C-terminal carboxyl group of Ub to a lysine residue of the targeted protein. The difference between both RING and HECT is the mechanism of the transfer of Ub to the substrate: The RING E3s mediate the transfer directly from E2 to the substrate, the HECT E3s mediate the transfer from E2 to E3 and then from E3 to the substrate. The substrate specificity is provided by the large variety of E3 enzymes which are responsible for a single one or only few different substrates.
Fig. 4.
The ubiquitination pathway. This figure shows the process of substrate labeling for terminal 26S proteasomal degradation. The first step is the “activation” of an Ubiquitin (Ub) by an E1 in an ATP-consuming manner. Then Ub is transferred by E1 to an E2 enzyme. Substrate specificity is provided by the large variety of available E3 enzymes that only target a small amount of substrates. There exist two types of E3 ubiquitin ligases: RING and HECT. In case of “RING” E3: both the substrate and the Ub-loaded E2 are bound by the E3, the Ub is directly transferred from E2 to the substrate. In case of “HECT” E3, the substrate is transferred from E2 to E3 and then from E3 to the substrate. After attachment of the first Ub to the substrate, a chain of Ub-molecules is attached to the first one by specialized E2/E4-enzymes that are only able to append Ub to another Ub-molecule. The substrate is then degraded by the 26S proteasome (a chain of Ub4 provides the strongest degradation signal), the Ub-chain is released into the cytosol and monomerized again for further loading of an E1 enzyme.
Thus, after attachment of the first Ub to a substrate, the label is elongated ‒ chains of 4 Ub-molecules attached to the substrate provide the strongest degradation signal for 26S mediated proteolysis [47]. In contrast, some proteins can be degraded by the proteasome following modification by a single ubiquitin (monoubiquitylation, like the proteins paired box 3 and syndecan 4) or multiple single ubiquitins (multiple monoubiquitylation, like the NF-κB precursor p105 and phospholipase D) [48]. Furthermore, also several proteins are known that can be degraded by the proteasome in an ubiquitin-independent manner: myeloid cell leukemia 1 (MCL1) [49], CCAAT/enhancer-binding protein δ (C/EBPδ) [50] and ornithine decarboxylase (ODC) [51].
How exactly a single Ub attached to a substrate is extended to a chain of Ub-molecules necessary for substrate recognition by 26S is still under discussion. A subset of the E2-pool is only capable to attach an Ub molecule to another Ub, resulting in the formation of Ub-chains [41]. This is possible for an Ub-loaded E2 without another partner (such as an E3 enzyme). Other authors state that elongation of the Ub-chain is mediated by so-called E4 enzymes [52], a special “subgroup” of the E3-pool. This discussion may be finally clarified by protein-phylogenetics. E2 enzymes are responsible for interaction with a special set of E3 enzymes or substrates: E1 attaches the C-terminal glycine76 of the activated Ub to a cysteine residue of E2, again via a thioester-bond.
Tough, even polyubiquitination of a substrate is not mandatorily a one-way street, because there are also so-called deubiquitinating enzymes (DUBs) which are able to remove an attached Ub-chain from a protein preventing it from proteolysis [53]. Those DUBs recently became a focus of research, since they represent another control instance of protein turnover/cellular proteostasis besides polyubiquitination [44]. Today, more than 95 genes expressing DUBs are known (about 80 are found in humans), involved in numerous cellular functions – some of those functions are essential, so that knockout-mutants are lethal. The number of DUBs is comparable to the number of E2-enzymes in mammals [41]. Three of them (PSMD14/POH1, the ubiquitin carboxyl-terminal hydrolase L5 (UCHL5/UCH37) and USP14) are associated with the lid (non-ATPase subunits) of the 19S regulator. Depletion of either UCHL5 or USP14 increases 26S proteasomal degradation of substrates, while a combined depletion of both decreases it. In studies, 11 of 66 human GFP-tagged DUBs have been found exclusively in the nucleus [54].
In contrast to the 20S proteasome, 26S in combination with the ubiquitin system enables the cells to degrade fully functional proteins – this is important for the quick adaptation of the cell to changed environmental conditions and for the removal of proteins that are no longer needed.
The whole proteome, both the functional one and damaged proteins are subject of a constant turnover, a steady state of de novo synthesis and proteolytic removal, which both depend on metabolic state, cell cycle, cell type, environmental conditions, pathologic changes, immune response, aging, even on circadian cycle [55] or melatonin mediated regulation of clock genes [56]. The need to remove a protein from the cell can have different causes – some proteins were not folded correctly during the de novo synthesis, a protein may have been damaged (by ROS), it may be fully functional but is no longer needed, or under starving conditions amino acids are needed for the constant de novo protein synthesis. In mammals, only few proteins such as the dentin of the teeth or the proteins of the eye lens are not constantly degraded and resynthesized during life span. Perturbation of this steady state (proteostasis) will result in reduced cellular function or even cell death [57].
2.2.1.2.2. 11S regulator and immunoproteasome (i20S)
Besides the already described “constitutive” 20S proteasome (sometimes also termed as “c20S”), an inducible form also exists in mammals, termed the “immunoproteasome” (i20S). Different regulatory proteins are co-expressed together with i20S, mainly the 11S regulator also named as “PA28” or “REG”. After induction of i20S subunits (i.e. of the respective subunits) via exposure of cells to interferon-gamma (IFN-γ), tumor necrosis factor alpha (TNF-α) or lipopolysaccharides (interpreted by the immune system as bacterial infection), in de novo synthesized proteasomes, the catalytic subunits β1, β2, and β5 are replaced by the respective inducible forms iβ1 (also termed PSMB9 or LMP2, for “low molecular weight protein 2”), iβ2 (PSMB10, LMP10, or MECL-1, for “multicatalytic endopeptidase complex-like 1”), and iβ5 (PSMB8 or LMP7). The inducible subunits have a higher affinity to the protein complexes involved in proteasome-assembly, and thus, within about 7 days of IFN- γ exposure, almost the whole proteasome pool of c20S is replaced by i20S [17]. Furthermore, i20S has a shorter half-life of about 27 h compared to the constitutive 20S (about 8–12 days). In the sum, i20S is both quickly formed in considerable amounts after induction and also quickly removed from the cell after decline of the inducing event.
Also, subunits of the proteasomal 11S regulator are co-expressed, forming heterohexameric, heteroheptameric or homoheptameric complexes. The contributing subunits are PA28α (REGα or PSME1) and PA28β (REGβ or PSME2), while the most stable complex seems to be the heteroheptameric PA28(α3β4)-combination, that contains a β-β-dimer. A third 11S subunit is known as PA28γ (also termed REGγ, 11Sγ, PSME3, or Ki antigen). PA28γ is found exclusively in the nucleus and is not inducible (in contrast to PA28α/β). It does not seem to play a role in the immune response, but in repair of damaged DNA, cell proliferation and tumorigenesis. PA28γ promotes proteasome-mediated degradation of regulatory proteins such as the “steroid receptor coactivator-3” (SRC-3) and the cyclin-dependent kinase inhibitors p16, p19 and p21 in an ATP- and ubiquitin-independent manner [58]. It may also play a role in cell cycle transition and proliferation, since REGγ-deficient mice show reduced body size and REGγ-deficient embryonic fibroblasts have an impeded transition from G- to S-phase during the cell cycle [58].
The immunoproteasome also plays an important role in the generation of short oligopeptides from pathogenic proteins, that show a hydrophobic C-terminus and are ideal for MHC-I-presentation (MHC-I, major histocompatibility complex class I) on the cell surface to CD8+-cells (cytotoxic T-lymphocytes). Interferon-γ also induces the formation of both immunoproteasomes and 11S regulator complexes. In case of a viral infection, this provides enhanced proteolysis of viral proteins and increased presentation of viral antigens on the cell surface, enhancing the immune-response especially in the infected tissue environment. Thus, cytotoxic T-lymphocytes antagonize intracellular infections (especially viral ones) by destroying infected cells. According to Mishto et al., the constitutive proteasome (20S) and the immunoproteasome (i20S) show only minor difference in the generated pool of oligopeptides [59], but differ in their degradation rates [60]. These results are still controversial, since other authors state that the immunoproteasome tends to cleave after hydrophobic and basic residues that are better suited for MHC-I-presentation [61].
Furthermore, in cells undergoing inflammation/inflammatory response especially in the phase of i20S de novo synthesis, also “mixed” proteasomes are formed which may have between one or five constitutive catalytic sites replaced by the inducible ones [62]. This may result in a broadened spectrum of MHC-I-compatible short antigenic oligopeptides available.
Despite of its role in production of presentable antigens, another supported function of the cytosolic PA28 is the degradation of oxidatively damaged proteins, since i20S shows a higher activity than c20S, especially when in complex with the 11S regulator which induces gate opening and thus increases proteolytic capacity [63], [64], [65]. In both inflammation and during aging the amount of i20S is increased, possibly due to the increased amount of damaged proteins [66], [67].
Recent findings point out, that some diseases are accompanied by a changed amount of immunoproteasomes, especially in pathologies associated with inflammation and/or infection [68], [69]. Whether the increase of i20S formation is a cause or a result of the pathology is still under discussion. Most likely, i20S is induced to cope with a shifted proteostasis, especially during and after phases of oxidative shifts/stress [70]. Diseases accompanied by increased formation of i20S are age-related macular degeneration (AMD), neurodegenerative pathologies such as Alzheimer’s, Huntington’s and Parkinson’s, as well as amyotrophic lateral sclerosis (ALS) – all of them showing inflammatory aspects, such as enhanced ROS formation, protein oxidation, lipid peroxidation, or formation of advanced glycation end products (AGEs).
Also several diseases are known that result from mutations in the PSMB8-gene, coding the iβ5-subunit: JMP (joint contracture, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy) [71], CANDLE (chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature) [72], NNS (Nakajo-Nishimura syndrome) [73] and JASL (Japanese autoinflammatory syndrome lipodystrophy). JMP is associated with a slight change in iβ5 activity [71] due to changes in its tertiary structure, resulting in mild metabolic disturbances, joint contractures, muscle atrophy, elevated liver enzymes and hypergammaglobulinemia; CANDLE, also showing decreased iβ5 activity, which manifests in skin lesions, slight fever, developmental delay and progressive lipodystrophy; NNS shows symptoms very similar to CANDLE; JASL is characterized by auto-inflammation and lipodystrophy [70].
Direct comparison of c20S and i20S revealed in most cases same efficiencies of both, while in certain cases i20S was slightly more active [70]. Binding of the 11S-regulator significantly enhances proteolysis by gate-opening and the selectivity of i20S towards oxidized substrates provides enhanced capacity during phases of oxidative stress. Also, in long-lived species (rodents and primates) a higher basal expression of i20S was shown compared to short-lived ones [74], while expression of the immunoproteasome as well increases with age (shown in rats [75]) and its induction via IFN-γ is reduced [76]. Since aging is also accompanied by both enhanced protein oxidation and thus, also an intracellular accumulation of oxidatively damaged proteins, expression of i20S may be a counteracting cellular antioxidative response in order to preserve proteostasis. Thus, the functions of i20S are beyond just the production of MHC-I-compatible antigens.
It is also important to mention the influence of oxidative stress and redox-shifts on the composition of the proteasomal system. Slight oxidative stress induces an antioxidative Nrf2-mediated response which also includes the expression of both the constitutive 20S proteasome as well as of the 11S regulator, but not of the immunoproteasome [77], while severe oxidative stress induces an inflammatory response (mediated by NF-κB) accompanied by expression of inducible proteasomal subunits [78]. A 20S proteasome which binds to both a 19S and an 11S regulator is termed “mixed type proteasome” or “hybrid proteasome”.
Another proteasomal subtype is the “thymoproteasome” (t20S), containing iβ1, iβ2 and tβ5 (PSMB11), a subunit, which is only found in cortical thymic epithelial cells (cTECs). In cTECS, t20S plays an important role in positive selection of T cells, while the expression of β5 or iβ5 cannot compensate the lack of tβ5 [79], [80], [81].
2.2.1.2.3. The protein activator “PA200”
The proteasomal regulator “protein activator 200 kDa” (“PA200”, in yeast termed “Blm10”) [82], is a protein which is only found in the nucleus of mammalian cells with a function that is still largely unknown.
In yeast, knockout mutants reveal hypersensitivity to DNA-damage and reduced respiratory capacity [83] while in mice the susceptibility to DNA-damaging agents is not changed. However, the male fertility was significantly decreased. The PA200/Blm10 protein is a monomeric dome-shaped regulator that binds to one of the alpha-rings of 20S and induces increased proteasomal activity via gate-opening, very similar to the 11S regulator. Both PA200 and Blm10 are ATP-independent and can be induced by ionizing radiation. Today, three different isoforms have been identified (PA200i, PA200ii, PA200iii), but only PA200i seems to actually bind to the 20S proteasome.
2.2.1.3. Kinetics of the 26S/20S mediates degradation
The characteristics and mechanisms of proteasomal degradation as well as the kinetic mechanism are of great interest. 26S proteasomal degradation is a process in the range of minutes as found with different substrates such as dihydrofolate dehydrogenase that is processed to oligopeptides within 5 min or the I27 domain of titin taking about 40 min. According to Henderson et al. [84], per minute of substrate degradation, a single 26S complex (19S-20S) consumes about 110 molecules of ATP. Considering an overall of 12 ATPases in a double 19S-capped 20S proteasome (19S-20S-19S), the authors estimated a consumption of about 300 molecules of ATP for complete unfolding of a single I27 domain [84].
Those rate-quantifying experiments were carried out in vitro using isolated 26S proteasomes from yeast in buffers preserving 26S functionality [84].
Thus, the I27 domain reveals high mechanical stability and may not be compared to the average substrate the 26S proteasome encounters in a cell – around 60% of the complete degradation time was spent unfolding I27. A V13P-mutant of I27 (I27V13P) reduced the overall degradation time to 9 min compared to the wildtype [84] in a substrate-saturated condition at steady state. Thus, at least a partial unfolding of the target protein reduces its “stability” and at the same time also its degradation time.
The 20S proteasome was also object of mathematical modeling and simulation (in silico). Most models of enzymatic activity assume Michaelis-Menten-type kinetics, but 20S for example revealed inhibition at high substrate concentrations (in this special case a fluorescent oligopeptide, which is activated by 20S mediated proteolysis); besides this, also the products seem to have enhancing effects: the proteolytic activity of 20S increased over time while the substrate Suc-LLVY-MCA was degraded [85]. Since the classical enzyme kinetic models fail to describe 20S mediated proteolysis, more complex models were established which also include effects such as substrate-inhibition (with feedback on substrate-binding or -hydrolysis), as well as enhancing or inhibiting regulatory sites at the outside of the proteasome or in its inside (proteolytic chamber). One of those models by Liepe et al. [85] suggests that a gate-opening site is located inside the proteasome as well as a transport-inhibiting site at its surface. This model is able to explain already known effects such as onset of reduced proteolysis before substrate-depletion, the substrate-inhibition at both early and late time points, as well as increased reaction velocity over time [85]. These effects are also dependent of the substrate.
2.2.1.4. Redox regulation of the UPS
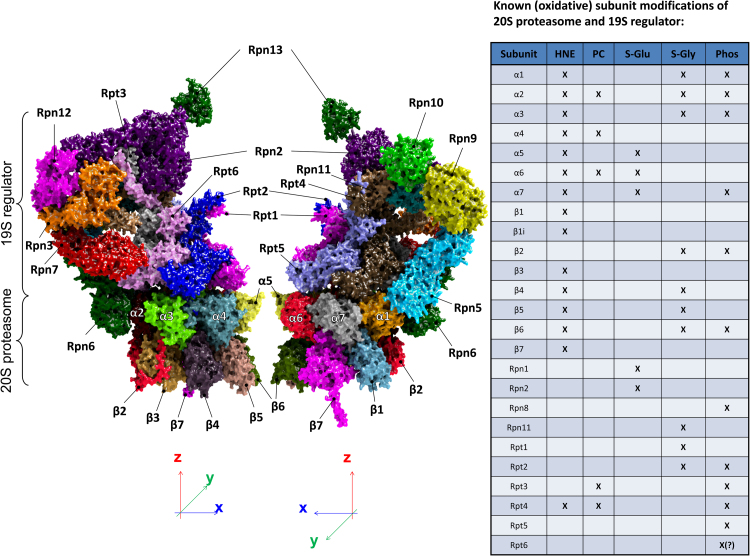
Since the formation of ROS and the resulting oxidative protein modification/damage is an inevitable process, the function of the UPS is also modulated in a redox-dependent manner. Such a redox-dependent regulation is mainly realized via redox-sensitive amino acids (such as cysteines) or residues that are modified by other proteins (for example kinases) which in turn are redox-regulated. Furthermore, also irreversible oxidation of 20S/26S is possible. Consequently, different 19S and 20S subunits were found to be modified/oxidized by formation of HNE-adducts or protein carbonyls, by S-glutathionylation, S-glycosylation or phosphorylation. In part, those modifications have direct impact on 20S activity. Our own work (unpublished data) revealed that S-glutathionylation of the mammalian 20S proteasome (according to [86], [87]) significantly increases the 20S proteasomal activity, while complete de-glutathionylation (using DTT, according to [88]) significantly reduces it. Fig. 5 shows a visualization of the 26S proteasome with the single subunits and their known modification(s) [89].
Fig. 5.
The structure of a half 30S proteasome and some important known posttranslational modifications. The single subunits of a half 30S proteasome - shown is the half of a 20S proteasome (one alpha- and one beta-ring) attached to the 19S regulator protein. The single subunits are color-coded, the structure is a 3D-reconstruction from cryo-electron microscopic data. The table on the right shows different posttranslational modifications (HNE=protein-adduct with the lipid peroxidation product trans-4-hydroxy-2-nonenal; PC= protein carbonyls; S-Glu=S-glutathionylation; S-Gly=S-glycosylation; Phos=phosphorylation) that have been shown already for the respective subunits [89].
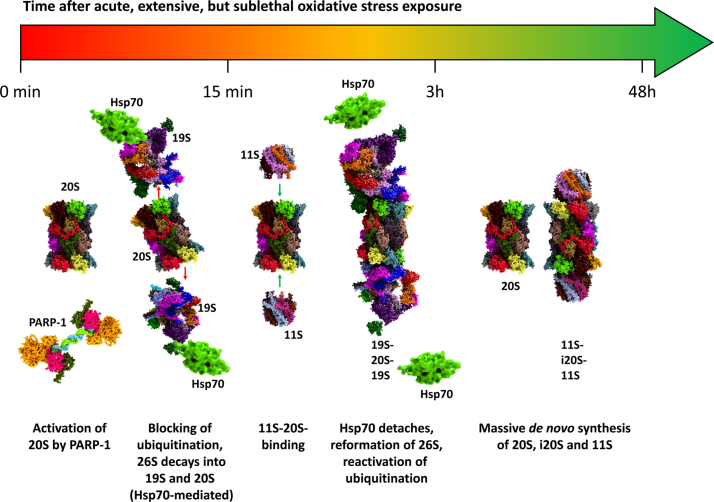
Besides 20S and 19S, other parts of the UPS and related proteins are affected by changing redox states. So, not only proteins are affected, since oxidative damage affects also lipids, carbohydrates and nucleotides. DNA-damage threatens the functionality of a cell. The nuclear poly(ADP-ribose)-polymerase-1 (PARP-1) is one of the first “detectors” of DNA-damage as single-(SSB) and double-strand breaks (DSB). PARP-1 binds to SSB/DSB and then actively produces poly-ADP-ribose, mainly targeting histones. Other targets are NF-κB, p53, DNA-topoisomerases, DNA-PKcs, also PARP-1 itself as well as the nuclear 20S proteasome. Most of those mentioned targets will be discussed below. It seems that this modification has a protective effect on histones, preserving the functional ones from degradation. PARP-1 is recognized as the linker between the DNA-damage-repairing machinery of the cell and the UPS removing damaged proteins, since PARP-1 is able to activate the 20S proteasome [90]. UPS-activation can be suppressed with both PARP-inhibitors as well as inhibitors of the proteasome (such as lactacystin or MG132). Besides this 20S proteasomal induction under conditions of oxidative stress, a rapid decline of 26S proteasomal activity is found, mainly driven by a Hsp70-mediated detaching of the 19S regulator [91], also resulting in a higher proteolytic capacity of 20S. Further redox-dependent modifications (such as S-glutathionylation and phosphorylation, while the latter one is not only redox-dependent) may also contribute to increased 20S activity while the 26S activity is reduced. Depending on the cell type, this response is induced within 5–30 min. Besides 19S detaching, enhanced binding of 20S to the 11S regulator takes place, also increasing 20S activity by gate-opening, thus providing more proteolytic capacity for the degradation of oxidized proteins [92]. The (poly)ubiquitinating machinery, formed of E1, E2, E3, and E4 enzymes is very susceptible to redox-shifts, mainly due to S-glutathionylation of active site-cysteines [93]. Within 3–24 h the bulk of oxidatively damaged proteins is removed from a mammalian cell mainly by the proteasomal system, since proteasomal inhibitors are able to prevent this removal to about 90% [21]. After the phase of oxidative stress and coping with the induced changes, 19S and 20S reassemble to 26S again, while Hsp70 is released. In the range from 12 to 72 h, massive de novo synthesis of 20S, i20S, and 11S subunits is induced. The described changes of the UPS during redox-shifts are depicted in Fig. 6.
Fig. 6.
Regulation of the proteasome after an oxidative event. The response of the proteasome and its different regulators to an event of acute, extensive, but sublethal oxidative stress over time is shown here. Within the first 15 min, PARP-1 activates the nuclear 20S proteasome. Ubiquitination is blocked, since the E-enzymes of the UPS are susceptible to oxidative stress (due to the functional cysteine in their active centers), while Hsp70 mediates the detachment of 19S from the 20S proteasome, reducing the amount of 26S proteolytic capacity. Within 15 min until 3 h after the stress-event, 11S attaches to the 20S proteasome, increasing its activity by gate-opening. The 26S proteasome is reformed (Hsp70 detaches) and the ubiquitinating machinery of the cell is recovering to a functional state. After 3–48 h a massive de novo synthesis of 20S, i20S and the 11S regulator is induced, strongly enhancing ATP-independent degradation of unfolded protein substrates.
This also explains the (for a short time) enhanced resistance of mammalian cells against oxidative stress after pre-treatment with low doses of oxidative agents, since the respective cellular systems are already induced [94]. Those cellular effects are termed as “adaptation” or “hormesis”. In toxicology, hormesis is a dose-response effect, characterized by low dose stimulation and a high dose inhibition, resulting in J- or inverted U-shaped dose responses. Though, both “hormesis” and “adaptation” share similar cellular mechanisms, they are not identical [95].
2.3. Autophagy-lysosomal system
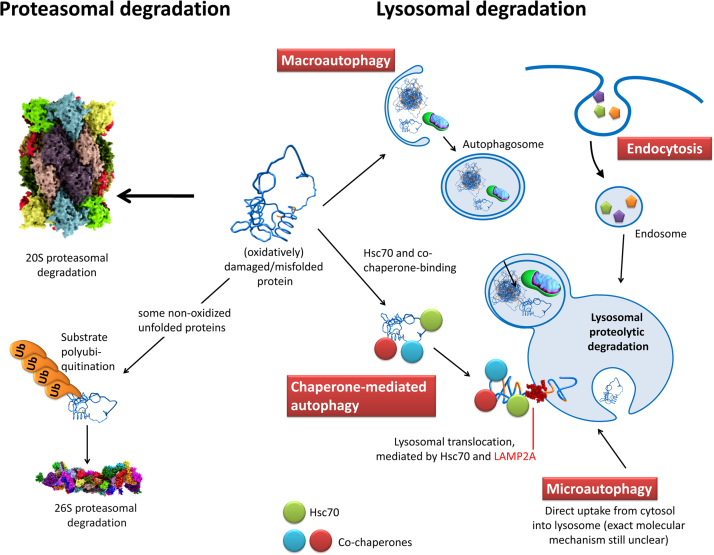
Oxidatively damaged or unfolded proteins can be removed from a cell via different pathways, as depicted in Fig. 7. The most important one is proteasomal degradation. Besides this, incorporation into the lysosomal system (autophagy) is possible. While the UPS is only able to degrade proteins, the lysosomal system incorporates macromolecules, protein aggregates as well as whole cellular organelles – the material can also be obtained from the extracellular space. Furthermore this evolutionary highly conserved system also plays a role in antigen processing, initiation of apoptosis, cholesterol homeostasis, signaling, differentiation, growth control, tissue remodeling, energy metabolism, degradation of the extracellular matrix and repair of the cellular plasma membrane [96], [97], [98].
Fig. 7.
The possible degradation-pathways of an (oxidatively) damaged/unfolded protein. The UPS-pathway - a damaged protein can be degraded by the 20S proteasome or the 26S proteasome. However, polyubiquitination is not a pathway for oxidatively damaged proteins, since those are not preferentially polyubiquitinated [198], [199]. Macroautophagy - a phagophore engulfs a cytosolic volume, that may also contain single damaged proteins, but mainly larger protein-aggregates (can be covalently cross-liked or not) or even whole organelles such as aged/dysfunctional mitochondria. The resulting autophagosome containing the lysosomal substrates fuses with a lysosome, exposing those substrates to a large variety of enzymes. Chaperone-mediated autophagy - a substrate protein is bound by different chaperones as Hsc70 and its co-chaperones and is translocated into the lysosomal system via interaction of Hsc70 and the “lysosome-associated membrane protein 2” (LAMP-2A). LAMP-2A translocates the substrate into the lysosomal volume where it is bound by a lysosomal form of Hsc70 (lys-Hsc70). Microautophagy - substrates are directly taken up from the cytosol via invagination by the lysosomal system. Endocytosis - substrates from the outside of the cell are taken up by the cell via an invagination of the cell membrane, forming an endosome that fuses with the lysosomal system, resulting in substrate degradation.
In mammalian cells, lysosomes are dynamic organelles with diameters from 0.1 up to 1.2 µm, filling about 0.5–5% of the intracellular space [99]. In the lysosome a large variety of acidic proteases, lipases and nucleases is found, often showing optimum activity at low pH-values (in lysosomes an acidic pH of 4.5–5.5 is found [100]) provided by membrane-bound ATP-consuming proton-pumps - thus, in case of lysosomal rupture, the released enzymes will not start to degrade cellular structures at the slightly basic cytosolic pH of about 7.2–7.4. The main proteases found in lysosomes are the serine proteases cathepsin A and G, the aspartic proteases cathepsin D and E (pepsin family A1), as well as the cysteine proteases cathepsin B, V, L, F, H, K, O, S, V, X and W (papain family C1A) [101]. Furthermore, the family C13 is found, including the cysteine protease asparaginyl endopeptidase, also termed legumain.
First, those organelles have been described by Christian de Duve in 1955, the term is composed of the Greek “lysis” (“destruction” or “dissolution”) and “soma” (“body”), thus describing a “lytic body”. Lysosomes are characterized by a single lipid double layer, containing large amounts of the lipophilic antioxidant α-tocopherol compared to other membranes [102]. The main function of α-tocopherol is disruption of lipid peroxidation chain reactions, what may be a hint to an increased amount of ROS formation within those organelles. Besides the first description, de Duve was also the first to observe a cellular self-eating process and coined the term “autophagy” as catabolic function of the lysosomal system in 1963 [103].
Also, different types of autophagy are possible for lysosomal substrates that depend on the pathways of delivery. One type is the so-called macroautophagy [104], [105] - in this case, a phagophore incorporates a cytosolic volume, and forms an autophagosome that fuses with a lysosome (Fig. 7, top left). This function can be induced by various cellular stresses such as limited nutrients [106], an accumulation of damaged proteins (aggrephagy) [107], dysfunctional/defective mitochondria (mitophagy) [108], [109], and finally also by invading pathogens (xenophagy) [110]. Until now, at least 36 genes are known (Atg1-36, Atg for “autophagy-related protein”) which are involved in this still poorly understood process. From those 36 genes, 17 form the “core machinery”, essential for most types of autophagy. The “mechanistic target of rapamycin complex 1“ (mTORC1) [111], is a complex of proteins, that functions as a sensor for nutrient supplementation, energy (via ATP) and redox state and controls protein synthesis. A large variety of factors impact mTORC1 signaling such as insulin, growth factors, phosphatidic acid, certain amino acids, hypoxia, mechanical and oxidative stress. One of the first upstream events in autophagy is the formation of the ULK1- (uncoordinated-51-like kinase 1, the mammalian homolog of Atg1) complex, formed of ULK1, ATG13, FIP200 (focal adhesion kinase family-interacting protein 200 kDa) and Atg101 [112].
Low amounts of glucose activate AMPK (5' AMP-activated protein kinase or AMPK or 5' adenosine monophosphate-activated protein kinase) by an altered ATP/ADP ratio. AMPK can, therefore, be considered as a sensor of AMP/ATP or ADP/ATP ratios determining the cellular energy level, and low amounts of glucose shift this ratio by a decrease of ATP in favor of ADP and AMP. Induced AMPK associates with ULK1, phosphorylates it at different sites, thus starting the formation of the ULK1 complex, required for autophagy. Autophagy can also be induced by a starvation of amino acids. In mammalian cells, mTORC1 binds the ULK1-complex under amino acid-rich conditions and inhibits induction of autophagy via phosphorylation of ULK1 at Ser757 and also via phosphorylation of ATG13 [112]. During starvation phases, mTORC1 detaches from the ULK1 complex that becomes active and induces autophagy. Insulin affects/regulates the ULK1-complex in an Akt (“protein kinase B”, also termed PKB)-mediated manner, amino acids take effect via mTORC1 and glucose via AMPK [112].
Thus, mTORC1 controls Atg1, one of the main inducers of autophagy. Atg1 regulates Atg9, a protein which induces the de novo formation of the phagophore at the so-called “preautophagosomal structure” (PAS) that engulfs the substrates to be degraded. In this process, also Atg6 and -14 are included, which form the class III PI3K complex I (containing Vps34, phosphoinositide 3-kinase, p150/Vps15, Atg6/Beclin1, and Atg14L) [113]; the exact mechanism of PAS assembly is still unclear. The activation of this Atg6-PI3K-complex produces PI3P (phosphatidylinositol 3-phosphate) providing a lipid signal recruiting other effectors such as DFCP1 (double FYVE domain-containing protein 1) as well as members of the WIPI (WD-repeat protein interacting with phosphoinositides) family that eventually form the phagophore [112].
Expansion and final closure of the forming membrane are regulated by the ubiquitin-like conjugation systems Atg8 and Atg12. Proteins of the Atg8-family are conjugated to phosphatidylethanolamine in a process involving Atg3 and -7, while Atg12 forms a complex with Atg5 which interacts with Atg16, also supporting the formation of the isolation membrane and its final closure around the substrates. The result is a vesicle (autophagosome), filled with substrates [114] - later the autophagosome fuses either directly with a lysosome or first with a “multivesicular body” (MVB, also termed “endosome”) forming an amphisome and then fuses with a lysosome. In both cases, the result is termed “autolysosome” [115]. However, an endosome is enclosed by only one lipid bilayer (formed of the invaginated cell membrane), while an autophagosome is engulfed by two lipid bilayers (formed by the phagophore) – the outer one fuses with the lysosome (the inner one is degraded together with the substrates it contains) to the resulting autolysosome [115]. The products of degradation are transported across the lysosomal membrane back into the cytosol for further use by the cellular metabolism. The mentioned MVBs contain nutrient transporters, complexes of ligands and growth-factors, lipids, extracellular material or pathogens. Most of this material is not degraded but recycled back to the cell membrane, while only a small amount of the MVBs ends up in the lysosomal system [114].
Another form of autophagic degradation is the microautophagy [116], [117] (the term was created in 1983), here, the lysosomal membrane directly engulfs a cytosolic volume by random membrane invagination, which forms an “autophagic tube” (mediated by Atg7) and finally buds into the lysosomal lumen [118], where it is degraded with its substrates (Fig. 7, bottom right).
The third type of autophagy is the so-called “chaperone-mediated autophagy” (CMA), discovered in 1981. CMA exclusively targets single proteins. Besides being recognized by the 20S proteasome, unfolded proteins can also be recognized by chaperones such as Hsc70 (“heat shock cognate 70”) triggering also chaperone-mediated autophagy (CMA) (Fig. 7, center). The basic mechanism involves Hsc70-binding to substrates in the cytosol which contain a specific KFERQ motif that is essential for CMA. About 30% of the cytosolic proteins in mammalian cells carry this motif [100]. Hsc70 recognizes this motif and transfers the substrate to the lysosome-associated membrane protein type 2A (LAMP-2A). LAMP-2A is a monomer which assembles into multimeric complexes after substrate binding to its monomeric form. After translocation of the substrate into the lysosome, the multimeric LAMP-2A-complex quickly disassembles. Hsc70 in complex with the co-chaperones Hsp90, Bag1, Hsp40, St13, and Stip1/Hop mediates recognition of the substrates’ KFERQ motif and translocation into the lysosome [119], [120]. This chaperone-complex seems also to be involved in substrate unfolding. Inside the lysosome, another variant of Hsc70 (lysosomal Hsc70, lys-Hsc70, with an isoelectric point of 5.3) is required for the translocation process of the substrate [121]. Lys-Hsc70 is stable in pH-ranges from 5.2 to 5.4, while it is degraded very quickly, if the lysosomal pH is shifted above 5.6, thus, reducing CMA significantly. Consequently, lysosomes are classified according to lys-Hsc70 in their lumen (or not) and without lys-Hsc70, CMA is not possible. However, the exact function of lys-Hsc70 is still unclear: it may be an active “pulling” process or a passive prevention of substrate retro-translocation into the cytosol. Furthermore it is still unknown, how exactly lys-Hsc70 enters the lysosomal lumen at all. Oxidative stress or starvation are also able to increase the amount of lys-Hsc70 in lysosomes [121], which is recently also known to be involved in the re-monomerization of LAMP-2A-clusters after substrate translocation [122].
Interestingly, Hsc70 may also play a role in microautophagy [123] and macroautophagy. Here, Hsc70 recognizes and binds protein aggregates and mediates their uptake in a process termed chaperone-assisted selective autophagy (CASA) [124], [125]. CASA was first shown for the protein filamin. In this case, the co-chaperone Bag3 binds both Hsc70 and HspB8 (heat shock protein beta 8), while the E3 ubiquitin-protein ligase Stub, that is bound to Hsc70 in this complex, mediates the ubiquitination of the substrate. However, the ubiquitinated form of filamin is not degraded by the 26S proteasome, but it is bound by the macroautophagy receptor sqstm1/p62. The sqstm1/p62-substrate complex [126] is then recognized by the autophagosomal membrane protein MAP1LC3 (microtubule-associated proteins 1A/1B light chain 3B, also termed LC3, the most commonly used marker-protein of autophagosomes) [127] which mediates uptake of the substrate into the autophagosome by a mechanism which is still under discussion.
2.4. Redox shifts regulating the UPS
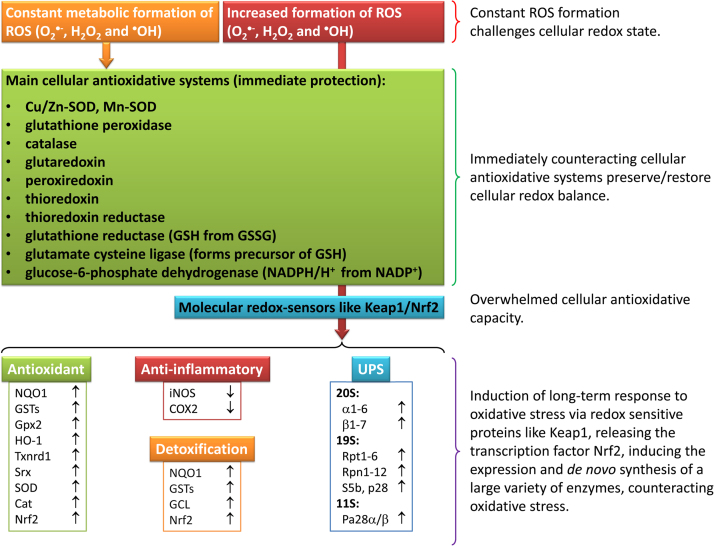
In order to counteract oxidative damage of cellular structures in redox-shifts, inflammation or oxidative stress, exceeding the “basic” amount of ROS produced in normal cellular function, there are powerful systems that can be induced, increasing the antioxidative capacity of the cell. In Fig. 8, the green box displays the most important cellular ROS-scavenging enzymes - if those systems are not sufficient to prevent a cellular redox shift, molecular redox-sensors such as Keap1/Nrf2 can be activated very quickly.
Fig. 8.
Overwhelming of antioxidative defenses leads to induction of defense mechanisms. Since the constant formation of reactive species is an inevitable by-product of life, mammalian cells are endowed with a remarkable asset of antioxidative systems. In case of an increase of ROS to above “normal” (physiological) level, an antioxidative response will be induced. The central redox-sensor Keap1/Nrf2 becomes activated and induces the expression of a variety of antioxidative (NQO1: NAD(P)H quinone dehydrogenase 1; GST: glutathione S-transferase; Gpx2: glutathione peroxidase 2; HO-1: heme oxygenase-1; Txnrd1: thioredoxin reductase 1; Srx: sulfiredoxin; SOD: superoxide dismutase; Cat: catalase; Nrf2: nuclear factor erythroid 2-related factor 2), anti-inflammatory (iNOS: inducible nitric oxide synthase; COX2: cyclooxygenase-2) and detoxifying enzymes (GCL: glutamate cysteine ligase), as well as parts of the ubiquitin-proteasome-system (UPS). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
2.4.1. Nrf2
Keap1 is the regulator of the “nuclear factor erythroid 2-related factor 2” (Nrf2), one of the central redox-sensitive messengers (Fig. 8). Nrf2 is bound to Keap1 in an inactive state under physiological conditions as Keap1/Nrf2-complex [128]. This complex binds the Cul3-Rbx1 holoenzyme, one of the (up to now) seven known Cullin-RING box (Cul-Rbx) E3-ubiquitin-ligases, mediating the polyubiquitination of Nrf2, resulting in its proteolytic degradation by the 26S proteasome. Thus, Nrf2 has only a very short intracellular half-life of about 10–20 min [129]. If Keap1 gets oxidized (its cysteine residues 151, 273, and 288 function as redox switches) and, therefore, its ability to bind Nrf2 is reduced, Nrf2 is released into the cytosol, where it becomes quickly phosphorylated mediated by a large variety of kinases such as protein kinase C (PKC), several mitogen-activated protein kinases (MAPKs), PKR-like endoplasmic reticulum kinase (PERK) or phosphatidylinositol 3-kinase (PIK3) [130]. After its phosphorylation [129], Nrf2 is translocated by the importins α5/β1 into the nucleus [131], where it forms a complex with the proteins sMaf, ATF4, JunD and PMF-1 which induces the expression of several antioxidant response elements/electrophile response elements (ARE/EpRE-elements), listed in Fig. 8 (bottom left). The amount (maximal nuclear accumulation) of Nrf2, as well as the accumulation time is affected by the intensity of oxidative stress: inducers of Nrf2 such as sulforaphane (SFN) increase in a dose dependent manner the maximal amount of Nrf2 translocated and reduce the necessary time [132]. The balance between Keap1-mediated proteasomal degradation of Nrf2 and its de novo synthesis provides a (very) small amount of Nrf2 that is found free in the cytosol in between synthesis and Keap1 binding. Pronounced oxidative conditions provide a reduced Nrf2-proteolysis and an increased synthesis, since increased Nrf2 expression is a part of Nrf2-mediated stress-response.
The nuclear fate of Nrf2 is still unclear: it can be translocated back into the cytosol, where it is degraded in a Keap1-mediated manner as described above or it can be degraded in the nucleus by the 26S proteasome, guided by “glycogen synthase kinase 3” (GSK-3) [133], which directs proteolysis of a variety of proteins via the SCFβ-TrCP1/2-complex which functions as an E3 ubiquitin ligase and mediates substrate-polyubiquitination. GSK-3 phosphorylates a cluster of Ser/Thr-residues in target proteins, which are then recognized by the SCFβ-TrCP1/2-complex, that also binds cullin-1 resulting in the formation of a complete E3-ligase after association with two other proteins (Skp1 and Rbx1) [134]. The resulting complex is able to recognize phosphorylated substrates and mediates their polyubiquitination, followed by 26S proteasomal degradation which may be an alternative pathway of Nrf2-proteolysis besides the Keap1-mediated one.
As mentioned above, oxidative stress is able to activate Nrf2, inducing the expression of both the 11S regulator and the (constitutive) 20S proteasome, while the immunoproteasome subunits are not induced by Nrf2. Nevertheless, H2O2 is able to induce expression of the immunoproteasome, but apparently by a mechanism, independent from Nrf2, since Nrf2 inducers, inhibitors, and the respective siRNA have only minimal effects on i20S induction mediated by H2O2[135].
Most of the proteins induced after Nrf2 activation are antioxidants, proteins with anti-inflammatory effects, detoxifying enzymes and subunits of the UPS (of 20S, 19S and 11S). This enables the cell to counteract both the increased amount of ROS, the possible sources of ROS and to remove already oxidatively damaged structures more efficiently. However, especially the induction of UPS subunits is still under discussion: One study using microarrays revealed an increase in the expression of the 20S subunits α1-2, α4-7, β1-6, as well as of the 19S subunits Rpt1/2/5, Rpn2/5/6/8-12, and S5b, but neither the inducible subunits of the immunoproteasome nor the ones of the 11S regulator were detected [136]. In contrast, Pickering et al. demonstrated induction of PA28α and -β by Nrf2 [135]. Other experiments on human fibroblasts detected induction of α4, β1, β2, and β5 [137]. The expression of POMP/Ump1 (a factor essential for proper 20S assembly) was also found to be induced [138]. Thus, the mRNA of Ump1 is significantly increased, the amount of Ump1 decreases and its half-life is reduced from 82 to 21 min, since Ump1 is the first substrate of a completely assembled proteasome and is thus quickly degraded with enhanced proteasome formation [17].
2.5. NF-κB
Another important regulator of cellular response to redox-changes is NF-κB which stands for the “nuclear factor 'kappa-light-chain-enhancer' of activated B-cells”. The most important inducers of NF-κB are pro-inflammatory cytokines such as tumor necrosis factor α (TNFα), interleukin-1β and bacterial lipopolysaccharides (LPS), also known to induce the expression of the immunoproteasome. NF-κB is a family of inflammatory transcription factors including RelA (p65), RelB, c-rel, p50 and p52 [139], also mediating immune responses to bacterial and viral infections, inflammation, aspects of development, cell proliferation and protects against UV radiation [139]. The canonical activation of NF-κB starts with the phosphorylation of IκBα, the negative regulator of NF-κB. IκBα-phosphorylation is mediated by the IKK kinase complex containing two kinases (IKK-α and IKK-β) as well as a regulatory subunit (IKK-γ, also termed NEMO) [140]. After binding the SCFβ-TrCP1/2-cullin-1-complex, phosphorylated IκBα is polyubiquitinated and degraded as 26S proteasomal substrate, releasing the NF-κB subunits which are translocated into the nucleus. In the nucleus NF-κB binds to target genes that include also IκBα, a protein chaperoning NF-κB back to the cytoplasm [141]. NF-κB induction also triggers increased mitochondrial activity, as well as expression of NADPH oxidase causing an increase in cellular ROS generation [142], [143]. At the same time, expression of inflammatory biomarkers as interleukin (IL)-β1, IL-6, TNF-α and the pro-inflammatory enzymes iNOS and COX2 are increased, which share a common induction via the NF-κB-pathway. The NF-κB-pathway is also involved in the regulation of the expression of several enzymes of the UPS, e.g. the muscle E3 ligase MUrf-1 [144].
It is known that induction of Nrf2, which has an inhibitory effect on NF-κB-induction, is able to reduce its pro-inflammatory response. In the same manner, NF-κB is able to suppress Nrf2 signaling at the transcriptional level by competing for the transcriptional co-activator “CREB binding protein” (CBP) and recruits the “histone deacetylase 3” (HDAC 3), causing a local hypoacetylation which reduces Nrf2-signaling [132], [145].
Both Nrf2 and NF-κB must maintain a balance between anti-oxidative and pro-inflammatory cellular response, since imbalances between both pathways are associated with a large number of diseases from neurodegeneration and autoimmune disorders to cancer [146]. Another interlink between Nrf2 and NF-κB (besides the above mentioned competition for CBP) is the ability of Keap1 to prevent the binding of IKKβ to “heat shock protein 90” (Hsp90) [139], inducing the autophagic degradation of IKKβ, as well as to prevent phosphorylation and thus, activation of IKKβ [147].
2.5.1. Nrf1
The “nuclear factor erythroid 2-related factor 1” (Nrf1) plays a role similar to that of Nrf2 [148]. It is an ER-bound transcription factor, regulating the expression of all 26S proteasomal subunits [149], of a variety of proteins necessary for proper assembly of the proteasome as well as for the so-called unfolded-protein-response (UPR) such as Np14, Ufd1, p47, p97, Usp14 and POMP/hUMP1 [17]. Furthermore, it induces expression of the proteasomal regulator PA200/Blm10 [82], while the inducible subunits of the immunoproteasome are suppressed. Slight amounts of proteasomal inhibitors are able to induce Nrf1 [150], while larger concentrations inhibit its activation. After induction, the respective mRNA of the proteasomal system and the mentioned co-factors are increased 2- to 4-fold within about 4 h.
Both Nrf1 (intracellular half-life of about 12 min) and Nrf2 (about 10–20 min) are degraded/regulated by the 26S proteasome [138], [151].
The induction of Nrf1 is probably regulated by a reduced proteasomal activity (also consequently inducible by proteasomal inhibitors) and thus indirectly mediated by redox regulation that affects the UPS. Normally polyubiquitinated Nrf1 is quickly degraded, but after reduction of UPS-mediated proteolysis, Nrf1 is released from the ER, enters the nucleus and promotes gene expression [82]. Furthermore, an accumulation of misfolded proteins in the ER (both induced by proteasome inhibition and ER stress) [152] is able to induce Nrf1. After proteasomal inhibition, Nrf1 restores UPS-function by induction of many genes encoding 26S subunits in mammalian cells [82]. Nrf1-/- mouse embryonic fibroblasts are unable to restore UPS activity after proteasomal inhibition by the covalent proteasome inhibitor YU101 [153]. Like its homolog (Nrf2), Nrf1 recognizes antioxidant response elements (AREs) in the promoters of many UPS-encoding genes [154]. Thus, Nrf1 is indirectly induced by the consequences of redox-shifts or oxidative stress.
2.5.2. PARP-1
Besides direct binding of regulatory complexes, other enzymes are also able to modulate 20S proteasomal activity. One example is the nuclear poly(ADP-ribose)polymerase 1 (PARP-1). PARP-2 and -3 fulfill similar functions in the cell, but PARP-1 shows the highest activity among the members of this family, responding to DNA-damage (both single and double strand breaks), that may result from increased cellular ROS-formation. PARP-1 poly(ADP-ribosyl)ates several protein targets such as (undamaged) histones, NF-κB, p53, DNA-topoisomerases, DNA-PKcs and also PARP-1 itself. PARP-1-mediated ADP-ribosylation enhances proteasomal activity about 18-fold within 15 min after an oxidative stress event [155].
In the nucleus of aged mammalian cells, the proteasomal activity is almost not affected by the aging process [156], [157], in contrast to the cytosolic proteasomal activity, where aggregates of damaged proteins are inhibiting the proteolysis (see below). However, the PARP-1 mediated activation of the nuclear 20S proteasome declines during cellular senescence [158], [159].
3. Lipofuscin and AGEs: the long-term consequence of disturbed proteostasis
The degradation of both, damaged proteins (mainly by 20S/26S) and dysfunctional organelles (autophagy) are essential for cellular function and survival. Loss of function of one or both systems will result in the accumulation of damaged proteins, forming aggregates as well as an increased formation of ROS. Highly covalent cross-linked protein aggregates are referred to as lipofuscin, containing cross-linked protein material, carbohydrates as well as peroxidized lipids. Lipofuscin is extremely resistant to mammalian proteases. Furthermore, lipofuscin is able to incorporate up to 2% of redox active transition metals such as copper and iron, both able to generate ROS via Fenton chemistry. The ROS formation in this case might be very low, but over years or decades, even slight increases will add up to detrimental effects and extensive accumulation of lipofuscin. Other products of oxidative modification of cell compounds are the so-called AGEs (advanced glycation end products), resulting from chemical reaction of proteins with reducing carbohydrates as well as the so-called ALEs (advanced lipid peroxidation end products), resulting from the reaction of proteins with products of lipid peroxidation.
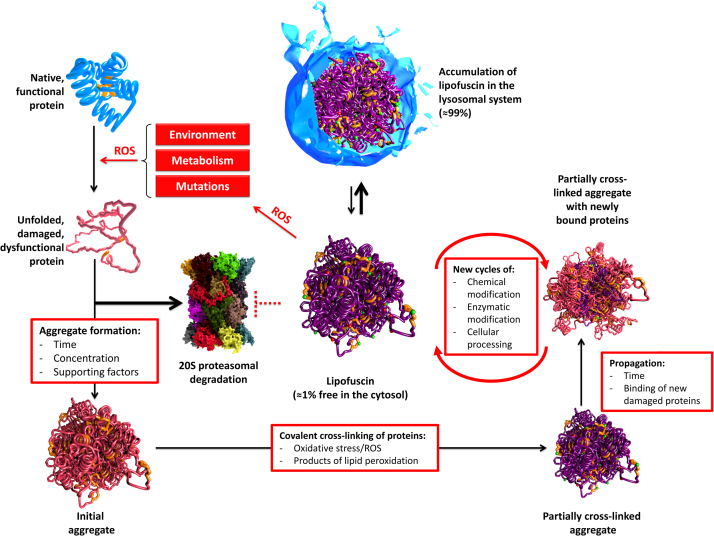
If a fully functional protein becomes oxidized, it may partially unfold and tend to form aggregates. However, proteasomal degradation considerably prevents aggregate formation. If the proteasomal degradation fails, the protein tends to aggregate with other unfolded proteins due to their exposed hydrophobic structures, driven by hydrophobic interactions (Fig. 9), forming initial aggregates (Fig. 9, bottom left), which become further oxidized during their life time. The result is a cross-linked aggregate (Fig. 9, bottom right). This aggregate can grow through binding of additional partially unfolded proteins over time that can also become further oxidized and covalently cross-linked. With time, a highly cross-linked, autofluorescent material – lipofuscin – is formed. Lipofuscin is taken up into the lysosomal system – more than 99% of lipofuscin is co-localized with lysosomes, only about 1% is found in the cytosol [160]. It is highly likely, that lipofuscin is released due to lysosomal membrane rupture that can be induced by oxidative damage. In the cytosol, lipofuscin can inhibit the proteasome [161], as well as form ROS by incorporation of transition metals as described above, thus providing the conditions for its own enhanced formation. While mitotic aging cells such as those in mucous tissues are able to “dilute” lipofuscin by ongoing cell division, it accumulates especially in postmitotic tissues during aging as observed in skeletal muscle and brain. Lipofuscin can fill up to 75% of the volume of the motor-neurons in centenarians and occupy up to 40% of the cytosolic volume in aged animals [162]. It was suggested that the remaining life span of a cell correlates negatively with the amount of lipofuscin, and that lipofuscin is one of the main life span limiting factors, not only resulting from oxidative stress/inflammation, but also from non-pathologic metabolism of the cell.
Fig. 9.
Ongoing protein oxidation leads to lipofuscin formation. Protein oxidation by various sources will lead to the unfolding of proteins, which are either degraded by the proteasome or start forming aggregates. Those initial aggregates of oxidized proteins may become further oxidized and covalently cross-linked. This is a multi-step procedure. Over time, such aggregates are incorporated into the lysosomal system. Because this material is resistant to proteases and lipases (also termed “lipofuscin”) it accumulates in the cell: about 99% are found in the lysosomal system, about 1% are found in the cytosol. Due to the ability of lipofuscin to incorporate redox-active transition metals, it is able to catalyze the Fenton reaction. Another aspect of lipofuscin’s cytotoxicity is its ability to inhibit the proteasome. The result is a decreased overall proteolytic capacity of the cell, an increased ROS formation that contributes both to lysosomal rupture, increased protein oxidation and covalent cross-linking of existing protein aggregates.
4. Aging and proteolytic systems
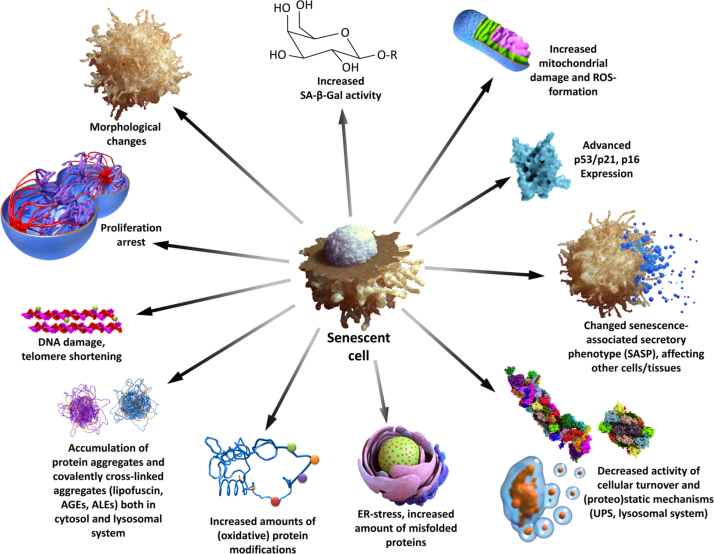
Aging is accompanied by a general decline of cellular functionality. Fig. 10 gives a short overview of changes that are induced by aging and may serve as markers.
Fig. 10.
Senescence of mammalian cells. This figure summarizes several important changes (some of them are used as experimental markers) during cellular senescence. Please note that it is not recommended using only one single marker of senescence, since every marker can be influenced by several pathologic changes.
There is no doubt about the decline of proteasomal activity in mammals during aging, although the exact reasons for this decrease are still discussed. This may be explained with changes in composition and/or structure of the proteasomal system [163], at least in part induced by modifications [164], both irreversible oxidative damage [165] and reversible (also redox-dependent) posttranslational modifications [89], [166]. Accompanying the decreased proteasomal functionality, an increase in both oxidatively damaged proteins (20S substrates) and polyubiquitinated proteins (26S substrates) was shown [167]. The increase of undegraded substrates may be due to decreased activity of the UPS or caused by an intracellular accumulation of heavily oxidized and covalently cross-linked protein aggregates (lipofuscin). Those structures are still recognized as proteasomal substrates and are found to be massively polyubiquitinated, but are very resistant to proteolytic degradation and thus, distract proteasomal activity from substrates that are still degradable. The decrease of proteolysis and the enhanced production of ROS in aged cells, result in increased formation of lipofuscin [168], [169], [170]. The age-related decline of proteasomal activity was found in several different mammalian tissues such as rat liver (− 50% of its peptidyl-glutamyl-peptide-hydrolyzing activity, located on the beta1 subunit) and rat brain: decrease of the chymotrypsin-like activity (located on beta5) in cortex, hippocampus and spinal cord of 12 month-old animals (accompanied by an increased amount of protein oxidation in those areas), but no change was found in brain stem or cerebellum [163]. In skeletal muscle, an upregulation of the immunoproteasome was detected, accompanied by accumulation of oxidized proteasomal subunits and also an induction of proteasomal activity mediated by the 19S regulator as well as a removal of polyubiquitinated substrates was found [171]. Another study revealed a 2‒3-fold increase in the levels of both beta1 and beta5 proteasomal subunits as well as of the 19S regulator subunits Rpt5 and Rpt6 in the hind limb muscles of 30 month-old Sprague-Dawley rats, the specific activity of the chymotrypsin-like site was increased by about 35% in aged animals compared to 4-month-old ones [172]. In contrast to those results, a study on F344 rats also revealed decreasing proteolytic activity (− 30% for the chymotrypsin-like one) in both 20S and 26S proteasomes, but without any change of the proteasomal amount compared to young animals [173], while in the hearts of aged rats a decrease of proteasomal mass and a loss of proteolytic capacity were detected [167]. In human BJ fibroblasts a decrease of all three proteasomal activities was found [174], [175] and detailed analysis of WI38 human fibroblasts came up with reduced expression of the active beta subunits as well as decreased assembly of functional proteasomes, since a large amount of alpha subunits was detected in a free state [176]. Also in aged retina cells, a reduced proteasomal activity was found that was reproducible by exposing young retina cells to N-ethyl-maleimide (NEM). NEM modifies the sulfhydryl-group of cysteine-residues and thus, the chymotrypsin-like activity decreased to about 65%, the peptidyl-glutamyl-peptide-hydrolyzing activity to about 80% compared to untreated samples [177].
Remarkably, the decline of the nuclear proteasomal fraction turned out to be smaller during aging, and no bulk amounts of aggregated proteins are found [21], [157], [159], [178].
In contrast, the effects of aging on the lysosomal system are quite incoherent. Induction of autophagy by caloric restriction or other interventions that affect autophagy, positively modulate life span in different organisms.
Both an increase and decrease of Beclin1 expression were found in aged tissues [179], and inconsistencies were also observed in LC3, Atg5 and-7 expressions and function. Also in lysosomes, lipofuscin is able to distract proteolytic capacity, significantly impairing lysosomal function. Changes in membrane-standing LAMP-2A-concentration were found, not because of reduced expression, but due to changes in the lysosomal membrane [180], [181]. Macroautophagy was found to be impaired in old murine brain tissue as well as in senescent human fibroblasts [170]. Moreover the degradation/turnover of certain proteins (in this case ferritin H) was found to be diminished in senescent human fibroblasts [182] which may also interfere with iron-metabolism, storage and transport, again contributing to increased oxidative stress (Fenton). Furthermore, the cross-talk between UPS and lysosomal system is affected in aged cells. Normally, macroautophagy becomes up-regulated when CMA is defective [183] and autophagy is induced by inhibition of the UPS [184]. Dysregulation of this cross-talk may result in changes of proteostasis or reduced turnover of organelles which could be responsible for alteration in the expression-pattern of autophagy-related proteins.
5. Future directions
The regulation of the cellular protein pool (proteostasis, mainly regulated via UPS) and turnover of organelles (mainly via lysosomal system and autophagy) are essential for cellular function. Aging is associated with a functional decline of those lytic systems (UPS and lysosomal system).
Especially the ability of a cell to preserve protein homeostasis (proteostasis) was recently shown to be essential in order to maintain physiological/organismal balance and functionality. Overexpression of the proteasomal β5 subunit in Caenorhabditis elegans resulted in both life span extension and increased resistance to oxidative stress (in this case mediated by the agents paraquat and juglone) [185]; similar results were found in human fibroblasts, where overexpression of β5 also increased the amount of 20S proteasomes, resulting in enhanced resistance to oxidative stress [186]. In nematode models of Alzheimer’s and Huntington’s disease, overexpression of β5 also induced resistance to proteotoxic stress pointing to a potential protective role of proteasomal activation in treatment of such disorders that result from proteostatic failure [185]. Accordingly, nematodes with decreased proteasomal activity were much more susceptible to proteotoxic insults [187].
In another Caenorhabditis elegans model, the 19S subunit Rpn6 also turned out to be a potent factor that can increase resistance to proteotoxic stress, since its upregulation was able to delay the according deleterious effects [188].
In Drosophila melanogaster, Tonoki et al. revealed that overexpression of Rpn11 (also a subunit of the 19S regulator) was able to alleviate age-related reduction of the 26S proteasomal activity, thus leading to extended life span and decreased levels of polyubiquitinated proteins [189].
Loss of Rpn11-function resulted in reduced 26S proteasomal activity and a premature age-dependent accumulation of polyubiquitinated proteins, combined with shorter life span and enhanced neurodegenerative phenotype. Thus, the authors concluded that maintaining 26S proteasomal activity with age may extend life span and also reduce the age-related progression of neurodegenerative diseases.
In humans, proteasome activities of fibroblasts of healthy centenarians (considered as examples of successful aging) were compared to the relative activities of fibroblasts derived from donors of different ages. Importantly, the cells from centenarians possessed 20S proteasome activities that were higher as compared to the ones found in cells from a 28 years old control donor. In contrast, the activity found in the 80 years old donor was the lowest one [190].
In early passage human embryonic fibroblasts, treatment with oleuropein reduced the intracellular levels of both ROS and oxidatively damaged proteins while it retained proteasome functionality during replicative senescence. In cell cultures that were treated with oleuropein throughout their lifespan, delay of senescent morphology was recorded and life span was extended by approximately 15% [191].
In a multicellular organism (in this case Caenorhabditis elegans), the compound 18α- glycyrrhetinic acid induces proteasomal activation and decelerates both aging and progression of Alzheimer’s disease [192].
Besides of the proteasomal system, the second main proteolytic machinery, the autophagy-lysosomal system, was also shown to be involved in longevity. Induced by caloric restriction or other interventions that affect autophagy, life span can be significantly increased in different organisms. Loss of IGF (insulin-like growth factor)-signaling due to mutated daf-2 elevated autophagy and extended life span by 70% (C. elegans), brain-specific overexpression of Atg8 (involved in autophagy) improved neuronal autophagy and extended life span by > 50% in female flies (D. melanogaster) [193]. Spermidine (a polyamine compound) treatment also induced autophagy levels, resulting in life span extension of about 30% (D. melanogaster) [194]. According to Seah et al. [195], in C. elegans lipoprotein production (an important process that mediates the transport of lipids between different tissues), autophagy and lysosomal lipolysis are linked in life span modulation in a conserved manner. They described a recently uncovered mechanism that links lipoprotein production and the transcriptional induction of both autophagy [196] and lysosomal lipolysis-related genes, guiding lipid remodeling and the according signaling that is associated with long-term survival. Though, the interaction of those mechanisms is still poorly understood.
All those above mentioned data point to the importance of a functional proteostasis for both healthy aging and even extension of life span (in some organisms).
Thus, better understanding of the two main proteostatic systems and especially their functional interaction may be the key for an extension of health-span and may become a powerful tool as therapeutic approach to various pathologies, in particular the ones associated with inflammation that can be considered as the most widespread future health-care problems with significant economic dimensions.
References
- 1.Sautin Y.Y., Johnson R.J. Uric acid: the oxidant-antioxidant paradox. Nucleosides Nucleotides Nucleic Acids. 2008;27:608–619. doi: 10.1080/15257770802138558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 3.Valko M., Jomova K., Rhodes C.J., Kuca K., Musilek K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016;90:1–37. doi: 10.1007/s00204-015-1579-5. [DOI] [PubMed] [Google Scholar]
- 4.La Penna G., Hureau C., Andreussi O., Faller P. Identifying, by first-principles simulations, Cu[amyloid-beta] species making Fenton-type reactions in Alzheimer's disease. J. Phys. Chem. B. 2013;117:16455–16467. doi: 10.1021/jp410046w. [DOI] [PubMed] [Google Scholar]
- 5.Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 6.Hoshi T., Heinemann S. Regulation of cell function by methionine oxidation and reduction. J. Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki S., Kodera Y., Saito T., Fujimoto K., Momozono A., Hayashi A., Kamata Y., Shichiri M. Methionine sulfoxides in serum proteins as potential clinical biomarkers of oxidative stress. Sci. Rep. 2016;6:38299. doi: 10.1038/srep38299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtman E.R., Van Remmen H., Richardson A., Wehr N.B., Levine R.L. Methionine oxidation and aging. Biochim. Biophys. Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Stadtman E.R. Protein oxidation and aging. Free Radic. Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson T.J., Lowther W.T. The peroxiredoxin repair proteins. Subcell. Biochem. 2007;44:115–141. doi: 10.1007/978-1-4020-6051-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huh J.Y., Kim Y., Jeong J., Park J., Kim I., Huh K.H., Kim Y.S., Woo H.A., Rhee S.G., Lee K.J., Ha H. Peroxiredoxin 3 is a key molecule regulating adipocyte oxidative stress, mitochondrial biogenesis, and adipokine expression. Antioxid. Redox Signal. 2012;16:229–243. doi: 10.1089/ars.2010.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handy D.E., Loscalzo J. Redox regulation of mitochondrial function. Antioxid. Redox Signal. 2012;16:1323–1367. doi: 10.1089/ars.2011.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinina E.V., Chernov N.N., Novichkova M.D. Role of glutathione, glutathione transferase, and glutaredoxin in regulation of redox-dependent processes. Biochemistry. 2014;79:1562–1583. doi: 10.1134/S0006297914130082. [DOI] [PubMed] [Google Scholar]
- 16.Mailloux R.J., Willmore W.G. S-glutathionylation reactions in mitochondrial function and disease. Front. Cell Dev. Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung T., Catalgol B., Grune T. The proteasomal system. Mol. Asp. Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Levine R.L., Berlett B.S., Moskovitz J., Mosoni L., Stadtman E.R. Methionine residues may protect proteins from critical oxidative damage. Mech. Ageing Dev. 1999;107:323–332. doi: 10.1016/s0047-6374(98)00152-3. [DOI] [PubMed] [Google Scholar]
- 19.Levine R.L., Mosoni L., Berlett B.S., Stadtman E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 21.Jung T., Engels M., Kaiser B., Poppek D., Grune T. Intracellular distribution of oxidized proteins and proteasome in HT22 cells during oxidative stress. Free Radic. Biol. Med. 2006;40:1303–1312. doi: 10.1016/j.freeradbiomed.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Jung T., Grune T. The proteasome and the degradation of oxidized proteins: Part I-structure of proteasomes. Redox Biol. 2013;1:178–182. doi: 10.1016/j.redox.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arendt C.S., Hochstrasser M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl. Acad. Sci. USA. 1997;94:7156–7161. doi: 10.1073/pnas.94.14.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jager S., Groll M., Huber R., Wolf D.H., Heinemeyer W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Biol. 1999;291:997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- 25.Orlowski M., Wilk S. Catalytic activities of the 20 S proteasome, a multicatalytic proteinase complex. Arch. Biochem. Biophys. 2000;383:1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- 26.Groll M., Bajorek M., Kohler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 27.Wenzel T., Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat. Struct. Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 28.Pacifici R.E., Salo D.C., Davies K.J. Macroxyproteinase (M.O.P.): a 670 kDa proteinase complex that degrades oxidatively denatured proteins in red blood cells. Free Radic. Biol. Med. 1989;7:521–536. doi: 10.1016/0891-5849(89)90028-2. [DOI] [PubMed] [Google Scholar]
- 29.Lasch P., Petras T., Ullrich O., Backmann J., Naumann D., Grune T. Hydrogen peroxide-induced structural alterations of RNAse A. J. Biol. Chem. 2001;276:9492–9502. doi: 10.1074/jbc.M008528200. [DOI] [PubMed] [Google Scholar]
- 30.Reeg S., Jung T., Castro J.P., Davies K.J., Henze A., Grune T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic. Biol. Med. 2016;99:153–166. doi: 10.1016/j.freeradbiomed.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karademir B., Bozaykut P., Kartal Ozer N. Heat shock proteins and proteasomal degradation in normal and tumor cells. Free Radic. Biol. Med. 2014;75(Suppl 1):S35. doi: 10.1016/j.freeradbiomed.2014.10.774. [DOI] [PubMed] [Google Scholar]
- 32.Rabl J., Smith D.M., Yu Y., Chang S.C., Goldberg A.L., Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luciani F., Kesmir C., Mishto M., Or-Guil M., de Boer R.J. A mathematical model of protein degradation by the proteasome. Biophys. J. 2005;88:2422–2432. doi: 10.1529/biophysj.104.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kish-Trier E., Hill C.P. Structural biology of the proteasome. Annu. Rev. Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wehmer M., Sakata E. Recent advances in the structural biology of the 26S proteasome. Int. J. Biochem. Cell Biol. 2016;79:437–442. doi: 10.1016/j.biocel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Bedford L., Paine S., Sheppard P.W., Mayer R.J., Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aizawa H., Kawahara H., Tanaka K., Yokosawa H. Activation of the proteasome during Xenopus egg activation implies a link between proteasome activation and intracellular calcium release. Biochem. Biophys. Res. Commun. 1996;218:224–228. doi: 10.1006/bbrc.1996.0039. [DOI] [PubMed] [Google Scholar]
- 38.Cascio P., Call M., Petre B.M., Walz T., Goldberg A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohn A., Konig J., Jung T. Metabolic Syndrome, Redox State, and the Proteasomal System. Antioxid. Redox Signal. 2016;25:902–917. doi: 10.1089/ars.2016.6815. [DOI] [PubMed] [Google Scholar]
- 40.Tsvetkov P., Myers N., Eliav R., Adamovich Y., Hagai T., Adler J., Navon A., Shaul Y. NADH binds and stabilizes the 26S proteasomes independent of ATP. J. Biol. Chem. 2014;289:11272–11281. doi: 10.1074/jbc.M113.537175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clague M.J., Heride C., Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 43.Schulman B.A., Harper J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ristic G., Tsou W.L., Todi S.V. An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Front. Mol. Neurosci. 2014;7:72. doi: 10.3389/fnmol.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzger M.B., Pruneda J.N., Klevit R.E., Weissman A.M. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashizawa A., Higashi C., Masuda K., Ohga R., Taira T., Fujimuro M. The ubiquitin system and Kaposi's sarcoma-associated Herpesvirus. Front. Microbiol. 2012;3:66. doi: 10.3389/fmicb.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao S., Ulrich H.D. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc. Natl. Acad. Sci. USA. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kravtsova-Ivantsiv Y., Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 49.Stewart D.P., Koss B., Bathina M., Perciavalle R.M., Bisanz K., Opferman J.T. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol. Cell. Biol. 2010;30:3099–3110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S., Dewille J.W. Proteasome-mediated CCAAT/enhancer-binding protein delta (C/EBPdelta) degradation is ubiquitin-independent. Biochem. J. 2007;405:341–349. doi: 10.1042/BJ20070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz A.L., Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev. Pharmacol. Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 53.Wolberger C. Mechanisms for regulating deubiquitinating enzymes. Protein Sci. 2014;23:344–353. doi: 10.1002/pro.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urbe S., Liu H., Hayes S.D., Heride C., Rigden D.J., Clague M.J. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol. Biol. Cell. 2012;23:1095–1103. doi: 10.1091/mbc.E11-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desvergne A., Ugarte N., Petropoulos I., Friguet B. Circadian modulation of proteasome activities and removal of carbonylated proteins. Free Radic. Biol. Med. 2014;75(Suppl 1):S18. doi: 10.1016/j.freeradbiomed.2014.10.631. [DOI] [PubMed] [Google Scholar]
- 56.Vriend J., Reiter R.J. Melatonin feedback on clock genes: a theory involving the proteasome. J. Pineal Res. 2015;58:1–11. doi: 10.1111/jpi.12189. [DOI] [PubMed] [Google Scholar]
- 57.Diaz-Villanueva J.F., Diaz-Molina R., Garcia-Gonzalez V. Protein folding and mechanisms of proteostasis. Int. J. Mol. Sci. 2015;16:17193–17230. doi: 10.3390/ijms160817193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao I., Liu J., Li X., Luo H. REGgamma, a proteasome activator and beyond? Cell Mol. Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanker D., Waithman J., Yewdell J.W., Chen W. Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. J. Immunol. 2013;191:52–59. doi: 10.4049/jimmunol.1300802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishto M., Liepe J., Textoris-Taube K., Keller C., Henklein P., Weberruss M., Dahlmann B., Enenkel C., Voigt A., Kuckelkorn U., Stumpf M.P., Kloetzel P.M. Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur. J. Immunol. 2014;44:3508–3521. doi: 10.1002/eji.201444902. [DOI] [PubMed] [Google Scholar]
- 61.Hulpke S., Tampe R. The MHC I loading complex: a multitasking machinery in adaptive immunity. Trends Biochem. Sci. 2013;38:412–420. doi: 10.1016/j.tibs.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Zanker D., Chen W. Standard and immunoproteasomes show similar peptide degradation specificities. Eur. J. Immunol. 2014;44:3500–3503. doi: 10.1002/eji.201445272. [DOI] [PubMed] [Google Scholar]
- 63.Dubiel W., Pratt G., Ferrell K., Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 64.Di Cola D. Human erythrocyte contains a factor that stimulates the peptidase activities of multicatalytic proteinase complex. Ital. J. Biochem. 1992;41:213–224. [PubMed] [Google Scholar]
- 65.Kuehn L., Dahlmann B. Proteasome activator PA28 and its interaction with 20 S proteasomes. Arch. Biochem. Biophys. 1996;329:87–96. doi: 10.1006/abbi.1996.0195. [DOI] [PubMed] [Google Scholar]
- 66.Ferrington D.A., Gregerson D.S. Immunoproteasomes: structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caniard A., Ballweg K., Lukas C., Yildirim A.O., Eickelberg O., Meiners S. Proteasome function is not impaired in healthy aging of the lung. Aging. 2015;7:776–792. doi: 10.18632/aging.100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarthy M.K., Weinberg J.B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Deventer S., Neefjes J. The immunoproteasome cleans up after inflammation. Cell. 2010;142:517–518. doi: 10.1016/j.cell.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 70.Johnston-Carey H.K., Pomatto L.C., Davies K.J. The Immunoproteasome in oxidative stress, aging, and disease. Crit. Rev. Biochem. Mol. Biol. 2015;51:268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agarwal A.K., Xing C., Demartino G.N., Mizrachi D., Hernandez M.D., Sousa A.B., Martinez d., V., dos Santos H.G., Garg A. PSMB8 encoding the beta5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am. J. Hum. Genet. 2010;87:866–872. doi: 10.1016/j.ajhg.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kunimoto K., Kimura A., Uede K., Okuda M., Aoyagi N., Furukawa F., Kanazawa N. A new infant case of Nakajo-Nishimura syndrome with a genetic mutation in the immunoproteasome subunit: an overlapping entity with JMP and CANDLE syndrome related to PSMB8 mutations. Dermatology. 2013;227:26–30. doi: 10.1159/000351323. [DOI] [PubMed] [Google Scholar]
- 73.Arima K., Kinoshita A., Mishima H., Kanazawa N., Kaneko T., Mizushima T., Ichinose K., Nakamura H., Tsujino A., Kawakami A., Matsunaka M., Kasagi S., Kawano S., Kumagai S., Ohmura K., Mimori T., Hirano M., Ueno S., Tanaka K., Tanaka M., Toyoshima I., Sugino H., Yamakawa A., Tanaka K., Niikawa N., Furukawa F., Murata S., Eguchi K., Ida H., Yoshiura K. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:14914–14919. doi: 10.1073/pnas.1106015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickering A.M., Lehr M., Miller R.A. Lifespan of mice and primates correlates with immunoproteasome expression. J. Clin. Investig. 2015;125:2059–2068. doi: 10.1172/JCI80514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gavilan M.P., Castano A., Torres M., Portavella M., Caballero C., Jimenez S., Garcia-Martinez A., Parrado J., Vitorica J., Ruano D. Age-related increase in the immunoproteasome content in rat hippocampus: molecular and functional aspects. J. Neurochem. 2009;108:260–272. doi: 10.1111/j.1471-4159.2008.05762.x. [DOI] [PubMed] [Google Scholar]
- 76.Stratford F.L., Chondrogianni N., Trougakos I.P., Gonos E.S., Rivett A.J. Proteasome response to interferon-gamma is altered in senescent human fibroblasts. FEBS Lett. 2006;580:3989–3994. doi: 10.1016/j.febslet.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 77.Pickering A.M., Linder R.A., Zhang H., Forman H.J., Davies K.J. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright K.L., White L.C., Kelly A., Beck S., Trowsdale J., Ting J.P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nitta T., Murata S., Sasaki K., Fujii H., Ripen A.M., Ishimaru N., Koyasu S., Tanaka K., Takahama Y. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 80.Xing Y., Jameson S.C., Hogquist K.A. Thymoproteasome subunit-beta5T generates peptide-MHC complexes specialized for positive selection. Proc. Natl. Acad. Sci. USA. 2013;110:6979–6984. doi: 10.1073/pnas.1222244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murata S., Sasaki K., Kishimoto T., Niwa S., Hayashi H., Takahama Y., Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 82.Sha Z., Goldberg A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fort P., Kajava A.V., Delsuc F., Coux O. Evolution of proteasome regulators in eukaryotes. Genome Biol. Evol. 2015;7:1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henderson A., Erales J., Hoyt M.A., Coffino P. Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 2011;286:17495–17502. doi: 10.1074/jbc.M110.212027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liepe J., Holzhutter H.G., Bellavista E., Kloetzel P.M., Stumpf M.P., Mishto M. Quantitative time-resolved analysis reveals intricate, differential regulation of standard- and immuno-proteasomes. Elife. 2015;4:e07545. doi: 10.7554/eLife.07545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Demasi M., Shringarpure R., Davies K.J. Glutathiolation of the proteasome is enhanced by proteolytic inhibitors. Arch. Biochem. Biophys. 2001;389:254–263. doi: 10.1006/abbi.2001.2332. [DOI] [PubMed] [Google Scholar]
- 87.Zmijewski J.W., Banerjee S., Abraham E. S-glutathionylation of the Rpn2 regulatory subunit inhibits 26 S proteasomal function. J. Biol. Chem. 2009;284:22213–22221. doi: 10.1074/jbc.M109.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Silva G.M., Netto L.E., Discola K.F., Piassa-Filho G.M., Pimenta D.C., Barcena J.A., Demasi M. Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. FEBS J. 2008;275:2942–2955. doi: 10.1111/j.1742-4658.2008.06441.x. [DOI] [PubMed] [Google Scholar]
- 89.Hohn T.J., Grune T. The proteasome and the degradation of oxidized proteins: part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ullrich O., Reinheckel T., Sitte N., Hass R., Grune T., Davies K.J. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc. Natl. Acad. Sci. USA. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szabo E., Kovacs I., Grune T., Haczku A., Virag L. PARP-1: a new player in the asthma field? Allergy. 2011;66:811–814. doi: 10.1111/j.1398-9995.2011.02551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jung T., Hohn A., Grune T. The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation. Redox Biol. 2014;2:99–104. doi: 10.1016/j.redox.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shang F., Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pickering A.M., Davies K.J. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012;523:181–190. doi: 10.1016/j.abb.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davies K.J. Adaptive homeostasis. Mol. Asp. Med. 2016;49:1–7. doi: 10.1016/j.mam.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Piao S., Amaravadi R.K. Targeting the lysosome in cancer. Ann. N.Y. Acad. Sci. 2016;1371:45–54. doi: 10.1111/nyas.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mony V.K., Benjamin S., O'Rourke E.J. A lysosome-centered view of nutrient homeostasis. Autophagy. 2016;12:619–631. doi: 10.1080/15548627.2016.1147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsukamoto S., Hara T., Yamamoto A., Ohta Y., Wada A., Ishida Y., Kito S., Nishikawa T., Minami N., Sato K., Kokubo T. Functional analysis of lysosomes during mouse preimplantation embryo development. J. Reprod. Dev. 2013;59:33–39. doi: 10.1262/jrd.2012-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li W., Yang Q., Mao Z. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell Mol. Life Sci. 2011;68:749–763. doi: 10.1007/s00018-010-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stoka V., Turk V., Turk B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res. Rev. 2016 doi: 10.1016/j.arr.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 102.Konig J., Besoke F., Stuetz W., Malarski A., Jahreis G., Grune T., Hohn A. Quantification of age-related changes of alpha-tocopherol in lysosomal membranes in murine tissues and human fibroblasts. Biofactors. 2016;42:307–315. doi: 10.1002/biof.1274. [DOI] [PubMed] [Google Scholar]
- 103.De Duve C. The lysosome. Sci. Am. 1963;208:64–72. doi: 10.1038/scientificamerican0563-64. [DOI] [PubMed] [Google Scholar]
- 104.Feng Y., He D., Yao Z., Klionsky D.J. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Esclatine A., Chaumorcel M., Codogno P. Macroautophagy signaling and regulation. Curr. Top. Microbiol. Immunol. 2009;335:33–70. doi: 10.1007/978-3-642-00302-8_2. [DOI] [PubMed] [Google Scholar]
- 106.Gallagher L.E., Williamson L.E., Chan E.Y. Advances in autophagy regulatory mechanisms. Cells. 2016;5 doi: 10.3390/cells5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Svenning S., Johansen T. Selective autophagy. Essays Biochem. 2013;55:79–92. doi: 10.1042/bse0550079. [DOI] [PubMed] [Google Scholar]
- 108.Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 109.Shaik A., Schiavi A., Ventura N. Mitochondrial autophagy promotes healthy aging. Cell Cycle. 2016;15:1805–1806. doi: 10.1080/15384101.2016.1181876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bauckman K.A., Owusu-Boaitey N., Mysorekar I.U. Selective autophagy: xenophagy. Methods. 2015;75:120–127. doi: 10.1016/j.ymeth.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheong H., Klionsky D.J. mTORC1 maintains metabolic balance. Cell Res. 2015;25:1085–1086. doi: 10.1038/cr.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gallagher L.E., Chan E.Y. Early signalling events of autophagy. Essays Biochem. 2013;55:1–15. doi: 10.1042/bse0550001. [DOI] [PubMed] [Google Scholar]
- 113.Kim J., Kim Y.C., Fang C., Russell R.C., Kim J.H., Fan W., Liu R., Zhong Q., Guan K.L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huber L.A., Teis D. Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 2016;39:8–14. doi: 10.1016/j.ceb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 115.Klionsky D.J., Eskelinen E.L., Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes... wait, I'm confused. Autophagy. 2014;10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xilouri M., Stefanis L. Chaperone mediated autophagy in aging: starve to prosper. Ageing Res. Rev. 2016 doi: 10.1016/j.arr.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 117.Morozova K., Clement C.C., Kaushik S., Stiller B., Arias E., Ahmad A., Rauch J.N., Chatterjee V., Melis C., Scharf B., Gestwicki J.E., Cuervo A.M., Zuiderweg E.R., Santambrogio L. Structural and biological interaction of hsc-70 protein with phosphatidylserine in endosomal microautophagy. J. Biol. Chem. 2016;291:18096–18106. doi: 10.1074/jbc.M116.736744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cell Mol. Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cuervo A.M., Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaushik S., Cuervo A.M. Chaperones in autophagy. Pharmacol. Res. 2012;66:484–493. doi: 10.1016/j.phrs.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stricher F., Macri C., Ruff M., Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. 2013;9:1937–1954. doi: 10.4161/auto.26448. [DOI] [PubMed] [Google Scholar]
- 122.Bandyopadhyay U., Kaushik S., Varticovski L., Cuervo A.M. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan Y., Pan S.S., Shen Y.J. Cardioprotection of exercise preconditioning involving heat shock protein 70 and concurrent autophagy: a potential chaperone-assisted selective macroautophagy effect. J. Physiol. Sci. 2016 doi: 10.1007/s12576-016-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ulbricht A., Gehlert S., Leciejewski B., Schiffer T., Bloch W., Hohfeld J. Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy. 2015;11:538–546. doi: 10.1080/15548627.2015.1017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jadhav T.S., Wooten M.W., Wooten M.C. Mining the TRAF6/p62 interactome for a selective ubiquitination motif. BMC Proc. 2011;5(Suppl 2):S4. doi: 10.1186/1753-6561-5-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Phadwal K. Analyzing the colocalization of MAP1LC3 and lysosomal markers in primary cells. Cold Spring Harb. Protoc. 2015;2015 doi: 10.1101/pdb.prot086272. (2015:pdb prot086272) [DOI] [PubMed] [Google Scholar]
- 128.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bryan H.K., Olayanju A., Goldring C.E., Park B.K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013;85:705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 130.Huang Y., Li W., Su Z.Y., Kong A.N. The complexity of the Nrf2 pathway: beyond the antioxidant response. J. Nutr. Biochem. 2015;26:1401–1413. doi: 10.1016/j.jnutbio.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Theodore M., Kawai Y., Yang J., Kleshchenko Y., Reddy S.P., Villalta F., Arinze I.J. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 2008;283:8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li W., Khor T.O., Xu C., Shen G., Jeong W.S., Yu S., Kong A.N. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008;76:1485–1489. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salazar M., Rojo A.I., Velasco D., de Sagarra R.M., Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 134.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pickering A.M., Linder R.A., Zhang H., Forman H.J., Davies K.J. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kwak M.K., Wakabayashi N., Greenlaw J.L., Yamamoto M., Kensler T.W. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jang J., Wang Y., Kim H.S., Lalli M.A., Kosik K.S. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32:2616–2625. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wardyn J.D., Ponsford A.H., Sanderson C.M. Dissecting molecular cross-talk between Nrf2 and NF-kappaB response pathways. Biochem. Soc. Trans. 2015;43:621–626. doi: 10.1042/BST20150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect. Biol. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 142.Mauro C., Leow S.C., Anso E., Rocha S., Thotakura A.K., Tornatore L., Moretti M., De Smaele E., Beg A.A., Tergaonkar V., Chandel N.S., Franzoso G. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Manea A., Manea S.A., Gafencu A.V., Raicu M. Regulation of NADPH oxidase subunit p22(phox) by NF-kB in human aortic smooth muscle cells. Arch. Physiol. Biochem. 2007;113:163–172. doi: 10.1080/13813450701531235. [DOI] [PubMed] [Google Scholar]
- 144.Rom O., Reznick A.Z. The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic. Biol. Med. 2016;98:218–230. doi: 10.1016/j.freeradbiomed.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 145.Liu G.H., Qu J., Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim. Biophys. Acta. 2008;1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 146.Ben-Neriah Y., Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 147.Kim J.E., You D.J., Lee C., Ahn C., Seong J.Y., Hwang J.I. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell. Signal. 2010;22:1645–1654. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 148.Schultz M.A., Abdel-Mageed A.B., Mondal D. The nrf1 and nrf2 balance in oxidative stress regulation and androgen signaling in prostate cancer cells. Cancers. 2010;2:1354–1378. doi: 10.3390/cancers2021354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Radhakrishnan S.K., den Besten W., Deshaies R.J. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vangala J.R., Sotzny F., Kruger E., Deshaies R.J., Radhakrishnan S.K. Nrf1 can be processed and activated in a proteasome-independent manner. Curr. Biol. 2016;26:R834–835. doi: 10.1016/j.cub.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 153.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Steffen J., Seeger M., Koch A., Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 155.Ullrich O., Reinheckel T., Sitte N., Hass R., Grune T., Davies K.J. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc. Natl. Acad. Sci. USA. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Merker K., Sitte N., Grune T. Hydrogen peroxide-mediated protein oxidation in young and old human MRC-5 fibroblasts. Arch. Biochem. Biophys. 2000;375:50–54. doi: 10.1006/abbi.1999.1657. [DOI] [PubMed] [Google Scholar]
- 157.Merker K., Ullrich O., Schmidt H., Sitte N., Grune T. Stability of the nuclear protein turnover during cellular senescence of human fibroblasts. FASEB J. 2003;17:1963–1965. doi: 10.1096/fj.03-0177fje. [DOI] [PubMed] [Google Scholar]
- 158.Bakondi E., Catalgol B., Bak I., Jung T., Bozaykut P., Bayramicli M., Ozer N.K., Grune T. Age-related loss of stress-induced nuclear proteasome activation is due to low PARP-1 activity. Free Radic. Biol. Med. 2011;50:86–92. doi: 10.1016/j.freeradbiomed.2010.10.700. [DOI] [PubMed] [Google Scholar]
- 159.Catalgol B., Wendt B., Grimm S., Breusing N., Ozer N.K., Grune T. Chromatin repair after oxidative stress: role of PARP-mediated proteasome activation. Free Radic. Biol. Med. 2010;48:673–680. doi: 10.1016/j.freeradbiomed.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 160.Hohn A., Sittig A., Jung T., Grimm S., Grune T. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radic. Biol. Med. 2012;53:1760–1769. doi: 10.1016/j.freeradbiomed.2012.08.591. [DOI] [PubMed] [Google Scholar]
- 161.Hohn A., Jung T., Grimm S., Catalgol B., Weber D., Grune T. Lipofuscin inhibits the proteasome by binding to surface motifs. Free Radic. Biol. Med. 2011;50:585–591. doi: 10.1016/j.freeradbiomed.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 162.Yin D. Biochemical basis of lipofuscin, ceroid, and age pigment-like fluorophores. Free Radic. Biol. Med. 1996;21:871–888. doi: 10.1016/0891-5849(96)00175-x. [DOI] [PubMed] [Google Scholar]
- 163.Baraibar M.A., Friguet B. Changes of the proteasomal system during the aging process. Prog. Mol. Biol. Transl. Sci. 2012;109:249–275. doi: 10.1016/B978-0-12-397863-9.00007-9. [DOI] [PubMed] [Google Scholar]
- 164.Schmidt M., Finley D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ferrington D.A., Kapphahn R.J. Catalytic site-specific inhibition of the 20S proteasome by 4-hydroxynonenal. FEBS Lett. 2004;578:217–223. doi: 10.1016/j.febslet.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 166.Demasi M., Netto L.E., Silva G.M., Hand A., de Oliveira C.L., Bicev R.N., Gozzo F., Barros M.H., Leme J.M., Ohara E. Redox regulation of the proteasome via S-glutathionylation. Redox Biol. 2013;2:44–51. doi: 10.1016/j.redox.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bulteau A.L., Szweda L.I., Friguet B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 168.Konig J., Ott C., Hugo M., Jung T., Bulteau A.L., Grune T., Hohn A. Mitochondrial contribution to lipofuscin formation. Redox Biol. 2017;11:673–681. doi: 10.1016/j.redox.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hohn A., Grune T. Lipofuscin: formation, effects and role of macroautophagy. Redox Biol. 2013;1:140–144. doi: 10.1016/j.redox.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ott C., Konig J., Hohn A., Jung T., Grune T. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 2016;10:266–273. doi: 10.1016/j.redox.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Husom A.D., Peters E.A., Kolling E.A., Fugere N.A., Thompson L.V., Ferrington D.A. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 172.Altun M., Besche H.C., Overkleeft H.S., Piccirillo R., Edelmann M.J., Kessler B.M., Goldberg A.L., Ulfhake B. Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J. Biol. Chem. 2010;285:39597–39608. doi: 10.1074/jbc.M110.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hayashi T., Goto S. Age-related changes in the 20S and 26S proteasome activities in the liver of male F344 rats. Mech. Ageing Dev. 1998;102:55–66. doi: 10.1016/s0047-6374(98)00011-6. [DOI] [PubMed] [Google Scholar]
- 174.Sitte N., Merker K., Von Zglinicki T., Davies K.J., Grune T. Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: Part II--aging of nondividing cells. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2000;14:2503–2510. doi: 10.1096/fj.00-0210com. [DOI] [PubMed] [Google Scholar]
- 175.Sitte N., Merker K., Von Zglinicki T., Grune T., Davies K.J. Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: Part I--effects of proliferative senescence. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2000;14:2495–2502. doi: 10.1096/fj.00-0209com. [DOI] [PubMed] [Google Scholar]
- 176.Chondrogianni N., Stratford F.L., Trougakos I.P., Friguet B., Rivett A.J., Gonos E.S. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 177.Kapphahn R.J., Bigelow E.J., Ferrington D.A. Age-dependent inhibition of proteasome chymotrypsin-like activity in the retina. Exp. Eye Res. 2007;84:646–654. doi: 10.1016/j.exer.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jung T., Hohn A., Catalgol B., Grune T. Age-related differences in oxidative protein-damage in young and senescent fibroblasts. Arch. Biochem. Biophys. 2009;483:127–135. doi: 10.1016/j.abb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 179.Carroll B., Hewitt G., Korolchuk V.I. Autophagy and ageing: implications for age-related neurodegenerative diseases. Essays Biochem. 2013;55:119–131. doi: 10.1042/bse0550119. [DOI] [PubMed] [Google Scholar]
- 180.Zhang C., Cuervo A.M. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kiffin R., Kaushik S., Zeng M., Bandyopadhyay U., Zhang C., Massey A.C., Martinez-Vicente M., Cuervo A.M. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J. Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 182.Ott C., Konig J., Hohn A., Jung T., Grune T. Reduced autophagy leads to an impaired ferritin turnover in senescent fibroblasts. Free Radic. Biol. Med. 2016;101:325–333. doi: 10.1016/j.freeradbiomed.2016.10.492. [DOI] [PubMed] [Google Scholar]
- 183.Kaushik S., Massey A.C., Mizushima N., Cuervo A.M. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Korolchuk V.I., Menzies F.M., Rubinsztein D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 185.Chondrogianni N., Georgila K., Kourtis N., Tavernarakis N., Gonos E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2015;29:611–622. doi: 10.1096/fj.14-252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chondrogianni N., Tzavelas C., Pemberton A.J., Nezis I.P., Rivett A.J., Gonos E.S. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J. Biol. Chem. 2005;280:11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 187.Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C.M., Xu C.F., Neubert T.A., Mor A., Philips M.R., Ron D. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2008;105:7094–7099. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P., Manning G., Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 189.Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chondrogianni N., Petropoulos I., Franceschi C., Friguet B., Gonos E.S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 2000;35:721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 191.Katsiki M., Chondrogianni N., Chinou I., Rivett A.J., Gonos E.S. The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Res. 2007;10:157–172. doi: 10.1089/rej.2006.0513. [DOI] [PubMed] [Google Scholar]
- 192.Papaevgeniou N., Sakellari M., Jha S., Tavernarakis N., Holmberg C.I., Gonos E.S., Chondrogianni N. 18alpha-glycyrrhetinic acid proteasome activator decelerates aging and alzheimer's disease progression in caenorhabditis elegans and neuronal cultures. Antioxid. Redox Signal. 2016;25:855–869. doi: 10.1089/ars.2015.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Madeo F., Zimmermann A., Maiuri M.C., Kroemer G. Essential role for autophagy in life span extension. J. Clin. Investig. 2015;125:85–93. doi: 10.1172/JCI73946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Eisenberg T., Knauer H., Schauer A., Buttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., Fussi H., Deszcz L., Hartl R., Schraml E., Criollo A., Megalou E., Weiskopf D., Laun P., Heeren G., Breitenbach M., Grubeck-Loebenstein B., Herker E., Fahrenkrog B., Frohlich K.U., Sinner F., Tavernarakis N., Minois N., Kroemer G., Madeo F. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 195.Seah N.E., de Magalhaes Filho C.D., Petrashen A.P., Henderson H.R., Laguer J., Gonzalez J., Dillin A., Hansen M., Lapierre L.R. Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy. 2016;12:261–272. doi: 10.1080/15548627.2015.1127464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lapierre L.R., Kumsta C., Sandri M., Ballabio A., Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–880. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 198.Kastle M., Reeg S., Rogowska-Wrzesinska A., Grune T. Chaperones, but not oxidized proteins, are ubiquitinated after oxidative stress. Free Radic. Biol. Med. 2012;53:1468–1477. doi: 10.1016/j.freeradbiomed.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 199.Kastle M., Grune T. Proteins bearing oxidation-induced carbonyl groups are not preferentially ubiquitinated. Biochimie. 2011;93:1076–1079. doi: 10.1016/j.biochi.2011.03.004. [DOI] [PubMed] [Google Scholar]