Abstract
The PROTAC (proteolysis-targeting chimera) ARV-825 recruits bromodomain and extraterminal (BET) proteins to the E3 ubiquitin ligase cereblon, leading to degradation of BET proteins, including BRD4. Whereas the BET-protein inhibitor (BETi) OTX015 caused accumulation of BRD4, treatment with equimolar concentrations of ARV-825 caused sustained and profound depletion (>90%) of BRD4 and induced significantly more apoptosis in cultured and patient-derived (PD) CD34+ post-MPN sAML cells, while relatively sparing the CD34+ normal hematopoietic progenitor cells. RNA-Seq, Reversed Phase Protein Array and mass cytometry ‘CyTOF’ analyses demonstrated that ARV-825 caused greater perturbations in mRNA and protein expressions than OTX015 in sAML cells. Specifically, compared to OTX015, ARV-825 treatment caused more robust and sustained depletion of c-Myc, CDK4/6, JAK2, pSTAT3/5, PIM1 and Bcl-xL, while increasing the levels of p21 and p27. Compared to OTX015, PROTAC ARV-771 treatment caused greater reduction in leukemia burden and further improved survival of NSG mice engrafted with luciferase-expressing HEL92.1.7 cells. Co-treatment with ARV-825 and JAK inhibitor ruxolitinib was synergistically lethal against the established and PD-CD34+ sAML cells. Notably, ARV-825 induced high levels of apoptosis in the in vitro generated ruxolitinib-persister or ruxolitinib-resistant sAML cells. These findings strongly support the in vivo testing of the BRD4-PROTAC based combinations against post-MPN sAML.
Keywords: Secondary AML, BRD4, PROTAC, protein degradation
INTRODUCTION
Hematopoietic stem/progenitor cells (HPCs) of BCR-ABL1-negative myeloproliferative neoplasms with myelofibrosis (MPN-MF) exhibit mutations in JAK2, c-MPL, or calreticulin (CALR) gene and display constitutive activation of JAK-STAT signaling 1,2. In MPN-MF, transformation to AML (sAML) occurs in up to 20% of patients3. Ruxolitinib (Rux), a type I, ATP-competitive, dual JAK1/2 inhibitor (JAKi), is currently used as the therapy of MPN-MF 4,5. Although treatment with ruxolitinib confers notable clinical benefit in MPN-MF, it exhibits only modest activity and does not significantly impact the clinical outcome in post-MPN sAML3,6–8. Prolonged exposure to ruxolitinib may also lead to a loss of response, causing the emergence of JAKi-resistant (JIR) sAML cells in patients 9,10. Although they lack additional JAK2 mutations, JIR cells exhibit reactivation of JAK-STAT signaling due to trans-phosphorylation of JAK2 by JAK1 or TYK2 tyrosine kinases (TKs)11. Sequential genomic assessments in pre- and post-sAML transformation have revealed mutations in TET2, ASXL1, IDH1&2, SRSF2, RUNX1, MYC, PTPN11, NRAS, SETBP1 and TP53 genes12,13. Because standard chemotherapy is also relatively ineffective as a therapy for sAML3,7 there is an urgent need to develop novel therapeutics for the treatment of this disease.
BET (bromodomain and extraterminal) family of proteins, including BRD4, are chromatin reader proteins14,15. They possess the N-terminal double tandem bromodomains, which bind to the acetylated lysine on nucleosomal histones and transcription factors14,15. Through their extra-terminal (ET) domain in the C-terminus, they interact and recruit co-regulatory chromatin modifying enzymes, remodeling factors and the mediator elements to the chromatin for regulating gene transcription15. The C-terminal domain (CTD) of BRD4 binds to P-TEFb (positive transcription elongation factor b); the heterodimer composed of cyclin dependent kinase 9 (CDK9) and its regulatory subunit cyclin T 16. The kinase activity of CDK9 in P-TEFb phosphorylates serine 2 in the heptad-repeats in the C-terminal domain (CTD) of RNA pol II (RNAP2), stimulating mRNA transcript elongation at the enhancers and promoters of oncogenes, especially those of c-Myc, Bcl-xL, PIM1 and CDK4/6, that are regulated by clustered or ‘super’ enhancers and are important for cell growth and survival of sAML cells 17–20. An RNAi screen identified BRD4 as an effective and promising target in AML cells 21. Several structure/activity-based BET protein small-molecule, acetyl-lysine-mimetic inhibitors (BETi) have been developed, including OTX015 and GSK525762 22,23. These agents displace BET proteins, along with the co-regulators of transcription chromatin, causing transcriptional repression of BCL-2, c-Myc, CDK4/6 22,24. Recently, we have demonstrated that treatment with BETi, e.g., JQ1 and I-BET 151, attenuates the expression of c-Myc, p-STAT5, PIM1 and CDK4/6, inhibits growth and induces apoptosis of cultured and patient-derived (PD), CD34+ sAML blast progenitor cells 25, including those that co-express JAK2 V617F and mutant TP53, e.g., HEL92.1.7 and SET2 cells 25,26. Co-treatment with BETi and ruxolitinib synergistically induced apoptosis of cultured and PD CD34+ sAML cells. As compared to treatment with vehicle control, or JQ1 or ruxolitinib as single agents, co-treatment with JQ1 and ruxolitinib significantly improved the median survival of the immune-depleted mice engrafted with HEL92.1.7 cells. Despite significant AML activity, BETi treatment leads to BRD4 protein accumulation over time27,28. Together with the reversible nature of the binding of BETi to BET proteins, this may account for sub-optimal BETi-mediated transcriptional repression of MYC, Bcl-xL, PIM1, CDK4/6 and other oncoproteins, as well as reduced apoptosis26,27. To circumvent this limitation, hetero-bifunctional PROTACs (proteolysis targeting chimera) with two recruiting ligands connected via a linker have been designed27–29. In the BET-PROTAC ARV-825 (Arvinas, Inc.), one ligand is the BRD4 binding moiety (OTX015) while the other moiety (pomalidomide) specifically hijacks and recruits the E3 ubiquitin ligase cereblon to polyubiquitylate and proteasomally degrade BETPs including BRD4 27. Unlike BETis, the BET-PROTAC can potentially act catalytically and facilitate multiple rounds of sub-stoichiometric catalysis 27–29, leading to efficient and prolonged depletion of the levels of BET proteins, including BRD4 (> 90%) in the sAML cells. Therefore, we compared the anti-sAML activity of ARV-825 with the BETi OTX015, against cultured and PD CD34+ sAML cells. Our findings demonstrate that at equimolar concentrations ARV-825 is significantly more potent than OTX015 in inducing apoptosis of sAML cells sensitive or resistant to ruxolitinib, while relatively sparing the CD34+ normal hematopoietic progenitor cells. Notably, whereas OTX015 treatment increased BRD4 levels, ARV-825 markedly depleted BRD4, as well as caused more robust attenuation of the levels of c-Myc, p-STAT5, Bcl-xL, PIM1 and CDK4/6, exerting synergistic lethality with ruxolitinib against sAML cells.
Materials and Methods
Cell lines and patient-derived (PD) post MPN-MF sAML cells
Post-MPN sAML SET2 and HEL 92.1.7 (HEL) cells expressing mutant JAK2-V617F and TP53 were obtained from the DSMZ (Braunschweig, Germany) and ATCC (Manassas, VA), respectively. UKE1 cells were obtained and characterized as previously described9. All experiments with the cell lines were performed within 6 months after thawing or obtaining from ATCC or DSMZ. HEL92.1.7 ruxolitinib-persister cells were generated by exposing the cells to 1.0 µM of ruxolitinib for 48 hours. Dead cells were removed by Ficoll Hypaque density gradient centrifugation. Live cells were expanded in drug-free media and then treated with additional rounds of identical ruxolitinib treatments and recovery. After this the viable, expanded cells were treated with the drugs for the described studies. Patient-derived (PD) peripheral blood and/or bone marrow aspirate samples were obtained with informed consent from patients with sAML and processed to separate the mononuclear and CD34+ sAML blast progenitor cells, as previously described9.
RNA purification, sequencing and quantitative polymerase chain reaction (QPCR) of sAML cells
The methods followed for RNA purification, sequencing (NGS) and qPCR analyses of RNA from the cultured and PD CD34+ sAML cells were as previously described30 and detailed in the Supplemental Methods.
Assessment of apoptosis by annexin-V staining
Untreated or drug-treated cells were stained with annexin-V (Pharmingen, San Diego, CA) and TO-PRO-3 iodide (Life Technologies, Carlsbad, CA) and the percentages of apoptotic cells were determined by flow cytometry as previously described31. For patient-derived sAML cells, loss of viability following drug treatment was determined by propidium iodide staining and flow cytometry, as previously described32.
Western blot analysis, Reverse Phase Protein Array (RPPA) and single cell mass cytometry (CyTOF) analysis
The methods followed for performing the Western blot, RPPA and CyTOF analyses were as previously described31–35, and are detailed in the Supplemental Methods.
In vivo sAML mouse model
Luciferase-expressing sAML HEL92.1.7 cells were engrafted into NSG mice and treated with OTX015 or ARV-771 as described in the Supplemental Methods. The survival of the mice is represented by a Kaplan-Meier plot.
Statistical Analysis
Significant differences between values obtained in a population of sAML cells treated with different experimental conditions were determined using a one-tailed, unpaired t-test. P values of less than 0.05 were assigned significance.
RESULTS
Compared to BETi, treatment with BET-PROTAC causes profound and sustained depletion of BRD4 in sAML cells
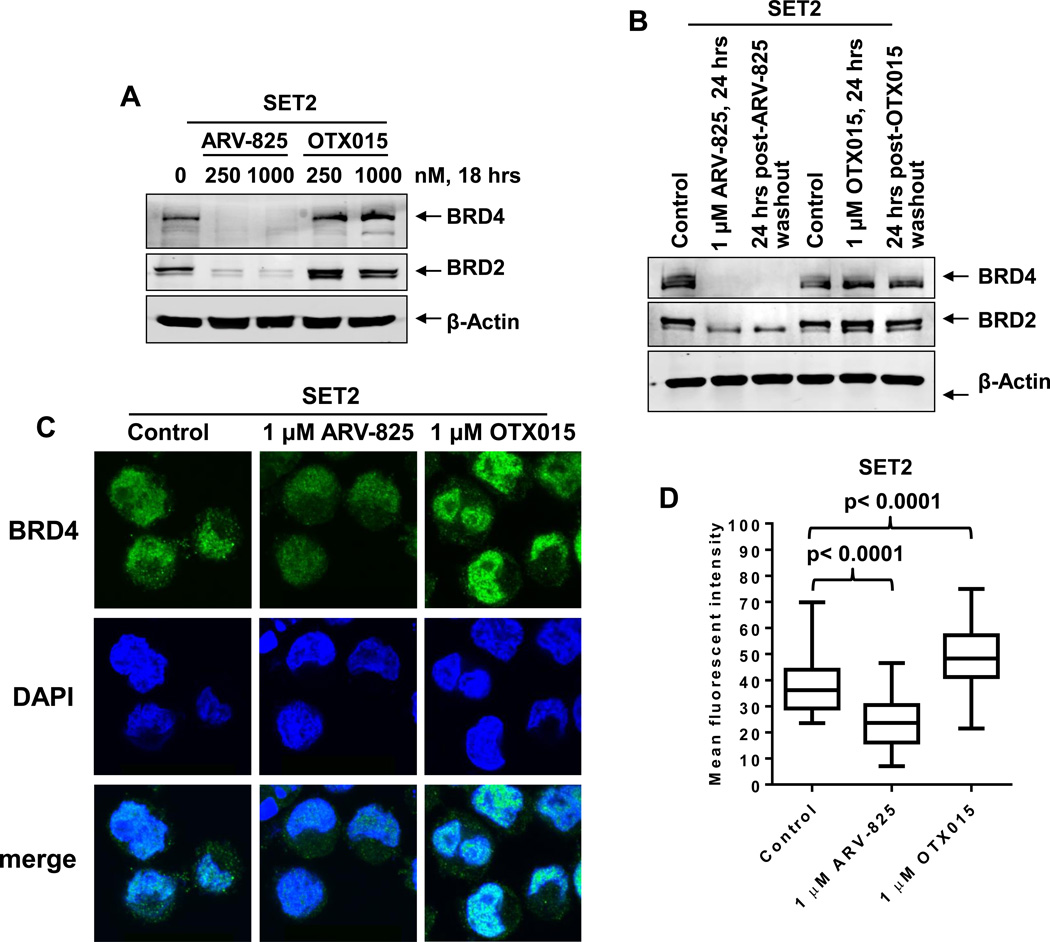
We first compared the effect of treatment with the BETi OTX015 versus ARV-825 on the levels of BET proteins BRD4 and BRD2 in sAML SET2 cells. As has been previously demonstrated in Burkitt’s Lymphoma (BL) cells lines27, in contrast to OTX015 which caused accumulation and increased levels of BRD4 protein, treatment with ARV-825 caused marked depletion of the levels of BRD4 and BRD2 in sAML SET2 cells (Figure 1A and 1B). The BETi JQ1 treatment also induced BRD4 levels in sAML HEL92.1.7 and SET2 cells (Supplemental Figure S1A). Confocal immunofluorescent microscopy also demonstrated that treatment with ARV-825 significantly depleted the nuclear expression of BRD4 in sAML cells, while OTX015 treatment increased BRD4 nuclear expression (Figure 1C and 1D). Whereas treatment with pomalidomide had no effect on the levels of BRD4 and BRD2, co-treatment with pomalidomide significantly restored the ARV-825-mediated depletion of BRD4 and BRD2 levels (Supplemental Figure S1B and S1C). We also assessed BRD4 and BRD2 levels in sAML cells treated with equimolar (1.0 μM) concentrations of ARV-825 or OTX015 for 24 hours, followed by compound washout, replenishment of the cells with fresh medium and incubating them for an additional 24 hours in drug-free medium. As shown in Figure 1B, compared to the high expression of BRD4 and BRD2 in the OTX015-treated post-washout cells, prior treatment with ARV-825 caused a sustained depletion of BRD4 and BRD2 in the post-washout cells. This implies that the sustained nuclear depletion of the levels of BRD4 mediated by exposure to ARV-825 would undermine the transcription-promoting effects of BRD4 at the enhancers and promoters.
Figure 1. Treatment with ARV-825 causes efficient and sustained depletion of BET protein expressions in sAML cells.
A. SET-2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 18 hours. Total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control. B. SET2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 24 hours. At the end of treatment, half of the cells were collected and snap frozen in liquid nitrogen. The remaining cells were washed three times in serum-free RPMI-1640 media to remove the drug (drug washout) and re-plated in complete media containing no drug for an additional 24 hours. Following this, total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control. C. SET2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 16 hours. Then, cells were cytospun onto glass slides and immunostained for BRD4 expression. Cells were counterstained with DAPI and images were acquired utilizing confocal microscopy. Original magnification is 100X. D. Median fluorescent intensity of nuclear BRD4 staining in SET2 cells. At least 20 cells in each condition were quantified. Statistical differences in the median fluorescent intensity are noted on the boxplot.
BET-PROTAC is more potent than BETi in inducing apoptosis of post-MPN cultured and patient-derived (PD) CD34+ sAML cells
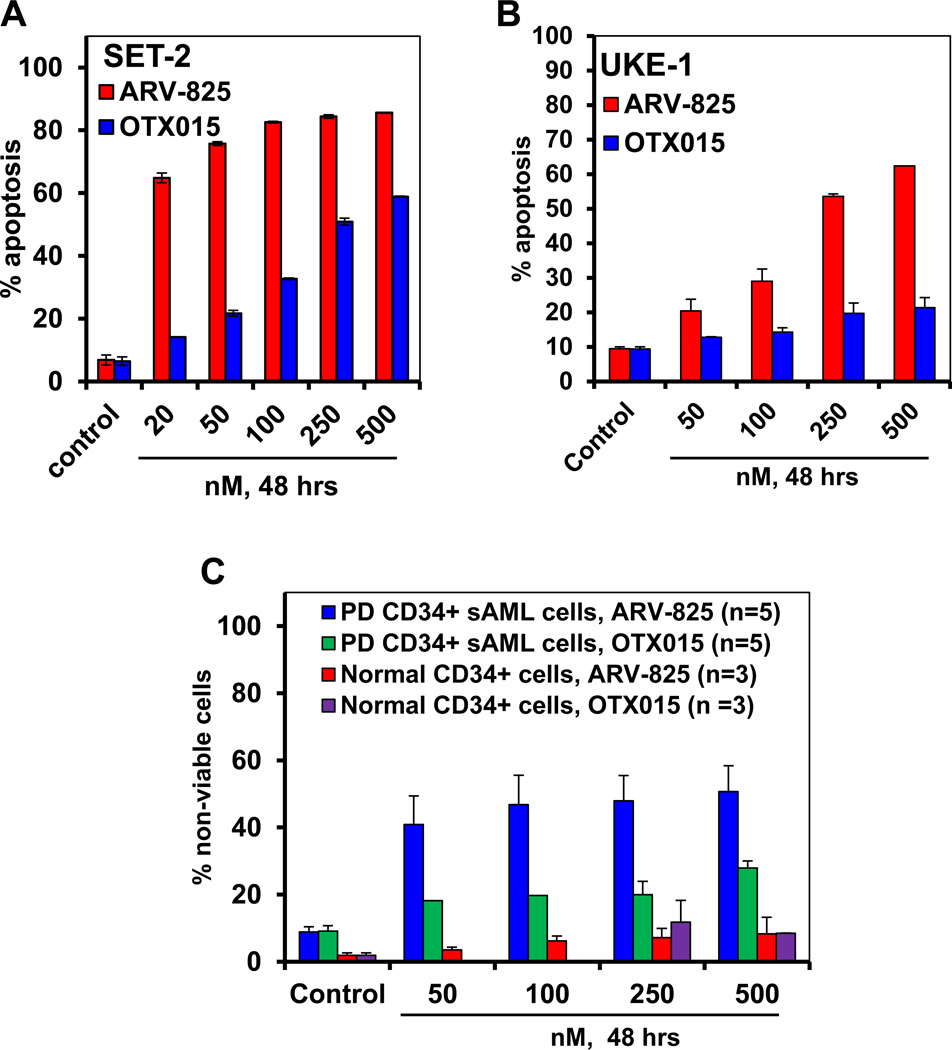
We next compared the effects of BET-PROTAC ARV-825 versus the BETi OTX015 in inducing apoptosis of cultured sAML SET2 and UKE1 cells. At equimolar concentrations, ARV-825 dose-dependently induced significantly more apoptosis than OTX015 in SET2 (ARV-825 IC50= 14.5 ± 0.2 nM; OTX015 IC50= 256 ± 21 nM) and UKE1 (ARV-825 IC50= 256 ± 12.45 nM; OTX015 IC50= >500 nM) cells (Figure 2A and 2B). SET2 cells were more sensitive than UKE1 cells to ARV-825 and OTX015-induced apoptosis. It is notable that compared to SET2, UKE1 cells are also endogenously less sensitive to ruxolitinib-induced apoptosis25. We next determined the apoptotic effects of ARV-825 versus OTX015 against CD34+ sAML cells procured from the bone marrow aspirates and/or peripheral blood of five patients with post-MPN sAML. As demonstrated in Figure 2C, again ARV-825 treatment dose-dependently induced more lethality than OTX015 in the PD CD34+ sAML cells. Notably, both ARV-825 and OTX015 induced relatively less apoptosis in the PD CD34+ sAML as compared to the cultured sAML cells types above. However, ARV-825 and OTX015 were significantly less effective in inducing apoptosis of the normal CD34+ cord blood-derived progenitors, as compared to the PD CD34+ sAML cells, indicating that the PROTAC and BETi exert in vitro selectivity against sAML cells (Figure 2C).
Figure 2. Treatment with BET PROTAC, ARV-825 induces more apoptosis and loss of cell viability than BET protein inhibitor, OTX015 in cultured and patient-derived (PD) CD34+ sAML cells.
A. SET-2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 48 hours. At the end of treatment, the % apoptotic cells were determined by flow cytometry. Columns, mean of three experiments; Bars, standard error of the mean. (The IC50 dose for ARV-825 is 14.5 ± 0.2 nM. The IC50 dose for OTX015 is 256 ± 21 nM. B. UKE1 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 48 hours. At the end of treatment, the percentages of annexin V-positive, apoptotic cells were determined by flow cytometry. Columns, mean of three experiments; Bars, standard error of the mean. (The IC50 dose for ARV-825 is 256 ± 12.45 nM. The IC50 dose for OTX015 is > 500 nM. C. Patient-derived (PD) CD34+ sAML cells and normal CD34+ cells were treated with the indicated concentrations of ARV-825 or OTX015 for 48 hours. Following this, the % of propidium iodide-positive, non-viable cells was determined by flow cytometry. Columns, mean loss of cell viability from five PD samples; Bars, standard error of the mean.
Compared to BETi treatment, BET-PROTAC induces more profound perturbation in mRNA transcript levels in sAML cells
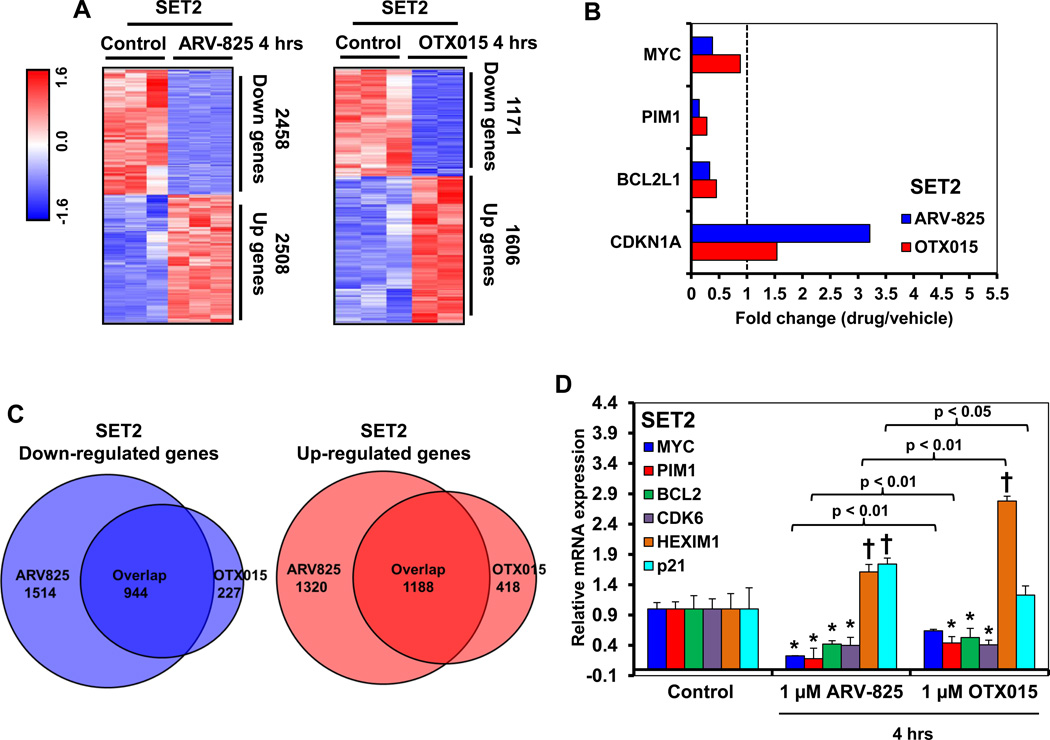
Utilizing RNA-Seq analysis, we compared the transcriptome changes induced by treatment with ARV-825 with those induced by OTX015 in the cultured sAML SET2 cells. Samples shown represent and were processed as biologic triplicates. Differential gene expression analysis showed that a larger number of genes were up- or down- regulated by the in vitro treatment of SET2 cells with ARV-825 compared to treatment with OTX015 (Figure 3A), at the same significance threshold. We also noted that for key oncogenes greater mRNA perturbations were induced by ARV-825, as tabulated in Figure 3B. Interestingly, some mRNA expressions such as MYC, PIM1 and Bcl2L1/Bcl-xL (Figure 3B), were downregulated more by ARV-825 than OTX015, whereas the mRNA of BCL2 and LMO2 were down-regulated approximately to a similar extent following treatment with ARV-825 or OTX015 (Supplemental Figure S2). LMO2 gene expression has been previously shown to be up regulated in advanced MPN and sAML cells36. In contrast, the mRNA levels of p21 were more up regulated by ARV-825 than OTX015, whereas β-catenin and HEXIM1 were induced to a similar level (Figure 3B and Supplemental Figure S2). HEXIM1 induction has been mechanistically linked to BETi-induced growth inhibition and apoptosis37, whereas Wnt/β-catenin induction has been shown to be an adaptive resistance mechanism in AML cells following treatment with BETi 38,39. The Venn diagram in Figure 3C shows that there was a significant overlap in the gene expressions that were up or down regulated, following treatment with ARV-825 and OTX015. Gene set enrichment analyses (GSEA) comparing the mRNA signature of ARV-825 with OTX015-treated SET2 cells with REACTOME pathways showed positively and negatively enriched pathways, which are presented in the tables in Supplemental Figures S3 and S4 as the normalized enrichment score for each pathway in the ARV-825 and OTX015 treated cells (all q-values are less than 0.25). As an example, the GSEA plot comparing the ARV-825 or OTX015-treated SET2 cells against the gene signature REACTOME pathway for RNA Pol II transcription showed that the q values for association were lower for ARV-825 (Supplemental Figure S3B and S3C). Notably, qPCR analyses confirmed that compared to OTX015, treatment with ARV-825 reduced MYC, PIM and BCL-2 and increased p21 mRNA expressions to a greater extent than OTX015 (Figure 3D). In contrast, HEXIM mRNA was induced more by OTX015 treatment. We also performed RNA-Seq and qPCR analyses, following treatment of PD CD34+ sAML cells with ARV-825 versus OTX015. ARV-825 treatment reduced MYC, IL7R, CDK6, LMO2, BCL2 and Bcl2L1/Bcl-xL, but induced p21 and CTNNB1 (β-catenin) mRNA levels to a greater extent than OTX015 in the PD CD34+ sAML cells (Supplemental Figure S5A to S5E). In PD CD34+ sAML cells, there was also a significant overlap in the gene expressions that were up or down regulated, following treatment with ARV-825 and OTX015 (Supplemental Figure S5C). Similar effects on the expression of MYC, BCL2, Bcl2L1/Bcl-xL, p21 and HEXIM1 were observed by qPCR analyses in additional PD CD34+ sAML samples (Supplemental Figure S5F to S5G).
Figure 3. Treatment with ARV-825 and OTX015 depletes the mRNA expression levels of c-Myc, PIM1, and BCL2L1/Bcl-xL but induces p21 protein expression levels in cultured and patient-derived sAML cells.
A. SET2 cells were treated with 1 µM of ARV-825 or OTX015 for 4 hours. Samples shown represent biologic triplicates. RNA-Seq analysis was performed on the isolated RNA. The heat map shows the number of mRNAs with a fold-change exceeding 1.33X in either direction (relative to the untreated control), and p-value less than 0.05, following treatment with ARV-825 or OTX015. B. The graph shows the fold change (drug-treated divided by control) of selected gene targets in ARV-825 and OTX015-treated cells. Vertical line at 1.0 indicates no fold change. C. Venn diagram representing the expression signature overlap in down and upregulated genes cells following treatment with ARV-825 and OTX015 in SET2 cells. D. SET-2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 4 hours. Data shown represent biologic triplicates. At the end of treatment, total RNA was isolated and reverse transcribed. The resulting cDNA was used for real-time, quantitative PCR analysis. The relative mRNA expression of each target was normalized to GAPDH and compared to the untreated control cells. * indicates mRNA values that are significantly depleted compared to the untreated control cells (p <0.05). † indicates mRNA values that are significantly induced compared to the untreated control cells (p <0.05).
Treatment of sAML cells with ARV-825 leads to more extensive protein-level perturbation compared to treatment with OTX015
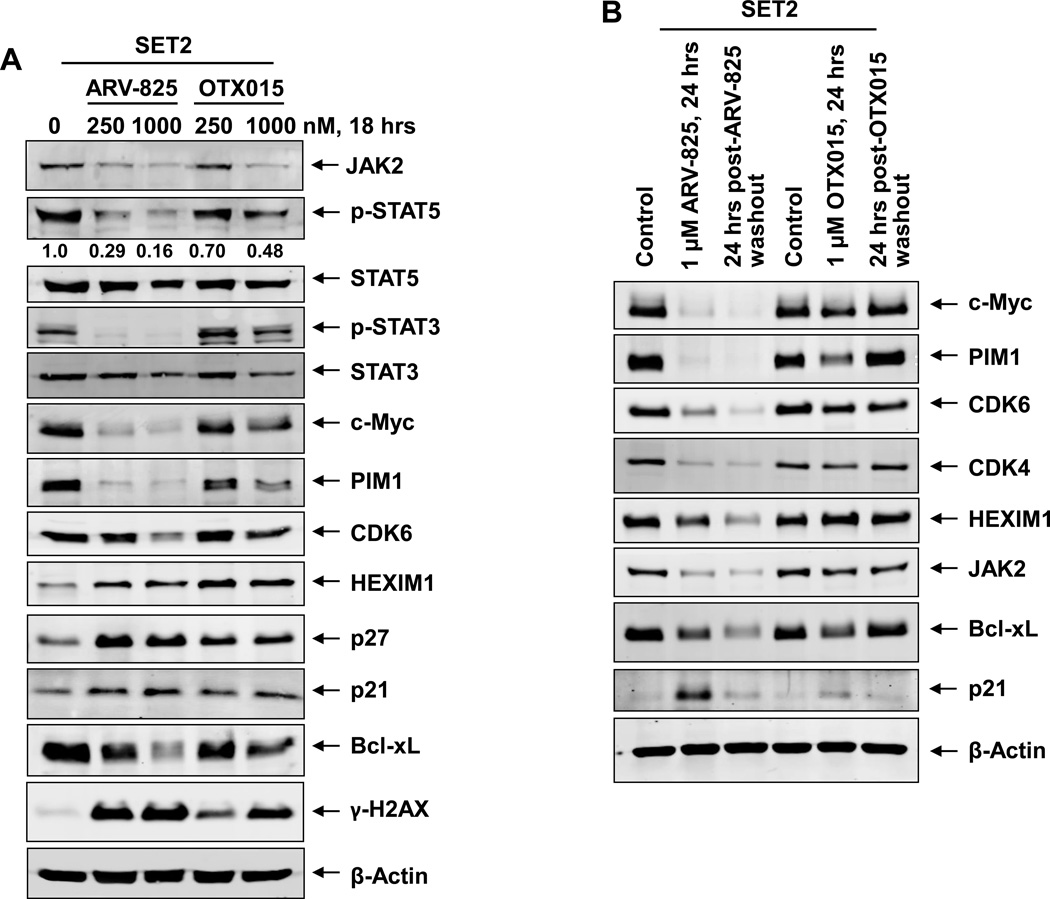
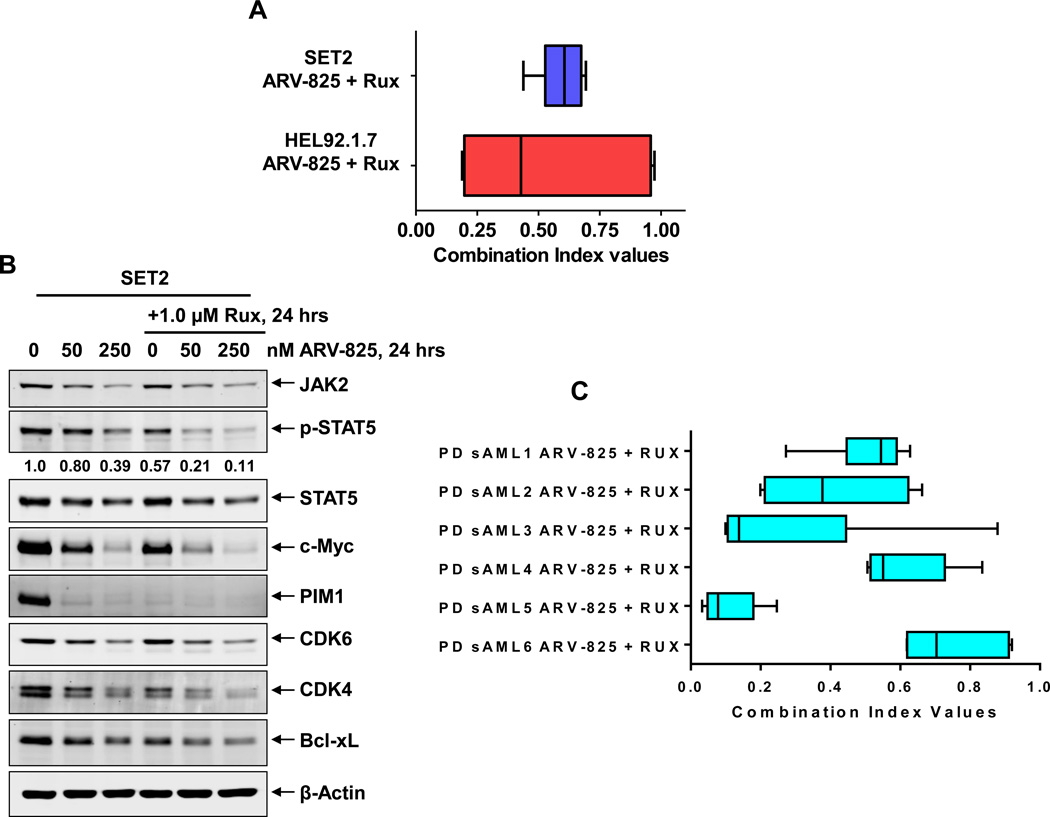
We also determined the effects of ARV-825 versus OTX015 treatment on protein expressions, utilizing specific and validated antibodies coupled to a reverse phase protein array (RPPA) 25,33. The heat maps in Supplemental Figure S6A and S6B show the changes in expression (in triplicate) of those proteins that exhibited a ≥ 1.25-fold increase or decrease in their expression and p < 0.05 (relative to the untreated cells) following treatment of SET2 cells with either 250 nM of ARV-825 or OTX015 for 18 hours, respectively. As expected, ARV-825 depleted BRD4 levels and OTX015 induced the levels of BRD4. In addition, ARV-825 treatment down and up regulated more proteins than OTX015 (Supplemental Table 1 and Supplemental Table 2). ARV-825 treatment also markedly down regulated pS6, pSTAT3, CHK1, p-Rb (surrogate for CDK4/6 down-regulation) JAK2, c-Myc and FOXM1, whereas the protein levels of DNA damage-associated γ-H2AX (H2AX pS140) as well as of the cleaved caspase 3 and 7 were up regulated by both ARV-825 and OTX015 (Supplemental Figure S6A and S6B; Supplemental Table 1 and Supplemental Table 2). Western analyses were conducted to further confirm the effects of ARV-825 versus OTX015 on the protein levels in the sAML SET2 cells. In comparison to OTX015, exposure to ARV-825 caused greater reduction in the levels of JAK2, p-STAT5, STAT5, p-STAT3, STAT3, c-Myc, PIM1, CDK6 and Bcl-xL (Figure 4A). Both ARV-825 and OTX015 treatment induced the protein expressions of HEXIM1, p27, p21 and γ-H2AX (Figure 4A). The latter represents increased DNA damage response due to loss of BRD4 function in the ARV-825 or OTX015-treated cells 40. Additionally, co-treatment with pomalidomide restored ARV-825-mediated depletion in the levels of c-Myc, PIM1, Bcl-xL and CDK4 levels, but reduced ARV-825-induced p21 levels in SET2 cells (Supplemental Figure S7A). This was associated with pomalidomide-mediated inhibition of ARV-825-induced apoptosis of SET2 cells (Supplemental Figure S7B). To determine whether treatment with ARV-825 has a lasting effect on the protein levels, following treatment of SET2 cells with equimolar concentrations of ARV-825 or OTX015 for 24 hours; cells were washed thrice with drug-free medium and incubated in media without drug for an additional 24 hours. Western analysis performed on the cell lysates following this incubation demonstrated that ARV-825–mediated depletions in the levels of c-Myc, PIM1, CDK4/6, JAK2 and Bcl-xL were sustained (Figure 4B). In contrast, after treatment with OTX015, incubation in the drug-free medium restored their levels, indicating a lack of sustained inhibition by OTX015 on the levels of BRD4 and of c-Myc, PIM1, CDK4/6, JAK2 and Bcl-xL (Figure 4B). Utilizing a recently described mass cytometry approach, we next determined the effects of equimolar concentrations of ARV-825 versus OTX015 on the PD CD34+ sAML cells that were immuno-phenotypically characterized as early stem/progenitor cells34, 35. A bioinformatics algorithm, SPADE (Spanning-tree Progression Analysis of Density-normalized Events; http://pengqiu.gatech.edu/software/SPADE/), was utilized on the mass cytometry data, using 4 cell surface markers, to define the cluster that represented the stem/progenitor cells, based on high expression of CD123, CD244, TIM3Fc+ and CD90 (Supplemental Figure S8A)41. As compared to the untreated control, ARV-825 treatment more potently reduced the expression of BRD4, c-Myc, and p-Rb while inducing the levels of p21 in the sAML stem/progenitor cells (Supplemental Figure S8B).
Figure 4. Treatment with ARV-825 causes efficient and sustained depletion of BRD4 target gene expressions in sAML cells.
A. SET-2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 18 hours. Total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control. B. SET-2 cells were treated with the indicated concentrations of ARV-825 or OTX015 for 24 hours. At the end of treatment, half of the cells were collected, washed with 1X PBS and snap frozen in liquid nitrogen. The remaining cells were washed three times in serum-free RPMI-1640 media to remove the drug (drug washout) and re-plated in complete media containing no drug for an additional 24 hours. Following this, total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control.
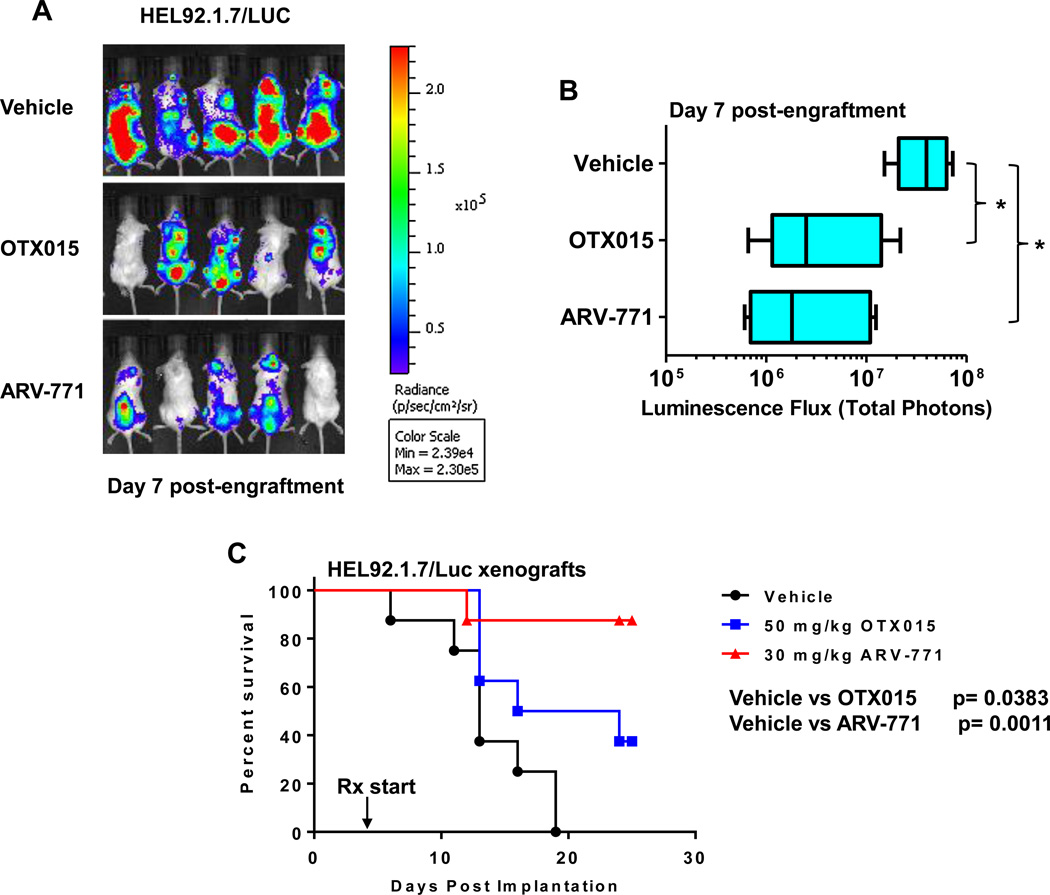
BET-PROTAC ARV-771 is more potent than BETi OTX015 in reducing the in vivo sAML burden and improving survival of NSG mice engrafted with luciferase transduced sAML cells
Following engraftment of luciferase-transduced sAML HEL92.1.7 cells into pre-irradiated NSG mice, we also compared the effects of treatment with ARV-771, a novel BET-PROTAC with superior in vivo pharmacology that recruits the E3 ligase VHL (Von Hippel-Lindau) to degrade BETPs, versus OTX015 or vehicle control on the sAML burden and survival of the mice. Figure 5A and 5B demonstrate that treatment with ARV-771 was more effective than OTX015 in reducing the bioluminescence in the NSG mice due to the sAML cells, as determined 7 days after engraftment of the sAML cells. Notably, compared to treatment with OTX015, ARV-771 was significantly more effective in improving the median survival of the NSG mice, as depicted in the Kaplan Meier plot in Figure 5C (p < 0.05). Whereas treatment with OTX015 appreciably reduced the weight of the NSG mice, treatment with ARV-771 had an insignificant effect on their weight (p = 0.16) (Supplemental Figure S9).
Figure 5. BET-PROTAC ARV-771 is more potent than BETi OTX015 in reducing the in vivo sAML burden and improving survival of NSG mice engrafted with luciferase transduced sAML cells.
A. NSG mice were implanted with luciferase-expressing HEL92.1.7 cells and monitored for 4 days. Mice were imaged to document engraftment of leukemia and then treated with vehicle, OTX015 or ARV771 as indicated for one week. At the end of treatment, mice were imaged with a Xenogen camera and total photon counts were recorded. B. Total photon counts in mice treated for one week with vehicle, OTX015 or ARV771 as determined by bioluminescent imaging. * indicates total photon counts that were significantly less in the OTX015 and ARV-771-treated mice compared to the vehicle-treated mice (p < 0.05). C. Kaplan-Meier plot of the in vivo activity of OTX015 or ARV771 against sAML HEL92.1.7 xenografts in NSG mice. Significance between OTX015 and ARV-771-treated versus vehicle-treated mice was determined by a Mantel-Cox Rank Sum test. P-values less than 0.05 were considered to be significant.
Co-treatment with BET-PROTAC and ruxolitinib exerts synergistic lethality in cultured and PD CD34+ sAML cells
Next, we determined the activity of co-treatment with ARV-825 and JAKi ruxolitinib against cultured SET2, HEL92.1.7 and PD CD34+ sAML cells expressing JAK2-V617F mutation. Figure 6A demonstrates that co-treatment with ARV-825 and ruxolitinib synergistically induced apoptosis of SET2, with combination indices below 1.0, utilizing the isobologram analyses (Dose and fraction tables presented in Supplemental Figure S10A and S10B) 42. As compared to treatment with each agent alone, co-treatment with ARV-825 and ruxolitinib caused greater attenuation of p-STAT5, c-Myc, CDK4/6, PIM1 and Bcl-xL in SET2 cells (Figure 6B). Similar synergy between ARV-825 and ruxolitinib was observed in six samples of patient-derived CD34+ sAML cells (Figure 6C; Dose and fraction tables presented in Supplemental Figure S10 C-H).
Figure 6. Synergistic lethal activity of co-treatment with ARV-825 and ruxolitinib in cultured and patient-derived (PD) CD34+ sAML cells.
A. SET2 and HEL92.1.7 sAML cells were treated with ARV-825 (dose range: 10–250 nM) and ruxolitinib (dose range: 50–1000 nM) at a constant ratio for 48 hours. Then, the % of annexin V- and TO-PRO-3-positive, apoptotic cells was determined by flow cytometry. Median dose effect and isobologram analyses were performed utilizing CompuSyn, assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination. B. SET2 cells were treated with the indicated concentrations of ARV-825 and/or ruxolitinib for 24 hours. Total cell lysates were prepared and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control. C. PD CD34+ sAML cells (n=6) were treated with ARV-825 (dose range: 2.5–500 nM) and ruxolitinib (dose range: 50–1000 nM) for 48 hours. Then, the % of propidium iodide positive, non-viable cells was determined by flow cytometry. Median dose effect and isobologram analyses were performed utilizing CompuSyn, assuming mutual exclusivity. Combination index (CI) values less than 1.0 indicate a synergistic interaction of the two agents in the combination.
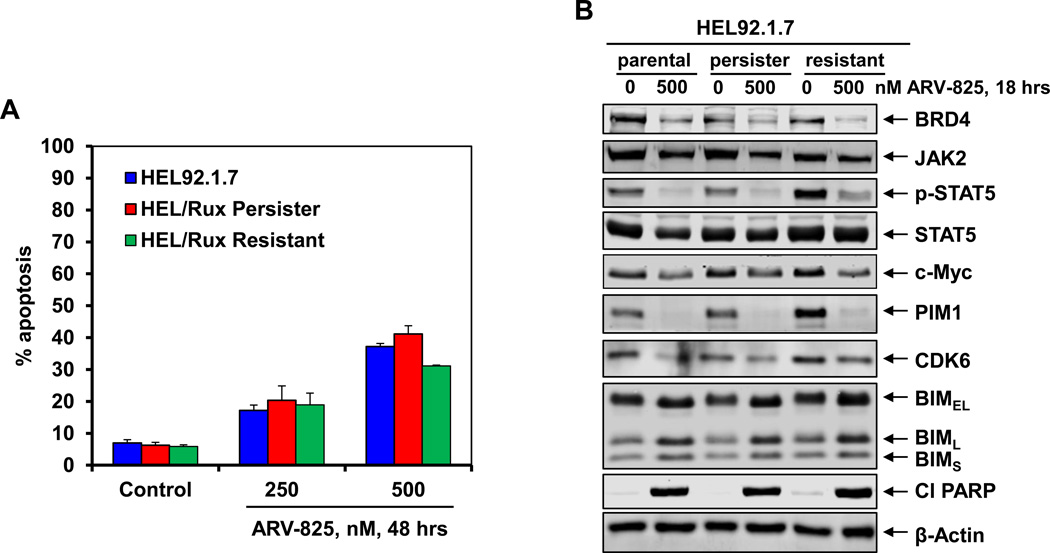
BET-PROTAC exerts lethality against in vitro generated ruxolitinib-persister and ruxolitinib-resistant cells
We had previously reported the isolation and characterization of JAKi-resistant (HEL/JIR) cells 9. Unlike the parental HEL92.1.7, HEL/JIR cells were relatively resistant to JAKi-induced apoptosis. It is notable that as compared to HEL92.1.7, UKE1 cells were endogenously less sensitive to ruxolitinib-induced apoptosis 25. As previously reported for EGFRi-resistant-persisters 43, we also developed ex vivo ruxolitinib-persister cells (HEL/Rux Persister), following repeated weekly exposure (1.0 µM for 48 hours) and recovery of the surviving HEL92.1.7 cells25. Figure 7A demonstrates that 48-hour exposure to ARV-825 dose-dependently induced apoptosis of not only the HEL92.1.7 cells, but also of the HEL/Rux Persister and HEL/Rux Resistant cells, although the latter cells are slightly less sensitive to higher levels of ARV-825. Treatment with 500 nM of ARV-825 also attenuated the protein levels of BRD4, JAK2, p-STAT5, c-Myc, PIM1, CDK6, while simultaneously inducing the levels of BIM and cleaved PARP (Figure 7B). These findings highlight that unlike ruxolitinib, ARV-825 retains lethal activity against HEL/Rux Persister and HEL/Rux Resistant cells, as well as against sAML UKE1 cells that are relatively resistant to ruxolitinib25.
Figure 7. Treatment with ARV-825 depletes BRD4 and its target proteins as well as induces apoptosis of sAML cells resistant to ruxolitinib.
A. HEL92.1.7 parental, HEL ruxolitinib-tolerant persister (HEL/Rux Persister) or ruxolitinib resistant (HEL/Rux Resistant) cells were treated with the indicated concentrations of ARV-825 for 48 hours. At the end of treatment, cells were stained with annexin V and TO-PRO-3 iodide and the % annexin V-positive, apoptotic cells was determined by flow cytometry. Columns, mean of three experiments; Bars, standard error of the mean. B. HEL92.1.7, HEL/ Rux Persister and HEL/Rux Resistant cells were treated with 500 nM of ARV-825 for 18 hours. Following this, cells were harvested and immunoblot analyses were conducted as indicated. The expression levels of β-Actin in the cell lysates served as the loading control.
DISCUSSION
In the present studies, we demonstrate unequivocally that, unlike treatment with BETi OTX015, which leads to intracellular and nuclear accumulation of BRD4 and BRD2, the BET-PROTAC ARV-825 treatment causes more profound and sustained depletion of the levels of BRD4 and BRD2 in the cultured and PD CD34+ sAML progenitor cells. Concomitantly, ARV-825 exerts significantly greater lethality against the cultured or PD CD34+ sAML cells, including those expressing JAK2-V617F and mutant TP53, as compared to the normal CD34+ progenitor cells.
As a transcriptional co-regulator, BRD4 recognizes and binds to the acetylated lysine residues on histone proteins and transcription factors at the enhancers and promoters of target genes 14,15,24. Recent studies have demonstrated that, in AML cells, hematopoietic transcription factors recruit BRD4 especially to the nucleosome-free clustered enhancers and to the promoters flanked by acetylated histones19,44,45. This co-occupancy is facilitated by the activity of the lysine acetyltransferase p300/CBP, which mediates the lysine acetylation of transcription factors and the histone proteins15, 19. BRD4 has also been demonstrated to bind through its ET domain to non-acetylated transcription factors 15, 19, the short form of the lysine methyl transferase NSD346, and to the mediator protein complex15, 47, whereas the C-terminal domain of BRD4 binds to P-TEFb14,15. BRD4 recruits these transcriptional co-regulators to the enhancers, which are known to loop over and make stable contact with the promoters to facilitate RNAP2-mediated mRNA transcript elongation48. Therefore, ARV-825 mediated depletion of BRD4 is likely to more profoundly disrupt the functions of the bound co-regulators than treatment with BETis, including the potential eviction of the mediator complex and P-TEFb, resulting in greater attenuation of the clustered enhancers-driven transcription of the sAML-relevant oncogenes, e.g., c-Myc, Bcl-xL, CDK6 and PIM119, 20,47,49. One of the most BRD4-occupied loci in AML is the cluster of 5 enhancers (super enhancer) of MYC that is located 1.8 mega-bases downstream of the promoter of c-Myc; all residing in a single large DNA TAD (topologically associated domain) 19, 47,48. Indeed, ARV-825 is more effective than OTX015 in reducing the levels of c-Myc in the cultured SET2 and PD CD34+ sAML cells. The serine–threonine kinase PIM1 is involved in phosphorylating and affecting the levels and/or activity of its target proteins, which include c-Myc, p21, p27, BAD and PRAS40 50, 51. PIM1 and c-Myc cooperate and synergistically increase cell growth, cellular biomass and survival 50, 51. Therefore, as demonstrated here, ARV-825-mediated simultaneous and greater depletion of PIM1 and c-Myc, as compared to OTX015, may also contribute to the greater induction of p21 and p27, as well as cause higher ARV-825-induced lethality in sAML cells. Mutations in JAK2, c-MPL or CALR constitutively activate JAK-STAT signaling, as well as promote survival and proliferation of MPN and sAML progenitor cells 2, 3, 10. Our findings also demonstrate that, compared to OTX015, treatment with ARV-825 causes greater depletion of JAK2, p-STAT3 and p-STAT5 levels, as well as of BCL2 and Bcl-xL, which further likely contributes to the higher lethal activity of ARV-825 in sAML cells. Treatment with both ARV-825 and OTX015 increased the expression of HEXIM1 and p21, of which p21 is known to be repressed by Myc 52, 53. Induction of HEXIM1 levels may also play a mechanistic role in inhibiting P-TEFb-mediated RNAP2 activity at and the expressions of AML-relevant oncogenes 17, 37. Up-regulation of BIM is also explained by ARV-825-induced Myc repression, since Myc induces miR-17-92 which inhibits BIM expression53. Utilizing mass cytometry coupled to SPADE algorithm clustering of the data 34, 35, 41, we also demonstrate for the first time that, compared to OTX015, treatment with ARV-825 caused marked attenuation of BRD4, c-Myc and p-Rb, while inducing more p21 in the CD34+ stem/progenitor cells.
Although inducing significant clinical benefit and improving the survival of patients with advanced post-MPN or primary MF, the JAKi ruxolitinib has modest and short-term clinical efficacy in patients with sAML3,7,8. This may be due to a lack of efficacy of ruxolitinib alone against sAML stem/progenitor cells. In contrast, findings presented here show that co-treatment with ARV-825 and ruxolitinib exerted synergistic lethality against cultured and PD CD34+ sAML cells. This may be at least partially explained by the observation that, as compared to treatment with each agent alone, co-treatment with ARV-825 and ruxolitinib caused greater attenuation of the levels of p-STAT5, c-Myc, CDK4, PIM1 and Bcl-xL in sAML cells. In post-MPN MF and sAML, resistance to ruxolitinib has been attributed to persistent JAK-STAT signaling due to trans-phosphorylation of JAK2 by either JAK1 or TYK211. Additional mechanisms of therapy-resistance in the post-MPN sAML stem/progenitor cells may be due to the co-occurring mutant TP53 and/or mutations in several of the chromatin modifying enzymes, including TET2, EZH2, ASXL1, IDH1/2 3,12,13. We hypothesized that the deregulated transcriptome due to these mutations would likely confer ruxolitinib-resistance in sAML cells. Since the deregulated transcriptome would still be BRD4-dependent, it would be undermined by treatment with ARV-825, thereby sensitizing ruxolitinib-resistant sAML cells to ARV-825. Indeed, our findings demonstrate that ARV-825 exerted lethality against ruxolitinib-persister/resistant sAML cells that also express mutant TP53 (Figure 7). Recent reports have demonstrated that in MLL-AF9-driven mouse AML and in human AML cells, resistance to BETi is not mediated through increased drug-efflux or metabolism but is, at least in part, due to increased activity of WNT-beta-catenin/TCF4 and the restoration of c-Myc expression, despite inhibition of chromatin-bound BRD4 38, 39. BETi resistance was also shown to emerge from leukemia stem cells both ex vivo and in vivo 38, 39. Although findings presented here demonstrate that treatment with ARV-825 transcriptionally induced the expression of CTNNB1 (β-catenin) in sAML cells, this did not compromise ARV-825-mediated depletion of c-Myc or affect ARV-825-induced lethality in sAML cells.
ARV-771 is a novel BET-PROTAC that recruits the E3 ligase VHL (Von Hippel-Lindau) to degrade BETPs, including BRD4 and BRD2. ARV-771 displays not only superior in vitro lethal effects but also exhibits superior in vivo pharmacology and was shown to exert anti-tumor efficacy when administered subcutaneously in the tumor-bearing mice 54. Our findings presented here clearly demonstrate for the first time a potent and superior anti-leukemia effect of ARV-771 compared to OTX015 in reducing the sAML burden in the NSG mice engrafted with the sAML HEL92.1.7 cells. This translated into a significantly improved survival of the NSG mice treated with ARV-771 versus OTX015, associated with greater tolerability for ARV-771. These findings strongly support future studies interrogating the in vivo efficacy of ARV-771 versus BETi, alone and in combination with ruxolitinib or other targeted therapies, utilizing patient-derived xenograft (PDX) models of sAML cells engrafted in immune-depleted mice.
Supplementary Material
Acknowledgments
The authors would like to thank the Flow Cytometry and Cellular Imaging (FCCI) Core Facility and the Functional Proteomics RPPA Core facility which are supported by MD Anderson Cancer Center Support Grant 5P30 CA016672-40. The heat maps were developed by the MD Anderson Cancer Center Department of Bioinformatics and Computational Biology, In Silico Solutions, Santeon and SRA International. This work was supported in part by U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, the Mary K. Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell), and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource). Additional support was also provided by CPRIT Metabolomics Core Facility Support Award RP120092 (CC) and a pilot grant from the Alkek Center for Molecular Discovery (CC). C.M.C acknowledges support from the National Institutes of Health (grant number R35CA197589). This research is supported in part by the MD Anderson Cancer Center Leukemia SPORE (P50 CA100632).
Footnotes
Conflict of Interest: C.M.C. is the founder and Chief Scientific Advisor of, and possesses an equity ownership stake in, Arvinas, Inc. Y.Q., K.R. K.G.C., A.P.C., A.S. are Arvinas employees and possess an equity ownership stake in Arvinas. N.P. serves on the scientific advisory board of Incyte Pharmaceuticals. He has also served as a consultant and received research funding from Novartis Pharmaceuticals. All other authors state that they have no conflict of interest to declare.
Supplementary information is available at Leukemia’s website.
References
- 1.Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118:1723–1735. doi: 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- 2.Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–e133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rampal R, Mascarenhas J. Pathogenesis and management of acute myeloid leukemia that has evolved from a myeloproliferative neoplasm. Curr Opin Hematol. 2014;21:65–71. doi: 10.1097/MOH.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 4.Keohane C, Mesa R, Harrison C. The role of JAK1/2 inhibitors in the treatment of chronic myeloproliferative neoplasms. Am Soc Clin Oncol Educ Book. 2013:301–305. doi: 10.14694/EdBook_AM.2013.33.301. [DOI] [PubMed] [Google Scholar]
- 5.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA, et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica. 2015;100:1139–1145. doi: 10.3324/haematol.2014.119545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kundranda MN, Tibes R, Mesa RA. Transformation of a chronic myeloproliferative neoplasm to acute myelogenous leukemia: does anything work? Curr Hematol Malig Rep. 2012;7:78–86. doi: 10.1007/s11899-011-0107-9. [DOI] [PubMed] [Google Scholar]
- 8.Eghtedar A, Verstovsek S, Estrov Z, Burger J, Cortes J, Bivins C, et al. Phase 2 study of the JAK kinase inhibitor ruxolitinib in patients with refractory leukemias, including post-myeloproliferative neoplasm acute myeloid leukemia. Blood. 2012;119:4614–4618. doi: 10.1182/blood-2011-12-400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiskus W, Verstovsek S, Manshouri T, Rao R, Balusu R, Venkannagari S, et al. Heat shock protein 90 inhibitor is synergistic with JAK2 inhibitor and overcomes resistance to JAK2-TKI in human myeloproliferative neoplasm cells. Clin Cancer Res. 2011;17:7347–7358. doi: 10.1158/1078-0432.CCR-11-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer SC, Levine RL. Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res. 2014;20:2051–2059. doi: 10.1158/1078-0432.CCR-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci. 2014;111:E5401–E5410. doi: 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;1 2:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, Vakoc CR. The mechanism behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itzen F, Greifenberg AK, Bosken CA, Geyer M. Brd4 activates P-TEFB for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–7590. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippakapoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 23.Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C, et al. The BET Bromodomain Inhibitor OTX015 Affects Pathogenetic Pathways in Preclinical B-cell Tumor Models and Synergizes with Targeted Drugs. Clin Cancer Res. 2015;21:1628–1638. doi: 10.1158/1078-0432.CCR-14-1561. [DOI] [PubMed] [Google Scholar]
- 24.Basheer F, Huntly BJ. BET bromodomain inhibitors in leukemia. Exp Hematol. 2015;43:718–731. doi: 10.1016/j.exphem.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Saenz DT, Fiskus W, Manshouri T, Rajapakshe K, Krieger S, Sun B, et al. BET protein bromodomain inhibitor-based combinations are highly active against post-myeloproliferative neoplasm secondary AML cells. Leukemia. 2016 doi: 10.1038/leu.2016.260. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiskus W, Verstovsek S, Manshouri T, Smith JE, Peth K, Abhyankar S, et al. Dual PI3K/AKT/mTOR inhibitor BEZ235 synergistically enhances the activity of JAK2 inhibitor against cultured and primary human myeloproliferative neoplasm cells. Mol Cancer Ther. 2013;12:577–588. doi: 10.1158/1535-7163.MCT-12-0862. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, et al. Phthalidimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toure M, Crews CM. Small-molecule PROTACS: New approaches to protein degradation. Agnew Chem Int Ed Engl. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 30.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;7:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 31.Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014;13:1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- 32.Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, et al. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther. 2014;13:2315–2327. doi: 10.1158/1535-7163.MCT-14-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113:154–164. doi: 10.1182/blood-2007-10-119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov. 2015;5:988–1003. doi: 10.1158/2159-8290.CD-15-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyspiańska BS, Bannister AJ, Barbieri I, Nangalia J, Godfrey A, Calero-Nieto FJ, et al. BET protein inhibition shows efficacy against JAK2V617F-driven neoplasms. Leukemia. 2014;28:88–97. doi: 10.1038/leu.2013.234. [DOI] [PubMed] [Google Scholar]
- 37.Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP, et al. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia. 2016;30:504–508. doi: 10.1038/leu.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathert P, Roth M, Neumann T, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong CY, Gilan O, Lam EY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qui P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 43.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanno T, Kanno Y, LeRoy G, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, et al. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013;85:629–643. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 51.Warfel NA, Kraft AS. PIM kinase (and Akt) biology and signaling in tumors. Pharmacol Ther. 2015;151:41–49. doi: 10.1016/j.pharmthera.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.