Abstract
Infections of immunocompromised patients with human adenoviruses (hAd) can develop into life-threatening conditions, whereas drugs with anti-adenoviral efficiency are not clinically approved and have limited efficacy. Small double-stranded RNAs that induce RNAi represent a new class of promising anti-adenoviral therapeutics. However, as yet, their efficiency to treat hAd5 infections has only been investigated in vitro. In this study, we analyzed artificial microRNAs (amiRs) delivered by self-complementary adeno-associated virus (scAAV) vectors for treatment of hAd5 infections in immunosuppressed Syrian hamsters. In vitro evaluation of amiRs targeting the E1A, pTP, IVa2, and hexon genes of hAd5 revealed that two scAAV vectors containing three copies of amiR-pTP and three copies of amiR-E1A, or six copies of amiR-pTP, efficiently inhibited hAd5 replication and improved the viability of hAd5-infected cells. Prophylactic application of amiR-pTP/amiR-E1A- and amiR-pTP-expressing scAAV9 vectors, respectively, to immunosuppressed Syrian hamsters resulted in the reduction of hAd5 levels in the liver of up to two orders of magnitude and in reduction of liver damage. Concomitant application of the vectors also resulted in a decrease of hepatic hAd5 infection. No side effects were observed. These data demonstrate anti-adenoviral RNAi as a promising new approach to combat hAd5 infection.
Keywords: adenovirus infection, RNA interference, antiviral therapy, AAV vector
Introduction
Human adenoviruses (hAds) belong to the genus Mastadenovirus and are divided into seven species (A–G).1 They usually induce mild, self-.limiting infections of the upper respiratory tract and the gastrointestinal tract. Children and adolescents are more often affected than adults.2, 3, 4 In immunocompromised patients, most notably those after transplantation of hematopoietic stem cells or solid organs, hAds can induce severe infections with fatal outcomes. Affected patients often suffer from disseminated viral infection and show a high and rapid increase of viral load in the blood serum.5, 6 Fulminant liver failure is the most frequent cause of death in such cases.7, 8 Morbidity rates in these patients are between 9% and 26%,9, 10 and the mortality can be as high as 80%.11, 12
Treatment options for immunocompromised patients with severe hAd infection are limited. Clinical protocols recommend intensive supportive care, reduction of immunosuppressive agents, administration of immunoglobulins, anti-hAd-adoptive T cell therapy, and application of antiviral drugs.13, 14 The most commonly used drug to treat hAd infections is cidofovir (CDV), a nucleoside analog that is phosphorylated by cellular kinases to become a deoxycytidine (dCTP) analog that specifically blocks viral DNA polymerase and interrupts viral DNA replication.15
The response rate to CDV is low (about 25%) and toxic side effects such as nephrotoxicity are widely observed.16 Brincidofovir (BCV) is a lipid-ester derivate of CDV. Due to its altered structure, bioavailability is increased, and its safety profile is improved.17 BCV was shown to be highly effective in a permissive animal model and in human patients with hAd infections.18, 19, 20, 21 However, fatal outcomes could not be prevented in several patients with severe hAd infections.22, 23
RNAi is an evolutionary conserved cellular mechanism of gene silencing, which is induced by small double-stranded RNAs.24 These RNAs can be delivered as synthetic short interfering RNAs (siRNAs) to the cells or expressed intracellularly from vectors as small hairpin RNAs (shRNAs) or artificial microRNAs (amiRs). In contrast to siRNAs, shRNAs and amiRs require intracellular processing, because of their hairpin structures, in order to form mature functional siRNAs.25 The siRNAs are incorporated into the RNAi silencing complex (RISC) and one strand is degraded, while the other guides the RISC to the target mRNA, where it binds to a complementary sequence and induces cleavage of the target sequence.26 The efficiency of anti-adenoviral siRNAs has been proven in several in vitro studies which have demonstrated that silencing of different adenoviral genes is suitable to inhibit hAd replication.27, 28, 29, 30 The most efficient inhibition (about two to three orders of magnitude in virus titer) was observed after the silencing of pre-terminal protein (pTP) and DNA Pol.27, 31 Both proteins are involved in adenoviral DNA synthesis, indicating that disturbing adenoviral DNA replication is a very potent approach to inhibit hAd infection. Other studies also found high anti-adenoviral activity of siRNAs directed against IVa2,29, 30 which is involved in transcriptional activation of the adenoviral major late promoter32 and is important for capsid assembly and for encapsidation of the viral genome,33, 34 and against the hexon protein,29, 30 which is the major protein of the viral capsid.35 E1A is the first viral gene to be transcribed and plays a key role in host cell metabolism modulation and transcription of other viral genes.35 siRNA-mediated silencing of E1A also resulted in inhibition of adenoviral replication, but depending on the study, the efficiency of this approach was very different.27, 28, 29, 30 Importantly, silencing of E1A was more potent in protecting cells from hAd-induced cell lysis compared to silencing of other adenoviral genes.27, 29 siRNAs can easily be delivered in vitro, reaching high intracellular concentrations which are sufficient to downregulate target gene expression. Certain cell lines and primary cells, however, are hard to transfect with siRNAs, and cell- and/or organ-specific delivery, as well as achievement of prolonged therapeutic levels in the target organ in vivo constitute a general problem for siRNA use.36, 37
In the present study, we investigated amiRs targeting different adenoviral genes in the context of self-complementary adeno-associated virus (scAAV) vectors. Based on in vitro evaluation, two scAAV9 vectors expressing amiRs targeting adenoviral genes pTP and E1A, or just pTP, were developed and analyzed for their antiviral efficiency in an animal model of immunosuppressed Syrian hamsters, which are permissive for hAd 5 (hAd5). We show that prophylactic application of the vectors resulted in a significant decrease of hAd5 titers in the liver, heart, and serum and led to a distinct reduction of liver injury. Moreover, inhibition of hAd5 replication in the liver was also detected when the vectors were applied concomitantly with hAd5.
Results
Evaluation of Anti-adenoviral amiRs
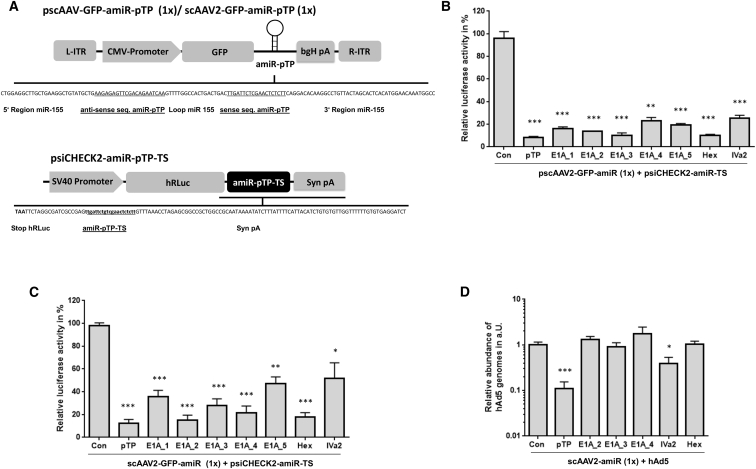
We generated eight different anti-adenoviral amiRs and inserted them into the 3′ UTR of a cytomegalovirus (CMV) promoter driven GFP reporter in AAV shuttle plasmids containing the sequence of scAAV vector genomes (Figure 1A). Five amiRs targeted different sequences of the adenoviral E1A gene, and one each was directed against target sequences of the adenoviral hexon, IVa2, and pTP genes (Table S1). To assess their silencing efficiencies, a multistep evaluation procedure was carried out. First, HeLa cells were co-transfected with the amiR-expression plasmids and with indicator plasmids containing fully complementary amiR target sequences (amiR-TS) in the 3′ UTR of a Renilla luciferase (hRLuc) reporter. All amiRs induced strong suppression of hRLuc expression of at least 70%, and amiR-pTP, amiR-E1A_3, and amiR-Hex even led to a reduction of luciferase activity of about 90% (Figure 1B). Next, we analyzed the amiRs in the context of scAAV2 vectors. HeLa cells were transduced with equal amounts of the vectors and transfected with corresponding amiR-TS containing hRLuc indicator plasmids. Again, hRLuc expression was inhibited by all amiRs. As before, amiR-pTP and amiR-Hex showed the strongest silencing effect, reaching a suppression of hRLuc activity of more than 80%. A similar efficiency was observed for amiR-E1A_2, whereas amiR-E1A_3, which was effective in the previous plasmid assay, showed distinctly lower silencing efficiency (Figure 1C). Finally, we investigated the ability of scAAV2 vector expressed amiRs to inhibit hAd5 replication. New scAAV2 vectors were constructed containing the sequences of the three most effective E1A-amiRs (amiR-E1A_2, amiR-E1A_3, and amiR-E1A_4), amiR-pTP, amiR-IVa2, and amiR-Hex, respectively. Compared to previously used scAAV2 vectors, the sequence encoding GFP was removed, as we observed negative crosstalk of GFP and amiRs when both were inserted in a single scAAV2 vector genome (data not shown). As shown in Figure 1D, the application of 1 × 104 vector genome equivalents (vge)/cell of the amiR-pTP-expressing vector scAAV2-amiR-pTP resulted in a strong decrease of hAd5 replication by about 89%. The amiR-IVa2-expressing scAAV2 vector also inhibited hAd5 replication, but only by about 61%. No inhibitory effect was seen for both scAAV2 vectors expressing the E1A targeting amiRs and amiR-Hex, respectively. In conclusion, the evaluation experiments demonstrated that among the considered amiRs, amiR-pTP showed the strongest anti-adenoviral efficiency if expressed from scAAV2 vectors. Therefore, it was selected for further investigation.
Figure 1.
Evaluation of Anti-adenoviral amiRs
(A) Schematic representation of the anti-adenoviral amiR and luciferase reporter vectors. One copy of anti-adenoviral amiR was inserted into a GFP-expression cassette of an AAV shuttle plasmid containing a self-complementary (sc) AAV2 vector genome (upper panel). One copy of amiR-target sequences (amiR-TS) was inserted downstream of a Renilla luciferase (hRLuc) gene in the reporter plasmid psiCHECK2-amiR-TS (lower panel). L-ITR/R-ITR = left and right inverted terminal repeat of AAV2, respectively; bGH pA - poly A signal of the bovine growth hormone; Syn pA - synthetic poly A signal. The silencing vector shows the amiR of the adenoviral pre-terminal protein (pTP). The reporter vector shows the corresponding amiR-pTP target sequence. (B) Silencing activity of anti-adenoviral amiRs in reporter gene assay. HeLa cells were transfected with anti-adenoviral amiR-expressing plasmids and co-transfected with hRLuc reporter plasmids containing the corresponding amiR-TS. The cells were harvested 48 hr after transfection and hRLuc activity was determined. As a control, each amiR-TS plasmid was co-transfected with a plasmid expressing an amiR against dsRed (con). The amiR-silencing activity was calculated as a percentage of hRLuc activity in samples treated with anti-adenoviral amiRs compared to samples treated with con. **p < 0.01 and ***p < 0.001 versus con. (C) HeLa cells were transduced with equal amounts of scAAV2 vectors containing one copy of the indicated amiR or the dsRed amiR (con) and 72 hr later transfected with hRLuc reporter plasmids containing the corresponding amiR-TS. Cells were harvested 96 hr after transduction, and the silencing activity of the anti-adenoviral amiRs was calculated as described under (B). *p < 0.05; **p < 0.01; and ***p < 0.001 versus con. (D) HeLa cells were transduced with 1 × 104 vge/cell of scAAV2 vectors containing one copy of the indicated amiRs concomitant to infection with hAd5 at an MOI of 0.05. The cells were lysed 48 hr after infection, and the number of adenoviral genomes was quantified by real-time PCR. The number of copies of hAd5 after treatment with amiRs expressing scAAV2 vectors is shown relative to the number of copies after treatment with con ( = 1). *p < 0.05 and ***p < 0.001 versus con. Note: in this panel, the scAAV2 vector genome lacks GFP cDNA, while GFP cDNA is present in (C). Results of (B)–(D) show mean values ± SEM.
Concatemerization of amiR-pTP and amiR-E1A_2 Increases Inhibition of hAd5 Infection
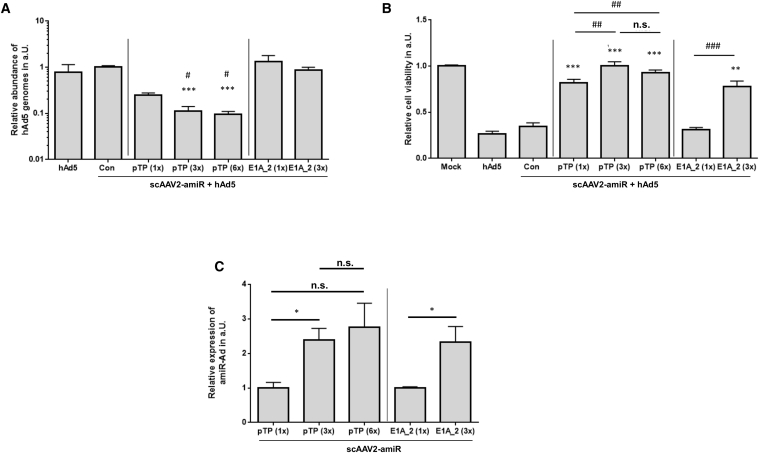
Earlier studies reported that use of concatemerized antiviral amiRs can result in higher inhibition of virus replication than the use of single amiRs.31, 38 To prove this in the context of amiR-pTP and AAV vectors, we generated scAAV2 vectors containing three and six copies of amiR-pTP and compared them with the scAAV2 vector containing only a single copy of amiR-pTP. The copy number of amiR-pTP was limited to six to ensure that the vector genome did not exceed the packaging capacity of the scAAV vectors. We also generated a scAAV2 vector expressing three copies of amiR-E1A_2 to compare it with the scAAV2 with a single copy of amiR-E1A_2. This was done because previous investigations had revealed that silencing of E1A can improve cell viability of hAd-infected cells much more efficiently than silencing of any other adenoviral gene.27, 29 Among the analyzed E1A-amiRs, amiR-E1A_2 showed the best silencing activity in the reporter gene assay when expressed from scAAV2 vectors, so it was selected (Figure 1C). Transduction of HeLa cells with 1 × 103 vge/cell of the scAAV2 vectors containing one, three, or six copies of amiR-pTP, concomitant with hAd5 (dose 0.05 MOI) led to inhibition of adenoviral replication by 76%, 89%, and 91%, as determined by real-time PCR, respectively (Figure 2A). In contrast, scAAV2 vectors with one or three copies of E1A_2 did not inhibit adenoviral replication. The same vectors were used to analyze cell viability of hAd5-infected cells. HeLa cells were transduced with 1 × 104 vge/cell of scAAV2 vector 5 hr before infection with 1 MOI of hAd5, and cell viability was determined 5 days later. Each of the scAAV2-amiR-pTP vectors significantly improved cell viability, but those with three or six copies were able to fully preserve it. There was no improvement for the vector containing one copy of amiR-E1A_2, but the vector with three copies of amiR-E1A_2 significantly protected cell viability (Figure 2B).
Figure 2.
Repetition of amiR-pTP and amiR-E1A Coding Units in the scAAV2 Vector Genome Increases Inhibition of hAd5
(A) Determination of hAd5 replication. HeLa cells were infected with hAd5 at an MOI of 0.05 and concomitantly transduced with 1 × 103 vge/cell of scAAV2-amiR vectors expressing the indicated amiRs. The cells were lysed 48 hr later, and the amount of viral DNA was quantified by real-time PCR. The copy number of each amiR in the vector genome is shown in parentheses. Cells, which were infected only with hAd5, are labeled “hAd5”. Con represents a scAAV2 control vector containing an array of six amiR sequences directed against dsRed. The genome copy number for each sample was related to con ( = 1). Significance was determined compared to con, ***p < 0.001. Significance compared to pTP (1×), #p < 0.05. (B) Determination of viability of hAd5-infected cells. HeLa cells were transduced with 1 × 104 vge/cell of indicated scAAV2-amiR vectors 5 hr prior to infection with hAd5 at an MOI of 1. Cell viability was determined 5 days after infection. Mock, untransduced/uninfected cells ( = 1). Con and hAd5, see under (A). Cell viability for each sample was related to mock infected. Significance compared to con, ***p < 0.001. Significance as shown, ##p < 0.01 and ###p < 0.001; n.s., not significant. (C) Relative expression of amiR-pTP and amiR-E1A_2 by scAAV2 vectors. HeLa cells were transduced with 1 × 103 vge/cell and analyzed 48 hr later for amiR-pTP and amiR-E1A_2 expression. The abundance of amiR-pTP and amiR-E1A_2 expression was analyzed with real-time RT-PCR and corrected for the expression of snU6RNA, *p < 0.05. Results of (A)–(C) show mean values ± SEM.
To determine the molecular basis leading to increased inhibition of hAd5 replication and reduction of cytotoxicity, we quantified the expression levels of amiR-pTP and amiR-E1A_2 48 hr after transduction of HeLa cells with 1 × 103 vge/cell of respective scAAV2 vectors. As shown in Figure 2C, the presence of three amiR copies in the vector genome increased the amiR levels of both amiR-pTP and amiR-E1A_2 by approximately 2.4-fold compared to the vectors containing only one copy of the individual amiRs. An increase of amiR-pTP expression was also seen for the scAAV2 vector containing six copies of amiR-pTP. However, the expression levels increased only 1.2-fold compared to the scAAV vector containing three copies of amiR-pTP. In conclusion, these data demonstrate that concatemerization of amiR-pTP and amiR-E1A_2 improves the effectiveness of the scAAV2 vectors.
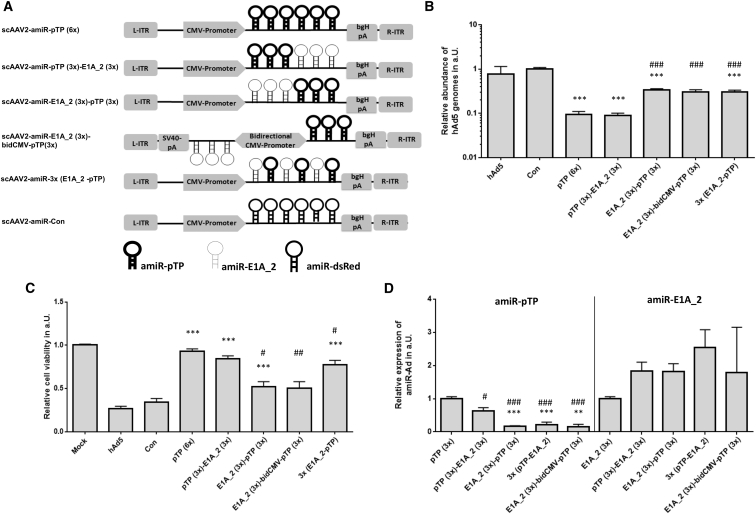
scAAV2 Vectors with Three Copies of amiR-pTP and Three Copies of amiR-E1A_2 Are Not More Efficient than scAAV2 Vector with Six Copies of amiR-pTP
Since both concatemerized amiR-pTP and amiR-E1A_2 showed an increased protective effect on hAd5 infections, we next investigated whether simultaneous expression of both amiRs could increase the anti-adenoviral efficiency. For this purpose, three copies of amiR-pTP and three copies of amiR-E1A_2 were inserted in different arrangements in the expression cassette of scAAV2 vectors (Figure 3A). Among the four investigated scAAV vectors with mixed amiRs, scAAV2-amiR-pTP (3×)-E1A_2 (3×) containing the amiR-pTP copies immediately downstream of the CMV promoter and amiR-E1A_2 copies downstream of the amiR-pTP showed the best performance. It inhibited viral replication in hAd5-infected cells by about 91% and increased cell viability by about 84%. Thus, its efficiency was in the range of scAAV2-amiR-pTP (6×), which was used as the control. The other vectors had significantly weaker responses in terms of adenoviral replication. Only scAAV2-3× (amiR-E1A_2-pTP) containing amiR-E1A_2 and amiR-pTP in alternating order increased cell viability similar to scAAV2-amiR-pTP (6×) and scAAV2-amiR-pTP (3×)-E1A_2 (3×) (Figures 3B and 3C). Analysis of amiR expression levels revealed that all four vectors expressed amiR-pTP at significantly lower levels than the control vector scAAV2-amiR-pTP (3×). However, scAAV2-amiR-pTP (3×)-E1A_2 (3×) showed significantly higher amiR-pTP expression than the other vectors with mixed amiR-pTP/amiR-E1A_2. In contrast, the expression of amiR-E1A_2 in the vectors with mixed amiR-pTP/amiR-E1A_2 was not significantly changed compared to the control vector scAAV2-amiR-E1A_2 (3×) (Figure 3D). These results demonstrate that co-expression of amiR-pTP and amiR-E1A_2 does not increase anti-adenoviral efficiency compared to positive control vectors expressing only amiR-pTP. Only scAAV2-amiR-pTP (3×)-E1A_2 (3×) showed a similar efficiency as scAAV2-amiR-pTP (6×). Therefore, this vector and scAAV-miR-pTP (6×), which showed best anti-adenoviral performance among all tested amiR-expressing scAAV vectors in previous experiments, were selected for further investigations.
Figure 3.
Configuration of the amiR pTP/amiR-E1A 2-Expression Cassette Affects Its Ability to Inhibit hAd5 Infection
(A) amiR-pTP- and amiR-E1A_2-expression cassettes of scAAV2-amiR vectors. Each scAAV2 vector contains an array of six amiR sequences in different configurations. scAAV2-amiR-Con (Con) contains six copies of amiR-dsRed. (B) Determination of hAd5 replication after treatment with amiR-pTP/amiR_E1A_2 scAAV2 vectors. HeLa cells were treated and analyzed as described in Figure 2A. Cells, which were infected only with hAd5, are labeled “hAd5”. Values are shown relative to Con ( = 1). Significance of anti-adenoviral amiR vectors compared to Con, ***p < 0.001. Significance of anti-adenoviral amiR vectors compared to scAAV2-amiR-pTP (6×), ###p < 0.001. (C) Determination of cell viability. HeLa cells were treated and analyzed as described under Figure 2B. Cell viability was related to mock ( = 1). hAd5 represents cells which were infected only with hAd5. Significance of anti-adenoviral amiR vectors compared to Con, ***p < 0.001. Significance of anti-adenoviral amiR vectors compared to scAAV2-amiR-pTP (6×), #p < 0.05 and ##p < 0.01. (D) Expression of amiR-pTP and amiR-E1A_2. HeLa cells were treated and analyzed as described under Figure 2C. Expression levels are shown relative to scAAV2-amiR-pTP (3×) (left side). Significance of amiR-pTP expression compared to scAAV2-amiR-pTP (3×), #p < 0.05 and ###p < 0.001. Significance of amiR-pTP expression compared to scAAV2-amiR-pTP (3×)-E1A_2 (3×), ***p < 0.001. Expression values are shown relative to scAAV2-amiR-E1A_2 (3×) ( = 1) (right side). There was no significant difference in the expression of amiR-E1A_2 levels between the different scAAV2 vectors. Results of (B)–(D) show mean values ± SEM.
Anti-adenoviral amiR-Expressing scAAV2 Vectors Inhibit Ongoing hAd5 Infections in the Terminally Differentiated Hepatic Cell Line HepaRG
hAds can infect the liver in immunosuppressed patients, which can lead to liver failure and death.7, 8 Therefore, we next investigated the inhibitory efficiency of scAAV2-miR-pTP (6×) and scAAV2-amiR-pTP (3×)-E1A_2 (3×) in terminally differentiated hepatic HepaRG cells. HepaRG cells were treated with 1 × 103 vge/cell of scAAV2-miR- TP (6×) and scAAV2-amiR-pTP (3×)-E1A_2 (3×) 5 hr before concomitant and 5 hr after infection with hAd5 and analyzed for abundance of hAd5 48 hr later by real-time PCR. HeLa cells were used as control in these experiments. Both vectors induced a marked inhibition of hAd5 replication at each time point in both cell lines (Figure S1). The results demonstrate the anti-hAd5 efficiency of both vectors in liver cells and furthermore in ongoing hAd5 infections.
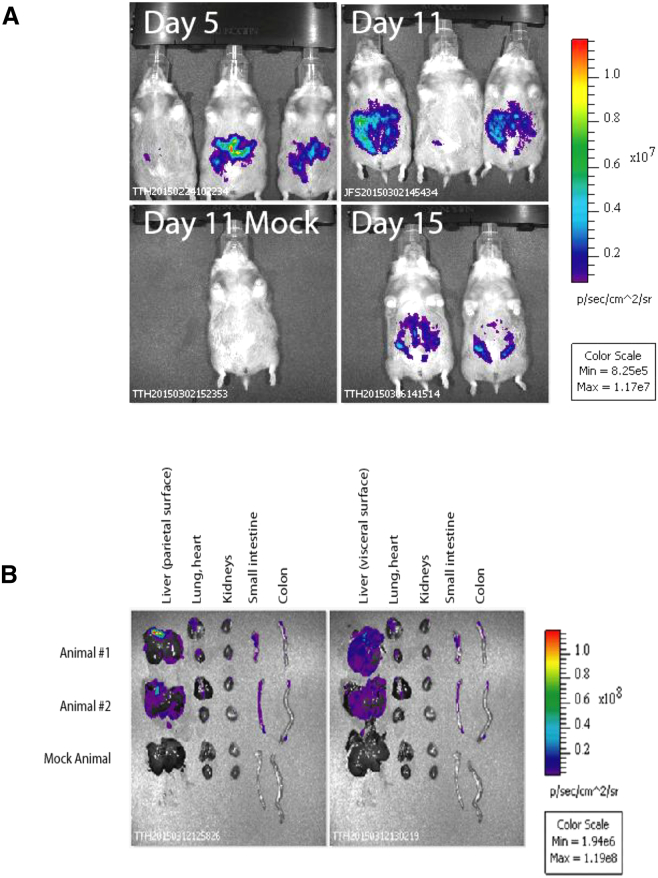
scAAV9 Vectors Transduce the Liver of Immunosuppressed Syrian Hamsters after Systemic Administration
Previous studies reported that AAV vectors containing the capsid of serotype 9 efficiently transduced the liver of rodents.39, 40, 41 The standard model for investigation of hAd5 infection in vivo makes use of immunosuppressed Syrian hamsters. Nothing is known, however, about the targeting of scAAV9 vectors and transgene expression in this system.18 Therefore, we injected Syrian hamsters intravenously (i.v.) with 2 × 1011 vge of scAAV9 vector-expressing hRLuc and immunosuppressed the animals with cyclophoshamide (CP). The animals were analyzed for hRLuc expression 5, 11, and 15 days later by in vivo imaging after intraperitoneal (i.p.) application of coelenterazine. Strong hRLuc activity was detected in the liver and in the small intestine of the investigated animals. Highest expression levels were found on day 11, but hRLuc was also abundantly expressed on days 5 and 15 (Figures 4A and 4B). On day 21, two hamsters were injected i.p. with coelenterazine, sacrificed, dissected, and the selected organs were imaged. Confirming the previous in vivo imaging data, the liver and small intestine showed the strong luminescent signals, whereas low or no expression was detected in the heart, the colon, the lung, and the kidney. The relatively low transduction of the heart was a surprising result, as a recent study found strong transgene expression after i.v. injection of AAV9 vectors into adult F1B hamsters.41 We observed that the luminescent signal intensity of explanted organs was stronger on the visceral surface of the liver, suggesting that the coelenterazine did not absorb well after i.p. injection. This may also have negatively affected the luminescence signal intensities of other organs, such as the heart.
Figure 4.
Transduction of scAAV9 Vectors after Intravenous Application in Immunosuppressed Syrian Hamsters
(A) Syrian hamsters were immunosuppressed by application of cyclophosphamide (CP). CP was administered i.p. at a dose of 140 mg/kg and then twice weekly at a dose of 100 mg/kg. Concomitant with the first CP treatment, all three animals were injected with 2 × 1011 virus genomes of scAAV9-hRLuc intravenously. On the indicated days, the hamsters were injected intraperitoneally (i.p.) with coelenterazine and 10 min later imaged in an IVIS Spectrum optical imaging platform. Images shown are normalized using the same color/intensity scale. The order of the animals is the same for day 5 and day 11, and the day 15 image has only animals #1 (left) and #2 (right). The mock infected animal shown at day 11 is a hamster that did not receive scAAV9-hRLuc, but was injected with coelenterazine i.p. (B) At 21 days post injection, the animals #1 (left) and #2 (right) and mock animal were injected i.p. with coelenterazine. At 20 min after the injection of coelenterazine, the animals were sacrificed, dissected, and selected organs were imaged. For the scAAV9-hRLuc-injected hamsters, strong luminescence was detected in the liver and small intestine. The luminescence was stronger on the visceral surface of the liver, suggesting that the coelenterazine substrate did not absorb well after i.p. injection. No luminescence was detected in the organs of the mock infected animal.
In summary, our data reveal that the liver was efficiently transduced by scAAV9 vectors. This is important, as the liver is the main target organ of hAd5 in the Syrian hamster animal model.18
Course of hAd5 Infection in Immunosuppressed Syrian Hamsters in a Moderate-Dose Infection Model
In the standard model for investigation of hAd5 infection, Syrian hamsters are immunosuppressed with cyclophosphamide (CP) for 2 weeks and then i.v. injected with 1.9 × 1012 vp/kg of hAd5.18 Animals treated in this manner developed a severe, disseminated hAd5 infection, which was accompanied by high mortality of about 50%.18 Applying the replace, reduce, or refine animal experiments (3R) principle42 aiming to specifically reduce the burden on experimental animals, we investigated whether immunosuppressed Syrian hamsters could also be infected with lower doses of hAd5. Therefore, the animals were immunosuppressed with CP, injected with 4 × 1011 vp/kg hAd5, and analyzed 3, 7, and 14 days later for hAd5 infection (Figure S2A). No deaths were observed in the investigation period. Animals showed a sustained reduction of body weight up to day 10 post infection. Thereafter, the body weight increased to day 14, reaching values similar to those determined immediately before hAd5 infection (Figure S2B). Liver tissues of the hAd5-infected animals showed minimal focal to moderately diffuse hepatocellular necrosis. The medians of the pathological scores were higher at day 7 and day 14 than at day 3. There was no tissue damage detected in the heart, lung, or kidney. One animal showed minimal pancreatitis (day 3) and another necroses of the pancreas islands at day 7 (Figures S2C and S2D). Replicating hAd5 was detected in the liver and at lower levels in the heart (about 100-fold) and the serum (about 10-fold). In the liver and heart, the virus titers increased from day 3 to day 7 and then dropped at day 14, whereas the hAd5 titers in the serum remained relatively constant over the whole investigation period (Figures S2E–S2H). These results demonstrate that injection of hAd5 into immunosuppressed Syrian hamsters at a moderate dose leads to hAd5 infection with significant changes to the liver, but without fatalities. As the highest hAd5 virus titers and pronounced liver damage were observed 7 days after hAd5 infection, we decided to choose this time point of analysis of the animals for follow up experiments.
Prophylactically Applied amiR-pTP and amiR-pTP/amiR-E1A_2-Expressing scAAV9 Vectors Inhibit hAd5 Infection and Reduce Liver Injury in Immunosuppressed Syrian Hamsters
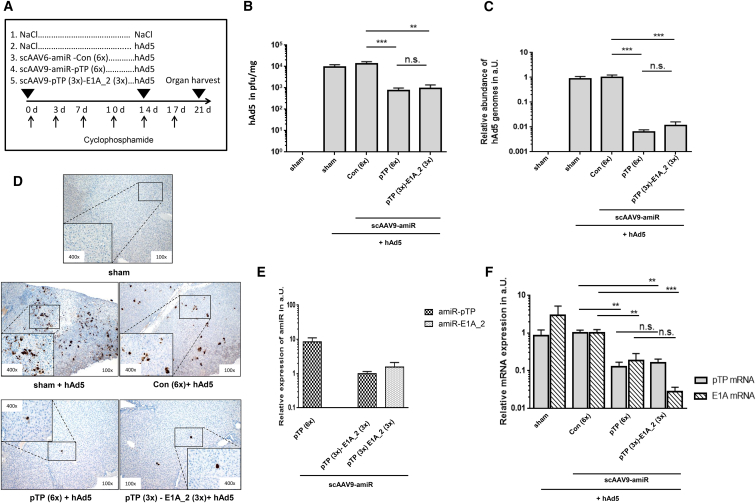
To assess the efficacy of scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×), Syrian hamsters were divided into five groups and immunosuppressed with CP. Concomitant with the first CP injection, each group was transduced with either 5 × 1013 vge/kg scAAV9-amiR-pTP (6×), scAAV9-amiR-pTP (3×)-E1A_2 (3×) or the control vector scAAV9-amiR-Con (6×), whereas two groups were sham operated and injected with physiological NaCl solution. At 2 weeks later, all animals, apart from one sham operated group, were injected i.v. with 4 × 1011 vp/kg hAd5 (Figure 5A). During the following investigation period of 7 days, only the uninfected animals and hAd5-infected animals that were treated with scAAV9-amiR-pTP (6×) did not lose weight after infection. Animals that were transduced with the scAAV9-amiR-pTP (3×)-E1A_2 (3×) and scAAV9-amiR-Con (6×) lost up to 3% of their body weight, but recovered, whereas untransduced hAd5-infected animals lost up 6% of their weight and did not recover (Figure S3).
Figure 5.
Prophylactic Application of scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3x)-E1A_2 (3x) to Immunosuppressed Syrian Hamsters Inhibits hAd5 Replication in the Liver
(A) Groups and time course of CP, scAAV9 vector, and hAd5 injection. Syrian hamsters were divided into five groups (n = 4 to 6 animals) and immunosuppressed by repetitive application of CP (as described in Figure 5). The first group (sham, n = 5) was injected with NaCl at two time points (0 day/14 day). The second group (sham + hAd5, n = 4) was injected with NaCl (0 day) and infected with hAd5 (14 day). The third (n = 4), fourth (n = 5), and fifth groups (n = 6) were transduced with scAAV9-amiR-Con (6×), AAV9-amiR-pTP (6×), and scAAV9-amiR-pTP (3×)-E1A_2 (3×), respectively, at 0 day and in each case infected with hAd5 at day 14. Animals were sacrificed and analyzed at day 21. The scAAV9 vectors were applied at a dose of 5 × 1013 vge/kg and hAd5 at a dose of 4 × 1011 vp/kg. (B) The titers of hAd5 measured in liver tissue. Titers of infectious hAd5 in liver tissue were determined by an in vitro hAd5 amplification/real-time PCR protocol, as described in the Materials and Methods. Significance, **p < 0.01 and ***p < 0.001; n.s., not significant. (C) hAd5 genome copy numbers in liver tissue. The hAd5 genome copy numbers were determined by real-time PCR and are shown relative to scAAV9-amiR-Con (6x). Significance, ***p < 0.001; n.s., not significant. (D) Immunohistochemical detection of adenoviral E1A protein expression in liver tissue using an E1A-specific antibody. Shown are representatives of each animal group, with the exception of the animal from the sham + hAd5 group, which shows a maximally positive stained liver. (E) amiR-pTP and amiR-E1A_2 mRNA expression in liver tissue. Abundance of amiR-pTP and amiR-E1A_2 expression was analyzed with real-time RT-PCR and corrected for the expression of snU6RNA. (F) pTP and E1A mRNA expression in liver tissue. Expression of pTP and E1A mRNA expression was determined by real-time RT-PCR and corrected for 18S rRNA expression. Expression is shown relative to scAAV9-amiR-Con (6×) ( = 1). Significance, **p < 0.01 and ***p < 0.001; n.s., not significant. Results of (B), (C), (E), and (F) show mean values ± SEM.
Determination of hAd5 titers in the liver revealed a significant reduction of virus burden in animals treated with scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) compared to animals treated with the control vector and untransduced, hAd5-infected animals. The hAd5 titer in the liver (determined by detection of infectious virus particles) in the scAAV9-amiR-pTP (6×) group and in the scAAV9-amiR-pTP (3×)-E1A_2 (3×) group was 94.5% and 92.8% lower compared to the control vector group, respectively (Figure 5B). Detection of hAd5 genome copies in the liver tissue by real-time PCR confirmed these data, but showed even stronger inhibition. Viral DNA abundance was significantly reduced about 99.4% in those hAd5-infected animals which were transduced with scAAV9-amiR-pTP (6×) and about 98.9% in those that were transduced with scAAV9-amiR-pTP (3×)-E1A_2 (3×) (Figure 5C). Immunohistochemical analysis revealed strong and diffuse distributed adenovirus E1A protein expression in the liver of hAd5-infected untransduced animals and in animals treated with the control vector, whereas in animals treated with the scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×), adenoviral E1A protein expression could only be found in small areas or single cells of the liver tissue, respectively (Figure 5D). We also investigated the expression of amiR-pTP and amiR-E1A_2 and the expression of adenoviral pTP- and E1A-mRNA in the liver. Both amiRs were specifically detected in animals treated with the respective amiR-expressing vector, but not in animals treated with the control vector. However, the expression of amiR-pTP was distinctly higher in the group treated with scAAV9-amiR-pTP (6×) than in the group treated with scAAV9-amiR-pTP (3×)-E1A_2 (3×) (Figure 5E). Both vectors induced a significant inhibition of pTP-mRNA expression by 87.4% (scAAV9-amiR-pTP 6×)) and 83.7% (scAAV9-amiR-pTP (3×)-E1A_2 (3×)) compared to the controls, respectively. A reduction of E1A-mRNA expression was also seen in both groups, but the reduction was higher in animals treated with scAAV9-amiR-pTP (3×)-E1A_2 (3×) (97.2%) than in animals treated with scAAV9-amiR-pTP (6×) (81.2%) (Figure 5F). The latter indicates that reduction of E1A expression in the scAAV9-amiR-pTP (3V)-E1A_2 (3×) treated animals was not only a result of reduced adenoviral replication induced by amiR-pTP, but also specifically resulted from E1A silencing.
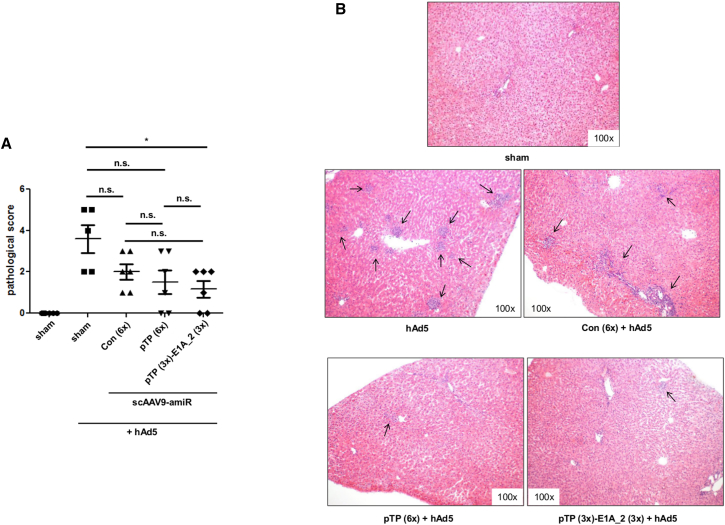
Hepatic pathological grading of the liver tissues revealed a reduction of liver injury in all animal groups which were transduced with scAAV9 vectors and infected with hAd5. Among these groups, there was no significant difference in pathological scores, but the reduction was more pronounced in animals treated with scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) than in animals treated with scAAV9-amiR-Con (6×). Only scAAV9-amiR-pTP (3×)-E1A_2 (3×) showed significantly lower liver injury compared to untreated hAd5-infected animals. Moreover, animals with no signs of liver pathology could be found only in the scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) groups (Figure 6A). Histological examination of the liver showed that in untreated hAd5-infected animals, the inflammation was evenly distributed throughout the tissue. In the scAAV9-amiR-Con (6×) group, distinct inflammation could only be detected in individual areas, whereas weak spots of inflammation were found in animals of the scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) groups (Figure 6B). We also measured activity of the alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in the serum of all investigated groups, but detected no differences among the groups. This indicates that liver damage induced by moderate hAd5 dose was below the level required to observe an increase in ALT and AST levels.
Figure 6.
Inhibition of Liver Injury after Prophylactic Application of scAAV9-amiR-pTP (6x) and scAAV9-amiR-pTP (3x)-amiR E1A_2 (3x) to hAd5-Infected Immunosuppressed Syrian Hamsters
Animals were treated and analyzed as shown in Figure 5A. (A) Severity of liver tissue damage. Liver damage was assessed and given as a pathological score presenting a scale from 0 (no damage) to 5 (severe damage). Significance of anti-adenoviral amiR vectors compared to scAAV9-amiR-Con (6×), *p < 0.05; n.s., not significant. (B) H&E staining of formalin-fixed liver slides. Shown are representatives of each group. Results of (A) show mean values ± SEM.
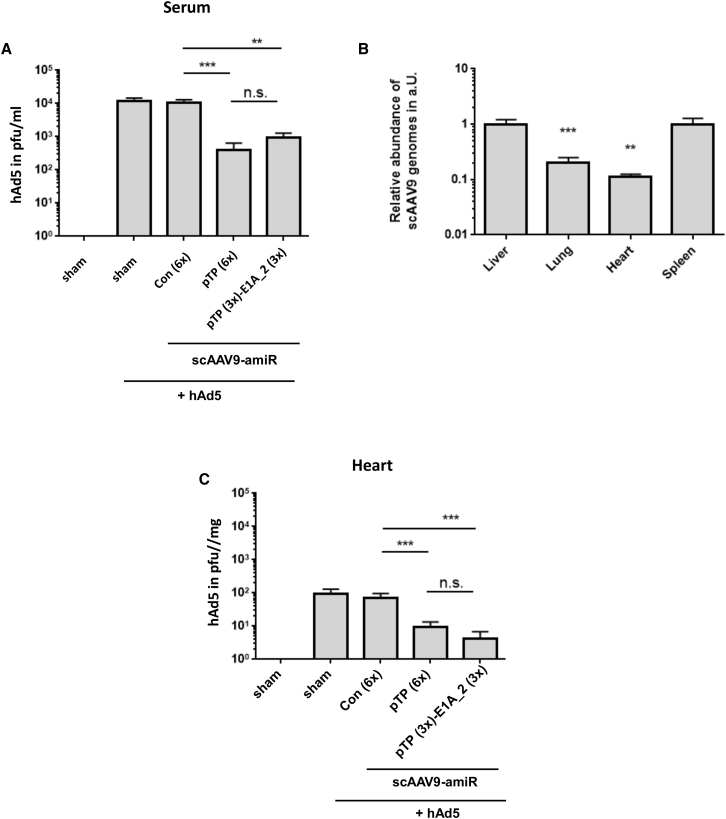
A significant reduction of hAd5 titers was also detected in the serum after application of scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) (Figure 7A), indicating that application of the vectors prevented viremia. To investigate whether hAd5 infection could also be inhibited in organs other than the liver, we investigated the heart, spleen, and lung, which were transduced by the scAAV9 vectors (Figure 7B). scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) treatment led to significant reduction of hAd5 titers in the heart (Figure 7C). hAd5 was also found in the spleen and the lung, but at very low levels and only sporadically in some animals (data not shown), making it impossible to assess the efficiency of the anti-adenoviral amiRs in these organs. These results indicate that prophylactic treatment with scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) inhibits hAd5 replication in the liver, decreases viremia, and inhibits development of disseminated hAd5 infection. More importantly, inhibition of replication was accompanied by reduced liver injury. Our data also show that there were no differences in anti-adenoviral efficiency between scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) relating to adenovirus replication, whereas slightly higher efficiency was seen for scAAV9-amiR-pTP (3×)-amiR-E1A_2 (3×) relating to the inhibition of hAd5-induced liver damage.
Figure 7.
Prophylactic Application of scAAV9-amiR-pTP (6x) and scAAV9-amiR-pTP (3x)-E1A_2 (3x) Reduces hAd5 Viremia and Decreases Virus Titers in the Heart
(A and C) The experimental procedure was the same as described in Figure 5A. The hAd5 titers were determined in the blood serum (A) and in the heart (C). Significance, **p < 0.01 and ***p < 0.001; n.s., not significant. (B) Relative amount of scAAV9-amiR-Con (6×) vector DNA in the lung, heart, and spleen compared to the liver ( = 1) at day 21 after vector application. Results of (A)–(C) show mean values ± SEM.
To determine whether hAd5 infection can also be inhibited by anti-hAd5 amiRs when a higher dose of hAd5 is used for infection, the experiments of the prophylactic approach were repeated with an about 10-fold higher hAd5 dose (5.58 × 1012 vp/animal). Unlike the moderate-dose model here, the treatment with scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) did not reduce the hAd5 levels in the liver. However, there was a slightly reduced liver pathology for animals treated with scAAV9-amiR-pTP (3×)-amiR-E1A (3×), as indicated by a distinctly lower activity of alanine aminotransferase (ALT) in the serum compared to scAAV9-amiR-Con (6×) (Figures S4A–S4C).
Concomitant Application of amiR-pTP- and amiR-pTP/amiR-E1A_2-Expressing scAAV9 Vectors Inhibit hAd5 Infection in the Liver of Immunosuppressed Syrian Hamsters
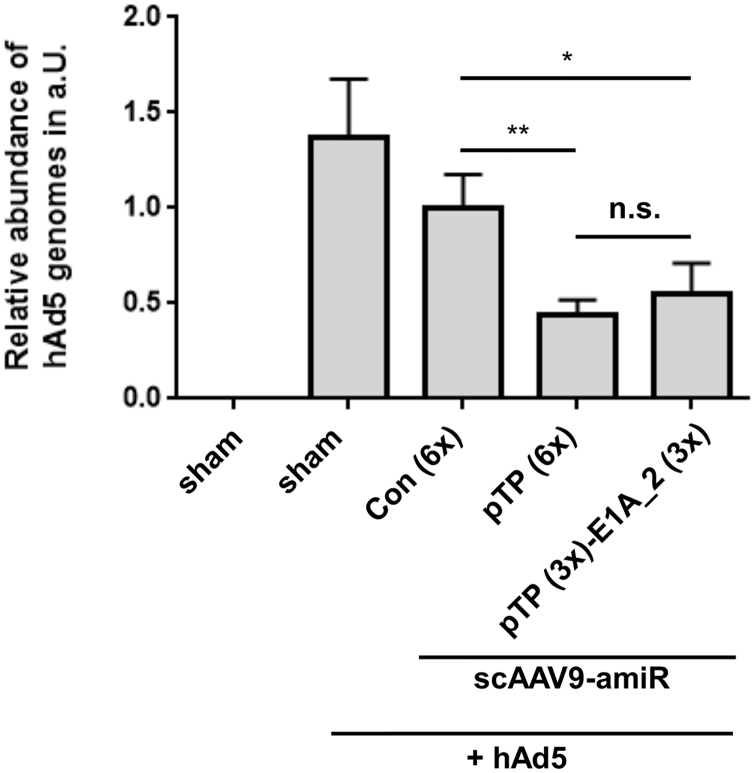
To investigate the employment of anti-adenoviral amiRs in a therapeutical approach, we repeated the above described in vivo experiments, but applicated the scAAV9 vectors concomitantly with hAd5. scAAV9 vectors have only low transgene expression in the first days after in vivo delivery.43 Therefore, in this experiment, the hAd5 dose was reduced 10-fold compared to the prophylactic approach to adapt it to an expected 10-fold lower amiR expression.43 Unfortunately, infectious hAd5 could not be detected in the liver of any of the animals 7 days after vector/hAd5 application, which probably resulted from a too low hAd5 dose. However, real-time qPCR revealed a significant reduction of hAd5 genome copy number of about 50% in animals treated with scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×) compared to the controls (Figure 8). These data indicate that anti-adenoviral amiRs can also inhibit hAd5 infection under therapeutic condition.
Figure 8.
Application of scAAV9-amiR-pTP (6x) and scAAV9-amiR-pTP (3x)-E1A_2 (3x) Concomitant with hAd5 in Immunosuppressed Syrian Hamsters Decreases hAd5 Infection of the Liver
The experimental procedure was the same as described in Figure 5A with two exceptions. The scAAV9 vectors were applied concomitantly with hAd5 at day 14 after first CP injection, and the hAd5 dose was reduced to 4 × 1010 vp/kg body weights. In each group, group n = 4, significance, *p < 0.05. Results show mean values ± SEM.
Discussion
We and others have previously demonstrated that hAd infections can be efficiently inhibited by anti-adenoviral RNAi in vitro.27, 28, 29, 30, 31, 44 However, the potential of RNAi inhibition of hAd infection in vivo has not been previously investigated. Thus, here we developed scAAV vectors expressing anti-adenoviral amiRs and analyzed their efficiency in the inhibition of hAd5 infection, first in vitro and then in vivo in immunosuppressed Syrian hamsters. This approach was chosen for several reasons: (1) scAAV vectors have a broad tissue tropism and enable rapid and strong transgene expression after transduction of the target cells in vitro and in vivo,40, 43, 45 (2) amiRs can efficiently silence target genes and can easily be delivered from viral vectors including AAV vectors in vitro and in vivo,46, 47, 48, 49 but were reported to have lower toxicity than shRNAs, which are more commonly used in the context of viral vector-based gene-silencing investigations,46, 48, 50, 51, 52 and (3) immunosuppressed Syrian hamsters represent an animal model that is permissive for hAd5.18, 53
Our initial in vitro evaluation experiments revealed that out of eight tested amiRs targeting the adenoviral genes E1A, IVa2, hexon, and pTP, only amiR-pTP inhibited the hAd5 replication with high efficiency. pTP is a component of the preinitiation complex which forms at the adenovirus origin of DNA replication and acts as the protein primer during DNA synthesis.54 Thus, our data confirm a previous assessment,31 that proteins of the adenoviral replication machinery represent the best targets for anti-adenoviral RNAi. The most plausible explanation for the lower inhibition of hAd5 replication by amiR-IVa2 seems to be the fact that it has a lower target gene-silencing activity than amiR-pTP. The reason for the failure of the amiRs directed against the E1A and hexon genes is less obvious, as their silencing activities were comparable with that of amiR-pTP. Moreover, previous investigations revealed efficient inhibition of hAd replication in vitro when E1A or hexon were silenced by siRNAs.27, 28, 29 It indicates that scAAV2 vector-mediated amiR delivery and/or intracellular amiR processing was too inefficient to generate amounts of active siRNAs which were sufficient to decrease E1A and hexon to a level that is low enough to prevent hAd5 replication.
The initial investigations cast a light on a comparable limited efficiency of scAAV2-delivered amiRs in vitro. This assessment is confirmed by the fact that amiR-pTP as the most efficient amiR only reduced adenoviral replication by about an order of magnitude, whereas in another study, a reduction of about two orders of magnitude was achieved with pTP-siRNAs.27 It has been shown that insertion of several copies of an amiR can increase amiR expression and improve the target gene silencing.31 In fact, here, we found that treatment with scAAV2 vectors containing three or six copies of amiR-pTP significantly increased amiR-pTP expression and hAd5 inhibition when compared with an scAAV2 vector containing only one copy of amiR-pTP. We did not investigate whether a further increase in amiR-pTP copy number would further enhance the performance of the vectors, however, anti-hAd vectors with three and six amiR-pTP copies had comparable anti-adenoviral efficiencies, so that it is unlikely that a further increase of the amiRs copy number will be advantageous. We also tested whether the anti-adenoviral efficiency of the amiR-pTP vector could be further increased by co-expression of amiR-E1A, as it has been shown previously that co-silencing of different adenoviral genes can reduce hAd-induced cytotoxicity, but only if one of the targeted genes is E1A.29 However, hAd5-induced in vitro cytotoxicity was not further reduced when amiR-E1A_2 was inserted into the vector genome in addition to amiR-pTP, although the concatemerized amiR-E1A_2 alone prevented the hAd5-induced cytotoxicity. One possible explanation for the failure of the co-silencing approach in vitro could be that the per se high efficiency of amiR-pTP could not be further improved by the addition of amiRs against E1A_2.
In consequence of the in vitro evaluation, we generated the amiR-pTP-expressing vector scAAV9-amiR-pTP (6×) for further in vivo investigations. We also generated an amiR-pTP/amiR-E1A-expressing vector scAAV9-amiR-pTP (3×)-E1A_2 (3×) in order to elucidate whether pTP/E1A co-silencing in vivo may perhaps still be advantageous compared to silencing of pTP alone. For the in vivo studies, we established a hAd5 infection model of immunosuppressed Syrian hamsters. This model is mainly characterized by strong replication of hAd5 in the liver and moderate liver pathology. Importantly, compared to a previously developed hAd5 infection model in immunosuppressed Syrian hamsters where a higher hAd5 dose is used,18 there were no mortalities of infected animals. The prophylactic application of scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-E1A_2 (3×), respectively, resulted in strong inhibition of hepatic hAd5 infection in infected animals. This was shown by up to two orders of magnitude fewer hAd5 in the liver and a substantial reduction of liver damage. In this regard, it should be noted that a certain, but not significant beneficial effect on liver pathology was observed with the amiR-Con vector (Figure 6A). Although the reason for this remains to be elucidated, we suggest that this effect was rather induced by the scAAV9 vector than by the expressed amiR-Con. This conclusion is supported by another experiment where coxsackievirus infections were treated by use of scAAV9 vectors. Also there, we observed a beneficial effect of the scAAV9 control vector, although the vector did not express amiRs (unpublished data). The antiviral effects correlated with the expression of amiR-pTP and amiR-E1A_2 in the liver and with silencing of the respective adenoviral genes pTP and E1A, demonstrating that inhibition of hAd5 infection was indeed a result of a specific amiR-induced RNAi mechanism. Even when applied concomitantly with hAd5, an anti-adenoviral efficiency of the vectors was detected, suggesting therapeutic potential of the amiR approach. Thus, our data demonstrate for the first time successful treatment of hAd5 infection by RNAi in vivo.
There were no significant differences between scAAV9-amiR-pTP (6×) and scAAV9-amiR-pTP (3×)-amiR-E1A_2 (3×) in reduction of hAd5 replication in the liver, but there was a trend toward reduced liver damage in animals treated with scAAV9-amiR-pTP (3×)-amiR-E1A_2 (3×) compared to those treated with scAAV9-amiR-pTP (6×). Interestingly, this effect was also seen in a further in vivo experiment with immunosuppressed Syrian hamsters treated with an about 10-fold higher hAd5 dose (Figure S4C). It suggests that co-silencing of pTP and E1A could possibly be beneficial to combat hAd5 infections in vivo, however, further investigations are necessary to verify it.
When comparing the efficiency of our anti-adenoviral RNAi approach to the efficiency of a new anti-adenoviral pharmacotherapy with the drug BCV in the animal model of immunosuppressed Syrian hamsters,18 BCV showed a distinctly higher efficiency. This is in some contrast to previous in vitro investigations, where a comparable efficiency of anti-adenoviral siRNAs and the BCV analog CDV has been observed.30, 31 Limited hepatic delivery of amiRs may be the main reason for this difference. An improvement of transduction by increasing vector dose, use of AAV vectors with modified capsids,55 or employment of small molecules56 may increase levels of expressed anti-adenoviral amiRs in the liver and increase the antiviral effect, but it may also increase the risk of side effects.52 In this regard, it should be noted that we did not find signs of liver pathology when the amiR-expressing vectors were applied to immunosuppressed hamsters (Figure S4C), indicating the safety of our approach.
Our study provides a first step in the translation of anti-adenoviral RNAi treatment from the bench to the clinic, but further developments will be necessary prior to its use in humans. In particular, it seems to be important to increase the amiRs levels in hAd5-infected cells. We suggest that for a substantial improvement of therapeutic efficiency for infections with moderate dose of hAd5, at least 10- to 100-fold higher scAAV9 vector doses are required. Although AAV vectors are currently the most promising vehicles for the delivery of transgenes and the first approved gene therapy is based on this type of viral vector,57 vector-free delivery systems may provide an appealing alternative. Relevant protocols using nanoparticle-mediated systemic delivery of stabilized siRNAs have been developed and were successfully used to treat hepatitis C virus infection in vivo recently.58 It may now be of interest to adapt these protocols to the specific requirements of hAd infection and analyze its efficacy in vivo and subsequently in humans.
In summary, here, we show that RNAi-induced gene silencing is a suitable approach to inhibit hAd5 infection in vivo. We found that amiRs delivered from scAAV9 vectors inhibit the development of disseminated hAd5 infections and reduce tissue damage of the liver in immunosuppressed Syrian hamsters. Moreover, the efficiency of anti-adenoviral RNAi was documented for the prophylactic and concomitant application of amiRs and no side effects were observed in our experiments. Thus, RNAi can be considered as a promising potential new approach to combat hAd infections in human patients.
Materials and Methods
Cell Culture
HEK293 cells and HeLa cell lines were cultured in high glucose DMEM (PAA Laboratories) supplemented with 10% fetal calf serum (FCS; c.c. pro and PAA Laboratories) and 1% each of penicillin and streptomycin (Sigma).
HepaRG cells were cultured in William’s E (GlutaMAX) supplemented with 5 μg/mL insulin, 50 μM hydrocortison, 10% FCS, and 1% penicillin and streptomycin for 14 days. The medium was changed twice a week. Thereafter, cells were treated with medium supplemented with 2% DMSO and cultured for an additional 14 days. Thereafter, the cells were used for transduction/infection experiments. After complete differentiation using DMSO, HepaRG resembled the liver cells in terms of metabolism in a stable, donor-independent way.
Design of amiRs
The amiRs amiR-E1A_1, amiR-E1A_2, amiR-E1A_3, amiR-Hex, and amiR-IVa2 were generated by embedding the previously described siRNA sequences siE1A_1, siE1A_2, siE1A_4, siHexon_4, and siIVa2_229 in an adapted form into the native environment of the miR-155 stem loop structure, respectively. For generation of amiRs-E1A_4 and amiR-E1A_5, the mature siRNA sequences of the amiR shR-E1A-73659 and shR-E1A-78559 were embedded in an adapted form into the miR-155 scaffold and named amiR-E1A_4 and amiR-E1_A5. The amiRs pTP-miR5 was used, as described previously31 and termed as amiR-pTP. Three different control amiRs were designed to target easily accessible areas of the red fluorescent protein drFP583 and referred to as amiR-Con_1, amiR-Con_2, and amiR-Con_3 (Figure 1A; Table S1).
Plasmids
The sequences of amiR-E1A_1, amiR-E1A_2, amiR-E1A_3, amiR-E1A_4, amiR-Hex, amiR-IVa2, and amiR-pTP (Table S1) were synthesized as a single segment of DNA and inserted into the plasmid pMK-RQ (kanR) by GeneArt. Each amiR was flanked by unique restriction sites for subcloning. As controls, amiR-Con_1 to 3 were synthesized and inserted as a cluster into pMK-RQ by GeneArt. The nine anti-adenoviral amiRs, as well as amiR-Con_1 to 3, were amplified by PCR, the DNA fragments digested with XhoI/ClaI, and inserted into the XhoI/ClaI-digested AAV shuttle plasmid pscAAV-CMV-GFP containing a self-complementary AAV vector genome and a GFP-expression cassette. The resulting plasmids were named pscAAV-CMV-GFP-amiR (9×) and pscAAV-CMV-amiR-Con (3×), respectively. pscAAV-CMV-GFP was generated from pscAAV-GFP60 by digestion with XbaI and religation. The AAV shuttle plasmid containing two stretches of amiR-Con_1 to 3 was constructed by insertion of a PCR product containing amiRs-Con_1 to 3 via BglII and ClaI into the plasmid pscAAV-CMV-GFP-amiR-Con (3×). The resulting plasmid was named pscAAV-GFP-amiR-Con (6×). Plasmids containing a GFP reporter and only one amiR were typically constructed by removal of the unnecessary amiRs from pscAAV-CMV-GFP-amiR (9×) and pscAAV-CMV-GFP-amiR-Con (3×) using restriction and cloning procedures. In plasmids expressing only the amiRs, but not the reporter, the GFP cDNA was cut out. These plasmids were named pscAAV-amiR-E1A_1, pscAAV-amiR-E1A_2, pscAAV-amiR-E1A_3, pscAAV-amiR-E1A_4, pscAAV-amiR-Hex, pscAAV-amiR-IVa2, pscAAV-amiR-pTP, pscAAV-amiR-Con, pscAAV-amiR-Con (3×), and pscAAV-amiR-Con (6×), respectively.
For generation of an AAV shuttle plasmid containing three copies of the amiR-pTP, the plasmid pMK-EcoRI-SpeI-amiR-pTP (3×)-NheI-ClaI (constructed by GeneArt) containing three copies of the amiR-pTP in tandem orientation was digested with EcoRI/ClaI, and the DNA fragment containing the pTP-amiRs was inserted into pscAAV-GFP via EcoRI/ClaI. The resulting plasmid was named pscAAV-CMV-amiR-pTP (3×). pscAAV-amiR-pTP (6×) contains six amiR-pTP. It was generated as follows: the plasmid pMK-EcoRI-SpeI-amiR-pTP (3×)-NheI-ClaI was digested with NheI/ClaI, and the DNA fragment containing the three amiR-pTP copies was inserted into NheI/ClaI-digested pscAAV-CMV-amiR-pTP (3×). The plasmid pAAV-amiR E1A_2 (3×) was generated from pscAAV-amiR-pTP (3×)-E1A_2 (3×) (see below) by restriction of the latter with EcoRI and religation.
Several plasmids were generated expressing three copies of amiR-pTP and amiR-E1A_2. These plasmids were constructed by a combination of gene synthesis of amiR sequences with plasmid cloning using pscAAV-CMV-amiR-pTP (3×) as the plasmid backbone. The pscAAV-amiR-E1A_2 (3×)-bidCMV-amiR-pTP (3×) contains a bidirectional CMV promoter with three copies of the amiR-pTP at one site and three copies of the amiR-E1A_2 at the opposite site of the bidirectional CMV promoter. pscAAV-amiR-E1A_2 (3×)-pTP (3×) contains a CMV promoter and three copies of the amiR-E1A_2 followed by three copies of the amiR-pTP. In pscAAV-amiR-pTP (3×)-E1A_2 (3×), the three copies of amiR-pTP and amiR-E1A_2 were inserted in reverse order compared to pscAAV-amiR-E1A_2 (3×)-pTP (3×). pscAAV-3× (amiR-E1A_2-pTP) contains three copies of amiR-E1A_2 and amiR-pTP in alternating order.
Plasmids containing miR-TS were generated by insertion of annealed miR-TS primers into the 3′ UTR of hRLuc reporter cDNA psiCheck2 (Promega) via XhoI and PmeI restriction sites.
To construct the AAV shuttle plasmid pscAAV-hRLuc containing hRLuc, hRLuc was amplified from a psiCHEK-2 vector using primers with EcoRI/XhoI linkers and inserted into pscAAV-GFP via EcoRI/XhoI. The correctness of all plasmids was confirmed by sequencing.
Construction of AAV Vectors
scAAV2 vectors were produced in 14.5 cm cell culture dishes by co-transfection of HEK293T cells with AAV shuttle plasmids and the AAV2 packaging plasmid pDG (kindly provided by Jürgen Kleinschmidt, Deutsches Krebsforschungszentrum, Heidelberg, Germany), as described.61 The luciferase-expressing vector scAAV9-hRLuc was produced in vented roller bottles by triple transfection of HEK293T cells by pscAAV-hRLuc- p5E18-VD2/9 (kindly provided by James M. Wilson, University of Pennsylvania, USA) and pHelper (Agilent Technologies), as described.62 At 2 days after transfection, vectors were released from the cells by three freeze-thaw cycles, treated with Benzonase (Merck; final concentration of 250 U/mL) for 1 hr at 37°C and the cell debris pelleted by centrifugation. The supernatants containing the scAAV2 vectors were stored at −80°C until use. The supernatants containing scAAV9-hRLuc were further purified by iodixanol gradient centrifugation, concentrated using Amicon Ultra-15 centrifugal filter devices (Merck Millipore) according to the instructions of the supplier, and stored at −80°C until use. AAV vector titers were determined by SYBR Green Real-Time PCR using the primers 5′-TGCCCAGTACATGACCTTATGG-3′ / 5′-GAAATCCCCGTGAGTCAAACC-3′ and SsoFast EvaGreen Supermix (Bio-Rad). The AAV vectors scAAV9-amiR-pTP (6×), scAAV9-amiR-pTP (3×)-E1A_2 (3×), and scAAV9-amiR-Con (6×) were constructed and the vector titers determined by Vigene Biosciences using the AAV shuttle plasmids pscAAV-amiR-pTP (6×), pscAAV-amiR-pTP (3×)-E1A_2 (3×) and pscAAV-amiR-Con (6×), respectively.
Production of hAd 5
hAd5 was a kind gift from Stefan Weger (Institute of Virology, Campus Benjamin Franklin, Charité - Universitätsmedizin Berlin, Berlin, Germany). It was reamplified on HEK293 cells, and the viral titers were determined by photometric measurement of the optical density at 260 nm to count virus particles ([vp]/μL) and by standard plaque assay to count plaque-forming units ([pfu]/μL) on HEK293 cells.
Luciferase Reporter Assays for Detection of amiR Activity
HeLa cells were seeded in 48-well plates. The next day they were transfected with plasmids expressing amiR and luciferase reporter plasmids containing the corresponding amiR-TS at a ratio of 7:1 using the transfection agent polyethylenimine. The cell cultures were incubated for 48 hr until analysis. Alternatively, HeLa cells were seeded in 48-well plates, transduced the next day with amiRs expressing scAAV2 vectors and incubated for an additional 72 hr. Thereafter, cells were trypsinized, plated out into 96-well plates, and transfected 24 hr later with luciferase reporter plasmids containing the corresponding amiR-TS for 24 hr until analysis. Firefly luciferase and hRLuc were analyzed using Dual-Luciferase Reporter Systems (Promega) in TriStar2 Multimode Reader LB 942 (Berthold Technologies), as recommended by the manufacturer.
Real-Time PCR for Detection of Adenoviral DNA
After discarding the supernatant of hAd5-infected cells, the cells were lysed in PBS by four freeze-thaw cycles. After centrifugation at 6,000 × g for 10 min, the supernatant was transferred to a fresh tube, an aliquot of 5 μL was diluted 1:10 in PBS, and inactivated at 95°C for 5 min. 2 μL of the solution were used directly in a real-time PCR for detection of hAd5 DNA using primers 5′-CACATCCAGGTGCCTCAGAA-3′ / 5′-AGGTGGCGTAAAGGCAAATG-3 directed against adenoviral hexon gene29 and the SsoFast EvaGreen Supermix (Bio-Rad). Alternatively, viral and genomic DNA were isolated from tissues using the Tissue DNA Mini Kit (PEQLAB) according to the manufacturer’s recommendations. 50 ng of isolated DNA were used in the real-time PCR reaction. Real-time PCR reaction was carried out in a C1000 Thermal Cycler and CFX96 Real-Time System (Bio-Rad) using the following program: 95°C, 3 min, 40 cycles with each: 95°C, 15 s, 55°C, 30 s, and 72°C, 30 s. 18S rRNA was analyzed as reference in a SYBR Green Real-Time PCR using the primer pair 5′-CCCCTCGATGCTCTTAGCTG-3′ / 5′-TCGTCTTCGAACCTCCGACT-3′ and same PCR program. The PCR reactions were carried out in duplicate. Relative hAd5 genome copy numbers were determined by the ΔΔc(t) calculation method.
Determination of Titers of Infectious hAd5 in Tissues
To release hAd5 from tissue, animal organs (<40 mg of tissue) were homogenized in 2 mL serum-free medium using a VDI 12 homogenizer (VWR International) followed by three freeze-thaw cycles. The debris was separated by centrifugation and the virus-containing supernatants stored at −80°C. Serum from animals was obtained by centrifugation of whole blood and stored at −80°C. To determine virus titers 6 × 105 HeLa cells per well were seeded in a 24-well cell culture plate. The next day the medium was removed, and cells were inoculated with 1:10 diluted virus solution in serum-free medium. The medium was changed 4 hr after infection, and the cells were harvested by trypsinization 48 hr after infection. The virus was released from the cells by three freeze-thaw cycles, and the amount of adenoviral DNA and 18S rRNA was detected by real-time PCR, as described above. Absolute quantification assays were done by infecting HeLa cells with 100, 500, 1,000, and 5,000 pfu of hAd5 ( = hAd5 standard) in parallel and the amount of adenoviral DNA after release of the virus from the cells 48 hr later, as described above. A standard curve was generated mapping ΔCt (Ct Hex − Ct 18S) over virus titer of the hAd5 standard and was used to calculate the hAd5 load in organs and cells. Alternatively to this procedure, virus titers were determined by use of the 50% tissue culture infective dose (TCID50), as described previously18 (only for data shown in the Figure S4B).
Measurement of ALT and AST
Serum analysis for ALT and AST activity was done by Advanced Veterinary Laboratories or carried out by use of Infinity ALT (GPT) Liquid Stable Reagent and Infinity AST (GOT) Liquid Stable Reagent (Thermo Fisher Scientific).
Cell Viability Assay
Cell viability was determined using the Cell Proliferation Kit II (XTT) (Roche) as recommended by manufacturer. Data were normalized to untreated mock infected cells.
Quantification of hAd5 mRNA Expression
Total RNA was isolated from tissue and cell cultures with TRIzol (Thermo Fisher Scientific) according to manufacturer’s recommendations. 5 μg of the RNA were treated with 2 U DNaseI (New England Biolabs) for 1 hr. 2 μg DNaseI-treated RNA were reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) in a 20 μL reaction. For determination of adenoviral pTP and E1A expression, 2 μL of this reaction were mixed with the SsoFast EvaGreen Supermix and the primer pairs 5′-CGGCGCAGGTCTTTGTAG −3′ / 5′-CACGCATGGGAGGAAGAG-3′ (pTP mRNA detection) and 5′-CTTGGGTCCGGTTTCTATGC-3′ / 5′-CCCGTATTCCTCCGGTGATA-3′ (E1A mRNA detection), respectively, and analyzed using the above described PCR program. Cellular 18S rRNA expression was used as reference and analyzed using the same PCR primers as mentioned above. The relative pTP and E1A mRNA expression was determined by the ΔΔc(t) calculation method.
Quantification of amiR Expression
Total RNA was isolated using TRIzol (Thermo Fisher Scientific), and the expression of mature amiR-pTP and amiR-E1A_2 was quantified using Custom Design TaqMan MicroRNA Assays (Thermo Fisher Scientific) according to the manufacturer’s recommendations. SnU6RNA served as internal standard (TaqMan MicroRNA Assay Number: 001973, Thermo Fisher Scientific).
Ethic Statement for In Vivo Experiments
All in vivo procedures involving the use and care of animals were performed according to the Guide for the Care and Use of Laboratory Animals published by the US NIH (NIH Publication No. 85-23, revised 1996) and the German animal protection code.
In Vivo Imaging of Luciferase Expression in Immunosuppressed Syrian Hamsters Transduced with scAAV9-hRLuc
Syrian hamsters were obtained from Envigo. Animals were immunosuppressed with CP according to a previously established protocol.18 Briefly, CP was administered i.p. at a dose of 140 mg/kg and then twice weekly at a dose of 100 mg/kg. At the time of the first injection, the animals weighed between 70 and 80 g. Concomitant with the first CP treatment, the animals were injected with 2 × 1011 vge of scAAV9-hRLuc i.v. percutaneously into the jugular vein. For detection of hRLuc expression, the hamsters were anesthetized with isoflurane to effect, injected with 150 μg/animal of coelenterazine (XenoLight RediJect Coelenterazine, PerkinElmer) i.p., and 10 min later imaged in an in vivo imaging system (IVIS) Spectrum Optical Imaging Platform (PerkinElmer). Images were normalized using the same color/intensity scale. In vivo imaging of luciferase activity was carried out 5, 11, and 15 days after vector transduction. At 21 days after vector injection, two scAAV9-hRLuc-transduced animals and one mock infected animal were injected i.p. with coelenterazine and sacrificed 20 min later. The animal was dissected and select organs were imaged for luciferase activity.
Course of hAd5 Infection in Immunosuppressed Syrian Hamsters
Male Syrian hamsters supplied from Charles River Laboratories were treated at an age of 5 weeks (weight between 100 and 135 g) with CP. CP treatment was repeated during the whole experiment, as described above. At 2 weeks after initial CP injection, the animals were narcotized with isoflurane, the left jugular vein was prepared, and hAd5 injected into the vein at a dose of 4 × 1011 vp/kg. The surgical step was accompanied by analgesic treatment with 5 mg/kg carprosol (CP-Pharma). Animals were sacrificed for organ harvest 3, 7, and 14 days after infection.
Determination of hAd5 Infection of Immunosuppressed Syrian Hamsters after Treatment with amiR-Expressing scAAV9 Vectors
Animals were supplied from Charles River Laboratories and treated at an age of 5 weeks with CP, as described above. In the prophylactic approach, concomitant to the onset of immunosuppression, the animals were transduced with 5 ×1013 vge/kg scAAV9-amiR-Con (6×), scAAV9-amiR-pTP (6×), or scAAV9-amiR-pTP (3×)-E1A_2 (3×) or were injected with physiological saline into the left jugular vein under isoflurane narcosis. At 2 weeks later, animals were infected with 4 × 1011 vp/kg hAd5 under isoflurane narcosis into the right jugular vein. One group that already had received saline was again injected with saline. Each surgical step was accompanied by analgesic treatment with carprosol, as described above. Animals were sacrificed 7 days after infection with hAd5, and the organs were dissected and rapidly frozen in liquid nitrogen or put in 4% formalin. In the therapeutic approach, Syrian hamsters were treated as described above with the following modification: scAAV9 vectors were injected concomitantly with hAd5 and hAd5 was used at a dose of 4 × 1010 vp/kg. Alternatively, 5-week-old hamsters were obtained from Envigo and treated with CP, as described above. Concomitant with the first CP injection, three groups of animals were injected via the jugular vein under ketamine-xylazine narcosis with one of the three scAAV9-amiR vectors at a dose of 5 × 1013 vge/kg and a fourth group received vehicle (PBS). At 14 days after vector administration, half of the animals in all groups were injected i.v. (into the jugular vein) with hAd5 at a dose of 5.58 × 1012 vp/kg, while the remaining animals received vehicle (PBS) injections. Animals were sacrificed 7 days later.
Immunohistological Examination
Tissues were formalin-fixed, paraffin-embedded (FFPE), cut, and stained with H&E, as previously described.63 For immunohistochemical staining, 4- to 5-μm sections of the FFPE hearts were cut and deparaffinized in xylene and rehydrated in graded alcohols.64 Endogenous peroxidases were blocked by incubating sections in methanol with 3% hydrogen peroxide for 20 min. Antigen heat retrieval was performed in citric buffer for 12 min, followed by a cooling period of 15 min. Slides were then incubated for 30 min with Roti-Immunoblock (Roth) and goat serum to block non-specific antibody binding. Sections were incubated overnight at 4°C with monoclonal mouse anti-adenovirus antibody (ab3648, Abcam). The antibody recognizes the hAd5 E1A protein.65 Slides were then incubated with a biotinylated secondary goat anti-mouse antibody (dilution 1:200 each; Vector Laboratories) for 30 min at room temperature. Immunolabeling was performed by an avidin-biotin-immunoperoxidase system (Vectastain Elite ABC Kit; Vector Laboratories). Diaminobenzidine tetrahydrochloride (DAB; Merck) was used for antigen visualization of viral protein.
Statistics
Results are expressed as means ± SEM. To test for statistical significance of in vitro data, an unpaired Student’s t test was applied. Statistical significance of in vivo data was determined using the Mann-Whitney U test.
Author Contributions
K.S. conducted in vitro and in vivo experiments with hAd5-infected immunosuppressed Syrian hamsters and contributed to writing the manuscript. A.G. performed in vitro experiments. M.K. designed amiRs and carried out in vitro experiments. S.P. and M.P. contributed to in vivo experiments with hAd5-infected immunosuppressed Syrian hamsters. R.K. conducted histological and immunohistological investigations. J.F.S., A.E.T., B.Y., W.S.W., and K.T. contributed by in vivo imaging of immunosuppressed Syrian hamsters after AAV9 vector transduction and by design of the study. J.K. contributed to the writing of the paper. H.F. designed the experiments and wrote the paper.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through grant FE785/4-1 to H.F. The project was also funded in part with funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health and Human Services (http://www.niaid.nih.gov), under contract no. HHSN272201000021.We thank Xiaomin Wang for help with hAd5 preparation and Erik Wade for critical reading of the manuscript and helpful comments.
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.07.002.
Supplemental Information
References
- 1.Kidd A.H., Jonsson M., Garwicz D., Kajon A.E., Wermenbol A.G., Verweij M.W., De Jong J.C. Rapid subgenus identification of human adenovirus isolates by a general PCR. J. Clin. Microbiol. 1996;34:622–627. doi: 10.1128/jcm.34.3.622-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echavarría M. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 2008;21:704–715. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong J.Y., Lee H.J., Piedra P.A., Choi E.H., Park K.H., Koh Y.Y., Kim W.S. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin. Infect. Dis. 2001;32:1423–1429. doi: 10.1086/320146. [DOI] [PubMed] [Google Scholar]
- 4.Rocholl C., Gerber K., Daly J., Pavia A.T., Byington C.L. Adenoviral infections in children: the impact of rapid diagnosis. Pediatrics. 2004;113:e51–e56. doi: 10.1542/peds.113.1.e51. [DOI] [PubMed] [Google Scholar]
- 5.Lion T., Baumgartinger R., Watzinger F., Matthes-Martin S., Suda M., Preuner S., Futterknecht B., Lawitschka A., Peters C., Potschger U., Gadner H. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood. 2003;102:1114–1120. doi: 10.1182/blood-2002-07-2152. [DOI] [PubMed] [Google Scholar]
- 6.Schilham M.W., Claas E.C., van Zaane W., Heemskerk B., Vossen J.M., Lankester A.C., Toes R.E., Echavarria M., Kroes A.C., van Tol M.J. High levels of adenovirus DNA in serum correlate with fatal outcome of adenovirus infection in children after allogeneic stem-cell transplantation. Clin. Infect. Dis. 2002;35:526–532. doi: 10.1086/341770. [DOI] [PubMed] [Google Scholar]
- 7.Vyas J.M., Marasco W.A. Fatal fulminant hepatic failure from adenovirus in allogeneic bone marrow transplant patients. Case Rep. Infect. Dis. 2012;2012:463569. doi: 10.1155/2012/463569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terasako K., Oshima K., Wada H., Ishihara Y., Kawamura K., Sakamoto K., Ashizawa M., Sato M., Machishima T., Nakasone H. Fulminant hepatic failure caused by adenovirus infection mimicking peliosis hepatitis on abdominal computed tomography images after allogeneic hematopoietic stem cell transplantation. Intern. Med. 2012;51:405–411. doi: 10.2169/internalmedicine.51.6432. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin A., Kingman H., Darville M., Foot A.B., Grier D., Cornish J.M., Goulden N., Oakhill A., Pamphilon D.H., Steward C.G., Marks D.I. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 2000;26:1333–1338. doi: 10.1038/sj.bmt.1702716. [DOI] [PubMed] [Google Scholar]
- 10.Howard D.S., Phillips G.L., II, Reece D.E., Munn R.K., Henslee-Downey J., Pittard M., Barker M., Pomeroy C. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 1999;29:1494–1501. doi: 10.1086/313514. [DOI] [PubMed] [Google Scholar]
- 11.Hierholzer J.C. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014;27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wy Ip W., Qasim W. Management of adenovirus in children after allogeneic hematopoietic stem cell transplantation. Adv. Hematol. 2013;2013:176418. doi: 10.1155/2013/176418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaar K., Röger C., Pozzuto T., Kurreck J., Pinkert S., Fechner H. Biological antivirals for treatment of adenovirus infections. Antivir. Ther. (Lond.) 2016;21:559–566. doi: 10.3851/IMP3047. [DOI] [PubMed] [Google Scholar]
- 15.De Clercq E., Holý A. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat. Rev. Drug Discov. 2005;4:928–940. doi: 10.1038/nrd1877. [DOI] [PubMed] [Google Scholar]
- 16.Plosker G.L., Noble S. Cidofovir: a review of its use in cytomegalovirus retinitis in patients with AIDS. Drugs. 1999;58:325–345. doi: 10.2165/00003495-199958020-00015. [DOI] [PubMed] [Google Scholar]
- 17.Ciesla S.L., Trahan J., Wan W.B., Beadle J.R., Aldern K.A., Painter G.R., Hostetler K.Y. Esterification of cidofovir with alkoxyalkanols increases oral bioavailability and diminishes drug accumulation in kidney. Antiviral Res. 2003;59:163–171. doi: 10.1016/s0166-3542(03)00110-4. [DOI] [PubMed] [Google Scholar]
- 18.Toth K., Spencer J.F., Dhar D., Sagartz J.E., Buller R.M.L., Painter G.R., Wold W.S. Hexadecyloxypropyl-cidofovir, CMX001, prevents adenovirus-induced mortality in a permissive, immunosuppressed animal model. Proc. Natl. Acad. Sci. USA. 2008;105:7293–7297. doi: 10.1073/pnas.0800200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voigt S., Hofmann J., Edelmann A., Sauerbrei A., Kühl J.S. Brincidofovir clearance of acyclovir-resistant herpes simplex virus-1 and adenovirus infection after stem cell transplantation. Transpl. Infect. Dis. 2016;18:791–794. doi: 10.1111/tid.12582. [DOI] [PubMed] [Google Scholar]
- 20.Florescu D.F., Pergam S.A., Neely M.N., Qiu F., Johnston C., Way S., Sande J., Lewinsohn D.A., Guzman-Cottrill J.A., Graham M.L. Safety and efficacy of CMX001 as salvage therapy for severe adenovirus infections in immunocompromised patients. Biol. Blood Marrow Transplant. 2012;18:731–738. doi: 10.1016/j.bbmt.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camargo J.F., Morris M.I., Abbo L.M., Simkins J., Saneeymehri S., Alencar M.C., Lekakis L.J., Komanduri K.V. The use of brincidofovir for the treatment of mixed dsDNA viral infection. J. Clin. Virol. 2016;83:1–4. doi: 10.1016/j.jcv.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Awosika O.O., Lyons J.L., Ciarlini P., Phillips R.E., Alfson E.D., Johnson E.L., Koo S., Marty F., Drew C., Zaki S. Fatal adenovirus encephalomyeloradiculitis in an umbilical cord stem cell transplant recipient. Neurology. 2013;80:1715–1717. doi: 10.1212/WNL.0b013e3182904f96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keyes A., Mathias M., Boulad F., Lee Y.J., Marchetti M.A., Scaradavou A., Spitzer B., Papanicolaou G.A., Wieczorek I., Busam K.J. Cutaneous involvement of disseminated adenovirus infection in an allogeneic stem cell transplant recipient. Br. J. Dermatol. 2016;174:885–888. doi: 10.1111/bjd.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 25.Poller W., Fechner H. Development of novel cardiovascular therapeutics from small regulatory RNA molecules--an outline of key requirements. Curr. Pharm. Des. 2010;16:2252–2268. doi: 10.2174/138161210791792813. [DOI] [PubMed] [Google Scholar]
- 26.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew. Chem. Int. Ed. Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kneidinger D., Ibrišimović M., Lion T., Klein R. Inhibition of adenovirus multiplication by short interfering RNAs directly or indirectly targeting the viral DNA replication machinery. Antiviral Res. 2012;94:195–207. doi: 10.1016/j.antiviral.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung Y.S., Kim M.K., Lee W.J., Kang C. Silencing E1A mRNA by RNA interference inhibits adenovirus replication. Arch. Virol. 2007;152:1305–1314. doi: 10.1007/s00705-007-0951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckstein A., Grössl T., Geisler A., Wang X., Pinkert S., Pozzuto T., Schwer C., Kurreck J., Weger S., Vetter R. Inhibition of adenovirus infections by siRNA-mediated silencing of early and late adenoviral gene functions. Antiviral Res. 2010;88:86–94. doi: 10.1016/j.antiviral.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Pozzuto T., Röger C., Kurreck J., Fechner H. Enhanced suppression of adenovirus replication by triple combination of anti-adenoviral siRNAs, soluble adenovirus receptor trap sCAR-Fc and cidofovir. Antiviral Res. 2015;120:72–78. doi: 10.1016/j.antiviral.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Ibrišimović M., Kneidinger D., Lion T., Klein R. An adenoviral vector-based expression and delivery system for the inhibition of wild-type adenovirus replication by artificial microRNAs. Antiviral Res. 2013;97:10–23. doi: 10.1016/j.antiviral.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing S.G., Byrd S.A., Christensen J.B., Tyler R.E., Imperiale M.J. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 2007;81:12450–12457. doi: 10.1128/JVI.01470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W., Imperiale M.J. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 2003;77:3586–3594. doi: 10.1128/JVI.77.6.3586-3594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyler R.E., Ewing S.G., Imperiale M.J. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 2007;81:3447–3454. doi: 10.1128/JVI.02097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell W.C. Update on adenovirus and its vectors. J. Gen. Virol. 2000;81:2573–2604. doi: 10.1099/0022-1317-81-11-2573. [DOI] [PubMed] [Google Scholar]
- 36.Juliano R., Alam M.R., Dixit V., Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe A., Arai M., Yamazaki M., Koitabashi N., Wuytack F., Kurabayashi M. Phospholamban ablation by RNA interference increases Ca2+ uptake into rat cardiac myocyte sarcoplasmic reticulum. J. Mol. Cell. Cardiol. 2004;37:691–698. doi: 10.1016/j.yjmcc.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Saha A., Bhagyawant S.S., Parida M., Dash P.K. Vector-delivered artificial miRNA effectively inhibited replication of Chikungunya virus. Antiviral Res. 2016;134:42–49. doi: 10.1016/j.antiviral.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisler A., Schön C., Größl T., Pinkert S., Stein E.A., Kurreck J., Vetter R., Fechner H. Application of mutated miR-206 target sites enables skeletal muscle-specific silencing of transgene expression of cardiotropic AAV9 vectors. Mol. Ther. 2013;21:924–933. doi: 10.1038/mt.2012.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inagaki K., Fuess S., Storm T.A., Gibson G.A., Mctiernan C.F., Kay M.A., Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L., Jiang J., Drouin L.M., Agbandje-McKenna M., Chen C., Qiao C., Pu D., Hu X., Wang D.Z., Li J., Xiao X. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. USA. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffelers M.J., Blaauboer B.J., Bakker W.E., Beken S., Hendriksen C.F., Koëter H.B., Krul C. Regulatory acceptance and use of 3R models for pharmaceuticals and chemicals: expert opinions on the state of affairs and the way forward. Regul. Toxicol. Pharmacol. 2014;69:41–48. doi: 10.1016/j.yrtph.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Röger C., Pozzuto T., Klopfleisch R., Kurreck J., Pinkert S., Fechner H. Expression of an engineered soluble coxsackievirus and adenovirus receptor by a dimeric AAV9 vector inhibits adenovirus infection in mice. Gene Ther. 2015;22:458–466. doi: 10.1038/gt.2015.19. [DOI] [PubMed] [Google Scholar]
- 44.Nikitenko N.A., Speiseder T., Lam E., Rubtsov P.M., Tonaeva Kh.D., Borzenok S.A., Dobner T., Prassolov V.S. Regulation of human adenovirus replication by rna interference. Acta Naturae. 2015;7:100–107. [PMC free article] [PubMed] [Google Scholar]
- 45.McCarty D.M., Fu H., Monahan P.E., Toulson C.E., Naik P., Samulski R.J. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Tang X., Zhu C., Song Y., Yin J., Xu J., Ertl H.C., Zhou D. Adenovirus-mediated artificial MicroRNAs targeting matrix or nucleoprotein genes protect mice against lethal influenza virus challenge. Gene Ther. 2015;22:653–662. doi: 10.1038/gt.2015.31. [DOI] [PubMed] [Google Scholar]
- 47.Ivacik D., Ely A., Ferry N., Arbuthnot P. Sustained inhibition of hepatitis B virus replication in vivo using RNAi-activating lentiviruses. Gene Ther. 2015;22:163–171. doi: 10.1038/gt.2014.94. [DOI] [PubMed] [Google Scholar]
- 48.Gröβl T., Hammer E., Bien-Möller S., Geisler A., Pinkert S., Röger C., Poller W., Kurreck J., Völker U., Vetter R., Fechner H. A novel artificial microRNA expressing AAV vector for phospholamban silencing in cardiomyocytes improves Ca2+ uptake into the sarcoplasmic reticulum. PLoS ONE. 2014;9:e92188. doi: 10.1371/journal.pone.0092188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoica L., Todeasa S.H., Cabrera G.T., Salameh J.S., ElMallah M.K., Mueller C., Brown R.H., Jr., Sena-Esteves M. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann. Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borel F., van Logtenstein R., Koornneef A., Maczuga P., Ritsema T., Petry H., van Deventer S.J., Jansen P.L., Konstantinova P. In vivo knock-down of multidrug resistance transporters ABCC1 and ABCC2 by AAV-delivered shRNAs and by artificial miRNAs. J. RNAi Gene Silencing. 2011;7:434–442. [PMC free article] [PubMed] [Google Scholar]
- 51.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 53.Toth K., Lee S.R., Ying B., Spencer J.F., Tollefson A.E., Sagartz J.E., Kong I.K., Wang Z., Wold W.S. Correction: Stat2 knockout syrian hamsters support enhanced replication and pathogenicity of human adenovirus, revealing an important role of type I interferon response in viral control. PLoS Pathog. 2016;12:e1005392. doi: 10.1371/journal.ppat.1005392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Botting C.H., Hay R.T. Role of conserved residues in the activity of adenovirus preterminal protein. J. Gen. Virol. 2001;82:1917–1927. doi: 10.1099/0022-1317-82-8-1917. [DOI] [PubMed] [Google Scholar]
- 55.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolson S.C., Li C., Hirsch M.L., Setola V., Samulski R.J. Identification and validation of small molecules that enhance recombinant adeno-associated virus transduction following high-throughput screens. J. Virol. 2016;90:7019–7031. doi: 10.1128/JVI.02953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon J.S., Lee S.H., Kim E.J., Cho H., Lee W., Kim G.W., Park H.J., Cho S.W., Lee C., Oh J.W. Inhibition of hepatitis c virus in mice by a small interfering RNA targeting a highly conserved sequence in viral ires pseudoknot. PLoS ONE. 2016;11:e0146710. doi: 10.1371/journal.pone.0146710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gürlevik E., Woller N., Schache P., Malek N.P., Wirth T.C., Zender L., Manns M.P., Kubicka S., Kühnel F. p53-dependent antiviral RNA-interference facilitates tumor-selective viral replication. Nucleic Acids Res. 2009;37:e84. doi: 10.1093/nar/gkp374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fechner H., Sipo I., Westermann D., Pinkert S., Wang X., Suckau L., Kurreck J., Zeichhardt H., Müller O., Vetter R. Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J. Mol. Med. (Berl.) 2008;86:987–997. doi: 10.1007/s00109-008-0363-x. [DOI] [PubMed] [Google Scholar]
- 61.Geisler A., Jungmann A., Kurreck J., Poller W., Katus H.A., Vetter R., Fechner H., Müller O.J. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011;18:199–209. doi: 10.1038/gt.2010.141. [DOI] [PubMed] [Google Scholar]
- 62.Sipo I., Knauf M., Fechner H., Poller W., Planz O., Kurth R., Norley S. Vaccine protection against lethal homologous and heterologous challenge using recombinant AAV vectors expressing codon-optimized genes from pandemic swine origin influenza virus (SOIV) Vaccine. 2011;29:1690–1699. doi: 10.1016/j.vaccine.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 63.Klopfleisch R., Lenze D., Hummel M., Gruber A.D. The metastatic cascade is reflected in the transcriptome of metastatic canine mammary carcinomas. Vet. J. 2011;190:236–243. doi: 10.1016/j.tvjl.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 64.Klose P., Weise C., Bondzio A., Multhaup G., Einspanier R., Gruber A.D., Klopfleisch R. Is there a malignant progression associated with a linear change in protein expression levels from normal canine mammary gland to metastatic mammary tumors? J. Proteome Res. 2011;10:4405–4415. doi: 10.1021/pr200112q. [DOI] [PubMed] [Google Scholar]
- 65.Harlow E., Franza B.R., Jr., Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.