Abstract
Both iron and lipids are involved in the progression of alcoholic fatty liver disease (AFLD), but the interaction between iron and lipids in AFLD is unclear. Here, we tested the hypothesis that iron regulates the expression of genes involved in lipid metabolism through iron regulatory proteins (IRPs), which interact with the iron-responsive elements (IREs) in the untranslated regions (UTRs) of genes, resulting in lipid accumulation. Using “RNA structure software”, we predicted the mRNA secondary structures of more than 100 genes involved in lipid metabolism to investigate whether the IRE structure exists in novel mRNAs. Cholesterol 7α-hydroxylase (Cyp7a1) has an IRE-like stem-loop, a noncanonical IRE structure, in its 3′-UTR. Cyp7a1 expression can be regulated by in vivo and in vitro iron treatment. In addition, the noncanonical IRE motif can efficiently bind both to IRP1 and IRP2. The results indicate that hepatic iron overloading in AFLD mice decreased Cyp7a1 expression and resulted in cholesterol accumulation, providing a new mechanism of iron-regulated gene transcription and translation through the interaction between iron and a noncanonical IRE structure in Cyp7a1 mRNA. This finding has significant implications in studying a proposed mechanism for the regulation of cholesterol homeostasis by an Fe/IRP/noncanonical IRE axis.
Keywords: alcoholic fatty liver disease, iron regulatory protein 1, iron regulatory protein 2, non-canonical IRE, mouse models, transcriptional regulation
Excessive alcohol consumption results in alcoholic liver disease (ALD), which refers to a spectrum of liver disorders ranging from alcoholic fatty liver (simple steatosis) to alcoholic steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma (1). Alcoholic fatty liver disease (AFLD) is a reversible condition and thus plays an important role in preventing the evolution of ALD toward severe stages (2). Therefore, AFLD has been a hot spot in research. The most commonly used laboratory models of alcohol abuse are intragastric infusion, the Lieber-DeCarli all liquid diet, and the alcohol agar block model. However, all of these methods have limitations. Therefore, we established a mouse model for AFLD by administering alcohol diluted in drinking water, which mimics human alcohol use behavior patterns. Recent evidence suggests that AFLD patients frequently display evidence of lipid accumulation (3) and iron overload (4–7). Even a slight alcohol intake has been shown to elevate the indices of iron storage (8). Our AFLD mouse model displayed apparent hepatic iron overloading and lipid accumulation. Iron is an essential element for living organisms that plays a vital role in many physiological events. Proteins, such as hemoglobin, may contain iron or iron-sulfur clusters in the prosthetic form. Such proteins frequently participate in the process of oxygen storage, catabolism, and redox reactions. Iron deficiency and iron overload can cause serious clinical consequences (9). Previous research has explored the subjects of iron and lipid metabolism. Most studies have focused on the oxidative stress caused by iron-mediated reactive oxygen species (ROS), which initiate lipid peroxidation. Several studies have shown that increased hepatic iron can increase plasma cholesterol (10). Also, Graham et al. (11) demonstrate that hepatic iron loading increases liver cholesterol synthesis. However, there are no findings to indicate that iron regulates gene expression, which participates in cholesterol metabolism directly. Therefore, further clarification of the potential role of iron in disordered lipid metabolism is required.
Cellular iron homeostasis is accomplished by the coordinated regulation of iron uptake, storage, and export by iron regulatory protein (IRP)1 and IRP2 (also known as ACO1 and IREB2) (12). IRPs bind iron-responsive elements (IREs) in the untranslated regions (UTRs) of mRNAs. When iron is in excess, IRPs do not generally bind to IREs. IRP1 acquires a [4Fe-4S] cluster and generates an aconitase, while IRP2 undergoes degradation. When cells are iron deficient, IRPs bind to IREs (13). If IREs are present in the 3′-UTR of target mRNAs, such as divalent metal transporter 1 (DMT1) (14) and transferrin receptor 1 (TFR1) (15), IRP binding can increase their expression by stabilizing the mRNAs. Conversely, if an IRE is located in the 5′-UTR of target mRNAs, such as in L- and H-ferritin (13) or ferroportin 1 (FPN1) (16), IRP binding can block the translation of these mRNAs. The canonical IRE consists of a stem-loop structure that is approximately 30 nucleotides long, with a terminal hexanucleotide CAGUG(N)/CAGAG(N) loop and an unpaired cytosine (C8) on the 5′ strand of the stem, as well as an additional variable length lower stem. Biochemical studies have indicated that the first (C14) and the fifth (G18) nucleotides of the terminal loop pair can form an AGU/AGA pseudo-triloop that is critical for the binding of IRPs (17). In addition, several IREs can present a noncanonical form of their structure, and so-called noncanonical IREs also exist (18–20). Noncanonical IREs can have a larger terminal loop (CAGUGUCA) (18) or have other nucleotides in the bulge region in place of the unpaired cytosine [e.g., in α-hemoglobin stabilizing protein (AHSP)] (19) and yet are still able to bind IRP1 and/or IRP2. In this report, we discovered a novel stem-loop structure in the 3′-UTR of cholesterol 7α-hydroxylase (Cyp7a1) mRNA that functions as a noncanonical IRE. Cyp7a1 is a cholesterol scavenger. Because excess cholesterol can be toxic like iron, there are several pathways that mammalian cells apply to clear excess cholesterol. The most vital pathway, accounting for approximately 90% of cholesterol breakdown, is the conversion of cholesterol into bile acids by more than 16 enzymes in the liver. Among these enzymes, Cyp7a1 catalyzes the first and rate-limiting step in the process (21, 22).
In this report, we verified the hypothesis that iron can regulate the expression of Cyp7a1 through the noncanonical IRE at the 3′-UTR of Cyp7a1 mRNA, which can interact with IRPs and thus result in cholesterol accumulation. The identification of a noncanonical IRE in Cyp7a1 mRNA represents a novel molecular mechanism connecting iron and lipid metabolism through the IRE/IRP regulatory system.
MATERIALS AND METHODS
RNA secondary structure predictions
RNA structure software was used to predict secondary structures of mRNAs. Cyp7a1 WT IRE and Cyp7a1 MUT IRE probes (Bioneer) for pulldown experiments are still predicted to form a stem loop structure, such as in the 3′-UTR of Cyp7a1 mRNA.
Mice
All animal experiments were approved by the Harbin Medical University Animal Care and Use Committee and were conducted according to National Institutes of Health guidelines. C57BL6/J mice were housed under a constant light/dark cycle in the SPF Mouse Barrier Unit. The control group was allowed free access to food and water. For chronic alcohol administration, male mice (8–10 weeks of age) were fed water containing 5% alcohol during the first week. The concentration was gradually increased every 2 weeks by 5% until the alcohol concentration reached 15% (v/v) (Fig. 1A). Mice were euthanized at 12 months of age. Livers were harvested and snap-frozen in liquid nitrogen. To create an iron deficiency model, approximately 400–500 μl of blood were drawn once a week for 4 weeks from control and AFLD mice. To create an iron loading model, mice were given iron-dextran (1 g/kg body weight; Sigma-Aldrich) or saline by an intraperitoneal injection once a week for 4 weeks.
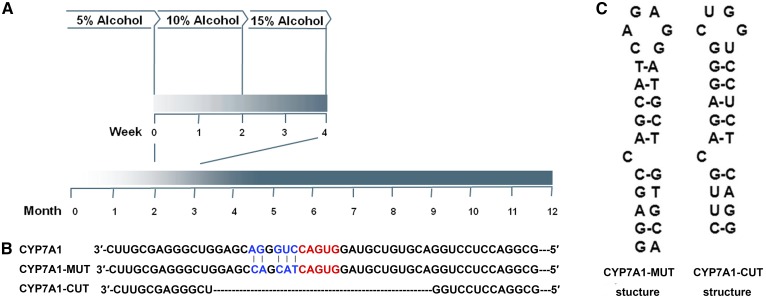
Fig. 1.
A: Strategy of alcohol administration in mice. B: The recombination construct of Luc/CYP7A1-3′-UTR-WT. The Luc/CYP7A1-3′-UTR-MUT sequence contains five mutations, which are marked in blue. Luc/CYP7A1-3′-UTR-CUT has a deletion in the region of the Cyp7a1 IRE-like sequence. C: The RNA secondary structure of the CYP7A1-3′-UTR-MUT and CYP7A1-3′-UTR-CUT was assessed by the software.
Cell culture
Mouse Hepa1-6 cells were cultured in Dulbecco’s modified essential medium (Invitrogen), and murine hepatocyte cell line AML12 cells (ATCC; CRL-2254) were cultured in complete growth medium that was prepared as follows: DMEM (1:1), Ham’s Nutrient Mixture F12 (1:1; Gibco, Life Technologies) with an insulin-transferrin-selenium mixture (Gibco, Life Technologies), and 40 ng/ml dexamethasone (Sigma-Aldrich), supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin at 37°C in 5% CO2.
Treatment of cells
To deplete Hepa1-6 cells of iron, the chelator, desferrioxamine mesylate (DFO) (0–100 μM, D9533; Sigma-Aldrich), was added to the cell culture medium. Iron loading of Hepa1-6 cells was achieved by incubating cells in the presence of iron-dextran (0–100 μg/ml; Sigma-Aldrich). Cells were collected 24 h after treatment and at 48 h after treatment for extraction of mRNA and protein, respectively. For alcohol treatment, Hepa1-6 cells were treated for 48 h with 0–200 mM alcohol (Sigma-Aldrich); then, cells were treated for 24 h with DFO (0–100 μM) in the presence of 200 mM alcohol.
Blood chemistry
Mice were fasted overnight before blood was drawn from the angular vein of the eye. Approximately 200 μl of serum was obtained after centrifugation 2,300 g for 5 min at 4°C) and was used for serum biochemical analysis by the automatic biochemistry analyzer (Sysmex Chemix-180; Sysmex, Japan).
Hepatic lipid accumulation analysis
The hepatic triglyceride (TG) levels were measured in liver homogenate using a commercial kit (Applygen, Beijing, China) according to the manufacturer’s instructions. The total cholesterol level in liver tissue was detected by HPLC, as described previously (23).
Quantitative real-time PCR
Total RNA was purified by homogenizing cells in Trizol reagent (Invitrogen). cDNA was obtained from 2 μg of total RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Gene expression was quantified by an Applied Biosystems 7500 real-time PCR system using SYBR Green reagent (Roche) and mRNA-specific primers, and the β-actin mRNA levels were used for normalization.
Western blot
Cells and tissues were lysed in ice-cold RIPA lysis buffer. After centrifugation of the lysates at 16,500 g for 15 min at 4°C, the supernatants were collected and stored at −80°C until analysis. Equal amounts of protein were separated on a 12% SDS polyacrylamide gel and transferred to a nitrocellulose membrane that was then preincubated with blocking solution (PBS containing 5% fat-free milk and 0.5%Tween-20) for 2 h, followed by a 2 h incubation with primary antibodies [CYP7A1 1:1,000, Abcam ab65596; ferritin light chain protein (Ft-L) 1:2,000, Abcam; IREB2 1:1,000, Abcam ab181153; aconitase 1 1:1,000, Abcam ab126595; β-actin 1:2,000, Haigenes M1501; GADPH 1:2,000, Abcam]. After washing with PBS containing 0.5%Tween-20, the blot was incubated with a 1:10,000 dilution of secondary antibody (HRP-conjugated goat anti-rabbit/mouse IgG, ZSGB-BIO, ZB2301/ZB2305) for 80 min and then washed and detected with an enhanced chemiluminescence detection kit (Applygen, Beijing, China).
Preparation of lysates
Cytoplasmic protein lysates (as a source for IRP1 and IRP2) were prepared for pulldown analysis. AML12 cells were cultured and, during the final 48 h, were treated with 200 μM DFO (20). Cells were lysed with ice-cold buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and protease inhibitors]. After centrifugation of lysates at 16,500 g for 15 min at 4°C, the supernatants were removed and stored at −80°C. The protein concentration was determined by a BCA protein assay kit (Applygen).
Hepatic histological grading
For histology, live tissues were fixed in 4% formalin for 24 h, dehydrated (Leica TP1020 automatic tissue hydroextractor; Leica, Germany) and embedded in paraffin (Leica EG1150C; Leica) for H&E and Oil Red O staining.
Construction of the Cyp7a1/TFR1 IRE expression vector
The luciferase expression construct designated as the pMir REPORT luciferase vector (Promega) encodes the Cyp7a1 mRNA 3′-UTR inserted between the SpeI and HindIII sites behind the luciferase gene in the pMir vector as Luc/CYP7A1-3′-UTR-WT. To generate the Luc/CYP7A1-3′-UTR-MUT, primers were used to obtain the mutant nucleic fragment: forward 5′-GCTTTTGTCTGCGAGGGCTGGAGCCAGCATCAGTGGATG-3′ and reverse 5′-CATCCACTGATGCTGGCTCCAGCCCTCGCAGACAAAAGC-3′. The five mutated nucleotides are underlined. In Luc/CYP7A1-3′-UTR-MUT, the IRE-like sequence can generate a canonical IRE (Fig. 1B, C). We also constructed a Luc/CYP7A1-3′-UTR-CUT, which had a deletion in the region of the Cyp7a1 IRE-like sequence. The Luc/TFR1-3′-UTR-WT sequence was cloned as a positive control. Because there are five canonical IREs in the 3′-UTR of mouse TFR1 mRNA, the IRE-IRP interaction in this mRNA is stronger than in others. Eventually, the sequence was confirmed by restriction enzyme digestion and sequencing.
Cell transfection
Hepa1-6 cells were seeded in 24-well culture plates for 24 h. DFO/iron-dextran was added to the cell culture medium before being cultured overnight. Transient transfection was performed with Lipofectamine 2000 (Invitrogen). Cells in serum-free antibiotic-free DMEM were transfected with 0.2 μg of total DNA per well, and growth medium was replaced after 6 h with fresh culture medium containing DFO/iron-dextran. The Renilla luciferase vector (pRL-TK) was used as an internal control to normalize for transfection efficiency.
Luciferase assays
Cells were harvested after 48 h of transfection to assay their luciferase activity. The cells were lysed with Promega passive lysis buffer at room temperature for 20 min and then assayed for firefly and Renilla luciferase activities by the Dual-Luciferase Reporter assay system (Promega, Madison, WI).
Biotin pulldown assay
RNA oligonucleotides: Cyp7a1 IRE-like (5′-GAGCAGGGUCCAGUGGAUGCUGUGCA-3′) and TFR1 IREb (5′-ACAUUAUCGGGAGCAGUGUCUUCCAUAAUGCAUA-3′) were purchased from Bioneer. To label target RNA, we used T4 RNA ligase to attach a single biotinylated nucleotide to the 3′ terminus of the RNA strands (Pierce RNA 3′ End Desthiobiotinylation kit). Fifty picomoles of biotinylated RNA was allowed to bind 50 μl of streptavidin magnetic beads for 30 min at room temperature; then, cell lysates (200 ng) were incubated with biotinylated oligonucleotides for 1 h at 4°C to bind with IRE. After four washes and elution of the RNA binding protein complexes, the proteins bound to the beads were analyzed by Western blotting targeting IRP1, IRP2, and GAPDH. IRPs were quantified using the Quantity One software (Bio-Rad).
Competition assay
Cytoplasmic protein lysate (20 ng) was incubated for 1 h at 4°C with 50 pmol of TFR1 IREb, Cyp7a1 IRE-like in the presence of increasing concentrations (0- to 50-fold) of nonbiotinylated competitor and analyzed by Western blotting.3
Measurement of Cyp7a1 and TFR1 mRNA stability
Hepa1-6 cells were grown in DMEM supplemented with 10% fetal bovine serum. DFO (200 μM) or iron-dextran (100 μg/ml) was added for 24 h prior to addition of 5,6-dichlorobenzimidazole riboside (DRB) (20 μg/ml; Sigma-Aldrich), which is an inhibitor of transcription. After DRB was added, cells were lysed by Trizol reagent (Invitrogen). RNA was isolated and the levels of Cyp7a1 and TFR1 mRNA were measured in real-time PCR. GAPDH was used as an internal standard.
Statistical analysis
All of the values are presented as the mean ± SD determined from three or more independent experiments. Student’s t-test was used to compare the values obtained from the two groups. Statistical significance was accepted at P < 0.05.
RESULTS
Establishment of an AFLD model in mice
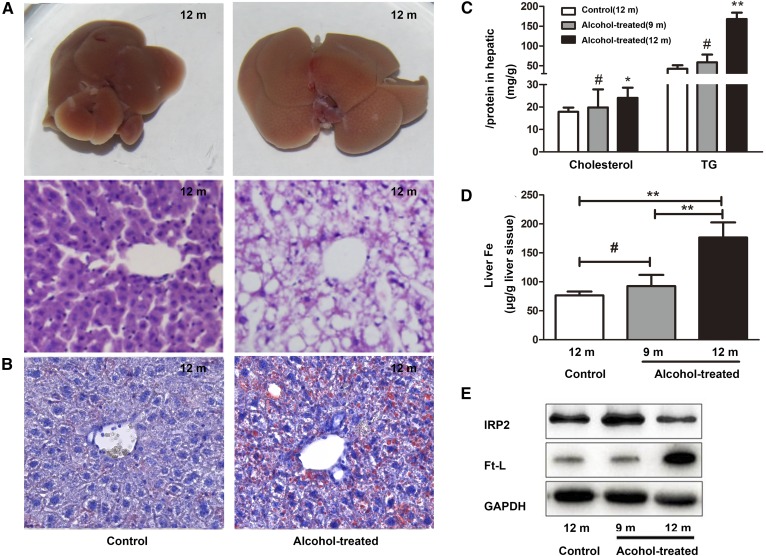
To simulate AFLD in patients with a history of alcoholism, 2- to 3-month-old mice were fed water containing 15% alcohol for approximately 9–10 months. As shown in the results, the body weights of 12-month-old mice and 9-month-old mice were significantly reduced compared to those of control mice. There were no obvious changes among alcohol-treated mice and control mice in daily food and fluid (water or alcohol) intake. Blood alcohol content in the serum was significantly elevated in 12-month-old alcohol-treated mice, and there were no significant changes in 9-month-old alcohol-treated mice compared with control mice (Table 1). The 12-month-old alcohol-treated mice exhibited increased concentration of alanine aminotransferase (ALT), aspartate aminotransferase (AST), cholesterol, TGs, and LDL cholesterol in serum (Table 2) and increased hepatic cholesterol and TGs (Fig. 2C) compared with the control group. However, 9-month-old alcohol-treated mice showed no significant change in serum ALT, AST, LDH, and TG, while cholesterol was slightly elevated in serum, but not statistically significantly different. Hepatic cholesterol and TGs showed no difference in 9-month-old alcohol-treated mice compared with the control group (Fig. 2C). The liver volumes of alcohol-treated 12-month-old mice increased significantly and appeared shallower compared with the livers of control mice. Histology (H&E) liver sections showed that the livers of the alcohol-treated 12-month-old mice had many cavities that were absent in the control mice (Fig. 2A) and alcohol-treated 9-month-old mice. Oil Red O stain of liver tissues showed that there were many lipid droplets present in the livers of alcohol-treated 12-month-old mice (Fig. 2B), whereas the alcohol-treated 9-month-old mice showed little change compared with the control group. Thus, chronic administration of 15% concentrations of alcohol caused AFLD in mice.
TABLE 1.
Changes in body weight, food intake, and fluid intake
| 9 Months | 12 Months | |||
| Control | Alcohol-Treated | Control | Alcohol-Treated | |
| Body weight (g) | 32.03 ± 1.87 | 30.08 ± 1.69a | 32.96 ± 1.72 | 29.88 ± 1.88b |
| Food intake (g/day) | 6.58 ± 0.74 | 5.89 ± 0.61 | 6.43 ± 0.50 | 5.79 ± 0.84 |
| Fluid intake (ml/day) | 6.19 ± 0.61 | 5.51 ± 0.48 | 6.08 ± 0.66 | 5.44 ± 0.46 |
| BCA (mM) | 1.42 ± 0.22 | 1.63 ± 0.31 | 1.73 ± 0.13 | 16.42 ± 8.1b |
Data are shown as the mean ± SD (n ≥ 8).
P < 0.05 versus control.
P < 0.01 versus control.
TABLE 2.
Serum parameters of control and chronic alcohol-fed mice
| Alcohol-Treated | |||
| Control (n = 8) | 9 Months (n = 9) | 12 Months (n = 8) | |
| ALT (U/l) | 28.84 ± 4.9 | 29.9 ± 1.22 | 35.56 ± 2.52a |
| AST (U/l) | 56.85 ± 6.02 | 59.92 ± 5.2 | 72.97 ± 6.44a |
| Cholesterol (mmol/l) | 1.89 ± 0.44 | 2.32 ± 0.49 | 2.68 ± 0.66b |
| TG (mmol/l) | 0.56 ± 0.23 | 0.51 ± 0.14 | 1.44 ± 0.49a |
| HDL-C (mmol/l) | 2.58 ± 0.16 | 2.29 ± 0.39 | 1.76 ± 0.45a |
| LDL-C (mmol/l) | 0.24 ± 0.03 | 0.23 ± 0.02 | 0.29 ± 0.05b |
Data are shown as the mean ± SD. HDL-C, HDL cholesterol; LDL-C, LDL cholesterol.
P < 0.01.
P < 0.05.
Fig. 2.
Establishment of an AFLD model in mice. A: Livers and H&E-stained liver sections of alcohol-treated mice and control mice (200× magnification). B: Oil Red O stain of liver sections of alcohol-treated mice and control mice (400× magnification). C: Total cholesterol and TGs were quantified in the livers of alcohol-treated mice and control mice. D: Hepatic iron content was measured using atomic absorption. E: Western blot analyses of Ft-L and IRP2 levels. #P > 0.05, *P < 0.05, **P < 0.01.
As reported previously, ALD patients frequently display evidence of iron overload (4, 7, 25, 26). Therefore, we assessed iron storage in mouse livers by measuring the iron content and expression of Ft-L and IRP2, which are regulated by the iron content. As shown in Fig. 2D, E, after chronic alcohol feeding for 8–9 months, an increase in the iron content and overexpression of Ft-L were observed in the AFLD mouse model compared with the control group. The expression of IRP2 protein was significantly decreased in the AFLD mouse model, and little change was observed in alcohol-treated 9-month-old mice compared with the control group. This result is consistent with the lipid levels, which had a positive correlation with the duration of alcohol exposure (Fig. 2C). Thus, alcohol-treated 12-month-old mice share similar causes and pathological processes as AFLD patients. Together, these results indicate there may be a correlation between hepatic lipid accumulation and the iron content.
Identification of a novel stem-loop structure in the 3′-UTR of Cyp7a1 mRNA
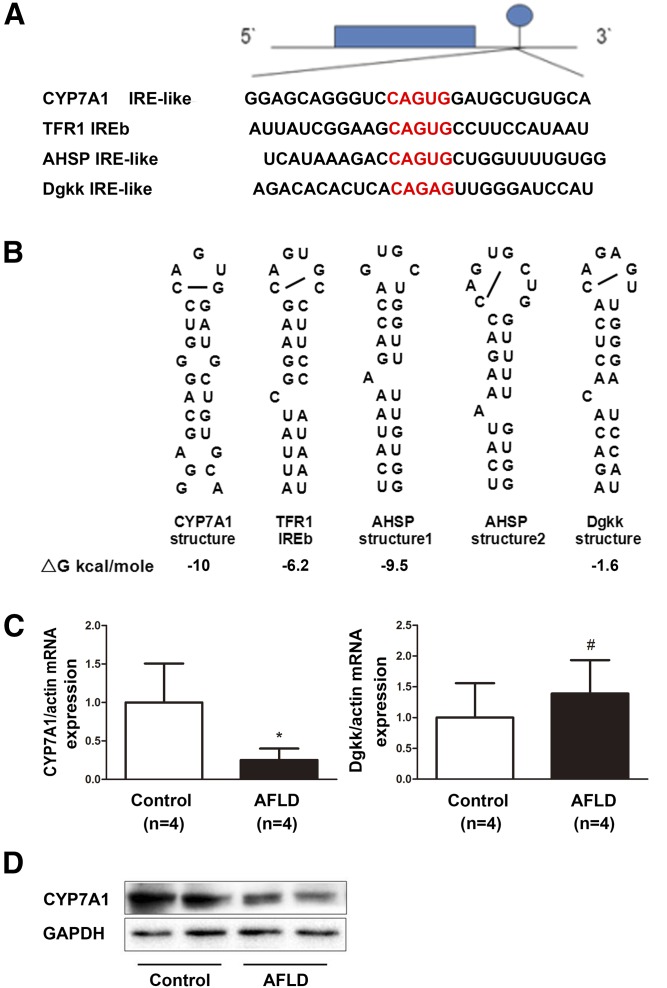
We hypothesized that iron might regulate the expression of lipid metabolism-related genes through interactions between IRPs and IREs in the UTR of these genes and, thus, result in lipid accumulation. Therefore, we used the RNA structure software’s most predicted secondary structures of more than 100 mRNAs of lipid metabolism-related genes to investigate to determine whether these mRNAs formed an IRE structure. We identified only two genes that were predicted to have structures similar to IREs in their 3′-UTR: Cyp7a1 and diacylglycerol kinase κ (Dgkk). The stem-loop structure in the 3′-UTR of the Dgkk mRNA (ΔG = −1.6 kcal/mol) has a canonical CAGAGU loop and a C8 in the bulge, but only one base pair in the downstream region. The mRNA of Cyp7a1 has a potential stem-loop structure in the 3′-UTR (ΔG = −10 kcal/mol; Fig. 3A, B). This stem-loop has a matching sequence, CAGUG, with no H and two unpaired Gs in place of the unpaired C8 (Fig. 3A, B). Because expression of Cyp7a1 mRNA and protein were significantly lower in the livers of AFLD mice without a significant change in the expression of Dgkk mRNA (Fig. 3C, D), we focused our study on Cyp7a1.
Fig. 3.
Sequence and predicted structures of an IRE-like stem-loop in Cyp7a1 mRNA. A: Sequence encoding the canonical IRE RNA stem loops in the 3′-UTR of TFR1 aligned to Cyp7a1 IRE, AHSP IRE, and Dgkk IRE. The CAGUG terminal loop of the IRE is shown in red letters. B: RNA secondary structure of the Cyp7a1 IRE-like, TFR1 IREb, AHSP, and Dgkk IRE was assessed by the software. C: Quantitative RT-PCR of hepatic Cyp7a1 and Dgkk mRNA levels in AFLD mice and control mice. D: Western blot analyses of CYP7A1 protein levels in AFLD mice and control mice. GAPDH blots represent tissue protein loading controls. #P > 0.05, *P < 0.05.
Iron removal or loading affects the expression of Cyp7a1 mRNA in vivo and in vitro
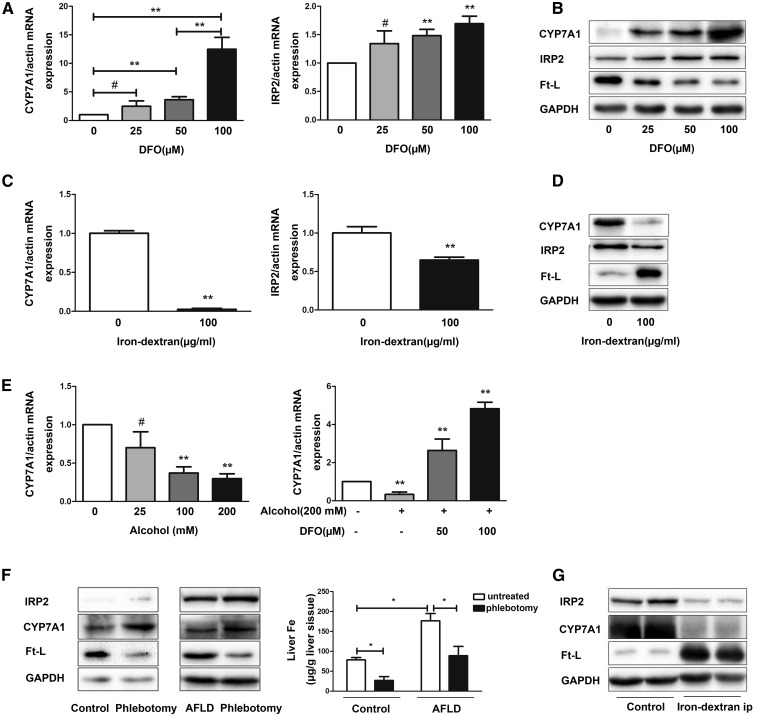
The results showed that the Cyp7a1 mRNA and protein levels were higher with increased concentrations of DFO in a dose-dependent manner in Hepa1-6 cells (Fig. 4A). Exposure of Hepa1-6 cells to 100 μg/ml iron-dextran resulted in lower Cyp7a1 mRNA and protein levels compared with controls. To determine the effectiveness of iron removal or loading, we tested the levels of the Ft-L and IRP2 mRNA and protein. The presence of DFO resulted in reduced Ft-L levels, whereas the Ft-L levels were elevated with exposure to iron-dextran. In addition, the levels of IRP2 mRNA and protein were shown to be higher with iron removal (Fig. 4B) and lower when iron loading was applied to the cells (Fig. 4C, D).
Fig. 4.
Iron regulates the intracellular and hepatic Cyp7a1 levels. A: DFO increased Cyp7a1 and IRP2 mRNA in a dose-dependent manner. Five independent experiments for each time point. B: Western blots of CYP7A1, IRP2, and Ft-L were performed. C: Cyp7a1 and IRP2 mRNA were detected by quantitative RT-PCR. D: Western blots of CYP7A1, IRP2, and Ft-L were performed. E: Hepa1-6 cells were treated with alcohol for the indicated concentrations. DFO was added after alcohol treated for 48 h, cells were harvested after 24 h, and Cyp7a1 mRNA levels were measured. F: Iron removal affects the expression of CYP7A1 in vivo. The expression of CYP7A1, IRP2, and Ft-L were measured, and hepatic iron content was measured. G: Iron loading affects the expression of CYP7A1 in vivo. The expression of CYP7A1, IRP2, and Ft-L were measured. #P > 0.05, *P < 0.05, **P < 0.01 (n = 3).
The CYP7A1 protein levels were markedly higher after phlebotomy, both in control mice and AFLD model mice, whereas the expression of Ft-L decreased and IRP2 increased in mice after phlebotomy. In contrast, iron-injection obviously reduced CYP7A1 expression in the livers of control mice, whereas the expression of Ft-L increased and IRP2 decreased (Fig. 4F, G).
Next, we assessed whether alcohol would stimulate Cyp7a1 expression in Hepa1-6 cells. As shown in Fig. 4E, the Cyp7a1 mRNA levels decreased in a dose-responsive pattern after alcohol exposure of approximately 48 h. To test whether the decrease of Cyp7a1 expression after alcohol treatment was due to increased cellular iron loading, DFO was added to cell cultures after alcohol treatment. The result showed that Cyp7a1 expression was induced by DFO in a pronounced dose-dependent manner at the mRNA level. Thus, we concluded that Cyp7a1 mRNA can be regulated by the iron levels.
IRE-like sequence in the 3′-UTR of mouse Cyp7a1 mRNA binds to IRP1 and IRP2
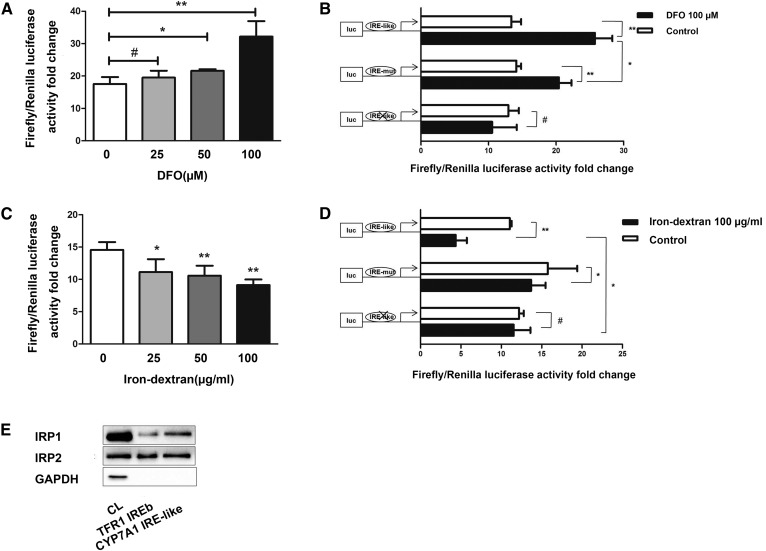
We used luciferase assays and a biotin pulldown assay to test for possible interactions between the 3′-UTR of mouse Cyp7a1 mRNA and IRPs. DFO chelated sufficient intracellular iron to promote Luc/TFR1-3′-UTR-WT luciferase activity, and iron-dextran inhibited Luc/TFR1-3′-UTR-WT luciferase activity in a dose-dependent manner (Fig. 5A, C). Luc/TFR1-3′-UTR-WT luciferase activity increased by 80% using 100 μM DFO. In contrast, treatment with 100 μg/ml iron-dextran resulted in 40% lower Luc/TFR1-3′-UTR-WT luciferase activity by increasing the intracellular iron content. The results suggested that the luciferase activity of Luc/CYP7A1-3′-UTR-WT and Luc/CYP7A1-3′-UTR-MUT were increased under low iron conditions and subject to decrease in response to high-iron content conditions. However, luciferase activity was unchanged in Luc/CYP7A1-3′-UTR-CUT transfectants under the same concentrations of DFO or iron-dextran. In addition, the Luc/CYP7A1-3′-UTR-WT group exhibited a stronger luciferase signal under treatment with DFO or iron-dextran than the Luc/CYP7A1-3′-UTR-MUT group (Fig. 5B, D). This suggests that the IRP-binding capacity of this noncanonical IRE on Cyp7a11 mRNA is stronger compared with the canonical IRE.
Fig. 5.
The IRE in the 3′-UTR of Cyp7a1 mRNA binds both to IRP1 and IRP2. A: TFR1 3′-UTR sequences inserted in the 3′-UTR of Luciferase reporter gene within the 3′-UTR HindIII/SpeI site. DNA from Luc/TFR1-3′-UTR-WT (100 ng) was transiently transfected into Hepa1-6 cells for 24 h as follows: untreated and DFO (0–100 μM) for 24 h prior. The TK plasmid was transfected to standardize. Luciferase activity of groups treated with DFO are higher than that of untreated group, and the highest activity was observed when 100 μM DFO was added. B: Luciferase reporter gene experiments performed in 100 μM DFO pretreated Hepa1-6 cells transfected with Luc/CYP7A1-3′-UTR-WT, Luc/CYP7A1-3′-UTR-MUT, and Luc/CYP7A1-3′UTR-CUT. The TK plasmid was transfected to standardize. C: DNA from Luc/TFR1-3′-UTR-WT (100 μg) was transiently transfected into Hepa1-6 cells for 24 h as follows: untreated and iron-dextran (0–100 μg/ml) for 24 h prior. The TK plasmid was transfected to standardize. Luciferase activity of groups treated with iron-dextran is lower than that of untreated group, and the lowest activity was observed when 100 μg/ml iron-dextran was added. D: Luciferase reporter gene experiments were performed in 100 μg/ml iron-dextran-pretreated Hepa1-6 cells transfected with Luc/CYP7A1-3′-UTR-WT, Luc/CYP7A1-3′-UTR-MUT, and Luc/CYP7A1-3′-UTR-CUT. The TK plasmid was transfected to standardize. E: Hepa1-6 cellular extracts were incubated with biotinylated RNA oligonucleotides of core IRE sequences from the TFR1 and Cyp7a1 mRNA 3′-UTRs. The protein-IRE complexes were precipitated with streptavidin-coated beads followed by Western blotting of IRP1/IRP2 proteins. GAPDH was used to exclude false positive reactions. #P > 0.05, *P < 0.05, **P < 0.01.
In pulldown assays, we used specific biotinylated RNA oligonucleotides to represent the core IRE sequences of the TFR1 and Cyp7a1. The data shown in Fig. 5E demonstrated that IRP1 and IRP2 both bound to the TFR1 IREb sequences. Under identical conditions, the Cyp7a1 IRE-like sequence bound to both IRP1 and IRP2 (Fig. 5E). To exclude false positive interactions, we concurrently detected GAPDH expression for reference. The pulldown assay results showed that the Cyp7a1 IRE-like sequence bound to both IRP1 and IRP2 and confirmed the results of our luciferase assays.
IRPs interacted with the Cyp7a1 IRE-like sequence in a similar binding affinity to the TFR1 in vitro
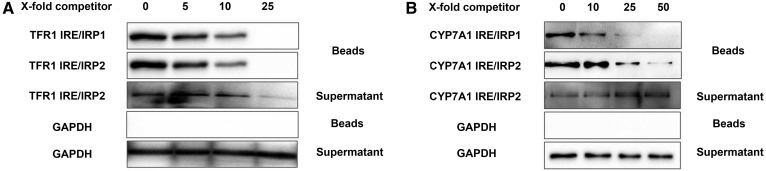
We next employed homologous competition assays in the pulldown experiments. As shown in Fig. 6A, B, the nonbiotinylated Cyp7a1 IRE probe efficiently competed with the labeled Cyp7a1 IRE probe, disrupting most of the biotinylated Cyp7a1 IRE-protein complex. We observed that the nonbiotinylated TFR1 IREb probe efficiently competed off approximately all of the labeled probe at approximately 25-fold. However, higher concentrations of nonbiotinylated Cyp7a1 probe were required. The result further demonstrated that the IRP-Cyp7a1 mRNA interaction is mediated by the IRE-like structure.
Fig. 6.
Homologous competition assays were employed. Cellular extracts were incubated with either 50 pmol biotinylated TFR1 IREb or Cyp7a1 IRE in the presence of a 0, 5, 10 or 25-fold TFR1 IRE (A) or in the presence of a 0, 10, 25 or 50-fold excess of nonbiotinylated Cyp7a1 IRE (B).
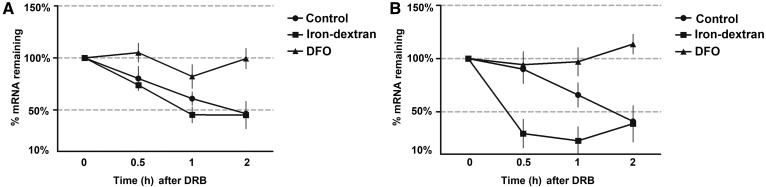
Cyp7a1 mRNA stability can be regulated by iron
We measured the effects of iron on Cyp7a1 mRNA stability. Cyp7a1 and TFR1 mRNA were stable under low-iron condition and degradation of these mRNAs was accelerated with iron-dextran. As shown in Fig. 7A, B, the half-life of Cyp7a1 and TFR1 mRNA are 90–100 min and 100–110 min, respectively. When iron-dextran was added to culture medium prior to DRB, both Cyp7a1 and TFR1 mRNA were more rapidly degraded (half-life 20–25 min and 50–60 min, respectively). In contrast, when DFO was added to culture medium prior, both Cyp7a1 and TFR1 mRNA became stable with a half-life of more than 2 h. The results suggest that the stability of Cyp7a1 mRNA can be regulated by iron levels.
Fig. 7.
Stability of Cyp7a1 and TFR1 mRNA. A: TFR1 mRNA was measured at the times indicated. TFR1 mRNA became stable after DFO treatment, the half-life of TFR1 decreased after iron-dextran treatment. B: Cyp7a1 mRNA was measured at the times indicated. Cyp7a1 mRNA became stable after DFO treatment, while the half-life of Cyp7a1 decreased four times in comparison with untreated controls after iron-dextran treatment. GAPDH was used as a control gene.
DISCUSSION
As an essential element for living organisms, iron plays a vital role in many crucial physiological events. It has also been shown to regulate many metabolic processes, such as lipid metabolism (11, 27–29). In this report, we present data indicating a correlation between the hepatic iron content and lipid metabolism in alcohol-induced hepatic steatosis using a mouse model. This study shows, for the first time, that the iron content can affect the expression of Cyp7a1 though a novel stem-loop structure in the 3′-UTR of Cyp7a1 mRNA, which we define as a noncanonical IRE sequence in this report.
Here, we established an AFLD model in mice. The alcohol in drinking water model most closely mimics the alcohol-dependent human drinking pattern (30). Several animal models have been established to study the pathological and clinical features of ALD, such as intragastric infusion, the Lieber-DeCarli all liquid diet (31), and the alcohol agar block model. The most commonly used animal model is the liquid diet model. However, the diet used in this model usually contains high fat, which made it difficult to isolate and study the influence of alcohol as a single factor in AFLD (32). The animals are forced to consume alcohol every time when both hungry and thirsty, so the model does not mimic human drinking. The liquid diet pair-feeding of control animals can also affect several aspects of the immune system (33). To develop a higher rate of alcohol breakdown, Tsukamoto et al. (34) developed the rodent intragastric ethanol infusion model. Blood alcohol concentration can be sustained at a desired level for long durations, but this model is not physiological. It is labor intensive and requires constant monitoring of animals and highly trained technical personnel to perform the catheter implantation surgery. Most importantly, these models are established in the late stage of AFLD. Our AFLD model resulted in early stages of steatosis, morphological changes, such as iron overload, inflammation (supplemental Figs. S1, S2), and oxidative stress (supplemental Table S1) in the livers. The blood alcohol content in the serum was significantly elevated in the 12-month-old alcohol-treated mice. However, this change was not observed in 9-month-old alcohol-treated mice (supplemental Table S1). So, we think it is important to increase the duration of alcohol feeding. Several studies have reported that alcohol in drinking water may cause dehydration due to the animals’ natural aversion to alcohol consumption (35). However, no dehydration occurred in the model used in this study. Several reasons were considered, such as the concentration and duration of alcohol in the period for adaptation. The concentration we used is the lowest concentration needed to lead to AFLD. Consistent with other reports (36), 9-month-old and 12-month-old alcohol-treated mice had lower body weight; it is well-known that growth hormone is reduced by alcohol in animals, as well as in humans (37). We also recorded the food and fluid (water or alcohol) intake of mice, and the results showed comparable levels of food intake and fluid intake in each group, so we hold that weight loss is not caused by anorexia. Furthermore, the AFLD model in this study is more practical, inexpensive, and maximally simulates the human disease process and, thus, may represent a more physiological model for alcoholism (38).
AFLD patients frequently display evidence of lipid accumulation and iron overload, which also appeared in the mouse model described in this report. In addition, we observed that lipid accumulation and iron overload positively correlated with the duration of alcohol consumption, which is a consistent feature in AFLD patients. We suspect that there may be a correlation between lipid accumulation and iron loading. Cellular iron metabolism is coordinately controlled through the IRE/IRP system (9). Most of the known genes that contain IREs in the UTRs of their mRNA are involved in iron metabolism, encoding proteins for iron absorption (DMT1), storage (L- and H-ferritin), transport (TFR1), utilization [EPAS1, hypoxia-inducible factor 2 alpha (HIF2a) (39), AHSP (19), 5′-aminolevulinate synthase 2 (ALAS2) (9)], and export [FPN1 (16)]. As previously described, these IREs form a canonical conserved secondary structure. However, a series of studies indicate that genes containing IREs and IRE-like elements are more diverse in function and structure, such as cell division cycle 14 (CDC14), a gene that is involved in dephosphorylation (40), and MRCKa, which is known to regulate the cytoskeleton (20). However, no IRE sequence has been found among lipid metabolism-related genes. The results described in this report open a new paradigm regarding the mechanisms of lipid metabolism regulation. In recent years, an increasing amount of literature has investigated IREs that are structurally distinct from canonical IREs, yet can mediate mechanistically similar responses (12, 19, 39). For example, the IRE-like stem-loop of AHSP (Fig. 3B) differs substantially from the canonical IRE, though there is strong evidence that IRP1 associates with AHSP mRNAs in cells. The IRE-like stem-loop of AHSP includes the CAGUGU loop consensus sequence and an A rather than a C at the stem bulge. In cytoplasmic extracts, AHSP mRNA coimmunoprecipitates with IRPs dependent on the IRE-like stem-loop structure (19). For the amyloid precursor protein (APP) IRE-like stem loop encoding a 13-base larger terminal loop (5′-GGCAGAGCAAGGA-3′) and a 6-base stem with two unpaired G residues, the CAGA-deleted version of the APP IRE abrogated IRP1 binding to APP IRE. Therefore, the terminal loop is the key for the IRP-IRE interaction (18, 41). The crystal structure of an IRP1-FTH1 IRE complex indicates that the protein binds a single IRE-RNA molecule through contacts at two spatially separate sites: the RNA terminal loop and C8 nucleotide. The IRP1 molecule adopts an L-shape with four domains. The IRE stem-loop terminal loops A15, G16, and U17 provide specificity and stability to interactions at domains 2 and 3, and eight bonds are formed between the C8 nucleotide and domain 4 of IRP1 (13, 42). Thus, noncanonical sequences that bind IRPs may include binding sites other than the ones described above.
Therefore, we used RNA structure software-predicted secondary structures to identify novel IREs from a group of more than 100 candidate mRNAs from lipid metabolism-related genes. The criteria for the evaluation were based on the common secondary structures of canonical and noncanonical IRE stem-loops: a) ∼28–30 nucleotide long-stem loops in the 3′- or 5′-UTRs; b) the apical loop has five conserved nucleotides, CAGU(A)GH (H is A, C, or U), of each IRE sequence, with the C forming a base pair with the second G to generate an AGU triplet at the apex; c) an unpaired base in the stem. We identified only two genes that had similar structures to the canonical IRE in the 3′-UTR, Cyp7a1 and Dgkk. These two genes were both predicted to have IRE-like structures in the 3′-UTR of their mRNA. We focused further analysis on Cyp7a1 because the expression of Dgkk mRNA showed no significant change, while the expression of Cyp7a1 mRNA and protein were significantly decreased in the AFLD mouse model, which showed significant iron deposition. Both the AHSP IRE and Cyp7a1 IRE sequences were predicted to fold into equally stable RNA structures with similar G values: −9.5 kcal/mol and −10 kcal/mol (Fig. 2B), whereas the G value of Dgkk was only −1.6 kcal/mol, so the predicted RNA structure of Dgkk in Fig. 2B may be not stable. Thus, we predicted a potential stem-loop structure to be a noncanonical IRE sequence in the 3′-UTR of the mouse Cyp7a1 mRNA. This stem-loop structure differs from the canonical IRE in that it contains a five-member loop (CAGUG) and two unpaired G residues in the stem instead of a six-member loop and C bulge. This is similar to the 75 kDa subunit of the mitochondrial complex I IRE, which has a five-member loop (CAGAG), and the noncanonical IRE of APP, which has unpaired G residues in the stem (18, 32).
Luciferase reporter experiments were performed to independently verify the effect of iron removal or loading on the expression of Cyp7a1 in Hepa1-6 cells. This study shows changes in luciferase activity upon transfection of cells with luciferase-3′-UTR sequences from Cyp7a1. The results suggest that when the IRE-like structure is disrupted, luciferase activity is decreased. This result indirectly shows that the IRP-Cyp7a1 mRNA interaction is mediated by the IRE-like structure. To confirm the functional role of this stem loop structure, we directly demonstrated that the 3′-UTR-specific Cyp7a1 IRE indeed bound IRP1 and IRP2 in pulldown assays. We also generated a Luc/CYP7A1-3′-UTR-MUT construct that contained five mutations to obtain the canonical IRE. Interestingly, we found that the IRP binding of the noncanonical IRE was stronger than that of the canonical IRE. In homologous competition assays, the Cyp7a1 probe exhibited a relatively low binding affinity to IRPs because a higher amount of nonbiotinylated probe was required in the assay compared with the TFR1 probe. It is possible that additional factors not present in the pulldown, such as Luc/CYP7A1-3′-UTR-WT, include extended sequences that may optimize the folding of an IRE-like stem-loop structure. The Cyp7a1 IRE, which only has a key AGU tri-loop, but binds IRPs, may include additional sites besides the two usual spatially separate sites (the RNA terminal loop and C8 nucleotide). More research is needed to study the interactions between the noncanonical IREs and IRPs, including binding site and spatial structure analyses.
We wanted to know whether the RNA stability could be regulated by iron content. In the RNA stability analysis, DRB was added to stop transcription, when DFO was added prior to DRB, both Cyp7a1 and TFR1 mRNA are stabilized with a half-life >2 h. On the other hand, under iron-rich condition, Cyp7a1 mRNA rapidly degraded (the half-life decreased approximately four times), while the half-life of TFR1 mRNA decreased two times from 100–110 min to 50–60 min. Consistent with the luciferase results, Cyp7a1 IRE showed more susceptive to iron content than TFR1 IREb in this result. Taken together, endogenous Cyp7a1 mRNA is destabilized, and its stability can be affected by intracellular iron levels via the IRE-like sequence in its 3′-UTR.
To understand how Cyp7a1 mRNA is degraded in low iron, we analyzed the 3′-UTR of Cyp7a1 mRNA. The endonucleolytic cleavage site is involved in TFR1 mRNA degradation (43). We were not able to identify the endonucleolytic cleavage site in the 3′-UTR of Cyp7a1 mRNA. However, the Cyp7a1 3′-UTR contains eight AREs (AUUUA), five of which are clustered within 300 nucleotides upstream of the noncanonical IRE. This situation is similar to that of the 3′-UTR of MRCKa mRNA (20), which also contains eight AREs that are complementary to miR16 through a UAAAUAUU sequence (44), which also seems to be essential in ARE-mediated mRNA degradation. Therefore, IRE/IRP stabilization of Cyp7a1 mRNA is perhaps associated with miR16/ARE binding. However, this hypothesis still needs to be further confirmed.
The present study has some limitations regarding the relationship between Cyp7a1 and iron. Cyp7a1 is a well-known cytochrome P450. It mediates cholesterol 7-hydroxylation reactions, binding cholesterol to ferric P450, thus forming a ferrous P450-cholesterol-O2 complex (45). However, no report has described a link between iron and Cyp7a1. This report proposes, for the first time, that Cyp7a1 mRNA is regulated by iron, most likely by IRP binding. IRP binding to the IRE-like structure can increase Cyp7a1 expression by stabilizing the mRNA.
Excessive consumption of alcohol could enhance ROS production and suppress the activity of antioxidants in vivo, such as superoxide dismutase (SOD) and GSH (46). CYP2E1 is one of the central pathways in the ability of alcohol to induce lipid peroxidation and oxidative stress. In our AFLD model, the ROS and CYP2E1 level in the liver were significantly elevated (supplemental Fig. S1). Increased cholesterol levels and decreased expression of Cyp7a1 can trigger inflammation (supplemental Fig. S2). An inflammatory/oxidative environment leads to a vicious circle, which can damage healthy neighboring epithelial and stromal cells to cause hepatocellular injury. Among various genes encoding for anti-oxidative and anti-inflammatory proteins, haem oxygenase-1 (HO-1) is particularly attractive. In a previous study, HO-1 expression could be induced by alcohol consumption in mice (47). HO-1 is an anti-inflammatory molecule that oxidizes heme to biliverdin, CO, and free iron (48–50). HO-1 overexpression leads to induction of iron deposition (51). We detected the activity of haem oxygenase; its activity was significantly increased in 12-month-old alcohol-treated mice. So we proposed that HO-1 expression was increased and its product iron subsequently overloaded. Then, IRP1 becomes an enzymatically active holoprotein with a 4Fe-4S cluster and aconitase activity, and IRP2 is rapidly targeted for proteasome-mediated degradation (52–54). Therefore, IRPs cannot bind to IRE-like motifs in the 3′-UTR of mouse Cyp7a1 mRNA. This reduces the mRNA stability and promotes the degradation of Cyp7a1 mRNA. The decreased expression of Cyp7a1 mRNA results in cholesterol accumulation in the liver. Several studies have shown that alcohol can give rise to bile acid production, but those studies all have one problem in common: the concentration of cellular alcohol was too low (7.7 or 50 mM) and time was too short (<24 h). In these studies, mice were administered alcohol feeding for only 5 weeks, and the concentration of alcohol in food was only 0.75–5% (1, 55). As the duration of alcohol administration was short, iron deposits had probably not accumulated. In a previously published article by our research group, we used a low dose alcohol concentration at 3.5% in drinking water fed to mice; the results showed that alcohol had positive effects on reverse cholesterol transport, a protective reaction, the expression and activity of CYP7A1 exhibited no changes (56). In another study published in 2013 (57), animals were administered Lieber-DeCarli liquid diet for a duration of 8 weeks, as the mice in our study were administered a moderate concentration of alcohol in the drinking water for approximately 8–9 months. The different duration between this study in 2013 (57) and our study may have caused some difference in the metabolism of the animals. There was no iron loading mentioned in the lecture. We believe that the short duration of massive alcohol may affect the expression of Cyp7a1 through other pathways. All of the above studies suggest that alcohol can regulate Cyp7a1 expression in a dose-responsive pattern and this regulation can be affected by multiple factors. It is well-known that overexpression of Cyp7a1 is induced by alcohol intake, perhaps due to reduced expression of FXR, which can be regulated by alcohol and upregulates expression of small heterodimer partner (SHP) in the liver and induces the transcription and secretion of fibroblast growth factor 15 (FGF15) from the intestines into the portal vein. FGF15 can downregulate Cyp7a1 as a target gene in the liver. Our AFLD model mice exhibited no changes in FXR and SHP expression, but showed a significant reduction in FGF15 expression. This reduction in FGF15 may relieve the inhibition and increase expression of CYP7A1 theoretically, but in our model CYP7A1 was reduced (supplemental Fig. S3). Therefore, it is possible that alcohol may regulate the expression of Cyp7a1 through multiple mechanisms. Under iron deposition conditions, it is possible that the inhibitory effect of iron is more dominant.
In the AFLD mouse model used in this study, massive cholesterol accumulation may have been partially caused by the decrease in Cyp7a1 expression. Certainly, the increase of cholesterol levels in the model cannot only result from the inhibition of Cyp7a1, but may also be the result of increased cholesterol synthesis. Some studies suggest that iron overloading in mice can increase cholesterol biosynthesis (11). While other studies suggest that alcohol intake can suppress expression of cholesterol synthesis genes (58), which can decrease cholesterol biosynthesis. In this model, the inhibition of the expression of these genes by alcohol may counteract the induction by iron, and results showed that HMGCR expression in mouse liver did not change. Of course, there are a number of factors unrelated to iron and IRE that take part in cholesterol synthesis and degradation pathways, such as HMGCR and CYP27A1 (supplemental Figs. S3, S4).
Our AFLD animal model displays evidence of both iron overload and cholesterol accumulation. Our data, for the first time, showed that Cyp7a1 mRNA contains a functional noncanonical IRE in the 3′-UTR that can bind to IRP1 and IRP2. Iron modulation of the Cyp7a1 noncanonical IRE points toward a possible link, previously unrecognized, between iron metabolism, the IRE/IRP regulatory system, and lipid metabolism. This finding has a significant implication in understanding the proposed mechanism for the regulation of cholesterol homeostasis by a Fe/IRP/noncanonical IRE axis.
Supplementary Material
Footnotes
Abbreviations:
- AFLD
- alcoholic fatty liver disease
- AHSP
- α-hemoglobin stabilizing protein
- ALD
- alcoholic liver disease
- ALT
- alanine aminotransferase
- APP
- amyloid precursor protein
- AST
- aspartate aminotransferase
- CYP7A1
- cholesterol 7α-hydroxylase
- DFO
- desferrioxamine mesylate
- Dgkk
- diacylglycerol kinase κ DRB, 5,6-dichlorobenzimidazole riboside
- FGF15
- fibroblast growth factor 15
- Ft-L
- ferritin light chain protein
- HO-1
- haem oxygenase-1
- IRE
- iron-responsive element
- IRP
- iron regulatory protein
- ROS
- reactive oxygen species
- TFR1
- transferrin receptor 1
- TG
- triglyceride
- UTR
- untranslated region
This work was supported by National Natural Science Foundation of China Grants 81200406 and 81570534, Scientific Research Foundation for the Returned Overseas Chinese Scholars Heilongjiang Province Grant C140202, and Graduate Innovation Foundation of Harbin Medical University Grant YJSCX2015-1HYD. Additional support was provided by Scientific Research Foundation of Heilongjiang Province Grant ZD201419 (Y.L.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
Many studies have shown that some elements structurally distinct from canonical IREs have the same functions as canonical IREs. These IRE-like elements are diverse in structure and many genes contain IRE-like elements such as AHSP a (24). “Cyp7a1 IRE-like” in this article refers to the IRE-like element in Cyp7a1 gene structure.
REFERENCES
- 1.Wu W., Zhu B., Peng X., Zhou M., Jia D., and Gu J.. 2014. Activation of farnesoid X receptor attenuates hepatic injury in a murine model of alcoholic liver disease. Biochem. Biophys. Res. Commun. 443: 68–73. [DOI] [PubMed] [Google Scholar]
- 2.Bieghs V., Hendrikx T., van Gorp P. J., Verheyen F., Guichot Y. D., Walenbergh S. M., Jeurissen M. L., Gijbels M., Rensen S. S., Bast A., et al. . 2013. The cholesterol derivative 27-hydroxycholesterol reduces steatohepatitis in mice. Gastroenterology. 144: 167–178.e1. [DOI] [PubMed] [Google Scholar]
- 3.Marí M., Morales A., Colell A., Garcia-Ruiz C., and Fernandez-Checa J. C.. 2014. Mitochondrial cholesterol accumulation in alcoholic liver disease: role of ASMase and endoplasmic reticulum stress. Redox Biol. 3: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison-Findik D. D. 2007. Role of alcohol in the regulation of iron metabolism. World J. Gastroenterol. 13: 4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong S., She H., Zhang A. S., Wang J., Mkrtchyan H., Dynnyk A., Gordeuk V. R., French S. W., Enns C. A., and Tsukamoto H.. 2008. Hepatic macrophage iron aggravates experimental alcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G512–G521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasselli E., Compalati A. D., Voci A., Vecchione G., Ragazzoni M., Gallo G., Borro P., Sumberaz A., Testino G., and Vergani L.. 2014. Altered oxidative stress/antioxidant status in blood of alcoholic subjects is associated with alcoholic liver disease. Drug Alcohol Depend. 143: 112–119. [DOI] [PubMed] [Google Scholar]
- 7.Tan T. C., Crawford D. H., Jaskowski L. A., Subramaniam V. N., Clouston A. D., Crane D. I., Bridle K. R., Anderson G. J., and Fletcher L. M.. 2013. Excess iron modulates endoplasmic reticulum stress-associated pathways in a mouse model of alcohol and high-fat diet-induced liver injury. Lab. Invest. 93: 1295–1312. [DOI] [PubMed] [Google Scholar]
- 8.Ioannou G. N., Dominitz J. A., Weiss N. S., Heagerty P. J., and Kowdley K. V.. 2004. The effect of alcohol consumption on the prevalence of iron overload, iron deficiency, and iron deficiency anemia. Gastroenterology. 126: 1293–1301. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson N., and Pantopoulos K.. 2014. The IRP/IRE system in vivo: insights from mouse models. Front. Pharmacol. 5: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields M., and Lewis C. G.. 1999. Cholesterol-lowering nature of unsaturated fat in rats may be due to its inability to increase hepatic iron. Metabolism. 48: 200–204. [DOI] [PubMed] [Google Scholar]
- 11.Graham R. M., Chua A. C., Carter K. W., Delima R. D., Johnstone D., Herbison C. E., Firth M. J., O’Leary R., Milward E. A., Olynyk J. K., et al. . 2010. Hepatic iron loading in mice increases cholesterol biosynthesis. Hepatology. 52: 462–471. [DOI] [PubMed] [Google Scholar]
- 12.Anderson C. P., Shen M., Eisenstein R. S., and Leibold E. A.. 2012. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim. Biophys. Acta. 1823: 1468–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walden W. E., Selezneva A. I., Dupuy J., Volbeda A., Fontecilla-Camps J. C., Theil E. C., and Volz K.. 2006. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 314: 1903–1908. [DOI] [PubMed] [Google Scholar]
- 14.Kayaaltı Z., Akyuzlu D. K., and Soylemezoglu T.. 2015. Evaluation of the effect of divalent metal transporter 1 gene polymorphism on blood iron, lead and cadmium levels. Environ. Res. 137: 8–13. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh M. C., Tong W. H., Zhang D., Ollivierre-Wilson H., Singh A., Krishna M. C., Mitchell J. B., and Rouault T. A.. 2008. Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc. Natl. Acad. Sci. USA. 105: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D. L., Hughes R. M., Ollivierre-Wilson H., Ghosh M. C., and Rouault T. A.. 2009. A ferroportin transcript that lacks an iron-responsive element enables duodenal and erythroid precursor cells to evade translational repression. Cell Metab. 9: 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campillos M., Cases I., Hentze M. W., and Sanchez M.. 2010. SIREs: searching for iron-responsive elements. Nucleic Acids Res. 38: W360–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho H. H., Cahill C. M., Vanderburg C. R., Scherzer C. R., Wang B., Huang X., and Rogers J. T.. 2010. Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J. Biol. Chem. 285: 31217–31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.dos Santos C. O., Dore L. C., Valentine E., Shelat S. G., Hardison R. C., Ghosh M., Wang W., Eisenstein R. S., Costa F. F., and Weiss M. J.. 2008. An iron responsive element-like stem-loop regulates alpha-hemoglobin-stabilizing protein mRNA. J. Biol. Chem. 283: 26956–26964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cmejla R., Petrak J., and Cmejlova J.. 2006. A novel iron responsive element in the 3′UTR of human MRCKalpha. Biochem. Biophys. Res. Commun. 341: 158–166. [DOI] [PubMed] [Google Scholar]
- 21.Li T., and Chiang J. Y.. 2015. Bile acids as metabolic regulators. Curr. Opin. Gastroenterol. 31: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50 (Suppl.): S120–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contreras J. A., Castro M., Bocos C., Herrera E., and Lasuncion M. A.. 1992. Combination of an enzymatic method and HPLC for the quantitation of cholesterol in cultured cells. J. Lipid Res. 33: 931–936. [PubMed] [Google Scholar]
- 24.dos Santos C. O., Dore L. C., Valentine E., Shelat S. G., Hardison R. C., Ghosh M., Wang W., Eisenstein R. S., Costa F. F., and Weiss M. J.. 2008. An iron responsive element-like stem-loop regulates alpha-hemoglobin-stabilizing protein mRNA. J. Biol. Chem. 283: 26956–26964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield J. B., Zhu G., Heath A. C., Powell L. W., and Martin N. G.. 2001. Effects of alcohol consumption on indices of iron stores and of iron stores on alcohol intake markers. Alcohol. Clin. Exp. Res. 25: 1037–1045. [PubMed] [Google Scholar]
- 26.De Feo T. M., Fargion S., Duca L., Cesana B. M., Boncinelli L., Lozza P., Cappellini M. D., and Fiorelli G.. 2001. Non-transferrin-bound iron in alcohol abusers. Alcohol. Clin. Exp. Res. 25: 1494–1499. [DOI] [PubMed] [Google Scholar]
- 27.Chitturi S., and George J.. 2003. Interaction of iron, insulin resistance, and nonalcoholic steatohepatitis. Curr. Gastroenterol. Rep. 5: 18–25. [DOI] [PubMed] [Google Scholar]
- 28.Turbino-Ribeiro S. M., Silva M. E., Chianca D. A. Jr., De Paula H., Cardoso L. M., Colombari E., and Pedrosa M. L.. 2003. Iron overload in hypercholesterolemic rats affects iron homeostasis and serum lipids but not blood pressure. J. Nutr. 133: 15–20. [DOI] [PubMed] [Google Scholar]
- 29.Pankow J. S., Boerwinkle E., Adams P. C., Guallar E., Leiendecker-Foster C., Rogowski J., and Eckfeldt J. H.. 2008. HFE C282Y homozygotes have reduced low-density lipoprotein cholesterol: the Atherosclerosis Risk in Communities (ARIC) study. Transl. Res. 152: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T. K., Lumeng L., McBride W. J., and Waller M. B.. 1979. Progress toward a voluntary oral consumption model of alcoholism. Drug Alcohol Depend. 4: 45–60. [DOI] [PubMed] [Google Scholar]
- 31.Sharda D. R., Miller-Lee J. L., Kanski G. M., Hunter J. C., Lang C. H., Kennett M. J., and Korzick D. H.. 2012. Comparison of the agar block and Lieber-DeCarli diets to study chronic alcohol consumption in an aging model of Fischer 344 female rats. J. Pharmacol. Toxicol. Methods. 66: 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Souza El-Guindy N. B., Kovacs E. J., De Witte P., Spies C., Littleton J. M., de Villiers W. J., Lott A. J., Plackett T. P., Lanzke N., and Meadows G. G.. 2010. Laboratory models available to study alcohol-induced organ damage and immune variations: choosing the appropriate model. Alcohol. Clin. Exp. Res. 34: 1489–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piano M. R., Artwohl J., Kim S. D., and Gass G.. 2001. The effects of a liquid ethanol diet on nutritional status and fluid balance in the rat. Alcohol Alcohol. 36: 298–303. [DOI] [PubMed] [Google Scholar]
- 34.Tsukamoto H., Towner S. J., Ciofalo L. M., and French S. W.. 1986. Ethanol-induced liver fibrosis in rats fed high fat diet. Hepatology. 6: 814–822. [DOI] [PubMed] [Google Scholar]
- 35.Mathews S., Xu M., Wang H., Bertola A., and Gao B.. 2014. Animals models of gastrointestinal and liver diseases. Animal models of alcohol-induced liver disease: pathophysiology, translational relevance, and challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 306: G819–G823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou H., Xie Q., Zhang M., Zhang C., Zhao G., Jin M., and Yu L.. 2009. Chronic alcohol consumption from adolescence-to-adulthood in mice–effect on growth and social behavior. Drug Alcohol Depend. 104: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tentler J. J., LaPaglia N., Steiner J., Williams D., Castelli M., Kelley M. R., Emanuele N. V., and Emanuele M. A.. 1997. Ethanol, growth hormone and testosterone in peripubertal rats. J. Endocrinol. 152: 477–487. [DOI] [PubMed] [Google Scholar]
- 38.Tan P., Liang H., Nie J., Diao Y., He Q., Hou B., Zhao T., Huang H., Li Y., Gao X., et al. . 2017. Establishment of an alcoholic fatty liver disease model in mice. Am. J. Drug Alcohol Abuse. 43: 61–68. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez M., Galy B., Muckenthaler M. U., and Hentze M. W.. 2007. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat. Struct. Mol. Biol. 14: 420–426. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez M., Galy B., Dandekar T., Bengert P., Vainshtein Y., Stolte J., Muckenthaler M. U., and Hentze M. W.. 2006. Iron regulation and the cell cycle: identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J. Biol. Chem. 281: 22865–22874. [DOI] [PubMed] [Google Scholar]
- 41.Rogers J. T., Randall J. D., Cahill C. M., Eder P. S., Huang X., Gunshin H., Leiter L., McPhee J., Sarang S. S., Utsuki T., et al. . 2002. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J. Biol. Chem. 277: 45518–45528. [DOI] [PubMed] [Google Scholar]
- 42.Goss D. J., and Theil E. C.. 2011. Iron responsive mRNAs: a family of Fe2+ sensitive riboregulators. Acc. Chem. Res. 44: 1320–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erlitzki R., Long J. C., and Theil E. C.. 2002. Multiple, conserved iron-responsive elements in the 3′-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J. Biol. Chem. 277: 42579–42587. [DOI] [PubMed] [Google Scholar]
- 44.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S. C., Gram H., and Han J.. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 120: 623–634. [DOI] [PubMed] [Google Scholar]
- 45.Shinkyo R., and Guengerich F. P.. 2011. Cytochrome P450 7A1 cholesterol 7alpha-hydroxylation: individual reaction steps in the catalytic cycle and rate-limiting ferric iron reduction. J. Biol. Chem. 286: 4632–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lijie Z., Ranran F., Xiuying L., Yutang H., Bo W., and Tao M.. 2016. Soyasaponin Bb protects rat hepatocytes from alcohol-induced oxidative stress by inducing heme oxygenase-1. Pharmacogn. Mag. 12: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palipoch S., Koomhin P., Punsawad C., Na-Ek P., Sattayakhom A., and Suwannalert P.. 2016. Heme oxygenase-1 alleviates alcoholic liver steatosis: histopathological study. J. Toxicol. Pathol. 29: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jais A., Einwallner E., Sharif O., Gossens K., Lu T. T., Soyal S. M., Medgyesi D., Neureiter D., Paier-Pourani J., Dalgaard K., et al. . 2014. Heme oxygenase-1 drives metaflammation and insulin resistance in mouse and man. Cell. 158: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D., Hui Y., Peng Y., Tang L., Jin J., He R., Li Y., Zhang S., Li L., Zhou Y., et al. . 2015. Overexpression of heme oxygenase 1 causes cognitive decline and affects pathways for tauopathy in mice. J. Alzheimers Dis. 43: 519–534. [DOI] [PubMed] [Google Scholar]
- 50.Lanceta L., Li C., Choi A. M., and Eaton J. W.. 2013. Haem oxygenase-1 overexpression alters intracellular iron distribution. Biochem. J. 449: 189–194. [DOI] [PubMed] [Google Scholar]
- 51.Song W., Su H., Song S., Paudel H. K., and Schipper H. M.. 2006. Over-expression of heme oxygenase-1 promotes oxidative mitochondrial damage in rat astroglia. J. Cell. Physiol. 206: 655–663. [DOI] [PubMed] [Google Scholar]
- 52.Iwai K., Drake S. K., Wehr N. B., Weissman A. M., LaVaute T., Minato N., Klausner R. D., Levine R. L., and Rouault T. A.. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA. 95: 4924–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cairo G., and Pietrangelo A.. 2000. Iron regulatory proteins in pathobiology. Biochem. J. 352: 241–250. [PMC free article] [PubMed] [Google Scholar]
- 54.Rouault T. A. 2009. Cell biology. An ancient gauge for iron. Science. 326: 676–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nilsson L. M., Sjovall J., Strom S., Bodin K., Nowak G., Einarsson C., and Ellis E.. 2007. Ethanol stimulates bile acid formation in primary human hepatocytes. Biochem. Biophys. Res. Commun. 364: 743–747. [DOI] [PubMed] [Google Scholar]
- 56.Li M., Diao Y., Liu Y., Huang H., Li Y., Tan P., Liang H., He Q., Nie J., Dong X., et al. . 2016. Chronic moderate alcohol intakes accelerate SR-B1 mediated reverse cholesterol transport. Sci. Rep. 6: 33032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie G., Zhong W., Li H., Li Q., Qiu Y., Zheng X., Chen H., Zhao X., Zhang S., Zhou Z., et al. . 2013. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 27: 3583–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein J. D., Sherrill J. B., Morello G. M., San Miguel P. J., Ding Z., Liangpunsakul S., Liang T., Muir W. M., Lumeng L., and Lossie A. C.. 2014. A snapshot of the hepatic transcriptome: ad libitum alcohol intake suppresses expression of cholesterol synthesis genes in alcohol-preferring (P) rats. PLoS One. 9: e110501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.