In this Review, Riera et al. review recent structural and biochemical insights that start to explain how specific proteins recognize DNA replication origins, load the replicative helicase on DNA, unwind DNA, synthesize new DNA strands, and reassemble chromatin.
Keywords: MCM2–7, DNA replication, pre-RC, CMG, replisome, cryo-EM
Abstract
DNA replication results in the doubling of the genome prior to cell division. This process requires the assembly of 50 or more protein factors into a replication fork. Here, we review recent structural and biochemical insights that start to explain how specific proteins recognize DNA replication origins, load the replicative helicase on DNA, unwind DNA, synthesize new DNA strands, and reassemble chromatin. We focus on the minichromosome maintenance (MCM2–7) proteins, which form the core of the eukaryotic replication fork, as this complex undergoes major structural rearrangements in order to engage with DNA, regulate its DNA-unwinding activity, and maintain genome stability.
Initiation of DNA replication is a multistep reaction that is carefully choreographed to promote replication fork assembly and regulated firing of replication origins (Costa et al. 2013; Riera et al. 2014; Tognetti et al. 2015; Bell and Labib 2016; Deegan and Diffley 2016; O'Donnell and Li 2016; Pellegrini and Costa 2016; Riera and Speck 2016; Bleichert et al. 2017). Moreover, this process is highly regulated in order to coordinate DNA synthesis with the cell cycle and the energy status of the cell. Checkpoints monitor the pathway and can halt DNA synthesis to overcome problems and safeguard the genome from damage (Alexander and Orr-Weaver 2016). In vivo analysis has identified most key players and addressed many regulatory principles, but many crucial mechanisms remain unknown. The recent reconstitution of budding yeast DNA replication using purified proteins now offers the chance for a detailed mechanistic and structural analysis of DNA replication initiation and DNA synthesis, which will help to advance the entire field (Yeeles et al. 2015, 2017; Devbhandari et al. 2017).
Initiation of DNA replication
The genomic sites where DNA replication is initiated are known as DNA replication origins (Marahrens and Stillman 1992; Hyrien 2016). In budding yeast, replication origins have conserved DNA sequences and always contain a binding site for the origin recognition complex (ORC) (Fig. 1A; Bell and Stillman 1992). This complex consists of six subunits and is organized in a C shape, with DNA being inserted into the central cleft, allowing for multiple protein–DNA contacts (Lee and Bell 1997; Speck et al. 2005; Sun et al. 2012; Yuan et al. 2017). During late M phase of the cell cycle, Cdc6 binding to an ORC/origin DNA complex starts a process of regulated protein assembly that culminates in the formation of the replication fork in S phase (Fig. 1B; Donovan et al. 1997; Rowles and Blow 1997; Weinreich et al. 1999). The ORC/Cdc6 complex contains four ATP-binding proteins: Cdc6, Orc1, Orc4, and Orc5. ATP binding by Orc1 and Cdc6 is required for ORC/Cdc6/DNA complex formation (Weinreich et al. 1999; Gillespie et al. 2001; Klemm and Bell 2001; Speck et al. 2005; Randell et al. 2006; Speck and Stillman 2007). Importantly, this ORC/Cdc6 complex is essential for the recruitment of the Cdt1/minichromosome maintenance 2–7 (MCM2–7) heptamer and the loading of MCM2–7 on dsDNA (Fig. 1C). MCM2–7 is the core of the replicative DNA helicase and consists of six subunits that have a spiral arrangement with a gap at the Mcm2/5 interface (Costa et al. 2011; Ticau et al. 2017; Zhai et al. 2017). During helicase loading, ORC, Cdc6, and Cdt1 insert DNA through the Mcm2/Mcm5 gate into the complex, and the MCM2–7 ring closes partially around dsDNA (Sun et al. 2013; Samel et al. 2014; Zhai et al. 2017). Crucially, the MCM2–7 recruitment process depends on an Mcm6–Cdt1 interaction, which alleviates an autoinhibitory activity of the Mcm6 C terminus (Fernandez-Cid et al. 2013). Upon ORC/Cdc6/Cdt1/MCM2–7 (OCCM) complex formation, ATP hydrolysis results in sequential Cdc6 and Cdt1 release and formation of an ORC/MCM2–7 (OM) intermediate (Fernandez-Cid et al. 2013; Coster et al. 2014; Kang et al. 2014; Ticau et al. 2015). Interestingly, Cdt1 release is associated with a structural change, as it promotes the closure of the MCM2–7 ring (Ticau et al. 2017). Following the recruitment of a second Cdc6, an ORC/Cdc6/MCM2–7 (OCM) complex is formed (Fig. 1D; Fernandez-Cid et al. 2013; Ticau et al. 2015, 2017). In contrast to the OCCM and the OM, the OCM complex is competent to rapidly recruit a second MCM2–7 hexamer, which also occurs in a Cdt1-dependent manner (Evrin et al. 2013, 2014; Fernandez-Cid et al. 2013; Sun et al. 2014; Ticau et al. 2015), resulting in a head-to-head MCM2–7 double hexamer (DH) that encircles dsDNA (Fig. 1E; Evrin et al. 2009; Remus et al. 2009; Gambus et al. 2011). Formation of the DH triggers Cdc6 release followed by simultaneous ORC and Cdt1 release and closure of the second MCM2–7 ring around DNA, resulting in a high-salt-stable complex (Evrin et al. 2009; Remus et al. 2009; Ticau et al. 2015, 2017). DH loading is also termed as prereplication complex (pre-RC) formation or DNA licensing. The large MCM2–7 DH is devoid of ATP hydrolysis (Sun et al. 2014) and DNA-unwinding activities (Evrin et al. 2009; Remus et al. 2009) but can slide on dsDNA in an ATP hydrolysis-independent manner (Evrin et al. 2009; Remus et al. 2009) to distribute DHs to non-origin-containing regions (Gros et al. 2015).
Figure 1.
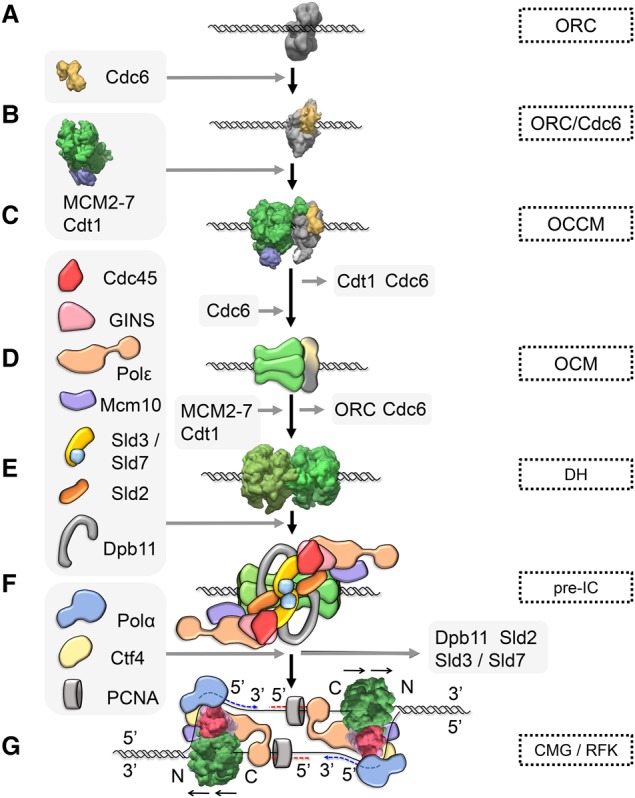
Eukaryotic initiation of DNA replication. Relevant complexes that have been characterized by electron microscopy are shown in surface view. (A) ORC is chromatin bound throughout the cell cycle (Electron Microscopy Data Bank [EMDB]: 1156). (B) ORC/Cdc6 is the landing platform MCM2–7/Cdt1 (EMDB: 5381). (C) The association of MCM2–7/Cdt1 (EMDB: 6671) with ORC/Cdc6 results in the OCCM (EMDB: 8540) formation with the dsDNA inserted into MCM2–7 hexamer. (D) Cdt1 and Cdc6 are released from the OCCM in an ATP hydrolysis-dependent reaction, and, upon recruitment of another Cdc6, the OCM the complex is formed. The OCM is an essential intermediate in the loading reaction and is responsible for the recruitment of a second MCM2–7/Cdt1 heptamer, although the details of the reaction are currently not known. (E) The final product of the loading reaction is a MCM2–7 DH embracing dsDNA (EMDB: 6338). This inactive complex is a stable DNA replication intermediate, which becomes activated only in S phase. (F) Preinitiation of DNA replication in S phase relies on Dbf4-dependent kinase (DDK)-dependent phosphorylation of the DH and a plethora of factors that interact with the DH. One of the landmarks of preinitiation complex formation is the binding of Cdc45 and GINS (from the Japanese go-ichi-ni-san, meaning 5-1-2-3, after the four related subunits of the complex: Sld5, Psf1, Psf2, and Psf3) to MCM2–7, resulting in formation of the replicative helicase: the Cdc45/MCM2–7/GINS (CMG) complex. (G) During the DNA-unwinding process, the CMG (EMDB: 8518) associates with both polymerases ε and α into a replication fork (RFK) to synthesize the leading and lagging strands. The helicase is propelled by the C-terminal AAA+ motor domains, and the unwinding takes place on the N-terminal face.
Activation of the MCM2–7 DH, termed preinitiation complex (pre-IC) formation, is a highly complex process and has been intensively studied in budding yeast. It depends on Dbf4-depedent kinase (DDK) Cdc7 and S-phase-specific cyclin-dependent kinase (CDK) and a large number of activation factors, including Sld3, Cdc45, Sld2, Dpb11, GINS (from the Japanese go-ichi-ni-san, meaning 5-1-2-3, after the four related subunits of the complex: Sld5, Psf1, Psf2 and Psf3), polymerase ε, and Mcm10 (Fig. 1F; Heller et al. 2011; Yeeles et al. 2015). During S phase, the MCM2–7 DHs become efficiently phosphorylated by DDK (Sheu and Stillman 2006, 2010; Francis et al. 2009; Sun et al. 2014), which in turn allows Sld3/Sld7 and Cdc45 recruitment to replication origins (Bruck et al. 2015; Herrera et al. 2015; Deegan et al. 2016; Fang et al. 2016). Moreover, CDK phosphorylates its essential targets, Sld2 and Sld3, which then allows these two phospho-proteins to interact with Dpb11 (Tanaka et al. 2007; Zegerman and Diffley 2007). Sld2, Dpb11, GINS, and polymerase ε form a loose complex in the cell (Muramatsu et al. 2010) and bind to Cdc45/Sld3/MCM2–7. Consequently, Sld2, Sld3, and Dpb11 are released, resulting in formation of the Cdc45/MCM2–7/GINS (CMG) complex (Kanemaki and Labib 2006; Heller et al. 2011; Yeeles et al. 2015). Interestingly, Mcm10 binding to the CMG is associated with a structural change in the CMG, rendering the complex high-salt-stable (Looke et al. 2017) and promoting origin firing (Kanke et al. 2012; van Deursen et al. 2012; Watase et al. 2012; Yeeles et al. 2015). The completely assembled CMG complex is highly active in ATP hydrolysis-driven 3′–5′ DNA unwinding, as seen first for the Drosophila variant, and forms the center of the eukaryotic DNA replication fork (Fig. 1G; Moyer et al. 2006; Ilves et al. 2010).
Crucially, during helicase activation, the MCM2–7 DH becomes extensively reorganized: Cdc45 and GINS bind to MCM2–7; the complex is split into two individual hexamers, potentially involving Mcm10 (Quan et al. 2015); one DNA strand becomes extruded from each hexamer; and this results in the two CMG complexes encircling ssDNA (Costa et al. 2014; Georgescu et al. 2017). Importantly, the mechanisms that lead to this MCM2–7 reorganization during initiation of DNA replication are largely unknown. DNA polymerases are known to associate with the CMG in part via Ctf4 to form a coupled DNA-unwinding and DNA synthesis assembly (Fig. 1G; Gambus et al. 2006; Simon et al. 2014; O'Donnell and Li 2016). Initial priming of DNA synthesis is carried out by DNA polymerase α, while leading and lagging strand DNA synthesis occurs mainly via polymerase ε (Pursell et al. 2007; Burgers et al. 2016) and polymerase δ (Nick McElhinny et al. 2008), respectively. However, some plasticity exists, with polymerase δ also playing a role during leading strand DNA synthesis, particularly during initiation of DNA replication and under conditions of replicative stress in budding yeast (Pavlov et al. 2001; Devbhandari et al. 2017; Yeeles et al. 2017) and after homologous recombination-dependent fork restart in Schizosaccharomyces pombe (Miyabe et al. 2015). Mcm10 acts also during elongation, as it travels with the replisome (Ricke and Bielinsky 2004; Gambus et al. 2006; Pacek et al. 2006), and as a specific mutation in its C terminus results in shorter replication products without affecting origin firing (Looke et al. 2017).
DNA synthesis is initiated at hundreds to thousands of replication origins in order to fully replicate the large eukaryotic genomes (Cvetic and Walter 2005; Mechali 2010). At each origin, one or more MCM2–7 DHs are loaded, but only a minority becomes transformed into active CMGs during S phase; with the remaining MCM2–7 DHs serving as “dormant origins,” which become activated only if a proximal replication fork becomes terminally arrested (Woodward et al. 2006; Ibarra et al. 2008).
Structural insights into key steps of DNA replication
The last few years have seen rapid progress for structural biology, in part due to the resolution revolution of cryo-electron microscopy (cryo-EM) (Kuhlbrandt 2014; Egelman 2016), and this has led to outstanding biology insights into DNA replication. Crystallography and cryo-EM have generated near-atomic resolution structures of several large protein complexes, including Drosophila melanogaster and Homo sapiens ORC (Bleichert et al. 2015; Tocilj et al. 2017); Saccharomyces cerevisiae MCM2–7 and MCM2–7/Cdt1 (Zhai et al. 2017), OCCM complex (Yuan et al. 2017), and MCM2–7 DH (Li et al. 2015); S. cerevisiae and D. melanogaster CMG (Abid Ali et al. 2016; Yuan et al. 2016; Georgescu et al. 2017); S. cerevisiae Ctf4 trimer (Simon et al. 2014), polymerase ε (Hogg et al. 2014), and polymerase δ (Swan et al. 2009); and H. sapiens Mcm2-H3/H4 (Huang et al. 2015; Richet et al. 2015). Here we concentrate on four key complexes involved in initiation of DNA replication; namely, S. cerevisiae ORC, OCCM, MCM2–7 DH, and CMG (Fig. 1A,C,E,G).
Origin recognition
Sequence-specific recognition of DNA replication origins by the S. cerevisiae ORC formed the biochemical basis for the discovery of this important DNA replication factor in 1992 (Bell and Stillman 1992), which started a 25-year journey toward the full reconstitution of budding yeast DNA replication (Yeeles et al. 2015, 2017; Devbhandari et al. 2017). The six-subunit Orc1–6 complex is well conserved from yeast to humans, and this homology extends even to archaea, where Orc monomers or dimers function in origin recognition (Li and Stillman 2012). Orc1–5, but not Orc6, have a conserved protein structure consisting of one N-terminal AAA+ domain and one C-terminal winged helix domain (WHD) (Fig. 2A; Bleichert et al. 2015; Tocilj et al. 2017; Yuan et al. 2017). Structural analysis showed that H. sapiens Orc6 has homology with transcription factor TFIIB (Liu et al. 2011).
Figure 2.
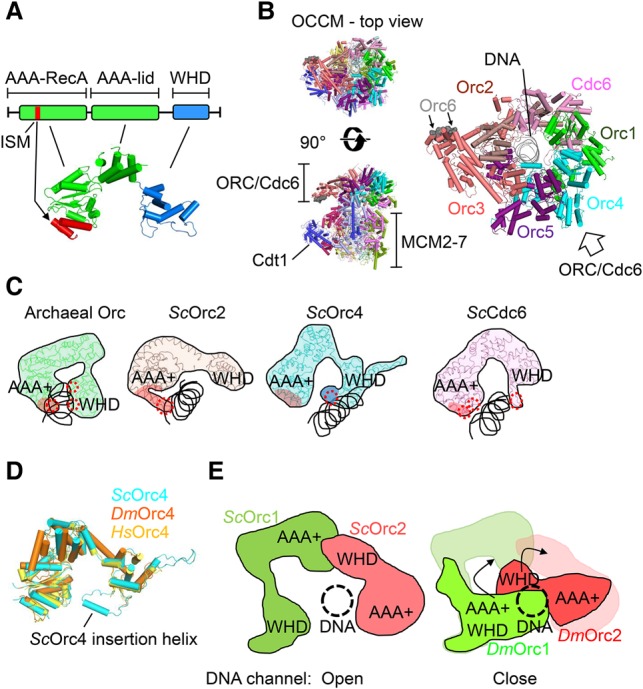
Origin recognition. (A) The domain and structural organization of Orc and Cdc6 proteins. The initiator-specific motif region of the AAA-RecA domain is indicated in red. (B, left) The OCCM structure in top view and side view (Protein Data Bank [PDB]: 5UDB). (Right) A top view of the ORC/Cdc6–DNA structure is shown enlarged. (C) Archaeal Orc (PDB: 2V1U) and the DNA-binding subunits of S. cerevisiae (Sc) ORC and Cdc6 are shown (PDB: 5UDB). The DNA-binding regions are indicated by red circles. The Orc4 insertion helix is shown in dark blue. (D) An overlay of S. cerevisiae Orc4 (PDB: 5UDB), D. melanogaster (Dm) Orc4 (PDB: 4XGC), and H. sapiens (Hs) Orc4 (PDB: 5UJM). (E) Structural comparison of D. melanogaster Orc1–Orc2 with S. cerevisiae Orc1–Orc2. The arrows show the rotation of D. melanogaster Orc1 AAA+ and Orc2 WHD to fit the S. cerevisiae Orc1 AAA+ and Orc2 WHD positions.
S. cerevisiae ORC recognizes specific DNA sequences within replication origins (Bell and Stillman 1992; Rao and Stillman 1995; Rowley et al. 1995; Dueber et al. 2007, 2011; Gaudier et al. 2007), while DNA structure and DNA-specific chromatin modifications appear more important for origin specification in metazoans (Remus et al. 2004; Eaton et al. 2011; Beck et al. 2012; Kuo et al. 2012; Cayrou et al. 2015). Interestingly, Cdc6 also consists of an N-terminal AAA+ domain and a C-terminal WHD, directly binds to ORC, and enhances the affinity and sequence specificity of budding yeast ORC (Speck et al. 2005; Speck and Stillman 2007).
However, the structural basis of origin recognition in eukaryotes was unknown for the longest time. Initially, crystal structures of archaeal ORC/DNA complexes revealed that both the β-hairpin wing and the helix–turn–helix (HTH) motif belonging to the WHD are deeply inserted into DNA grooves of the origin DNA. In addition, an initiator-specific motif (ISM) within the AAA+ domain contacts the DNA (Fig. 2A; Dueber et al. 2007; Gaudier et al. 2007). Both the ISM and WHD induce DNA bending, while the ISM also has a role in DNA sequence recognition (Dueber et al. 2007, 2011; Gaudier et al. 2007). Recently, the first high-resolution cryo-EM structure of DNA-bound ORC/Cdc6 in complex with Cdt1/MCM2–7 showed that S. cerevisiae Orc1–5 encircles DNA (Yuan et al. 2017). Here, the C-shaped structure of the Orc1–5 proteins is arranged in the order of Orc1–Orc4–Orc5–Orc3–Orc2, and Cdc6 fills the gap between Orc1 and Orc2, consistent with the previous low-resolution structure of S. cerevisiae ORC/Cdc6 (Chen et al. 2008) and the D. melanogaster ORC crystal structure (Fig. 2B; Bleichert et al. 2013). In this conformation, DNA makes multiple contacts with ORC/Cdc6 and is topologically trapped by the ring-shaped complex (Speck et al. 2005; Bleichert et al. 2015). Although Orc6 was not well resolved, a conserved C-terminal α helix was found to interact with Orc3 (Fig. 2B). Remarkably, this Orc6 helix is mutated in Meier-Gorlin syndrome. Indeed, this disease results in primordial dwarfism in humans, which is also caused by mutations in Orc1, Orc4, Cdt1, and Cdc6 (Bicknell et al. 2011; Tocilj et al. 2017). Consistently, in the case of Orc6, the mutation results in a defective Orc3 interaction, which in turn was shown to reduce MCM2–7 loading in Drosophila (Bleichert et al. 2013).
In the OCCM structure, the AAA+ domains and the WHDs of Orc1–5/Cdc6 form a central channel. In this context, Orc2, Orc4, and Cdc6 make direct DNA contacts, while Orc1, Orc3, and Orc5 do not touch the DNA (Fig. 2B; Yuan et al. 2017). Within the Orc2 protein, only the ISM of the AAA+ domain interacts with DNA, mostly contacting the phosphate backbone (Fig. 2C). Compared with most other species, budding yeast Orc4 contains an α-helix insertion, which is directly involved in DNA interaction. Unlike other Orc/Cdc6 proteins, Orc4 appears responsible for sequence-specific DNA interactions in the OCCM, as it uniquely makes base-specific contacts in the major groove of the DNA (Fig. 2D; Yuan et al. 2017). However, the functional relevance of these interactions is still outstanding. Moreover, site-specific DNA binding of the S. pombe ORC is also determined by the same Orc subunit (Chuang and Kelly 1999; Kong and DePamphilis 2001). Here, multiple AT-hook domains at the N terminus of S. pombe Orc4 are used for binding to replication origin sequences, highlighting the Orc4 protein as the most important module for sequence-specific DNA recognition in yeast. In contrast, D. melanogaster Orc4 and H. sapiens Orc4 are lacking the insertion α-helix and AT-hook domains. Intriguingly, metazoan replication origins share no common DNA sequence (Cayrou et al. 2015; Hyrien 2015). However, H. sapiens Orc1 has affinity for G-quadruplex ssDNA (Hoshina et al. 2013), shown to act as an origin-positioning motif (Valton et al. 2014), while DNA topology has been shown to be an important determinant for Drosophila ORC–DNA interactions (Remus et al. 2004). Moreover, epigenetic modifications, in particular H4K20 dimethylation and trimethylation, promote chromatin recruitment of H. sapiens ORC via an Orc1 bromo-adjacent homology (BAH) domain and DNA licensing (Beck et al. 2012; Kuo et al. 2012).
Besides S. cerevisiae, Cdc6 also plays a role in DNA binding in the OCCM (Yuan et al. 2017), with its ISM and WHD making prominent contacts with the phosphate backbone (Fig. 2A,C). Unlike archaeal ORC/Cdc6, S. cerevisiae Cdc6 uses only the WHD β-hairpin wing to contact DNA. Basically, S. cerevisiae Cdc6 contributes toward DNA binding at two levels: S. cerevisiae Cdc6 interaction with S. cerevisiae ORC traps DNA in the central channel, and, in addition, its two nucleic-acid binding surfaces contact the DNA directly, providing a structural explanation of why S. cerevisiae Cdc6 results in increased affinity of S. cerevisiae ORC for DNA (Mizushima et al. 2000). However, how the Cdc6 ATPase-dependent regulation of sequence specificity works (Speck and Stillman 2007) is still unknown at a structural level. The crystal structure of the D. melanogaster ORC core revealed that D. melanogaster Orc3–4–5 has a configuration similar to that of S. cerevisiae and H. sapiens Orc3–4–5, but the whole structure adopts an autoinhibited conformation that is incompatible with DNA and Cdc6 binding (Bleichert et al. 2015; Tocilj et al. 2017; Yuan et al. 2017). Aligning the D. melanogaster ORC structure to the S. cerevisiae ORC structure shows that the D. melanogaster Orc1 AAA+ domain and the D. melanogaster Orc2 WHD cover the central channel of the ORC DNA passage (Fig. 2E; Bleichert et al. 2015; Yuan et al. 2017). Thus, the conformational changes of those regions must be essential for DNA interaction and are also required for Cdc6 contacting Orc1 and Orc2 in Drosophila. With Orc1, Orc4, and Orc5 binding to ATP and ATP binding to Orc1 being essential for ORC/Cdc6/DNA complex formation (Weinreich et al. 1999; Gillespie et al. 2001; Klemm and Bell 2001; Speck et al. 2005; Randell et al. 2006; Speck and Stillman 2007), it appears possible that the interaction with ATP may result in strong conformational changes in the D. melanogaster Orc1 AAA+ domain and the D. melanogaster Orc2 WHD.
In summary, the recent structural insights reveal decreasing DNA sequence specificity during evolution. Archaeal ORC/Cdc6 proteins are forming multiple DNA sequence-specific contacts, and S. cerevisiae Orc/Cdc6 proteins are making fewer, mostly phosphate backbone and some base-specific contacts with DNA, while H. sapiens ORC is missing the sequence-specific DNA recognition motifs observed in yeast. The structural basis of topology-specific DNA recognition in D. melanogaster ORC and G-quadruplex-specific ssDNA binding in H. sapiens ORC is still outstanding and will offer further insights into the definition of replication origins in metazoan.
General organization of MCM
The MCM2–7 proteins are very well conserved from yeast to humans, while, in archaea, highly related homologs exist that assemble in homohexameric complexes. The six distinct subunits that make up MCM2–7 are dominated by a two-part domain structure (Fig. 3A–C): (1) the N-terminal protein interaction and DNA-binding domain and (2) the highly conserved C-terminal AAA+ ATPase motor domain (Fig. 3A,B; Costa and Onesti 2009). The two domains shape the characteristic dumbbell silhouette of the Mcm subunits, generating double ring structures in the context of the MCM2–7 hexamer (Fig. 3B,D).
Figure 3.
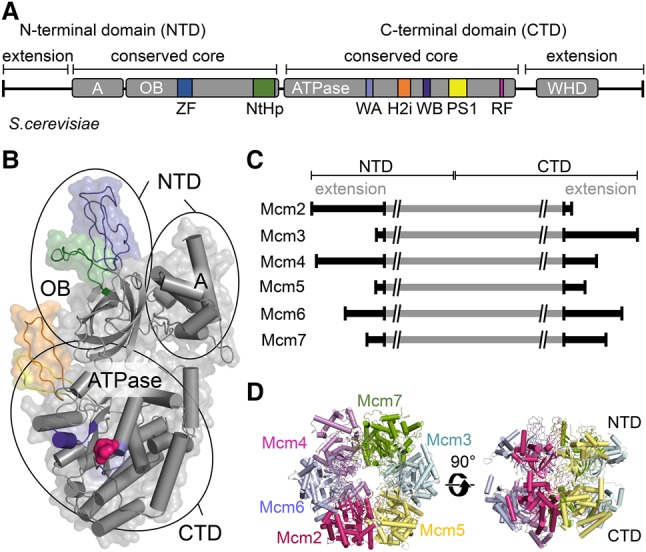
MCM2–7 helicase function arises from its architecture. (A) Domain structure of an Mcm protein with its four domains: A, oligonucleotide binding (OB), ATPase, and WHD. The color-coding for specific motifs—zinc finger (ZF), N-terminal hairpin (NtHp), Walker A (WA), Walker B (WB), presensor-1 (PS1), helix 2 insertion (H2i) β hairpin, arginine finger (RF), and WHD—is identical in A and B. (B) Atomic model of an exemplary Mcm subunit (PDB: 5U8S; Mcm2 of CMG bound to a replication fork) outlining its domain organization, with the WHD not shown. (C) Mcm subunits show different N-terminal and C-terminal extensions. (D) The MCM2–7 hexameric ring structure extracted from the CMG bound to a replication fork viewed from the top and side (PDB: 5U8S).
The N-terminal domain (NTD) can be divided roughly into two subdomains. (1) α Helices in the regulatory subdomain A form a compact bundle on the outside of the helicase ring. This domain has been suggested to regulate MCM helicase activity and function as a conformational switch in archaea (Slaymaker and Chen 2012; Miller and Enemark 2015). Its role in eukaryotes is less clear. (2) The second subdomain exhibits an oligonucleotide-binding (OB)-fold. This fold is involved in making contacts with ssDNA, as seen in the context of Pyrococcus furious MCM, with a potential role in initial DNA unwinding during the DH activation (Froelich et al. 2014). In addition, this domain also contributes via hydrophobic interactions to oligomerization of MCM2–7 into the characteristic hexamer structure (Bochman and Schwacha 2009). The OB-fold is interrupted by a DNA-binding N-terminal hairpin (NtHp) and a zinc finger (ZF). One of the main functions of the ZFs is to form the DH interface, which gives rise to the extraordinary salt stability of the complex (Li et al. 2015; Zhai et al. 2017). N-terminal extensions (NTEs) (Fig. 3C) distinguish eukaryotic Mcms from their archaeal counterparts. The large NTEs in Mcm2, Mcm4, and Mcm6 are mostly disordered and regulate both initiation and fork progression in a DDK-dependent manner (Sheu and Stillman 2006, 2010; Randell et al. 2010; Sheu et al. 2014, 2016), while the Mcm2 NTE is also important for replication-coupled assembly of chromatin (Huang et al. 2015; Richet et al. 2015).
The C-terminal domain (CTD) of Mcm proteins contains the motor of the helicase that has several key features: the ATP-binding motif Walker A (WA) and ATP hydrolysis motifs Walker B (WB) and arginine finger (RF), with the latter being localized at each of the intersubunit interfaces. In addition, the CTD contributes to DNA binding using the DNA-binding presensor-1 (PS1) and helix 2 insertion (H2i) β-hairpin loops, which protrude into the inner channel. Furthermore, most of the Mcm proteins include beyond the ATPase domain C-terminal extensions (CTEs) (Fig. 3C), which are composed of WHDs and additional sequences representing protein interaction motifs with various functions during DNA replication (Li et al. 2015; Abid Ali et al. 2016; Yuan et al. 2016, 2017).
MCM2–7 conformations in the OCCM, DH, and CMG reveal MCM2–7 ring-opening mechanisms
Although MCM2–7 is at the core of the replication fork, the complex is unable to unwind or even associate with dsDNA on its own. Instead, MCM2–7 requires different sets of cofactors in order to carry out each of these reactions (Takeda et al. 2005; Evrin et al. 2009; Remus et al. 2009; Fernandez-Cid et al. 2013; Frigola et al. 2013). Indeed, MCM2–7 is known to form complexes with various factors during the different stages of DNA replication initiation and DNA synthesis (Fig. 1). Three of the complexes, representing prominent stages of DNA replication, have been structurally characterized recently by cryo-EM at high resolution: the OCCM, the MCM2–7 DH, and the CMG (Fig. 4A–C; Costa et al. 2011, 2014; Sun et al. 2013, 2014; Li et al. 2015; Yuan et al. 2016, 2017; Georgescu et al. 2017). Interestingly, the Mcm subunit conformations are different in these three complexes, reflecting distinct functional states. In order to visualize these differences, we performed a structural alignment of the CTDs of each Mcm subunit of the three complexes. Here we took advantage of a highly regular Mcm conformation observed across the six subunits in the MCM2–7 DH. Thus, the DH served as a unique reference point (Fig. 4D; labeled with a green dot) to understand the alternative Mcm conformations present in the OCCM and CMG. This analysis revealed that, in the OCCM, all MCM2–7 NTDs are left-twisted by various degrees, while, in the CMG, half of the hexamer has left-twisted NTDs (Mcm2/3/5), and the other half has right-twisted NTDs (Mcm4/6/7). In the following, we discuss the three different complexes and their functions in the context of these structural changes.
Figure 4.
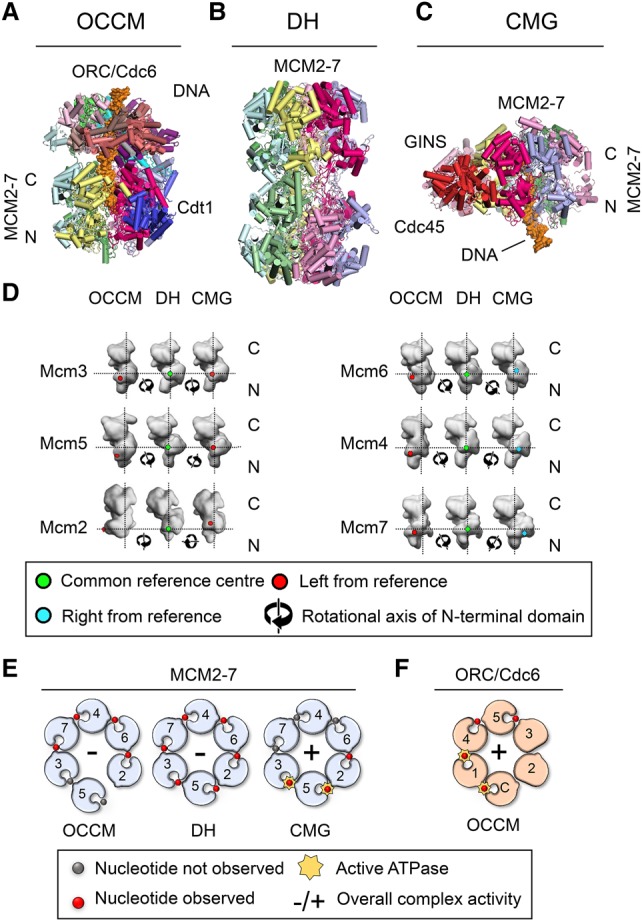
Structural changes in Mcm subunits dictate their function. Cartoon-style atomic models of OCCM (A; PDB: 5UDB), MCM2–7 DH (B; PDB: 3JA8), and CMG bound to a replication fork (C; PDB: 5U8S). (D) Comparison of the conformation of the MCM2–7 subunits in the OCCM, DH, and CMG. The CTDs of each Mcm subunit in the OCCM/DH/CMG conformations were aligned with the CTD of Mcm6 in the DH conformation, and this was taken as a fixed common position that was then used to generate the common reference center in the NTD (marked in green in the DH). This common reference point was used to detect the movement of the Mcm NTD in the OCCM and CMG relative to the DH, considering the Mcm NTD as a unit. Movement of the NTD toward the left is shown in red, and movement toward the right in is shown blue; the rotational axis relative to the common reference point is shown as a symbol. The alignment used the atomic structures of the proteins, but the figure depicts 10 Å surface view representations for improved clarity. (E) Schematic representation of the ATPase pockets of MCM2–7 in the OCCM, DH, and CMG. (F) ATPase pockets of ORC/Cdc6 in context of the OCCM.
The OCCM complex represents a highly transient intermediate prior to ATP hydrolysis-driven MCM2–7 DH formation (Fig. 4A). In the OCCM, the helicase is associated with its DNA loader (ORC/Cdc6), and the dsDNA is already inside the central channel of MCM2–7, although the DNA entry gate between the Mcm2 and Mcm5 interface still remains partially open (Sun et al. 2013; Yuan et al. 2017). Crucially, while, in the MCM2–7/Cdt1 precursor, the six Mcm proteins are arranged in a spiral structure, in the OCCM complex, they are arranged in a plane similar to those in the MCM2–7 DH and CMG (Yuan et al. 2017; Zhai et al. 2017). Importantly, Cdt1, which is essential for ORC/Cdc6 and MCM2–7 association, interacts with both the core helicase and the DNA loader (Chen et al. 2007; Fernandez-Cid et al. 2013; Sun et al. 2013; Ticau et al. 2015; Yuan et al. 2017). Surprisingly, in the OCCM, the Mcm subunits exhibit a very different conformation compared with the DH or the CMG. Here, the Mcm2 NTD is dramatically left-twisted, which extends to the neighboring Mcm6 and Mcm4 subunits and a more minor left twist in Mcm3, Mcm5, and Mcm7 (Fig. 4D). This is likely due to the presence of Cdt1 and a network of interactions between the WHDs of MCM2–7 and ORC/Cdc6 (Yuan et al. 2017). The extended conformation allows Cdt1 to make multiple contacts with MCM2–7. Its N-terminal region is bound to the Mcm2 NTD, directly touching the twisted Mcm2 domain. A long loop links the Cdt1 N-terminal region to its C-terminal region. This C-terminal region itself interacts with Mcm6 but also with the Mcm4 NTD. Therefore, Cdt1 embraces half of the hexamer (Mcm2, Mcm4, and Mcm6). Interestingly, most of these Cdt1–MCM2–7 interactions have been also observed in the context of the Cdt1–MCM2–7 complex, with the exception of the highly conserved Cdt1–Mcm6 CTD interaction (Wei et al. 2010; Liu et al. 2012; Fernandez-Cid et al. 2013; Yuan et al. 2017; Zhai et al. 2017), suggesting that this interaction has an important role in OCCM formation and could contribute toward the structural changes in the Mcm NTDs (Fig. 4D). Intriguingly, it has been suggested that Cdt1 acts to stabilize Mcm2, Mcm4, and Mcm6, potentially allowing the movement of the other half (Mcm5, Mcm3, and Mcm7) (Yuan et al. 2017). Indeed, Cdt1 release from the OCCM is associated with MCM2–7 ring closure (Ticau et al. 2017). In sum, these data support the concept that Cdt1 is important for remodeling of MCM2–7 and closing of the twisted Cdt1/MCM2–7 ring during OCCM formation, but much more work is needed to directly link these events to Cdt1.
The MCM2–7 DH represents a very stable DNA replication intermediate. In this complex, the Mcm2/5 gate is tightly locked, and no DNA-unwinding activity can be detected until the DH becomes activated during the G1/S transition. Indeed, the two MCM2–7 hexamers are stacked via the NTDs in an interaction largely mediated by their ZFs. These tight interactions stabilize the two hexamers and hinder MCM2–7 ring opening (Sun et al. 2014). Moreover, in this head-to-head configuration, the interaction modes of each of the six ZFs are completely different, causing a visible tilt and twist between the two hexamers of the complex (Remus et al. 2009; Sun et al. 2014; Li et al. 2015). Although the functional relevance of the imperfect stacking of the two hexamers has not been demonstrated, it could have a role in the helicase activation process (Li et al. 2015). Interestingly, within the DH, the individual Mcm subunits are twisted between the NTD and CTD rings (Sun et al. 2014; Li et al. 2015). In fact, the NTDs of all six subunits display right-handed twists of varying degrees when compared with their conformation in the OCCM (Fig. 4D). This twist could represent an additional mechanism of sealing the weak Mcm2/5 interface (Samel et al. 2014; Sun et al. 2014). Furthermore, the distances between neighboring CTDs are different, with three tightly (Mcm4/7, Mcm7/3, and Mcm5/2) and three loosely (Mcm6/4, Mcm3/5, and Mcm2/6) packed pairs (Li et al. 2015). Again, the functional relevance is not known, but it could be possible that the loosely packed subunits could participate in MCM2–7 ring opening during helicase activation.
Finally, the CMG is the active form of the replicative helicase (Fig. 4C). This complex represents the center of the replication fork and actively unwinds dsDNA, exposing DNA single strands to the polymerases and allowing their replication (Langston et al. 2014). In this configuration, the leading strand passes through the MCM2–7 ring in a 3′–5′ direction, while the lagging strand is excluded from it, and the gate between Mcm2 and Mcm5 is sealed by tight interactions with GINS and Cdc45 (Costa et al. 2011; Georgescu et al. 2017). When the MCM2–7 DH becomes transformed into the CMG, the NTDs become reorganized (Fig. 4D). Here, the NTDs of half of the MCM2–7 ring (Mcm3, Mcm5, and Mcm2) are twisted to the left, while the NTDs in the other half (Mcm6, Mcm4, and Mcm7) are twisted to the right (Fig. 4D). This process involves a range of complex rearrangements in the three dimensions of space, and we propose that these structural changes reconfigure the central channel of the helicase to allow interaction with ssDNA.
MCM2–7 ATP hydrolysis activities of the OCCM, DH, and CMG
MCM2–7 uses ATP binding and hydrolysis to unwind dsDNA during DNA replication. ATP hydrolysis is a coordinated process and requires the hexameric ring to be in a closed circular conformation (Schwacha and Bell 2001). The active ATPase sites are located within the Mcm dimer interfaces and form from conserved motifs of adjacent subunits; i.e., residues from both flanking protomers coordinate ATP binding and hydrolysis (Davey et al. 2003). The intersubunit nature of the active ATPase sites allows cooperative interactions that can be transmitted through the MCM2–7 ring. Indeed, incorporation of a single ATPase mutant into the MCM2–7 ring can result in a noticeable reduction in the ATP hydrolysis activity of the entire complex (Schwacha and Bell 2001; Davey et al. 2003; Ilves et al. 2010). However, some subunits, such as Mcm4, Mcm5, and Mcm7, appear to have a greater contribution toward the overall MCM2–7 ATP hydrolysis rate than others, as mutations in their WB motifs result in stronger ATP hydrolysis defects (Bochman et al. 2008; Bochman and Schwacha 2010; Bell and Botchan 2013). Many other ring-shaped hexameric motors show high cooperativity between active sites and sequential order of ATP hydrolysis (ATP hydrolysis in one subunit at the time following the order in the ring) (Abrahams et al. 1994; Liao et al. 2005; Adelman et al. 2006; Thomsen and Berger 2008; Eckert et al. 2012) but not all of them. For instance, studies on the hexameric protein unfoldase ClpX revealed that ATP hydrolysis in the hexamer is probabilistic. In ClpX, ATP hydrolysis in a given subunit depends on nucleotide binding of adjacent subunits, structural constraints, and substrate interactions but does not rely heavily on intersubunit cross-talk (Martin et al. 2005). Thus, ClpX ATP hydrolysis does not occur in the regular order of its six subunits but is fine-tuned for its specific function in unfolding proteins. In the context of MCM2–7, the data support a cooperative model of ATP hydrolysis. However, whether MCM2–7-dependent DNA unwinding at the replication fork follows a purely sequential order of ATP hydrolysis is still not clear.
MCM2–7 ATP hydrolysis during helicase loading has been analyzed by in vitro reconstituted reactions using a battery of ATP-binding and ATP hydrolysis mutants and a limited number of direct ATP hydrolysis assays (Fernandez-Cid et al. 2013; Coster et al. 2014; Kang et al. 2014). Here we discuss these data in the context of recent structural insights. Crucially, due to cryo-EM structures and fluorescence resonance energy transfer (FRET) data, we know that the MCM2–7 spiral becomes transformed into a nearly closed planar ring during OCCM formation, which then becomes further closed during OCM formation (Ticau et al. 2017; Yuan et al. 2017; Zhai et al. 2017). It is important to consider that the spiral-to-ring transformation introduces a tension into the complex, which puts the MCM2–7 complex under serious strain once it reaches the OCCM/OM/OCM stages. Consequently, mutations affecting the intersubunit interface geometry could block the ring closure, which in turn could affect (1) ATP hydrolysis-dependent Cdt1 release, (2) enclosure of DNA by the MCM2–7 ring, or (3) the ability of the complex to establish the correct hexamer–hexamer interface. Interestingly, MCM2–7 ATP-binding mutants are known to impair complex stability in the presence of low salt and elevated temperatures and display strong defects in the OCCM-to-DH transition (Coster et al. 2014; Kang et al. 2014). We suggest that the underlying reason for the pre-RC formation defect of Mcm ATP-binding mutants is the reduced complex stability, particular during the spiral-to-ring transition—a concept that could be tested using cryo-EM.
Furthermore, Mcm subunits are interconnected by RF mutants, which function for both complex stability and ATP hydrolysis (Coster et al. 2014). Two studies showed recently that mutation of the conserved Mcm arginine affects helicase loading, particularly when introduced in Mcm5 and Mcm6 but also in Mcm2, Mcm3, and Mcm7 (Coster et al. 2014; Kang et al. 2014). Moreover, arginine mutations in Mcm2 and Mcm5 specifically affect Cdt1 release. As Cdt1 release is a hallmark of pre-RC ATP hydrolysis, it was suggested that MCM2–7 ATP hydrolysis is essential for pre-RC formation. Interestingly, a different study showed that the RF mutation in Mcm3 does not have an impact on ATP hydrolysis rates during pre-RC formation (Fernandez-Cid et al. 2013). Direct ATP hydrolysis measurements for Mcm2 and Mcm5 RF mutants are still outstanding; therefore, it is not entirely clear whether these mutations affect only Mcm ATP hydrolysis (Coster et al. 2014; Kang et al. 2014) or may affect the intersubunit geometry and hence indirectly impair closing of the Mcm2–7 ring and/or Cdt1 release.
The cryo-EM analysis of the OCCM has provided further insights into the role of MCM2–7 ATP binding and hydrolysis during helicase loading. Here it was shown that ATP is at the interface of Mcm3/7, Mcm7/4, Mcm4/6, and Mcm6/2, where it possibly helps to stabilize intersubunit interactions. No nucleotide was detected at the Mcm3/5 or Mcm5/2 interfaces, consistent with the observation that the Mcm5/2 interface is broken (Yuan et al. 2017). Considering a cooperative MCM2–7 ATP hydrolysis model, the data would suggest that MCM2–7 ATPase activity is blocked or much reduced in the OCCM (Fig. 4E). However, Orc1 and Cdc6 are bound to ATP within the OCCM and appear primed for ATP hydrolysis (Fig. 4F; Yuan et al. 2017). Indeed, Cdc6 has been strongly implicated for ATP hydrolysis-dependent removal of failed helicase loading intermediates. Although the removal mechanism is not yet identified, Cdc6 ATPase activity is known to be induced during pre-RC formation (Fernandez-Cid et al. 2013), and Cdc6 ATPase mutants affect the release of Mcm3 (Coster et al. 2014), of MCM2–7 ATP-binding and ATP hydrolysis mutants that cause defective pre-RC formation (Coster et al. 2014; Kang et al. 2014), and of MCM2–7 loaded by Orc1–5 (missing Orc6) (Coster et al. 2014).
The role of Orc1 ATP hydrolysis during pre-RC formation is more complicated. An S. cerevisiae Orc4 RF mutant (Orc4R), which blocks ORC ATP hydrolysis (Bowers et al. 2004; Fernandez-Cid et al. 2013), showed no influence on ATP hydrolysis during pre-RC formation (Fernandez-Cid et al. 2013) or on MCM2–7 DH formation using a reconstituted pre-RC assay (Fernandez-Cid et al. 2013; Coster et al. 2014; Kang et al. 2014) but is lethal in vivo and allows only a single round of helicase loading using an extract-based pre-RC assay (Bowers et al. 2004). Furthermore, a Orc1 WB mutant (ORC-d1), which has defects in ATP binding and ATP hydrolysis (Klemm and Bell 2001), was analyzed under conditions of saturating ATP concentrations in order to test the role of Orc1 ATP hydrolysis during pre-RC formation. It was found that this mutant reduced helicase loading, Cdt1 release, and pre-RC-dependent ATP hydrolysis (Fernandez-Cid et al. 2013), suggesting that Orc1 ATP hydrolysis works in an ORC4R-independent manner during pre-RC formation. In summary, the role of ATP hydrolysis during pre-RC formation is complicated, as none of the mutants studied so far produces the same type of arrest in complex formation as has been observed with the slowly hydrolyzable ATP analog ATPγS.
As stated above, loading of multiple DHs at many replication origins is a way to store large amounts of the inactive replicative helicase before DNA synthesis takes place (Alver et al. 2014). Consistent with this notion, the DH is very stable even in the presence of high salt concentrations. Cryo-EM analysis revealed that ATP can be found at all of the Mcm subunit interfaces (Fig. 4E; Li et al. 2015). However, the strongly tilted conformation of MCM2–7 subunits resulted in structural changes in crucial ATP hydrolysis motifs, in particular the RFs localized at the intersubunit interfaces. Consistently, the DH was found to display minimal ATPase activity (Sun et al. 2014). Indeed, blocking MCM2–7 ATP hydrolysis represents a powerful mechanism to restrict MCM2–7 helicase activity in G1 phase, prior to its activation in S phase (Sun et al. 2015).
Progress has been made recently to better understand the fork organization (Abid Ali et al. 2016; Yuan et al. 2016; Georgescu et al. 2017). The very first cryo-EM structure of the S. cerevisiae CMG in complex with a forked DNA allowed Georgescu et al. (2017) to propose a new model of the replisome architecture, generating an improved framework to understand DNA synthesis. Most importantly, it was observed that, in the S. cerevisiae CMG, the NTD is at the leading edge of DNA unwinding (Georgescu et al. 2017). In contrast, a previous model suggested that the CTD is in front, which was based on FRET experiments with archaeal Mcm on a forked DNA and cryo-EM structures of the D. melanogaster CMG in complex with ssDNA (McGeoch et al. 2005; Costa et al. 2014). The new model, with the NTD near the fork, makes sense, as it places the leading and lagging strand polymerases in the correct locations for DNA synthesis, but additional studies would be useful to confirm these data and fully understand the DNA path through the replication fork (Sun et al. 2015; Miller and Costa 2017). In order to unwind the DNA, ssDNA needs to be actively propelled through the central channel of the CMG in a process that requires ATP hydrolysis (Moyer et al. 2006). Surprisingly, in S. cerevisiae, CMG ATP binding was detected at only three of the six Mcm interfaces; namely, Mcm3/5, Mcm5/2, and Mcm2/6 (Fig. 4E). Two of these (Mcm3/5 and Mcm5/2) correspond to the interfaces that are most important for helicase activity, as determined by analyzing the impact of ATPase mutants in the context of the D. melanogaster CMG (Ilves et al. 2010). Therefore, these studies suggest a differential contribution of the Mcm subunits toward the ATP hydrolysis and helicase activities of the CMG, with Mcm3, Mcm5, and Mcm2 being the key players. This suggests that MCM2–7 ATP hydrolysis does not follow the rotary model as observed in the E1 helicase or F1 ATPase (Enemark and Joshua-Tor 2008; Junge and Nelson 2015) but may have a substrate-specific activity similar to ClpX (Stinson et al. 2015).
Evolutionary conservation of the MCM helicase DNA channel
The Mcms hydrolyze ATP to translocate on DNA, with the six subunits forming a strongly positively charged DNA channel (Kumar and Remus 2016). All Mcm proteins are conserved throughout evolution and are highly similar in their three-dimensional structure, especially in key residues that are necessary for DNA binding or translocation through the central channel (see Fig. 5A). Superimposition of published structures of Mcms from archaea (Sulfolobus solfataricus) and eukaryotes (S. cerevisiae) highlight their structural similarity (Brewster et al. 2008; Yuan et al. 2016). This is especially the case in regions such as the N-terminal ZF, PS1 loop, H2i loop, and NtHp (Fig. 5A), which are important for DNA binding and dsDNA unwinding. Indeed, archaeal Mcm mutants that lack either the PS1 loop or the H2i loop motif are still competent for binding DNA but are devoid of dsDNA-unwinding activity (McGeoch and Bell 2005; Jenkinson and Chong 2006). Although flexibility in these loops and structures may occur, their general structure and position within the helicase are very well conserved (Fig. 5A), suggesting a universal binding mode and DNA path. Even more, the structure of the archaeal Mcm PS1 loop is similar to the corresponding loops found in viral helicases, such as the E1 helicase or the SV40 T-antigen, hinting at a universal mechanism of DNA unwinding (Li et al. 2003; Abbate et al. 2004).
Figure 5.
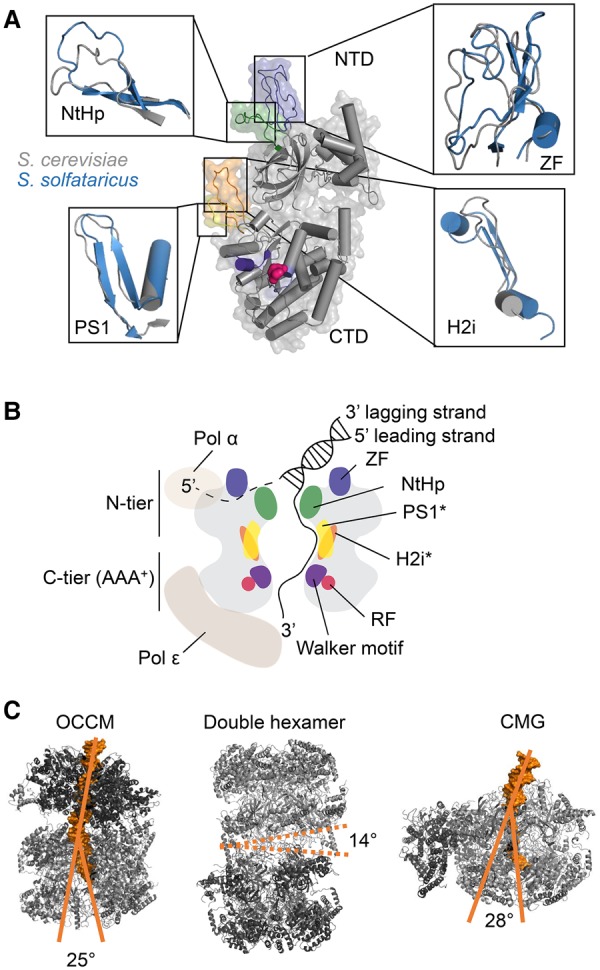
MCM2–7 DNA interactions within the OCCM, DH, and CMG complexes. (A) Detailed structural overlays of selected budding yeast (gray; PDB: 5U8S; Mcm2 of CMG bound to a replication fork) and S. solfataricus (blue; PDB: 3F9V) Mcm regions are shown. A superimposition of the PS1 loop, H2i loop, NtHp, and ZFs is depicted. (B) A schematic cut-through of the CMG with adjacent polymerases is shown with important domains labeled and color-coded as in A. The asterisk denotes that the hairpins of Mcm2/3/5/6 gather on one side of the central channel to interact with the passing ssDNA. (C) Structures of the budding yeast OCCM (PDB: 5UDB), DH (PDB: 3JA8), and CMG (PDB: 5U8S; CMG bound to a replication fork) are depicted. Furthermore, the central channels of the OCCM and CMG and its DNA bending are indicated. In the context of the DH, the interhexamer angle is shown.
Models for ORC-dependent MCM loading
Crucial MCM2–7 DNA interactions take place during ORC-dependent helicase loading, generating the critical topological link between the DNA and the helicase. Previous studies observed MCM2–7 in different conformations: The S. cerevisiae MCM2–7 hexamer was reported to be a closed ring, as observed by low-resolution negative-stained samples (Bochman and Schwacha 2007; Samel et al. 2014), which is also true for its archaeal and bacterial counterparts (Enemark and Joshua-Tor 2008; Miller et al. 2014). On the other hand, hexamers from Drosophila and the microsporidian parasite Encephalitozoon cuniculi were shown to adopt spiral-shaped or open-ringed conformations (Costa et al. 2011; Lyubimov et al. 2012). However, a recent study showed that S. cerevisiae MCM2–7 forms a left-handed spiral shape, with a 10–15 Å gap between Mcm2 and Mcm5 that is too narrow for DNA insertion (Zhai et al. 2017). In addition, it was found that the central channel of MCM2–7 was partially occluded by the CTEs of Mcm5 and Mcm6. This half-open complex is stabilized by Cdt1 binding to the N-terminal regions of Mcm2, Mcm4, and Mcm6 (Zhai et al. 2017). As MCM2–7 in this conformation does not allow DNA insertion, this suggests that the initial contact of Cdt1/MCM2–7 with ORC/Cdc6 needs to widen the Mcm2/5 gate to allow DNA entry. In the context of the OCCM, the MCM2–7 ring remains partially open (Sun et al. 2013; Yuan et al. 2017). The sealing of the Mcm2/5 gate requires ATP hydrolysis and removal of Cdt1 (Ticau et al. 2017), but the necessary structural rearrangements are still unknown. It will be fascinating to uncover the molecular mechanisms that allow controlled MCM2–7 ring opening, DNA insertion, and ring closure.
A ssDNA path through the budding yeast MCM DNA channel
As discussed, the apparent DNA path through the S. cerevisiae CMG complex has been observed recently by using an artificial DNA fork substrate (Georgescu et al. 2017). Here, the S. cerevisiae MCM2–7 helicase travels in a 3′–5′ polarity with the N-terminal tier ahead of the C-terminal tier (Rothenberg et al. 2007; Costa et al. 2014; Sun et al. 2015; Georgescu et al. 2017). These data allow us to understand how DNA traverses through the CMG (Fig. 5B): The dsDNA enters the N-terminal tier of the helicase and is unwound. The investigators suggest that the lagging strand leaves the helicase sandwiched between the ZFs and the NtHps on the N-terminal surface of MCM2–7, where the polymerase α can directly prime lagging strand synthesis (Georgescu et al. 2017).
Upon strand separation, the leading strand is then passed through the positively charged central channel, where it interacts with the PS1 loop or the H2i loop of Mcm2/3/5/6 and adopts a right-handed spiral B-DNA form before being handed to polymerase ε for leading strand synthesis (Georgescu et al. 2017). Interestingly, earlier structural studies from archaea suggested that the ssDNA binds in the plane of the ring rather than perpendicular (Froelich et al. 2014). It will be interesting to investigate whether the new structural data of the CMG in complex with an artificial replication fork (Georgescu et al. 2017) will be supported by additional biochemical or in vivo data to verify the direction of DNA translocation of the CMG. Moreover, additional structures of replication fork intermediates captured in complex with DNA, which have been largely elusive so far, will be important to fully understand the path of dsDNA and ssDNA through the CMG.
MCM and DNA channel flexibility during DNA replication
When comparing structures that depict different MCM2–7 states in DNA replication initiation in the order of their occurrence (OCCM → DH → CMG) (Fig. 5C), several differences are noticeable. Despite the overall unaffected structure of the two-tiered MCM2–7 helicase, internal rearrangements occur. In the OCCM, after the first MCM2–7 is loaded, DNA is bent by ∼25° at the ORC–MCM2–7 hexamer interface, which could represent a mechanism of DNA insertion (Yuan et al. 2017). Loading of the second hexamer and dissociation of the ORC leads to the MCM2–7 DH formation, in which hexamers are tilted by 14°, which may have a role in initial DNA unwinding (Li et al. 2015). Finally, dissociation of the DH and association of Cdc45 and GINS with the MCM2–7 rings lead to the assembly of the CMG. After activation, this complex continuously unwinds dsDNA to prepare it for replication. Through this, the dsDNA strand is bent again by ∼28° to the right of the vertical axis (Georgescu et al. 2017). This kink could be introduced by the necessity of efficiently unwinding the relatively rigid dsDNA. The designated lagging strand is proposed to leave the CMG on the surface of the Mcm3/Mcm5 interface (Georgescu et al. 2017).
Protein interactions remodel MCM2–7 during different stages of DNA replication initiation
Unlike bacteria, eukaryotes coordinate their DNA synthesis with the cell cycle and organize their replisome around the replicative helicase (Stillman 2005). Clearly, the MCM2–7 proteins evolved significantly from their prokaryotic precursor and gained additional regulatory and functional features. Protein–protein interactions are at the core of this new functionality, and we discuss their roles in the context of MCM2–7 structure in this section.
Two centers of the MCM2–7 interactome: the C-terminal and N-terminal protein-binding hubs
C-terminal interactions
Cryo-EM structures of the OCCM complex (Sun et al. 2013; Yuan et al. 2017) and biochemical experiments (Fernandez-Cid et al. 2013; Frigola et al. 2013) have shown the important role of the Mcm C termini in pre-RC formation (Fig. 6A). In the following section, we discuss the progress in understanding the protein interaction interface of ORC/Cdc6 and Cdt1/MCM2–7. Indeed, Cdt1, which is essential for chromatin binding of MCM2–7 (Maiorano et al. 2000; Nishitani et al. 2000), interacts with the Mcm6 C terminus (Jee et al. 2010; Wei et al. 2010). Nuclear magnetic resonance (NMR) analysis of the human and budding yeast proteins identified the structure of this important interaction surface, highlighting the Mcm6 WHD for Cdt1 binding (Wei et al. 2010; Liu et al. 2012). Mutation of conserved amino acids in this domain affects the cell in two ways, blocking MCM2–7 nuclear import and DNA synthesis (Liu et al. 2012). In vitro analysis revealed that the Mcm6 WHD adopts an autoinhibited conformation that blocks the binding of MCM2–7 to ORC/Cdc6 (Fernandez-Cid et al. 2013). The structural basis of the Mcm6 WHD autoinhibition was observed recently (Yuan et al. 2017; Zhai et al. 2017). In the absence of Cdt1, the ORC/Cdc6 interaction with MCM2–7 is blocked due to a clash between Orc4 and the Mcm6 WHD. Cdt1 overcomes this block by reorganizing the Mcm6 WHD, which prevents the steric clash and enables tight ORC/Cdc6 interactions with Cdt1/MCM2–7. Other studies reported that the conserved C terminus of S. cerevisiae Mcm3 is also essential for the initial recruitment of Cdt1/MCM2–7 to ORC/Cdc6 (Frigola et al. 2013; Sun et al. 2013). Moreover, in the same studies, it was observed that Mcm3 interacts directly with Cdc6 and that the Mcm3 C terminus induces ORC/Cdc6 ATP hydrolysis with a role in quality control of complex assembly. Finally, the Mcm3 C-terminal region also contains a WHD that interacts with Cdc6 and Orc2, but the function of this interaction is currently unknown (Yuan et al. 2017).
Figure 6.
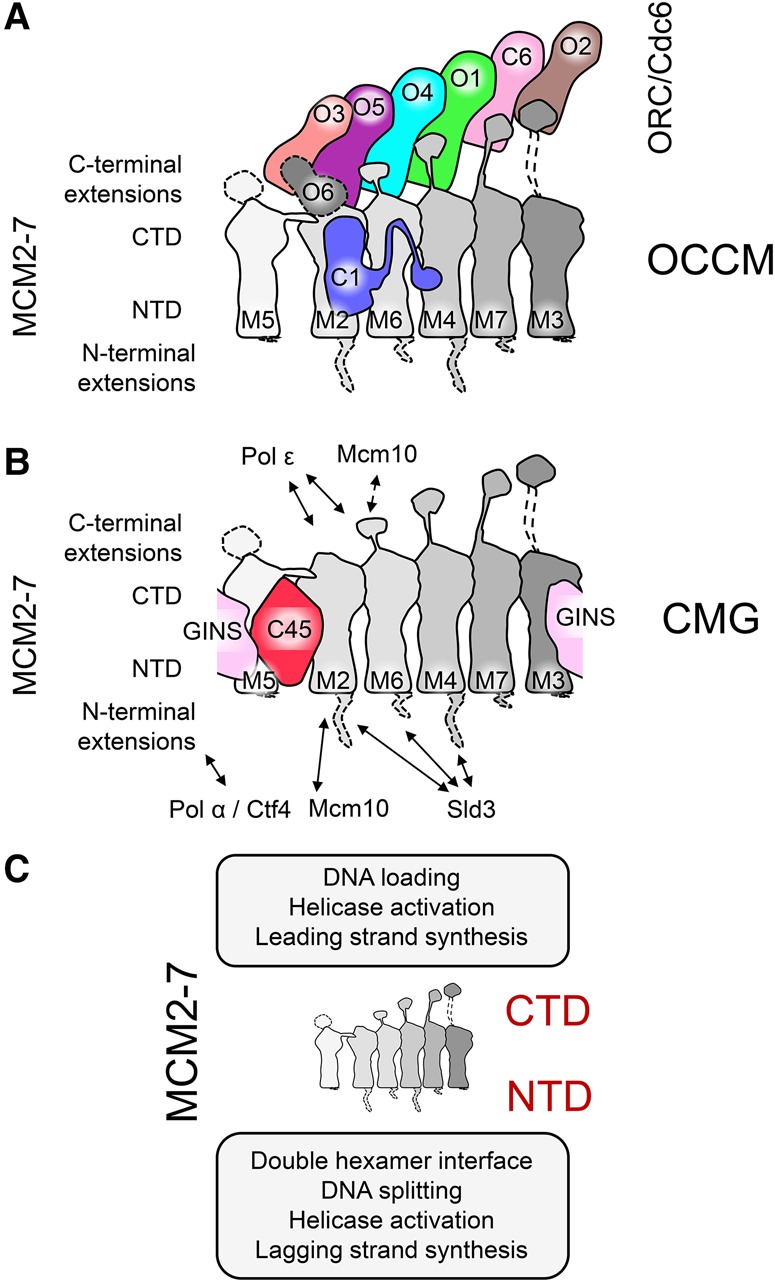
MCM2–7 protein–protein interactions in the context of OCCM and CMG. Interactions between the Mcm subunits (in gray gradient) and the Orc subunits (Orc1 in green, Orc2 in brown, Orc3 in salmon, Orc4 in cyan, Orc5 in purple, and Orc6 in gray), Cdc6 (light pink), and Cdt1 (blue) in the OCCM (A) and GINS (pale pink) and Cdc45 (red) in the CMG (B). The dashed lines indicate flexible regions not solved in the structures. (C) Summary of the different functions in which the MCM2–7 complex is involved though interactions with other proteins.
In S. cerevisiae Mcm2, unlike the other Mcm subunits, the CTE is missing (Fig. 3C). Interestingly, Mcm2 in higher eukaryotes contains a C-terminal WHD. Furthermore, these eukaryotes also possess geminin, a crucial inhibitor of pre-RC formation (Lee et al. 2004). Interestingly, the N-terminal part of H. sapiens Cdt1 interacts with geminin; due to this proximity, geminin is well placed to interact with the Mcm2 CTE. We suggest that the Mcm2 WHD may have coevolved as an additional geminin-anchoring point and in this way could block pre-RC formation by interfering with direct Cdt1–MCM2–7 interactions.
Within the MCM2–7/Cdt1–ORC/Cdc6 interface (Yuan et al. 2017), the two complexes are tethered together using flexible links. These connections are made up from the Mcm CTEs and contain a flexible linker and a Mcm WHD, which latches onto ORC/Cdc6. In addition, the CTDs of Mcm2, Mcm6, and Mcm4 are connected directly to the WHDs of Orc5, Orc4, and Orc1, respectively, linking one half of the complex more tightly to ORC/Cdc6, while the other half remains more mobile. Although the functional relevance of these interactions is not yet entirely clear, this built-in flexibility has been suggested to be involved in the process of DNA loading (Yuan et al. 2017).
In contrast to the OCCM, the Mcm CTEs appear mostly flexible in the MCM2–7 DH (Li et al. 2015). However, in the CMG, the CTEs of Mcm4, Mcm5, and Mcm6 are more rigid. Two CMG cryo-EM structures—an Apo form and a variant bound to a fork DNA—have highlighted that the CTEs can adopt different conformations. Without DNA, the CTEs of Mcm5 and Mcm6 partially occlude the DNA channel, but, in the context of the replication fork, these are repositioned to allow ssDNA passage through the helicase, with the Mcm4 CTE touching the DNA itself, suggesting a complex mechanism of DNA sensing (Georgescu et al. 2017). In addition, the Mcm6 CTE makes contacts with Mcm10, but this is not essential for Mcm10 recruitment or DNA replication initiation (Fig. 6B; Douglas and Diffley 2016). At the C-terminal face, the CMG expels ssDNA from its central channel. Recently, polymerase ε was found to be localized in this position, contacting the CTDs of Mcm2 and Mcm6, ideally placed to carry out leading strand DNA synthesis (Fig. 6B; Sun et al. 2015). In summary, the MCM2–7 C-terminal face has a major role in MCM2–7 loading, helicase activation, and leading strand DNA synthesis (Fig. 6C).
N-terminal interactions
During MCM2–7 DH formation, the N-terminal face of the two hexamers bind to each other, resulting in a highly stable complex. Within this complex, the long NTEs of Mcm2, Mcm4, and Mcm6 likely adopt a very flexible conformation, as they were not well resolved in the cryo-EM structure (Li et al. 2015). Once the cell enters S phase, DDK phosphorylates the MCM2–7 DH (Sun et al. 2014) to promote CMG formation and replication fork assembly. Specifically, the Mcm2, Mcm4, and Mcm6 NTEs are major sites for DDK phosphorylation and serve as binding sites for Sld3 (Fig. 6B; Sheu and Stillman 2006; 2010; Randell et al. 2010; Bruck and Kaplan 2015; Herrera et al. 2015; Deegan et al. 2016; Fang et al. 2016). In the CMG cryo-EM structures, the NTEs are only poorly resolved, again suggesting a flexible conformation (Yuan et al. 2016; Georgescu et al. 2017). However, upon interaction with specific partners, these flexible NTEs are likely to adopt a specific structure. In the case of Mcm2, a section of the NTE was crystallized together with an H3/H4 dimer and assumed a defined structure encircling the histones. Accordingly, this Mcm2 NTE has been suggested to play an important role in nucleosome recycling (Huang et al. 2015; Richet et al. 2015). Within the CMG, the NTDs are kept in a stable ring conformation through multiple interactions with Cdc45 and GINS. Here, Cdc45 interacts with the NTDs of Mcm2 and Mcm5 (Costa et al. 2011; Abid Ali et al. 2016; Simon et al. 2016). Cdc45 also closely interacts with the four proteins that constitute the GINS complex, which in turn binds the NTDs of Mcm3 and Mcm5. This broad system of interactions between GINS, Cdc45, Mcm2, Mcm3, and Mcm5 has been hypothesized to serve as a mechanism that keeps the Mcm2/5 gate in a closed state (Fig. 6B; Costa et al. 2011). Similarly, Cdt1 can contact Mcm2, Mcm6, and Mcm4 due to its striking extended conformation (Fig. 6A). However, this alternative network of interactions induced by Cdt1 has been suggested to have a different function, potentially allowing opening and closing the Mcm2/5 gate during OCCM formation (Ticau et al. 2017; Yuan et al. 2017; Zhai et al. 2017). Interestingly, although both Cdc45 and Cdt1 bind the Mcm2 NTD, they contact mutually exclusive surfaces of Mcm2, with Cdc45 binding the Mcm5-proximal part, while Cdt1 contacts the Mcm6-proximal section. Therefore, the two proteins might affect Mcm2 in alternative ways, with Cdc45 closing the gate to support helicase activity, and Cdt1 allowing opening and closing of the gate during MCM2–7 loading. Moreover, Mcm10 might affect these surfaces as well, as it contacts the Mcm2 NTD (Apger et al. 2010; Lee et al. 2010; Looke et al. 2017) in order to stabilize the CMG complex (Looke et al. 2017). At an in vitro assembled replication fork, ssDNA appears from the N-terminal MCM2–7 face during DNA unwinding (Georgescu et al. 2017). This ssDNA is well positioned to serve as a template for the lagging strand DNA synthesis. Indeed, DNA polymerase α/primase and Ctf4 were found to colocalize at the N-terminal MCM2–7 face in a low-resolution negative stain EM study (Fig. 3B; Sun et al. 2015). Ctf4 is a homotrimer that acts as a hub of protein interactions, linking several factors involved in DNA synthesis, chromatin remodelling, rDNA stability, DNA recombination, and sister chromatid cohesion to the CMG (Simon et al. 2014; Villa et al. 2016), while polymerase α primes DNA synthesis prior to extension by processive DNA polymerases (Zhang and O'Donnell 2016). These recent structural findings start to reveal a picture of the eukaryotic replisome assembly process as a whole; namely, the N-terminal MCM2–7 face has a major role in DH organization and stability, helicase activation, controlled opening and closing of the Mcm2/5 gate, and lagging strand DNA synthesis (Fig. 6C).
Outlook
Structural biology has provided major biological insights into replisome assembly and function from the architectural point of view and also by providing detailed mechanistic insights. In the future, these data will stimulate the design of sophisticated biochemical and single-molecule experiments, facilitating the measurement of novel activities and revealing specific mechanisms and their dynamics. The replisome consists of 50 or more factors and many more associated factors; the integration of these into a structural and functional network will be an exciting task for the years to come. Translating this knowledge into the context of disease has already started (for example, in the case of Meier-Gorlin syndrome), but thousands of mutations, as observed in the context of cancer, have not yet been analyzed. Importantly, the structural and mechanistic insight into DNA replication initiation will help the development of novel inhibitors, which are aimed to block DNA replication before it even starts (Shreeram et al. 2002; Blow and Gillespie 2008). However, as the focus of drug development is considerably directed toward enzymes, it may still take some years before major progress is achieved.
Acknowledgments
We thank Indiana Magdalou and Almut Caspary for critical reading of the manuscript. We thank the Biotechnology and Biological Sciences Research Council (BB/M003760/1 and BB/N000323/1), the Medical Research Council (MC_U120085811), and the Wellcome Trust (107903/Z/15/Z) for funding.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.298232.117.
Freely available online through the Genes & Development Open Access option.
References
- Abbate EA, Berger JM, Botchan MR. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev 18: 1981–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid Ali F, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC, Rueda D, Costa A. 2016. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun 7: 10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams JP, Leslie AG, Lutter R, Walker JE. 1994. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628. [DOI] [PubMed] [Google Scholar]
- Adelman JL, Jeong YJ, Liao JC, Patel G, Kim DE, Oster G, Patel SS. 2006. Mechanochemistry of transcription termination factor Rho. Mol Cell 22: 611–621. [DOI] [PubMed] [Google Scholar]
- Alexander JL, Orr-Weaver TL. 2016. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev 30: 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver RC, Chadha GS, Blow JJ. 2014. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair 19: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apger J, Reubens M, Henderson L, Gouge CA, Ilic N, Zhou HH, Christensen TW. 2010. Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics 185: 1151–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DB, Burton A, Oda H, Ziegler-Birling C, Torres-Padilla ME, Reinberg D. 2012. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev 26: 2580–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Botchan MR. 2013. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 5: a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Labib K. 2016. Chromosome duplication in Saccharomyces cerevisiae. Genetics 203: 1027–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, et al. 2011. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM. 2013. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife 2: e00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Botchan MR, Berger JM. 2015. Crystal structure of the eukaryotic origin recognition complex. Nature 519: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Botchan MR, Berger JM. 2017. Mechanisms for initiating cellular DNA replication. Science 355: eaah6317. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ. 2008. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer 8: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2007. Differences in the single-stranded DNA binding activities of MCM2–7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem 282: 33795–33804. [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73: 652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. 2010. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2–7 ‘gate’. Nucleic Acids Res 38: 6078–6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, Schwacha A. 2008. Subunit organization of Mcm2–7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol 28: 5865–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell 16: 967–978. [DOI] [PubMed] [Google Scholar]
- Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci 105: 20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I, Kaplan DL. 2015. Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase. Proc Natl Acad Sci 112: 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I, Perez-Arnaiz P, Colbert MK, Kaplan DL. 2015. Insights into the initiation of eukaryotic DNA replication. Nucleus 6: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM, Gordenin D, Kunkel TA. 2016. Who is leading the replication fork, Pol ε or Pol δ? Mol Cell 61: 492–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Mechali M. 2015. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25: 1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. 2007. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev 21: 2897–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. 2008. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci 105: 10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci 96: 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Onesti S. 2009. Structural biology of MCM helicases. Crit Rev Biochem Mol Biol 44: 326–342. [DOI] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. 2011. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Hood IV, Berger JM. 2013. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem 82: 25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM. 2014. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife 3: e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster G, Frigola J, Beuron F, Morris EP, Diffley JF. 2014. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell 55: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetic C, Walter JC. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol 16: 343–353. [DOI] [PubMed] [Google Scholar]
- Davey MJ, Indiani C, O'Donnell M. 2003. Reconstitution of the Mcm2–7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem 278: 4491–4499. [DOI] [PubMed] [Google Scholar]
- Deegan TD, Diffley JF. 2016. MCM: one ring to rule them all. Curr Opin Struct Biol 37: 145–151. [DOI] [PubMed] [Google Scholar]
- Deegan TD, Yeeles JT, Diffley JF. 2016. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J 35: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. 2017. Chromatin constrains the initiation and elongation of DNA replication. Mol Cell 65: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci 94: 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Diffley JF. 2016. Recruitment of Mcm10 to sites of replication initiation requires direct binding to the minichromosome maintenance (MCM) complex. J Biol Chem 291: 5879–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM. 2007. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Dueber EC, Costa A, Corn JE, Bell SD, Berger JM. 2011. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res 39: 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert T, Link S, Le DT, Sobczak JP, Gieseke A, Richter K, Woehlke G. 2012. Subunit Interactions and cooperativity in the microtubule-severing AAA ATPase spastin. J Biol Chem 287: 26278–26290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. 2016. The current revolution in cryo-EM. Biophys J 110: 1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. 2008. On helicases and other motor proteins. Curr Opin Struct Biol 18: 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. 2009. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci 106: 20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C. 2013. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2–7 hexamer dimerization. Nucleic Acids Res 41: 3162–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Riera A, Zech J, Clarke P, Herrera MC, Tognetti S, Lurz R, Speck C. 2014. The ORC/Cdc6/MCM2–7 complex facilitates MCM2–7 dimerization during prereplicative complex formation. Nucleic Acids Res 42: 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Cao Q, Lou H. 2016. Sld3–MCM interaction facilitated by Dbf4-dependent kinase defines an essential step in eukaryotic DNA replication initiation. Front Microbiol 7: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. 2013. An ORC/Cdc6/MCM2–7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell 50: 577–588. [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. 2009. Incorporation into the prereplicative complex activates the Mcm2–7 helicase for Cdc7–Dbf4 phosphorylation. Genes Dev 23: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JF. 2013. ATPase-dependent quality control of DNA replication origin licensing. Nature 495: 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. 2014. A conserved MCM single-stranded DNA binding element is essential for replication initiation. Elife 3: e01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366. [DOI] [PubMed] [Google Scholar]
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. 2011. MCM2–7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 286: 11855–11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. 2007. Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216. [DOI] [PubMed] [Google Scholar]
- Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, O'Donnell ME. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci 114: E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ. 2001. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Kumar C, Lynch G, Yadav T, Whitehouse I, Remus D. 2015. Post-licensing specification of eukaryotic replication origins by facilitated Mcm2–7 sliding along DNA. Mol Cell 60: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. 2011. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera MC, Tognetti S, Riera A, Zech J, Clarke P, Fernandez-Cid A, Speck C. 2015. A reconstituted system reveals how activating and inhibitory interactions control DDK dependent assembly of the eukaryotic replicative helicase. Nucleic Acids Res 43: 10238–10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, Johansson E. 2014. Structural basis for processive DNA synthesis by yeast DNA polymerase ε. Nat Struct Mol Biol 21: 49–55. [DOI] [PubMed] [Google Scholar]
- Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, Nakatsuka A, Kunichika T, Obuse C, Waga S. 2013. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem 288: 30161–30171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Stromme CB, Saredi G, Hodl M, Strandsby A, Gonzalez-Aguilera C, Chen S, Groth A, Patel DJ. 2015. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol 22: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. 2015. Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol 208: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. 2016. How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000Res 5: 2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J. 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci 105: 8956–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. 2010. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee J, Mizuno T, Kamada K, Tochio H, Chiba Y, Yanagi K, Yasuda G, Hiroaki H, Hanaoka F, Shirakawa M. 2010. Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins. J Biol Chem 285: 15931–15940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson ER, Chong JP. 2006. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci 103: 7613–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge W, Nelson N. 2015. ATP synthase. Annu Rev Biochem 84: 631–657. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Labib K. 2006. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25: 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Warner MD, Bell SP. 2014. Multiple functions for Mcm2–7 ATPase motifs during replication initiation. Mol Cell 55: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. 2012. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J 31: 2182–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Bell SP. 2001. ATP bound to the origin recognition complex is important for preRC formation. Proc Natl Acad Sci 98: 8361–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, DePamphilis ML. 2001. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 21: 8095–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. 2014. Biochemistry. The resolution revolution. Science 343: 1443–1444. [DOI] [PubMed] [Google Scholar]
- Kumar C, Remus D. 2016. Eukaryotic replication origins: strength in flexibility. Nucleus 7: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. 2014. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci 111: 15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Bell SP. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17: 7159–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Kim Y, Cho Y. 2004. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature 430: 913–917. [DOI] [PubMed] [Google Scholar]
- Lee C, Liachko I, Bouten R, Kelman Z, Tye BK. 2010. Alternative mechanisms for coordinating polymerase α and MCM helicase. Mol Cell Biol 30: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stillman B. 2012. The origin recognition complex: a biochemical and structural view. Subcell Biochem 62: 37–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423: 512–518. [DOI] [PubMed] [Google Scholar]
- Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N. 2015. Structure of the eukaryotic MCM complex at 3.8 A. Nature 524: 186–191. [DOI] [PubMed] [Google Scholar]
- Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G. 2005. Mechanochemistry of t7 DNA helicase. J Mol Biol 350: 452–475. [DOI] [PubMed] [Google Scholar]
- Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y. 2011. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci 108: 7373–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wu R, Zhou B, Wang J, Wei Z, Tye BK, Liang C, Zhu G. 2012. Structural insights into the Cdt1-mediated MCM2–7 chromatin loading. Nucleic Acids Res 40: 3208–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looke M, Maloney MF, Bell SP. 2017. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev 31: 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. 2012. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci 109: 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404: 622–625. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. 2005. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437: 1115–1120. [DOI] [PubMed] [Google Scholar]
- McGeoch AT, Bell SD. 2005. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell 120: 167–168. [DOI] [PubMed] [Google Scholar]
- McGeoch AT, Trakselis MA, Laskey RA, Bell SD. 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol 12: 756–762. [DOI] [PubMed] [Google Scholar]
- Mechali M. 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738. [DOI] [PubMed] [Google Scholar]
- Miller TC, Costa A. 2017. The architecture and function of the chromatin replication machinery. Curr Opin Struct Biol 47: 9–16. [DOI] [PubMed] [Google Scholar]
- Miller JM, Enemark EJ. 2015. Archaeal MCM proteins as an analog for the eukaryotic Mcm2–7 helicase to reveal essential features of structure and function. Archaea 2015: 305497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JM, Arachea BT, Epling LB, Enemark EJ. 2014. Analysis of the crystal structure of an active MCM hexamer. Elife 3: e03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe I, Mizuno K, Keszthelyi A, Daigaku Y, Skouteri M, Mohebi S, Kunkel TA, Murray JM, Carr AM. 2015. Polymerase δ replicates both strands after homologous recombination-dependent fork restart. Nat Struct Mol Biol 22: 932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T, Takahashi N, Stillman B. 2000. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev 14: 1631–1641. [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. 2006. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci 103: 10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Polε, and GINS in budding yeast. Genes Dev 24: 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. 2008. Division of labor at the eukaryotic replication fork. Mol Cell 30: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404: 625–628. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Li H. 2016. The eukaryotic replisome goes under the microscope. Curr Biol 26: R247–R256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Kunkel TA. 2001. In vivo consequences of putative active site mutations in yeast DNA polymerases α, ε, δ, and ζ. Genetics 159: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Costa A. 2016. New insights into the mechanism of DNA duplication by the eukaryotic replisome. Trends Biochem Sci 41: 859–871. [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. 2007. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 317: 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y, Xia Y, Liu L, Cui J, Li Z, Cao Q, Chen XS, Campbell JL, Lou H. 2015. Cell-cycle-regulated interaction between Mcm10 and double hexameric Mcm2–7 is required for helicase splitting and activation during S phase. Cell Rep 13: 2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39. [DOI] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. 2010. Mec1 is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol Cell 40: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Stillman B. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci 92: 2224–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J 23: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet N, Liu D, Legrand P, Velours C, Corpet A, Gaubert A, Bakail M, Moal-Raisin G, Guerois R, Compper C, et al. 2015. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res 43: 1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK. 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 16: 173–185. [DOI] [PubMed] [Google Scholar]
- Riera A, Speck C. 2016. Licensing of replication origins. In The initiation of DNA replication in eukaryotes (ed. Kaplan DL), pp. 189–211. Springer International Publishing, New York. [Google Scholar]
- Riera A, Tognetti S, Speck C. 2014. Helicase loading: how to build a MCM2–7 double-hexamer. Semin Cell Dev Biol 30: 104–109. [DOI] [PubMed] [Google Scholar]
- Rothenberg E, Trakselis MA, Bell SD, Ha T. 2007. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem 282: 34229–34234. [DOI] [PubMed] [Google Scholar]
- Rowles A, Blow JJ. 1997. Chromatin proteins involved in the initiation of DNA replication. Curr Opin Genet Dev 7: 152–157. [DOI] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J 14: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel SA, Fernandez-Cid A, Sun J, Riera A, Tognetti S, Herrera MC, Li H, Speck C. 2014. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2–7 onto DNA. Genes Dev 28: 1653–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Bell SP. 2001. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell 8: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. 2006. Cdc7–Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Kinney JB, Lengronne A, Pasero P, Stillman B. 2014. Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc Natl Acad Sci 111: E1899–E1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Kinney JB, Stillman B. 2016. Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res 26: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ. 2002. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21: 6624–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, et al. 2014. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510: 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Sannino V, Costanzo V, Pellegrini L. 2016. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun 7: 11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Chen XS. 2012. MCM structure and mechanics: what we have learned from archaeal MCM. Subcell Biochem 62: 89–111. [DOI] [PubMed] [Google Scholar]
- Speck C, Stillman B. 2007. Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem 282: 11705–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. 2005. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12: 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. 2005. Origin recognition and the chromosome cycle. FEBS Lett 579: 877–884. [DOI] [PubMed] [Google Scholar]
- Stinson BM, Baytshtok V, Schmitz KR, Baker TA, Sauer RT. 2015. Subunit asymmetry and roles of conformational switching in the hexameric AAA+ ring of ClpX. Nat Struct Mol Biol 22: 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. 2012. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure 20: 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. 2013. Cryo-EM structure of a helicase loading intermediate containing ORC–Cdc6–Cdt1–MCM2–7 bound to DNA. Nat Struct Mol Biol 20: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. 2014. Structural and mechanistic insights into Mcm2–7 double-hexamer assembly and function. Genes Dev 28: 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O'Donnell ME. 2015. The architecture of a eukaryotic replisome. Nat Struct Mol Biol 22: 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. 2005. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 19: 2827–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. 2008. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol 69: 1071–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. 2015. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Champasa K, Correa IR Jr, Gelles J, Bell SP. 2017. Mechanism and timing of Mcm2–7 ring closure during DNA replication origin licensing. Nat Struct Mol Biol 24: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocilj A, On KF, Yuan Z, Sun J, Elkayam E, Li H, Stillman B, Joshua-Tor L. 2017. Structure of the active form of human origin recognition complex and its ATPase motor module. Elife 6: e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti S, Riera A, Speck C. 2015. Switch on the engine: how the eukaryotic replicative helicase MCM2–7 becomes activated. Chromosoma 124: 13–26. [DOI] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. 2014. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J 33: 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. 2012. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J 31: 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, Wirthensohn D, Wightman M, Matak-Vinkovic D, Pellegrini L, Labib K. 2016. Ctf4 is a hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol Cell 63: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase G, Takisawa H, Kanemaki MT. 2012. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45–Mcm–GINS. Curr Biol 22: 343–349. [DOI] [PubMed] [Google Scholar]
- Wei Z, Liu C, Wu X, Xu N, Zhou B, Liang C, Zhu G. 2010. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem 285: 12469–12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. 1999. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci 96: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. 2006. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell 65: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Bai L, Sun J, Georgescu R, Liu J, O'Donnell ME, Li H. 2016. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol 23: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. 2017. Structural basis of Mcm2–7 replicative helicase loading by ORC–Cdc6 and Cdt1. Nat Struct Mol Biol 24: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Cheng E, Wu H, Li N, Yung PY, Gao N, Tye BK. 2017. Open-ringed structure of the Cdt1–Mcm2–7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol 24: 300–308. [DOI] [PubMed] [Google Scholar]
- Zhang D, O'Donnell M. 2016. The eukaryotic replication machine. Enzymes 39: 191–229. [DOI] [PubMed] [Google Scholar]


