Abstract
Replacement of canonical histones with specialized histone variants promotes altering of chromatin structure and function. The essential histone variant H2A.Z affects various DNA‐based processes via poorly understood mechanisms. Here, we determine the comprehensive interactome of H2A.Z and identify PWWP2A as a novel H2A.Z‐nucleosome binder. PWWP2A is a functionally uncharacterized, vertebrate‐specific protein that binds very tightly to chromatin through a concerted multivalent binding mode. Two internal protein regions mediate H2A.Z‐specificity and nucleosome interaction, whereas the PWWP domain exhibits direct DNA binding. Genome‐wide mapping reveals that PWWP2A binds selectively to H2A.Z‐containing nucleosomes with strong preference for promoters of highly transcribed genes. In human cells, its depletion affects gene expression and impairs proliferation via a mitotic delay. While PWWP2A does not influence H2A.Z occupancy, the C‐terminal tail of H2A.Z is one important mediator to recruit PWWP2A to chromatin. Knockdown of PWWP2A in Xenopus results in severe cranial facial defects, arising from neural crest cell differentiation and migration problems. Thus, PWWP2A is a novel H2A.Z‐specific multivalent chromatin binder providing a surprising link between H2A.Z, chromosome segregation, and organ development.
Keywords: chromatin, H2A.Z, mitosis, neural crest, PWWP2A
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Post-translational Modifications, Proteolysis & Proteomics
Introduction
H2A.Z, an evolutionary highly conserved histone H2A variant (Iouzalen et al, 1996), is implicated in fundamental biological processes, such as transcriptional regulation, cell cycle control, and DNA repair (Santisteban et al, 2000; Meneghini et al, 2003; Rangasamy et al, 2004; Kalocsay et al, 2009; Weber et al, 2010; Zovkic et al, 2014), as well as cancer initiation and progression (Svotelis et al, 2010; Dryhurst et al, 2012; Valdes‐Mora et al, 2012; Kim et al, 2013). In flies, frogs, and mice, H2A.Z is required for early development and survival (van Daal & Elgin, 1992; Iouzalen et al, 1996; Faast et al, 2001). Vertebrates contain two non‐allelic H2A.Z‐encoding genes (H2AFZ and H2AFV), whose protein products (H2A.Z.1 and H2A.Z.2) differ in only three amino acids (Eirin‐Lopez et al, 2009). Alternative splicing of H2A.Z.2 mRNA in primates gives rise to H2A.Z.2.1 (for simplicity reasons referred to as H2A.Z.2) and the nucleosome‐destabilizing H2A.Z.2.2 isoform (Bonisch et al, 2012; Wratting et al, 2012). Interestingly, we recently found H2A.Z.2 to contribute to metastatic melanoma progression through regulation of E2F‐target gene expression promoting cell proliferation (Vardabasso et al, 2015). Despite progress in recognizing H2A.Z's contributions to DNA‐based processes, little is known about the underlying mechanisms. H2A.Z‐ and H2A‐containing nucleosome stabilities seem to be rather similar (Bonisch & Hake, 2012), but their molecular surfaces differ (Suto et al, 2000). The acidic patch, a C‐terminal region that is further extended in H2A.Z in comparison with H2A, is important for H2A.Z chromatin deposition (Latrick et al, 2016) and is hypothesized to also play a role in the recruitment of chromatin context‐dependent readers that in turn differentially modify surrounding chromatin regions (Suto et al, 2000).
In order to identify such an H2A.Z network, we utilized a label‐free quantitative mass spectrometry approach using two different human cell lines. We found > 35 proteins constituting readers, writers, and erasers of histone marks, as well as novel chromatin interactors to be significantly enriched on H2A.Z‐ versus H2A‐containing nucleosomes. One of those is PWWP2A, a functionally uncharacterized vertebrate‐specific protein containing a conserved PWWP domain. PWWP2A is found at H2A.Z‐containing transcriptional start sites of highly expressed genes and an internal amino acid stretch is sufficient to confer H2A.Z‐specificity, as well as nucleosome binding in vivo. Strong chromatin affinity in vivo depends on both the internal stretch and the highly conserved PWWP domain, which functions as a DNA interaction module, revealing a multivalent binding mode. H2A.Z chromatin occupancy does not depend on PWWP2A, while H2A.Z via its C‐terminal tail is one important feature for strong PWWP2A chromatin association. Depletion of PWWP2A in human cells results in impaired cellular proliferation due to a block in metaphase‐anaphase transition. While chromosomal passenger complex members are unaffected, loss of PWWP2A results in the deregulation of many genes involved in metabolism and morphology. In X. tropicalis, PWWP2A is expressed early during development in the neuroectoderm and neural crest. Knockdown of frog PWWP2A results in severe defects in head development, arising from neural crest cell migration and differentiation problems. This phenotype can be rescued by human PWWP2A protein variants that recognize H2A.Z. In summary, our study identifies PWWP2A as an H2A.Z‐specific multivalent chromatin binder that participates in regulating cell‐specific crucial gene expression programs.
Results
Identification of PWWP2A as novel H2A.Z‐nucleosome interactor
Previously, we employed a label‐free quantitative mass spectrometry approach and identified H2A.Z mononucleosome binders in the metastatic melanoma cell line SK‐mel147 stably expressing GFP and GFP‐tagged H2A variants (Vardabasso et al, 2015). Further experiments in this system as well as in HeLa Kyoto (HeLaK) cells (Fig EV1A and B) allowed us to quantitate candidate protein binding and to determine cell type‐specific H2A.Z network differences (Fig EV1C–E). Upon comparison between SK‐mel147 biological replicates, we identified 44 proteins that were significantly enriched on H2A.Z.1 or H2A.Z.2 nucleosomes compared to H2A‐containing ones (Figs 1A and EV1C). Almost all members of the H2A.Z‐specific SWR1‐related chaperone complex SRCAP (Ruhl et al, 2006) were found, confirming the sensitivity and specificity of our approach (Fig 1A and B). In contrast to SRCAP, only EP400 and TRRAP, the largest subunits of the second multicomponent H2A.Z deposition complex p400/TIP60 (Cai et al, 2005), were precipitated, suggesting that this complex either binds only transiently to chromatin or requires larger chromatin domains than just mononucleosomes. We also found BRD2, which has previously been shown in 293T cells to preferentially bind hyperacetylated H2A.Z nucleosomes (Draker et al, 2012) to be a specific interactor, further verifying our method (Vardabasso et al, 2015). In concordance with our hypothesis of H2A.Z being a recruiter of many chromatin‐altering proteins, we identified repressive as well as activating histone‐modifying complexes. Members of the H3K4me3‐catalyzing MLL complex as well as the H3K9me2 and H3K36me3 demethylases PHF2 and KDM2A, respectively, were specific interactors, likely accounting for the characteristic histone posttranslational modification (PTM) profile of H2A.Z nucleosomes (Fig 1C). Together, the identification of complexes involved in diverse DNA‐based processes supports our hypothesis of H2A.Z being a general but selective recruitment factor of chromatin‐modifying proteins.
Figure EV1. Mononucleosome generation and LFQ‐MS analysis workflow.
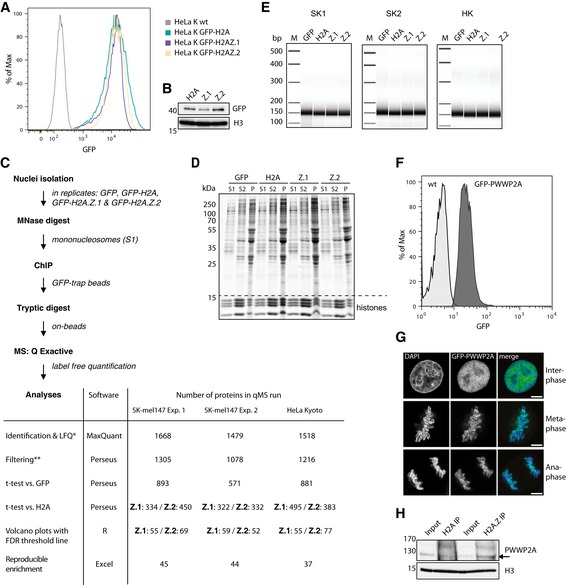
- Flow cytometry analysis of HeLaK cells either non‐transfected (wt, gray histogram) or stably transfected with GFP‐H2A (green histogram), GFP‐H2A.Z.1 (blue histogram), and GFP‐H2A.Z.2 (orange histogram). Notice that expression levels of tagged histones are similar.
- Immunoblots of nuclear extracts derived from HeLaK cells stably expressing GFP‐tagged histones (see also A). Anti‐H3 antibody was used as loading control.
- Schematic workflow of LFQ‐MS experiment (see also Fig 1A and Materials and Methods). Table depicts the number of proteins identified by indicated method in each step of the procedure.
- Coomassie staining of proteins found in S1, S2, and pellet (P) fractions after MNase digest of chromatin derived from SKmel‐147 cells stably expressing GFP, GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.2), or GFP‐H2A.Z.2 (Z.2). Notice that all fractions contain histones, but remaining protein content is different.
- Bioanalyzer separated DNA sizes of S1‐containing nucleosomes after MNase digest of two independent SKmel‐147 replicates (SK1 and SK2) or HeLaK (HK) cells stably expressing GFP, GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.2), or GFP‐H2A.Z.2 (Z.2) used in LFQ‐MS experiments shown in Fig 1A. Notice that S1 fractions that were used for LFQ‐MS IP experiments contain > 90% pure mononucleosomes as seen by DNA sizes of around 150 bp.
- Flow cytometry analysis of HeLaK cells either non‐transfected (control, light gray histogram) or stably transfected with GFP‐PWWP2A (dark gray histogram). Notice that a pure population of HeLaK cells expressing GFP‐PWWP2A at low level was obtained.
- Confocal microscopy images of HeLaK cells stably expressing GFP‐PWWP2A in interphase, metaphase, or anaphase. DNA was visualized by DAPI staining (blue), and GFP‐PWWP2A is seen in green. Scale bars = 10 μm. Notice that some GFP‐PWWP2A remains stably associated with condensed mitotic chromatin.
- Immunoblotting of endogenous PWWP2A upon mono‐IPs using antibodies against endogenous H2A or H2A.Z. Arrow indicates endogenous PWWP2A.
Source data are available online for this figure.
Figure 1. PWWP2A is a novel H2A.Z mononucleosome binder.
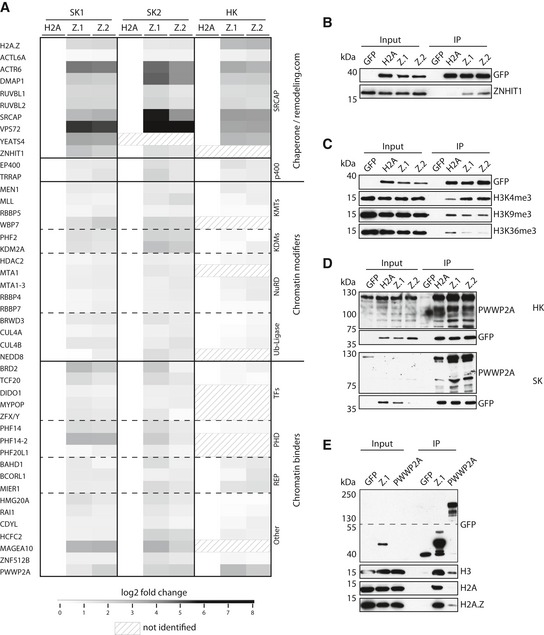
-
AHeatmap of significant outliers from pull‐downs analyzed by label‐free MS‐based proteomics with mononucleosomes derived from Skmel‐147 (two independent experiments: SK1 and SK2; see also Vardabasso et al, 2015 for first replicate) or HeLaK (HK) cells stably expressing GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.1), or GFP‐H2A.Z.2 (Z.2), normalized to H2A. Scale bar: log2‐fold. See also Fig EV1A–E for details on experimental procedure and verification of results, and Datasets EV1, EV2 and EV3 for detailed lists of H2A.Z‐binders.
-
BImmunoblotting of SRCAP complex‐specific member ZNHIT1 upon SK‐mel147 GFP, GFP‐H2A, GFP‐H2A.Z.1, or GFP‐H2A.Z.2 mono‐IPs. GFP served as control.
- C, D
-
EImmunoblots of GFP‐H2A.Z.1 or GFP‐PWWP2A mono‐IPs detecting endogenous H3, H2A, or H2A.Z. Notice enrichment of H2A.Z in comparison with H2A in GFP‐PWWP2A pull‐down. See also Fig EV1F–H for generation and characterization of GFP‐PWWP2A cell lines and endogenous pull‐down.
Source data are available online for this figure.
We chose to further investigate the so far functionally uncharacterized vertebrate‐specific PWWP2A protein because of its interesting domain structure and its presence in previous H2A.Z‐pull‐down experiments (Draker et al, 2012). PWWP2A belongs to a group of proteins containing a PWWP domain that was first identified in Wolf‐Hirschhorn syndrome candidate 1 (WHSC1) (Stec et al, 2000). First, we confirmed PWWP2A interaction with H2A.Z nucleosomes by immunoblots applying native GFP‐variant mononucleosome IPs (mono‐IPs; Fig 1D). Next, we cloned PWWP2A from SKmel‐147 cDNA and generated HeLaK cell lines stably expressing N‐terminally GFP‐tagged PWWP2A (Fig EV1F). As suspected, GFP‐PWWP2A was predominantly found in the nucleus (Fig EV1G). Although most of the protein was relieved from chromosomes during mitosis, a fraction of it remained stably associated with chromatin, assuming a strong chromatin binding ability of a sub‐pool of PWWP2A. Native mono‐IPs using these cells independently confirmed PWWP2A's specificity for H2A.Z nucleosomes (Fig 1E). Additionally, binding of endogenous PWWP2A to endogenous H2A.Z and not to H2A nucleosomes confirmed our findings (Fig EV1H). In summary, we have identified PWWP2A as a novel and functionally uncharacterized H2A.Z‐nucleosome interactor.
Multivalent binding mode of PWWP2A enables H2A.Z‐nucleosome binding specificity as well as strong chromatin interaction
Intrigued by PWWP2A's specific and strong H2A.Z interaction, we used recombinant GST‐tagged PWWP2A and mononucleosomes isolated from HeLaK cells for in vitro pull‐down assays, which efficiently pulled‐down tagged and untagged H2A.Z‐containing nucleosomes and reproduced our previous results (Fig 2A and B). Further, GST‐PWWP2A was able to interact with recombinant nucleosomes, thereby showing a higher affinity to H2A.Z than to H2A (Fig 2C) verifying a direct H2A.Z‐nucleosome binding specificity. Human PWWP2A contains two N‐terminal proline‐rich stretches (P1, P2) and a C‐terminal serine‐rich region (S) followed by a conserved PWWP domain (Appendix Fig S1A). Since the highly conserved PWWP domain plays a role in chromatin recognition (Qin & Min, 2014), we generated a PWWP domain deletion mutant (ΔPWWP) and a construct entirely consisting of the PWWP domain and the last C‐terminal amino acid stretch (PWWP) (Fig 2D, top). Interestingly, both deletion mutants were able to pull‐down HeLaK‐derived nucleosomes (Fig 2D, bottom), suggesting that at least two separate domains participate in chromatin interaction. IPs with additional recombinant GST‐PWWP2A deletion proteins (Fig 2E and Appendix Fig S1B) determined the domain sufficient for H2A.Z‐nucleosome interaction to be an internal (I) stretch between the P2 and S regions. Surprisingly, this domain could be further divided into an N‐terminal region (IN) necessary for nucleosome binding and a C‐terminal part (IC) mediating H2A.Z‐specificity (Fig 2F and G, and Appendix Fig S1C and D). This is a unique and so far undescribed feature as BLAST searches did not find any sequence homology of this internal stretch in other proteins.
Figure 2. Two separate internal regions of PWWP2A confer nucleosome binding and H2A.Z‐specificity.
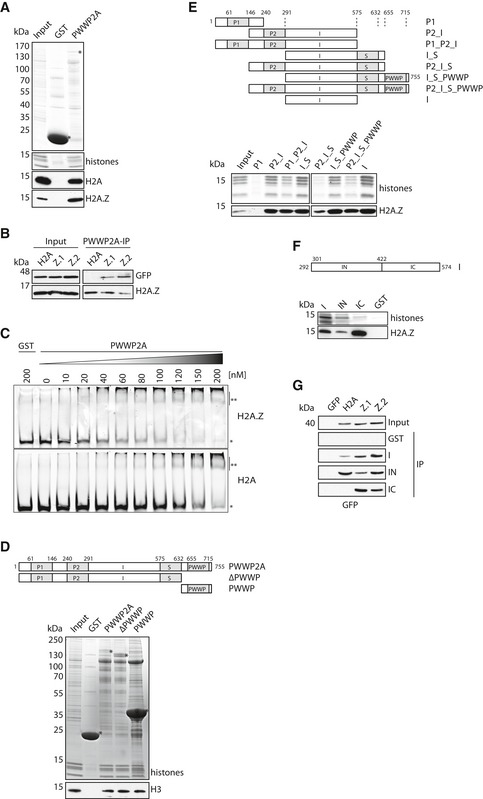
- Pull‐downs of GST or GST‐PWWP2A with mononucleosomes (input) derived from HeLaK cells. Precipitated recombinant GST proteins and histones are detected with Coomassie blue staining and H2A and H2A.Z with specific antibodies in immunoblots. * indicates right sizes of purified and precipitated GST and GST‐PWWP2A.
- Immunoblots of GST‐PWWP2A IPs with mononucleosomes derived from HeLaK cells stably expressing GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.1), or GFP‐H2A.Z.2 (Z.2).
- Representative competitive EMSA using recombinant H2A (bottom)‐ or H2A.Z (top)‐containing nucleosomes incubated with indicated increasing concentrations of GST‐PWWP2A. GST alone served as negative control. * indicates nucleosome; ** indicates nucleosome‐GST‐PWWP2A complex.
- Schematic representation of recombinant GST‐tagged PWWP2A and PWWP domain deletion (ΔPWWP) and PWWP domain only (PWWP)‐containing constructs (top) used in cell‐derived mono‐IPs followed by Coomassie staining and immunoblotting (bottom). * indicates respective GST proteins. Notice that both the PWWP domain alone as well as the PWWP‐deletion protein are able to interact with nucleosomes, indicating at least two independent nucleosome binding sites within PWWP2A.
- Schematic representation of recombinant GST‐PWWP2A deletions (top) used in cell‐derived mono‐IPs followed by Coomassie and immunoblotting (bottom). See also Appendix Fig S1B for protein purification.
- Schematic representation of recombinant GST‐PWWP2A internal deletions (top) used in cell‐derived mono‐IPs followed by Coomassie and immunoblotting (bottom). See also Appendix Fig S1C and D for protein purification and further IPs.
- IPs as described in (F) with mononucleosomes derived from HeLaK cells stably expressing GFP‐H2A and GFP‐H2A.Z isoforms detected with anti‐GFP antibody.
Source data are available online for this figure.
Next, we wondered how the PWWP domain contributes to further chromatin association. Recently, PWWP domains have been shown to be DNA and/or histone PTM binders (Chen et al, 2004; Yang & Everett, 2007). Based on the PWWP domain solution structure of PWWP2B (Qin & Min, 2014), a close homolog of PWWP2A, we modeled the 3D structure of the PWWP domain of PWWP2A (Fig 3A). The electrostatic potential of the solvent‐accessible surface revealed an overall enrichment in basic residues, suggestive of DNA‐binding potential (Fig 3A). Indeed, PWWP2A's PWWP domain bound DNA with low affinity (Fig 3B), but did not confer H2A.Z‐specificity directly (Appendix Fig S2A). Thus, PWWP2A interacts with chromatin via a multivalent binding mode with separated nucleosome interaction, H2A.Z‐specificity, and DNA‐binding domains.
Figure 3. Multivalent binding mode of PWWP2A enables high‐affinity chromatin interaction.
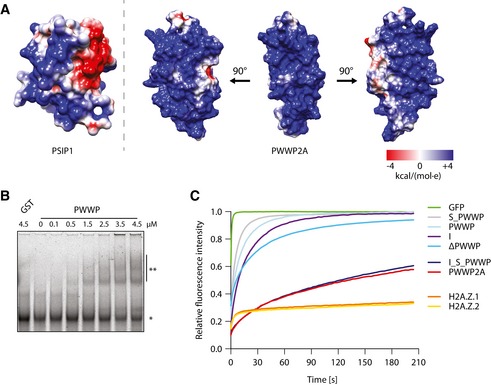
- The electrostatic surface potential (ESP) of the published structure of the PSIP DNA‐binding PWWP domain (Eidahl et al, 2013) was calculated (left), and the PWWP2A‐PWWP domain (right) was modeled based on published PWWP2B 3D structure (Qin & Min, 2014). ESP color values are in units of kcal/(mol*e) at 298 K. Three representative views of the ESP of the PWWP2A PWWP domain (right).
- Representative EMSA using Cy‐5 labeled 75‐bp dsDNA and indicated increasing concentrations of GST‐PWWP domain. *: free DNA; **: shifted PWWP‐DNA complex. See also Appendix Fig S2A for H2A.Z‐independence of PWWP domain in nucleosome interaction.
- FRAP quantification curves of average GFP signal relative to fluorescence signal prior to bleaching from transiently expressed GFP (negative control), GFP‐tagged histones, and GFP‐tagged PWWP2A mutants (n = 5–14). See also Appendix Fig S2B for FRAP experiments comparing recovery signals of different chromatin binding proteins to GFP‐PWWP2A, as well as Appendix Fig S2C–E for FRAP analyses of stable and transient GFP and GFP‐tagged proteins expressing HeLaK cells.
Next, we determined PWWP2A's chromatin association in vivo by Fluorescence Recovery After Photobleaching (FRAP) experiments. GFP‐PWWP2A showed an extremely slow recovery curve, suggesting that it is stably bound to chromatin (Fig 3C and Appendix Fig S2B–E). Constructs containing either the internal or the PWWP domain or a deletion of the PWWP domain (ΔPWWP) featured much faster recovery kinetics than wild‐type PWWP2A (Fig 3C, Appendix Fig S2E), further supporting a multivalent chromatin‐binding mode. PWWP2A deletion protein consisting only of the internal, serine‐rich and PWWP domains (I_S_PWWP), showed an identical recovery curve as full‐length PWWP2A (Fig 3C). Thus, concerted low‐affinity interactions of the internal region and the PWWP domain are needed for an exceptionally strong chromatin association paired with H2A.Z‐nucleosome specificity.
PWWP2A predominantly localizes to the promoter region of highly transcribed genes
To determine PWWP2A's chromatin occupancy in comparison with H2A.Z‐enriched sites genome‐wide, we performed native chromatin immunoprecipitation followed by high‐throughput sequencing (nChIP‐seq) using stable cell lines expressing GFP‐tagged H2A.Z isoforms or GFP‐PWWP2A and found that PWWP2A was particularly enriched at promoters and 5′‐UTR sites (Figs 4A and EV2A). PWWP2A overlapped to 77 and 66% with H2A.Z‐containing peaks at promoter or non‐promoter regions, respectively (Fig 4B). Interestingly, both H2A.Z.1 and H2A.Z.2 peaks showed an enrichment of PWWP2A at promoter regions, but only little overlap with PWWP2A at non‐promoter sites (Fig 4C), suggesting that H2A.Z is one main but not the sole determinant for PWWP2A's site specificity. Heatmap alignment revealed a strong correlation of PWWP2A and H2A.Z variants with highly transcribed genes in euchromatic H3K4me3‐positive regions (Figs 4D and EV2B). Both H2A.Z variants and PWWP2A were located at the −1 and +1 nucleosomes at transcriptional start sites (TSSs; Fig 4E). Interestingly, PWWP2A also accumulated at the nucleosome‐depleted region (NDR), possibly recognizing free DNA via its PWWP domain and protecting it from MNase digestion. We conclude that PWWP2A interacts specifically with H2A.Z nucleosomes at the TSS of highly transcribed genes.
Figure 4. PWWP2A binds H2A.Z nucleosomes at TSS of actively transcribed genes.
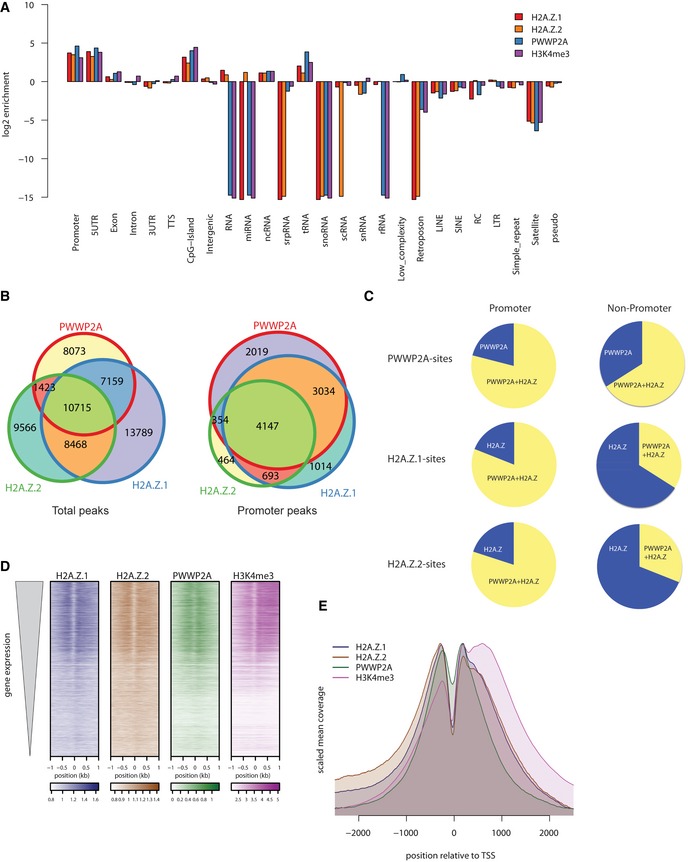
- Log2‐enrichment plot representing genomic regions after GFP‐PWWP2A (blue), GFP‐H2A.Z.1 (red), GFP‐H2A.Z.2 (orange), and H3K4me3 (purple) nChIP‐seq. Shown are two biological replicates for each nChIP. See also Fig EV2A for representative genome browser captures.
- Venn diagrams displaying total (left) or promoter‐occupying (right) peaks of HeLaK GFP‐PWWP2A, GFP‐H2A.Z.1, and GFP‐H2A.Z.2. Promoters are defined as −3 kb < TSS < +1 kb.
- Distribution of overlapping peaks between GFP‐PWWP2A (top), GFP‐H2A.Z.1 (middle), and GFP‐H2A.Z.2 (bottom) nChIP‐seq data of promoter (left) and non‐promoter (right) regions according to (B).
- Heatmap of nChIP‐seq peaks at transcriptional start sites (TSS) sorted for expression level (top: high expressed, bottom: low expressed genes).
- Correlation of GFP‐PWWP2A (green), ‐H2A.Z.1 (blue), ‐H2A.Z.2 (brown), and H3K4me3 (purple) mean coverage signals at TSS of expressed genes. See Fig EV2B for GFP‐PWWP2A localization at euchromatic regions.
Figure EV2. PWWP2A localizes to euchromatic regions.
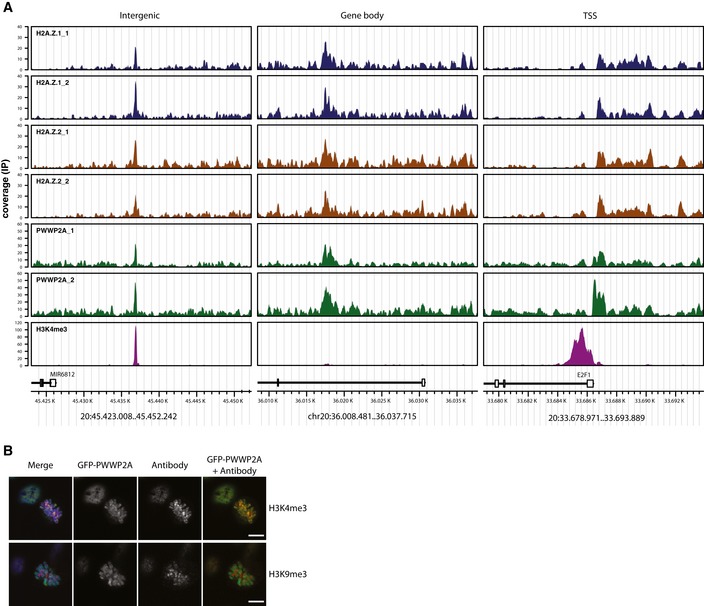
- Genome browser snap shot of a representative region in chromosome 20 displaying two independent replicates of HeLaK GFP‐H2A.Z.1, GFP‐H2A.Z.2, GFP‐PWWP2A, and H3K4me3 nChIPs signals at intergenic, gene body, and TSS regions. Annotated genes are displayed below. See also Fig 4 for nChIP‐seq analyses.
- Representative confocal IF pictures of HeLaK cells stably expressing GFP‐PWWP2A (green), co‐stained with DAPI (blue) and either H3K4me3 antibody (top, red) or H3K9me3 (bottom, red). Scale bars = 10 μm.
PWWP2A depletion leads to deregulation of gene expression and results in a proliferation defect caused by a metaphase‐anaphase block
Next, we investigated PWWP2A's biological function by RNA interference (RNAi) in HeLaK cells using two independent siRNAs that efficiently downregulated PWWP2A mRNA (Fig 5A) as well as protein (Fig 5B). Having PWWP2A found predominantly at the TSS of actively transcribed genes, we wanted to know whether it is involved in the regulation of gene expression. Indeed, RNA‐seq of PWWP2A‐depleted cells identified ~600 genes to be deregulated (Fig 5C). Gene ontology (GO) analysis revealed an enrichment of genes involved in developmental process regulation and cell morphogenesis (Appendix Fig S3). With so many genes deregulated and important cellular processes possibly affected, we wondered about phenotypic and functional consequences of PWWP2A reduction. Interestingly, we noticed that fewer cells (Fig 5B) with overall slightly enlarged nuclei (Fig EV3A) were present upon knockdown, correlating with a reduction in cell proliferation (Fig 5D).
Figure 5. PWWP2A depletion in HeLaK cells leads to alterations in gene expression programs and a proliferation defect.
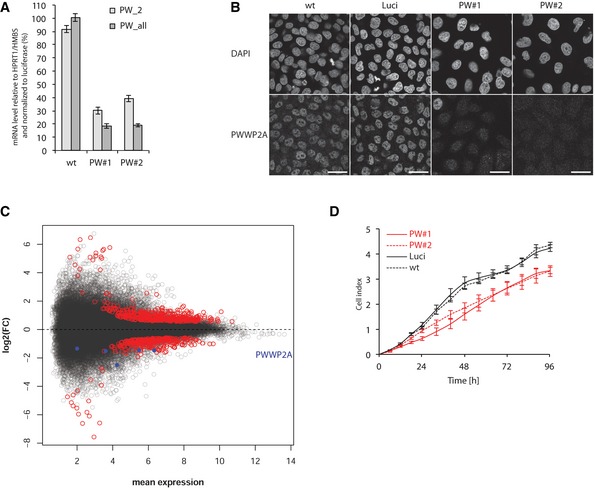
- PWWP2A expression analysis by qPCR 2 days after knockdown with two independent siRNAs (PW#1, PW#2) using two different primer pairs (PW_2: recognizes main splice product; PW_all: recognizes all predicted splice forms). Luciferase siRNA (Luci) and non‐transfected wild‐type (wt) cells were used as controls. All data were normalized to HPRT1/HMBSS expression and depicted as % of luci transfectants. Error bars indicate SEM of four biological replicates.
- IF microscopy analysis of cells treated as described in (A). Cells were stained with DAPI to visualize DNA (top) and anti‐PWWP2A antibody (bottom). Scale bars = 10 μm. Notice slight increase in nuclear size upon PWWP2A depletion (quantification in Fig EV3A).
- Scatter plot summarizing genes up‐ (top red) or down‐regulated (bottom red) upon PWWP2A depletion (blue dots indicate different PWWP2A RNA isoforms) as determined by RNA‐seq. See also Appendix Fig S3 for GO term analysis of deregulated transcripts.
- Growth curve of HeLaK cells after RNAi (red: PWWP2A siRNAs, black: wt and luciferase control siRNAs). Error bars indicate SEM of three independent biological replicates.
Figure EV3. Role of PWWP2A during mitosis.
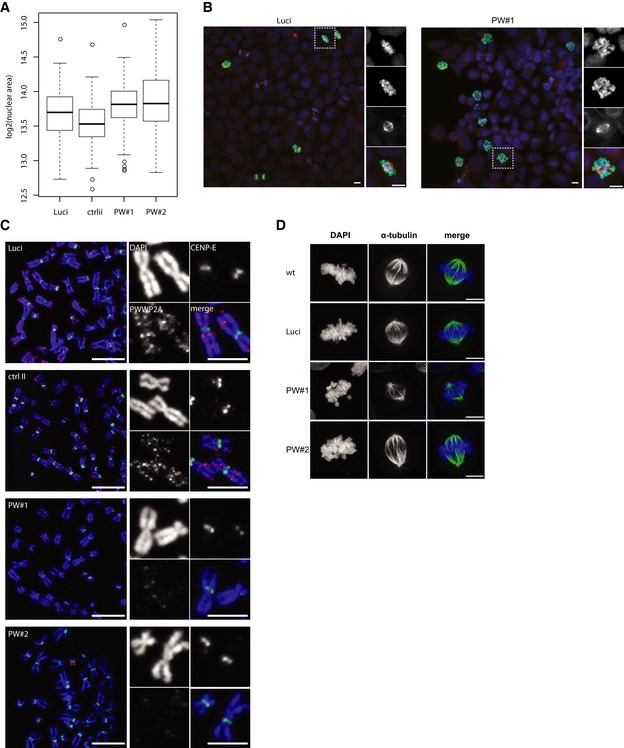
- Standard Tukey boxplots of nuclear sizes of control (ctrlii, luci) and PWWP2A‐depleted (PW#1, PW#2) cells (n = 3). Horizontal line = median, box = interquartile range.
- Confocal IF microscopy analysis of HeLaK RNAi cells to visualize mitotic phases. Two days after RNAi cells were stained with DAPI (blue), H3S10ph (green), and tubulin (red) antibodies. Scale bars = 10 μm. See also Fig 6E for quantitation of mitotic stages.
- Representative confocal IF pictures of chromosomes of control (ctrlii, Luci) and PWWP2A‐depleted (PW#1, PW#2) HeLaK cells co‐stained with DAPI (blue), CENP‐E (green), and PWWP2A (red). Scale bars = 10 μm (enlarged chromosomes in boxes; scale bars = 5 μm).
- Representative confocal IF pictures of mitotic HeLaK cells two days after control (wt, Luci) and PWWP2A (PW#1, PW#2) siRNA transfections. HeLaK cells were co‐stained with DAPI (blue) and tubulin (green) antibodies. Scale bars = 10 μm.
Analysis of cell cycle phases by flow cytometry of propidium iodide (PI)‐stained cells revealed an accumulation in G2/M phase cells upon PWWP2A knockdown (Fig 6A and B). Subsequent co‐staining with PI and H3S10ph antibody showed that PWWP2A depletion leads to an increase in mitotic cells (Fig 6C and D), especially enriching for prometaphase and depleting anaphase cells as demonstrated by immunofluorescence microscopy of H3S10ph‐positive cells (Figs 6E and EV3B). Real‐time‐lapse microscopy imaging confirmed a halt of PWWP2A‐depleted cells in mitosis for up to 10 h, while control cells traversed through mitosis in < 1.5 h (Fig 6F and Movies EV1, EV2, EV3 and EV4). PWWP2A RNAi cells repeatedly shuffled back and forth between prometaphase and metaphase unable to keep all chromosomes at the equatorial plate. In agreement with the observation that chromosomes were able to move, we did not find any changes in centromere and kinetochore formation (Fig EV3C). Further, although tubulin spindles were generated and contributed to the moving motion of chromosomes, they showed a slight abnormal morphology (Fig EV3D). Interestingly, this mitotic phenotype strongly copies the previously described, but mechanistically not understood, mitotic defect observed in H2A.Z double knockout vertebrate cells (Kusakabe et al, 2016), thus identifying PWWP2A as a possible mediator of the H2A.Z‐dependent cell cycle progression phenotype.
Figure 6. PWWP2A is crucial for proper mitosis progression.
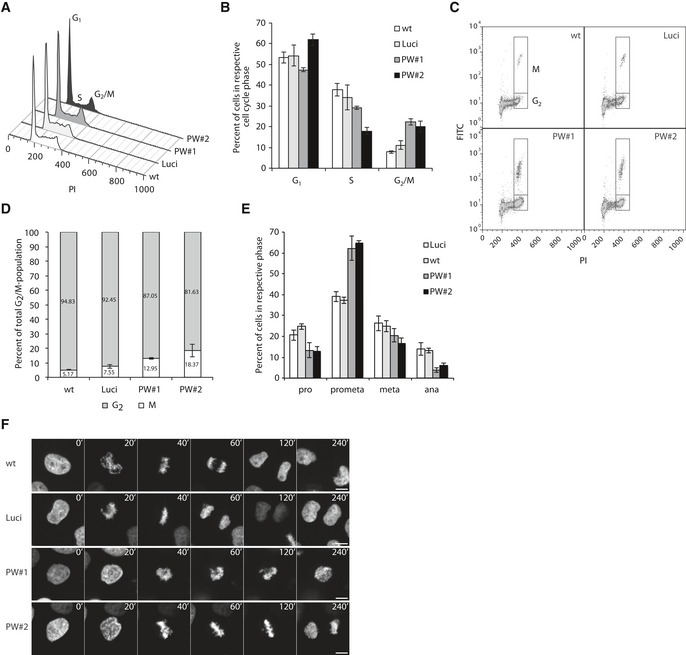
- Cell cycle analysis of PI‐stained HeLaK RNAi cells using flow cytometer.
- Quantification of experiments described in (A). Percentages of cells in G1, S, or G2/M phases 3 days after RNAi. Error bars indicate SEM of five independent biological replicates.
- Flow cytometry dot‐plots of HeLaK cells co‐stained with PI and H3S10ph antibody 3 days after RNAi to distinguish between G2 and M phase cells.
- Quantification of experiments described in (C). Percentages of cells in G2 or M phases 3 days after RNAi. Error bars indicate SEM of three independent biological replicates.
- Quantification of mitotic phases 2 days after RNAi by visually distinguishing morphological characteristics of H3S10ph‐positive cells. Error bars indicate SEM of four independent biological replicates. See also Fig EV3B–D for stainings and chromosome spreads of control and RNAi cells.
PWWP2A does not influence H2A.Z occupancy, while H2A.Z's C‐terminal tail contributes to PWWP2A chromatin recruitment
One possible explanation for the strong cell cycle progression phenotype upon PWWP2A loss could be the result of changes in H2A.Z occupancy due to either defects in its deposition or an increase in chromatin destabilization. However, H2A.Z chromatin binding ability was not affected upon PWWP2A depletion (Fig 7A, Appendix Fig S4A). Accordingly, GFP‐H2A.Z.1 nChIP‐seq upon PWWP2A knockdown revealed only small differences in H2A.Z peak heights (Figs 7B and EV4A and B) and no effect on global H2A.Z levels (Fig EV4C). Given that the observed reduction was found on PWWP2A‐bound as well as non‐bound sites, it is highly likely that general cellular changes, possibly due to defects in mitosis, and not a specific H2A.Z‐depositioning defect, are the cause of this phenomenon. Likewise, overexpression of Cherry‐tagged PWWP2A (Ch‐PWWP2A) in GFP‐H2A.Z expressing HeLaK cells (Appendix Fig S4B) did not influence H2A.Z occupancy (Fig EV4D). Based on these observations, we speculate that PWWP2A is not needed for H2A.Z targeting, while on the other hand H2A.Z might be one important requirement for PWWP2A's recruitment to specific chromatin regions.
Figure 7. PWWP2A does not influence H2A.Z occupancy but C‐terminal tail of H2A.Z mediates PWWP2A nucleosome binding.
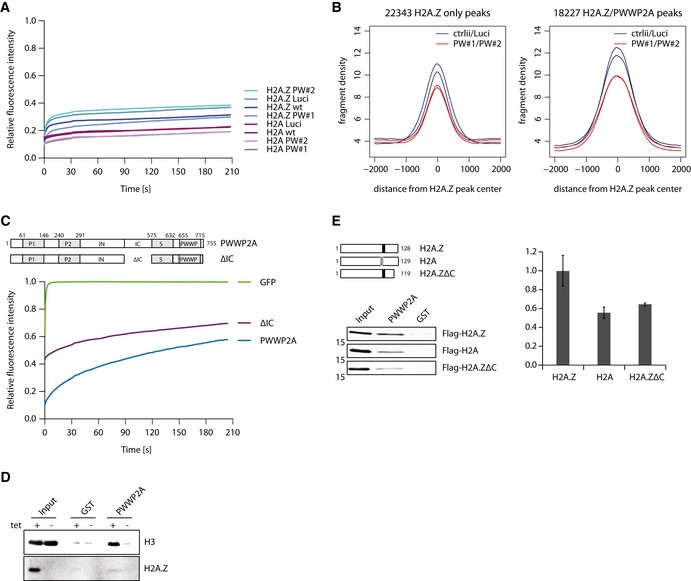
- FRAP quantification curves of average GFP signal relative to fluorescence signal prior to bleaching from HeLaK stably expressing GFP‐H2A or GFP‐H2A.Z.1 2 days after control (wt, Luci) or PWWP2A (PW#1, PW#2) knockdown (n = 9–19) (see Appendix Fig S4A for FRAP experiments).
- Venn diagram displaying sole GFP‐H2A.Z.1 (left, 22,343 peaks) or overlapping with PWWP2A (left, 18,227 peaks) nChIP‐seq signals in control (ctrlii, Luci; dark blue) or PWWP2A‐depleted (PW#1, PW#2; red) background (see also Fig EV4A and B).
- Schematic representation of PWWP2A and IC region deletion (ΔIC) (top). FRAP quantification curves (bottom) as shown and described in Fig 3C with the addition of GFP‐ΔIC (n = 16) (see also Appendix Fig S4C for FRAP IF pictures).
- Immunoblotting of endogenous H3 and H2A.Z (verification of tetracycline (tet)‐induced knockout efficiency) upon IP of mononucleosomes derived from WT or H2A.Z DKO DT40 cells with recombinant GST (negative control) or GST‐PWWP2A. Notice that less H3 is pulled down with PWWP2A when H2A.Z is depleted.
- Schematic representation of Flag‐tagged chicken H2A, H2A.Z, and H2A.Z C‐terminal deletion mutant (H2A.ZΔC) (top, left). Note indication of acidic patch in H2A (gray box) and extended acidic patch in H2A.Z (black box). After transient transfection of HK cells with Flag‐constructs, derived mononucleosomes were incubated with recombinant GST or GST‐PWWP2A and binding efficiency and variant‐specificity was tested in immunoblots with anti‐Flag antibody (left). Right: Quantification of signal intensities of immunoblots using Image Studio Lite Ver 5.2 (LI‐COR). Error bars indicate SEM of four independent replicates. See Fig EV4E for partial growth rescue of Flag‐H2A.ZΔC in DKO cells.
Source data are available online for this figure.
Figure EV4. PWWP2A does not affect H2A.Z genomic localization.
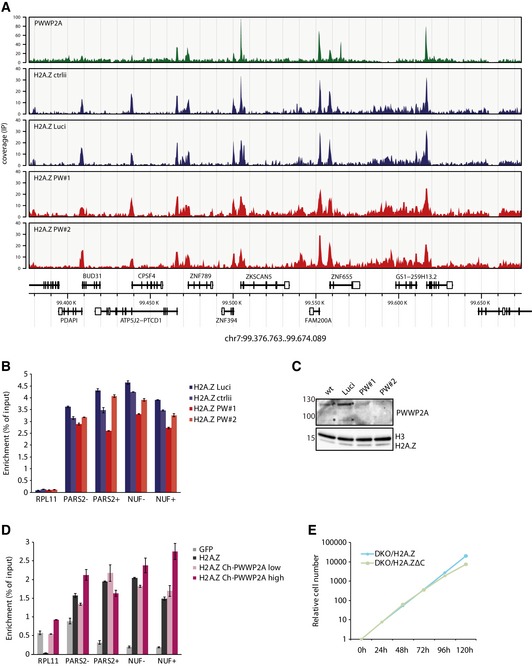
- Genome browser snap shot of a representative region in chromosome 7 displaying GFP‐PWWP2A (green, taken from Fig 4) and GFP‐H2A.Z nChIP‐seq signals 2 days after control (ctrlii, Luci, blue) or PWWP2A depletion (PW#1, PW#2, red). Annotated gene structures are shown below. Notice that H2A.Z peaks do not significantly change upon PWWP2A knockdown and in comparison with PWWP2A‐containing or PWWP2A‐missing sites. See also Fig 7B for Venn diagrams.
- nChIP‐qPCR of GFP‐H2A.Z to verify results obtained in (A) and Fig 7B. RPL11 represents a gene body site missing H2A.Z nucleosomes (negative control), while all other regions show H2A.Z‐containing promoter sites (− = −1 nucleosome, + = +1 nucleosome). Error bars indicate SEM of two to three technical replicates. Notice that PWWP2A depletion does not lead to a significant change in H2A.Z occupancy.
- Immunoblot of cell extracts 2 days after control (wt, Luci) or PWWP2A (PW#1, PW#2) knockdown with antibodies against PWWP2A, H3, and H2A.Z. Notice that PWWP2A reduction does not influence global H2A.Z levels.
- nChIP‐qPCR of GFP (negative control, light gray), GFP‐H2A.Z without exogenous Cherry‐PWWP2A (Ch‐PWWP2A) expression (dark gray) or upon sorting of low (light red) or high (dark red) Ch‐PWWP2A expression levels (see Appendix Fig S4B for FACS levels) using primer pairs shown in Fig EV4B. Error bars indicate SEM of two to three technical replicates. Notice that PWWP2A overexpression does not significantly affect H2A.Z occupancy.
- Growth curve of DKO cells rescued with either wt Flag‐H2A.Z (red, see also Kusakabe et al, 2016) or Flag‐H2A.ZΔC (green). Notice that although Flag‐H2A.ZΔC transfected cells grow slower than cells expressing wt Flag‐H2A.Z, they are able to partially rescue the severe mitotic defect observed in DKO cells implying chromatin deposition took place (see also Fig 7E).
Indeed, depletion of the IC region in PWWP2A (ΔIC), which mediates H2A.Z‐specificity (Fig 2F and G), resulted in increased mobility in FRAP (Fig 7C, Appendix Fig S4C), suggesting that H2A.Z is one important but not the sole mediator of PWWP2A chromatin association. Accordingly, performing IPs with recombinant GST‐PWWP2A and mononucleosomes derived from an inducible H2A.Z double knock out (DKO, both H2A.Z.1 and H2A.Z.2 isoforms are targeted) DT40 chicken cell line (Maruyama et al, 2012; Kusakabe et al, 2016) resulted in a diminished, but due to its multivalent binding mode not abolished, mononucleosome‐binding ability of PWWP2A (Fig 7D).
Next, we determined the region in H2A.Z that mediates PWWP2A specificity. In particular the C‐terminal acidic patch of H2A.Z has been proposed to provide a variant‐specific binding platform for nuclear proteins (Suto et al, 2000). We therefore incubated recombinant PWWP2A with mononucleosomes derived from HeLaK cells transiently expressing Flag‐tagged H2A, H2A.Z.1, or C‐terminal deleted H2A.Z.1 (H2A.ZΔC) (see Fig EV4E for rescue experiments implying proper chromatin incorporation). Indeed, depletion of the nine unique C‐terminal amino acids in H2A.Z keeping the extended acidic patch intact led to a reduction in PWWP2A binding, resembling the binding ability of canonical H2A (Fig 7E). In conclusion, PWWP2A is not needed for proper H2A.Z occupancy, while H2A.Z via its C‐terminal tail is partially required for PWWP2A chromatin binding.
PWWP2A is crucial for neural crest cell differentiation and migration during Xenopus laevis development
As PWWP2A depletion also affected gene programs of developmental processes in HeLaK cells (Appendix Fig S3), we next asked whether PWWP2A plays any role in vivo during organismal development. We chose the African clawed frogs X. laevis and X. tropicalis as model systems. Both species contain a PWWP2A gene whose protein sequence is strongest conserved in the internal, the serine‐rich region, and the PWWP domain (Appendix Fig S5). Whole‐mount RNA in situ hybridization in various developmental stages indicated endogenous X. tropicalis PWWP2A (XtPWWP2A) mRNA to be maternally expressed and to increase during gastrula stages (Fig EV5A), similarly to the H2A.Z mRNA profile (Ridgway et al, 2004). Subsequently, XtPWWP2A transcripts became enriched in the neuroectoderm, notably in neural folds, retina, and cranial neural crest. In tailbud stages, XtPWWP2A remained broadly expressed with a clear anterior bias. We investigated the biological impact of XtPWWP2A on development by protein‐knockdown with a specific PWWP2A translation‐blocking Morpholino oligonucleotide (pwMO) (Fig EV5B) by injecting control (CoMO) or pwMO into one blastomere of 2‐cell stage Xenopus laevis embryos. All embryo cohorts developed normally throughout neurulation. However, during tadpole stages a strong delay in head differentiation, most visible in the eye, appeared in the pwMO‐injected side (Fig 8A). The lack of retinal tissue combined with a reduced head size was observed in nearly 80% of the pwMO‐injected embryos (Table EV1). This phenotype was specific and PWWP2A's function evolutionary conserved, because coinjection of the full‐length human GFP‐PWWP2A mRNA, which was not affected by pwMO, restored wild‐type head morphology in the majority of animals (65%) (Fig 8A, Table EV1). Interestingly, the_I_S_PWWP and ΔPWWP protein variants restored normal head development with nearly the same efficiency as the full‐length protein, while the ΔIC variant did not revert the pwMO phenotype. These findings suggest that PWWP2A exerts its functions in head development through interaction with H2A.Z. Furthermore, injection of pwMO into embryos of the closely related X. tropicalis caused a comparable morphological phenotype (Fig EV5C). As development proceeded, the majority of the PWWP2A morphant embryos managed to form small eyes with lenses, but they failed to recover a bilaterally symmetric head and remained bent toward the injected body‐halve.
Figure EV5. Analysis of Xenopus pwwp2a expression and function.
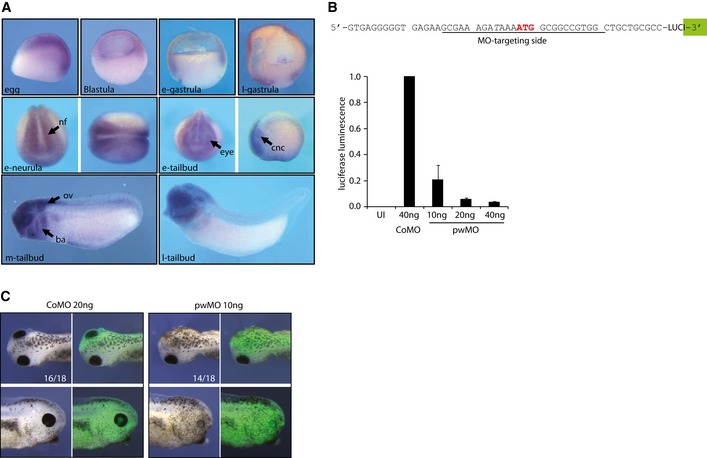
- Expression pattern of endogenous Xenopus tropicalis pwwp2a mRNA during early development. Pictures show whole‐mount RNA in situ hybridization patterns (purple color) for the following developmental stages: egg, blastula and early (e‐)/late (l‐) gastrula stages in lateral views of sagittal sections; e‐neurula and e‐tailbud stages show anterior views (left picture) or dorsal views (right picture, anterior to the left). Mid (m‐) and l‐tailbud stages are shown in lateral views (anterior left). Abbreviations: ba, branchial arches; cnc, cranial neural crest; eye, retinal Anlage; nf, neural folds; ov, otic vesicle.
- Top: Targeting region of the pwMO oligonucleotide on the X. laevis pwwp2a mRNA. The translational start site is highlighted in red. To determine the translational blocking efficiency of pwMO in vivo, a ˜275‐bp fragment of pwwp2a 5′ cDNA sequence, including the AUG, was cloned in frame upstream of the luciferase coding region. Bottom: The above depicted luciferase construct was injected in CoMO‐ or pwMO‐loaded embryos and chemiluminescence measured at gastrula stage. Error bars indicate SEM of three independent biological replicates. UI, uninjected embryos.
- Morphology of X. tropicalis embryos injected with CoMO and pwMO together with lineage‐tracer Alexa‐488; numbers indicate penetrance of major morphological phenotype over total embryos inspected (n = 3 experiments).
Figure 8. PWWP2A is essential for Xenopus eye, head, and brain development.
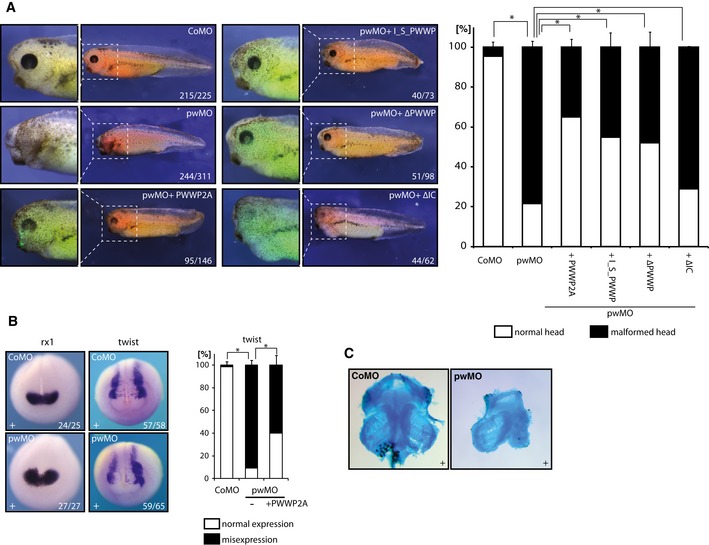
- Left side: One‐cell of two‐cell stage Xenopus laevis embryos was injected with either control (CoMO) or pwwp2a‐specific (pwMO) morpholino. For rescue experiments, pwMO morphant embryos were coinjected with mRNAs encoding either GFP‐tagged full‐length or variant human PWWP2A proteins, as indicated. Injected body sites were identified by Alexa 594 red fluorescence, while GFP fluorescence indicates synthesis of coinjected human PWWP2A protein variants. Panels display side views of representative embryos from the indicated conditions; numbers indicate penetrance of the major morphological phenotype over total embryos inspected. Right side: Quantification of the percentage of the observed phenotype, that is, malformation of head structures and eyes. Error bars indicate SEM of at least three independent biological replicates (see also Table EV1). *P ≤ 0.05 (Student's t‐test, two‐sided, unpaired).
- Whole‐mount RNA in situ hybridization assays with probes against rx1 (anterior view) and twist (dorsoanterior view) in either CoMO‐ or pwMO‐injected embryos at neurula stage. The rx1 gene is induced normally; the cranial neural crest marker twist is diminished on the pwMO‐injected side (91% affected), which is partially restored by coinjection of full‐length PWWP2A mRNA (60% affected). Numbers indicate penetrance of the depicted molecular phenotype over total embryos inspected. Right: Quantification of the percentage of misexpression of twist mRNA in controls compared to pwMO morphant and rescue condition. Error bars indicate SEM of three independent biological replicates. *P ≤ 0.05 (Student's t‐test, two‐sided, unpaired).
- Representative images of dissected facial cartilage from CoMO‐ or pwMO‐injected embryos visualized by Alcian blue staining. (+) indicates injected body side.
Since the morphological phenotype coincided with the region of high expression of PWWP2A mRNA, we investigated the expression of eye and cranial neural crest (CNC) markers in half‐injected morphant embryos. At mid‐neurula stages, the retinal homeobox gene rx1 is expressed in a symmetric anterior domain of the prospective eye field (Casarosa et al, 1997). Rx1 mRNA staining was indistinguishable between CoMO‐ and pwMO‐injected embryos (Fig 8B), indicating that the observed defect in eye‐formation occurs downstream of eye field induction. During neurulation, cranial neural crest cells are induced at the border of the anterior neural plate, migrate ventrally shortly after, and differentiate ultimately into cartilage and bones (LaBonne & Bronner‐Fraser, 1998; Theveneau & Mayor, 2012). At the tadpole stage, the number of cells expressing the CNC regulatory gene twi was reduced in pwMO‐injected embryos, a phenotype that was partially rescued by coexpression of the human full‐length PWWP2A protein (Fig 8B). To corroborate this result, we performed Alcian blue staining of the cranial cartilage at the tadpole stage. In contrast to control morphants, the cartilage in the PWWP2A‐morphant side of embryos was strongly reduced in mass and disorganized, most obviously within the branchial arches (Fig 8C). We conclude that the reduced CNC formation in PWWP2A‐depleted embryos translates into a permanent defect in cartilage formation. These results identify an essential role for PWWP2A in cranio‐facial morphogenesis.
Discussion
H2A.Z is involved in all DNA‐related processes by so far not well‐understood mechanisms. We and others (Hu et al, 2013) hypothesized that this evolutionary conserved histone variant serves as a binding platform for distinct chromatin‐modifying complexes whose recruitment depends on additional chromatin surroundings. In our native quantitative MS approach, we used high‐stringency settings to enrich for variant‐specific binders and excluded proteins that also interacted strongly with H2A nucleosomes. Accordingly, we identified some previously found proteins, like BRD2 (Draker et al, 2012) and members of the SRCAP complex, as well as several transcription factors and histone‐modifying proteins/complexes While we were able to pull‐down the H2A.Z‐specific chaperone complex SRCAP, we only managed to identify the two largest subunits of the other H2A.Z chaperone complex p400/TIP60 (EP400 and TRRAP), but not other unique members. This finding raises the question why these two chaperone complexes that evolved from the yeast SWR1 complex and have been suggested to both participate in H2A.Z nucleosome exchange bind so differently to chromatin. It is possible that they function in distinct processes, as the p400/TIP60 complex has been shown to participate in DNA damage repair (reviewed in Jacquet & Cote, 2014), while SRCAP is important for H2A.Z deposition in gene regulation, as, for example, in muscle differentiation (Cuadrado et al, 2010). Alternatively, the p400/TIP60 complex together with ANP32E rather mediates H2A.Z removal and is therefore not stably associated with H2A.Z nucleosomes (Mao et al, 2014; Obri et al, 2014), or it requires larger chromatin domains than just one nucleosome for binding. The identification of histone‐modifying complexes involved in gene activation (e.g., MLL) and repression (e.g., HDAC1/2), supports the idea of H2A.Z being a recruitment platform for many proteins that participate in different aspects of gene regulation. We suspect that these complexes are not recruited to the same regions but are found at distinct H2A.Z sites in the genome, but where they are located and how they are recruited to these sites needs to be determined in future experiments.
Our studies identified the uncharacterized vertebrate‐specific PWWP2A protein to be a novel H2A.Z‐nucleosome binder that interacts via the variant's C‐terminus. Surprisingly, the extended acidic patch required for proper targeting and deposition and being proposed to be an interaction hub (Suto et al, 2000) is not sufficient for PWWP2A nucleosome binding. Interestingly, the short nine amino acid long C‐terminal stretch that is required for PWWP2A binding is a flexible region that extends out of the nucleosome core particle, it is different from H2A and apparently only conserved between vertebrates that also contain PWWP2A. As PWWP2A interacts with both H2A.Z isoforms that differ in the last residue (valine versus alanine) PWWP2A's interaction sequence appears to be “GKKGQQKT” and includes residues that can be ubiquitylated (Sarcinella et al, 2007). Whether this modification that marks transcriptionally silent facultative heterochromatin has any effect on PWWP2A binding and function remains to be elucidated.
PWWP2A might act as a “mediator” or “adapter” between H2A.Z nucleosomes and further chromatin‐modifying proteins, because it apparently does not contain any enzymatic active domains. In line with this hypothesis is our finding that PWWP2A is not needed for proper H2A.Z deposition and chromatin occupancy, while H2A.Z is at least partially required for PWWP2A nucleosome binding. PWWP2A specifically recognizes H2A.Z nucleosomes at the TSS of active genes, while most H2A.Z at non‐promoter sites are depleted of PWWP2A. These data suggest that PWWP2A needs an additional so far unknown signal that either keeps it at the TSS or prevents its recruitment to most H2A.Z non‐promoter regions. It is possible that the NDR region is recognized by the PWWP domain and is thereby one of the specificity determinant(s) as such a long stretch of free DNA is characteristic for TSS regions and might not occur at non‐promoter intergenic sites. Alternatively or additionally, another not yet identified chromatin feature, maybe histone PTMs that can be recognized via a conserved aromatic cage in the PWWP domain, confers extra specificity.
We have shown that PWWP2A binds to chromatin utilizing at least three different domains that confer H2A.Z variant‐specificity, nucleosome interaction, and DNA binding. Such a combinatorial binding mode might enable the recognition of different sites with different affinities and therefore distinct modes of regulation. Although PWWP2A is found predominantly at H2A.Z genomic sites, non‐promoter regions containing H2A.Z are mostly devoid of PWWP2A. This suggests that H2A.Z is a primary determinant for PWWP2A recruitment, but additional signals (possible free DNA and/or PTMs) confer a stable, high‐affinity association. As PWWP2A is predominantly found at the TSS of highly expressed genes, it is not surprising that its depletion affects gene expression programs and has profound consequences on cellular processes. We speculate that depending on the cell type and developmental stage, PWWP2A is involved in ensuring proper gene expression levels and any interference with this process results in cell type‐specific defects. HeLaK cells, which are immortal, aneuploid tumor cells with a high proliferation capacity might therefore be strongly affected in mitotic processes. Although we were not able to identify the original cause of the observed chromosome congression defect (reviewed in Auckland & McAinsh, 2015), it is possible that global metabolic alterations or the deregulation of a combination of factors involved in chromosome migratory mechanisms affect the cell's ability to properly align chromosomes at the spindle equator and synchronously pull them to the opposite poles. Accordingly, changes in transcriptional programs might be the driving parameter for the developmental defects observed in PWWP2A‐depleted X. laevis and X. tropicalis embryos. Our analyses revealed that PWWP2A's function in neural crest cell differentiation and/or migration during early development is evolutionary conserved, as a rescue with human PWWP2A RNA was overly successful. Indeed, PWWP2A's rescuing activity depends on the IC domain, suggesting that it exerts its developmental function in the context of H2A.Z‐containing chromosomal sites.
We propose that PWWP2A modulates expression of many H2A.Z‐containing genes, and speculate that the combined deregulation of metabolism and morphogenesis causes defects in mitotic spindle pulling and sliding as well as neural crest stem cell migration and differentiation. The latter alterations are hallmarks of human cranio‐facial diseases, such as neurocristopathies (Zhang et al, 2014) affecting facial features and include eye abnormalities, as seen in DiGeorge, Treacher‐Collins, or Waardenburg syndromes (reviewed in Mayor & Theveneau, 2013). Although the genetic causes of some of these diseases have been identified, for example, the ATP‐dependent chromatin remodeler CHD7 (Bajpai et al, 2010) is mutated in CHARGE syndrome (Van Nostrand et al, 2014), the genetic origin of many other neural crest‐associated diseases is not known. Future studies will elucidate whether mutations in PWWP2A might contribute to the pathogenesis of human neurocristopathies.
Materials and Methods
Cell culture and transfections
HeLa Kyoto (HeLaK) cells and metastatic melanoma Skmel‐147 cells and were grown in DMEM medium (PPA), supplemented with 10% FCS (Sigma‐Aldrich) and 1% penicillin/streptomycin (PAA) at 37°C and 5% CO2. DT40 cells were cultured and H2A.Z expression suppressed as described (Kusakabe et al, 2016). HeLaK and SKmel‐147 cells expressing GFP‐tagged histone variants are culture as described in (Bonisch et al, 2012; Vardabasso et al, 2015). GFP‐PWWP2A plasmids were transfected into HeLaK cells using X‐tremeGENE HP (Roche) according to the manufacturer's instructions and then selected in medium containing 400–600 μg/ml G‐148 sulfate liquid (PAA). Cell populations of 80–100% GFP‐positive stable cells were aspired after 3 weeks of selection and employed for further experiments. Expression levels of GFP‐proteins were measured using a FACSCanto machine (BDI Bioscience). Establishment and analysis of conditional H2A.Z DKO cells expressing Flag‐H2A.ZΔC was done as described previously (Kusakabe et al, 2016).
Antibodies
The following primary antibodies were used in this study: α‐PWWP2A (Novus (Acris), NBP2‐13833), α‐H2A.Z (Abcam, ab4174), α‐H2A (Abcam, ab13923), α‐H3 (Abcam, ab1791), α‐H3K4me3 (Diagenode, C15410003), α‐H3K9me3 (Diagenode, C15410056), α‐H3K36me3 (Active Motif, 61101), α‐H3S10ph (Active Motif, 39253 and 39636), α‐GST (clone 6G9; gift from E. Kremmer, Helmholtz Munich), α‐GFP (Roche, 11814460001), α‐CENP‐E (Active Motif, 39620), α‐ZNHIT1 (Sigma, HPA019043), α‐alpha‐Tubulin (Active Motif, 39527), α‐Flag (Sigma, F1804).
The following secondary antibodies were used: α‐mouse HRP (GE Healthcare, NA931), α‐rabbit HRP (GE Healthcare, NA934), and α‐rat HRP (GE Healthcare, NA935), α‐rabbit AlexaFluor 488 (Dianova, 711‐545‐152), α‐mouse AlexaFluor 488 (Dianova, 715‐545‐151), α‐mouse AlexaFluor 555 (Life Technologies, A21424), α‐rabbit AlexaFluor 555 (Life Technologies, A31572), α‐rabbit AlexaFluor 594 (Dianova, 711‐585‐152), α‐mouse AlexaFluor 594 (Dianova 715‐585‐151), α‐rabbit IRDye 800CW (LI‐COR Biosciences, 926‐32211), or α‐mouse IRDye 680RD (LI‐COR Biosciences, 926‐68070).
Cloning
Total RNA from Skmel‐147 cells was isolated using the RNeasy mini kit (QIAGEN) and cDNA synthesized with the ProtoScript First Strand cDNA Synthesis kit (NEB) according to the manufacturer's instructions and as described previously (Wiedemann et al, 2010). A DNA fragment containing the PWW2A coding sequence and parts of the 5′ and 3′ UTRs was amplified with a gene specific primer pair (Fwd: 5′‐GGAGTTGGAGGAGGGAGAAG‐3′, Rev: 5′‐TTCCAATGGTCTTGCCTACC‐3′) and Phusion DNA Polymerase (Biolabs). PCR product was subcloned into the shuttle vector pT7blue3. The full‐length PWWP2A fragment was subsequently used in Gateway cloning to create a N‐terminally GFP‐tagged PWWP2A fusion protein construct (pIRESneo‐eGFP‐PWWP2A) for in vivo studies and a GST‐tagged PWWP2A fusion construct (pGEX6P1‐PWWP2A) for recombinant bacterial expression. In addition, several PWWP2A truncation constructs for eukaryotic and bacterial expression were created using Gateway and Gibson (Gibson et al, 2009) cloning strategies. Flag‐H2A and Flag‐H2A.Z constructs are described in Kusakabe et al (2016). Flag‐H2A.ZΔC was prepared by inserting a chemically synthesized H2A.Z.1 cDNA (Eurofins Genomics) lacking Gly120 to Val128 into the p3xFLAG‐CMV10 plasmid (Sigma‐Aldrich).
Generation of S1 mononucleosomes
Mononucleosomes for IP experiments (qMS, ChIP‐seq as well as in vitro binding studies) were isolated as described previously (Sansoni et al, 2014) and the S1 fraction used for subsequent experiments.
Generation of mononucleosomes from DKO cells was performed as described in (Kusakabe et al, 2016) with one modification: At the last step of the preparation, the nucleosome pellet was solubilized with a buffer containing 150 mM NaCl instead of 500 mM NaCl. The mono‐nucleosome fraction was checked by electrophoresis on 2% agarose gel.
Label‐free quantitative mass spectrometry (LFQ‐MS) and data analysis
Mononucleosome immunoprecipitations, LC‐MS/MS analysis, and MS Raw Data Analysis were basically carried out as previously described (Vardabasso et al, 2015). For more details see the Appendix Supplementary Methods.
nChIP‐seq
Immunoprecipitations were carried out with S1 mononucleosomes derived from HeLaK cells stably expressing GFP or GFP‐tagged H2A, H2A.Z.1, H2A.Z.2, or PWWP2A from 4 × 107 cells in 1.5 ml low‐binding tubes. For more details, see the Appendix Supplementary Methods.
RNA‐seq and data analysis
Total RNA from HeLaK cells subjected to PWWP2A siRNAs and control siRNAs was isolated utilizing the RNeasy mini kit (Qiagen) according to manufacturer's instructions. The quality of the RNA (amount and integrity) was assessed using the BioAnalyzer and a RNA 6000 pico kit (Agilent). mRNA isolation, fragmentation, first‐ and second‐strand cDNA synthesis and end repair were carried out using the NEBNext Ultra RNA library prep kit for Illumina (NEB) following the manufacturer's instructions. cDNA sequencing libraries were established with the MicroPlex Library Preparation Kit (Diagenode).
Next generation sequencing (50 bp, single end) was performed by Dr. Stefan Krebs at the Laboratory of Functional Genome Analysis (LAFUGA) in Munich (Gene Center).
Raw sequencing reads were aligned to the human genome (GRCh38) using kallisto (v 0.42.4; Bray et al, 2016) with parameters “‐t 8 ‐b 100 –single ‐l 190 ‐s 10”. Differential expression was tested using an R/sleuth model including the experimental batch as random effect (preprint: Pimentel et al, 2016). Responders were defined by a q‐value cutoff < 5%. Gene Ontology group enrichment for differentially expressed genes was defined using the “goana” function in R/limma. GO groups were kept for visualization using REVIGO (Supek et al, 2011), if group size was larger than 19 genes and the P‐value smaller than 5%.
Recombinant expression and purification of GST proteins and in vitro binding studies
Expression and purification of GST proteins are described in detail in the Appendix Supplementary Methods.
Fluorescence recovery after photobleaching and live cell imaging
Fluorescence recovery after photobleaching analyses were performed essentially as described in Schneider et al (2013). For more details, see the Appendix Supplementary Methods.
Electrophoretic mobility shift assay
Details on all electrophoretic mobility shift assays (EMSAs) can be found in the Appendix Supplementary Methods.
RNAi, cell cycle and proliferation analysis
siRNAs were designed using siDESIGN Center (http://dharmacon.gelifesciences.com/design-center/?redirect=true) and synthesized (MWG‐Biotech AG). siRNAs were prevalidated by BLAST searches (NCBI) to confirm their targeting specificity to PWWP2A and to reduce the chance of off‐target effects. The following double‐stranded siRNAs were used: Luciferase, 5′‐CUUACGCUGAGUACUUCGA‐3′; ctrlII, 5′‐UAAGGCUAUGAAGAGAUACTT‐3′; PWWP2A#1, 5′‐GGACAGAAGUCAAGUGUGAUUTT‐3′; PWWP2A#2, 5′‐GCUAUUAAACUACGACCCAUUTT‐3′; cells were transfected with siRNA using oligofectamine (Invitrogen) according to the manufacturer's instructions. Two to three days after transfection, cells were used for various assays that are described in detail in the Appendix Supplementary Methods.
IF microscopy (confocal, chromosome spreads)
IF stainings were performed as previously described (Wiedemann et al, 2010; Mulholland et al, 2015). Metaphase Spreads were prepared as described previously (Jeppesen, 2000). For details, see the Appendix Supplementary Methods.
Modeling of PWWP domain structure
A potential structure of the PWWP domain of PWWP2A was modeled employing the web browser‐based tool iTASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/; Roy et al, 2010) and visualized with the freely available software Chimera (1.8.0). Template was the already published structure of the PWWP2B PWWP domain (Qin & Min, 2014), Protein Data Bank (PDB) code 4LD6. The predicted PWWP2A structure was compared to the published structure of the DNA‐binding PWWP domain of PSIP1 (Eidahl et al, 2013), PDB code 4FU6). The Electrostatic Surface Potential (ESP) of PSIP1 and PWWP2A was calculated utilizing the Coulombic Surface Coloring algorithm, which is part of the Chimera (1.8.0) software package, using default settings.
Xenopus experiments
Expression constructs and in vitro transcription
For rescue experiments, the human full‐length pwwp2a sequence (NM_001130864.1) was cloned into the pCS2 + expression vector with a N‐terminal GFP‐tag. For testing morpholino targeting efficiencies, the cDNA region from −121 to +154 of X. laevis pwwp2a was fused in frame to the luciferase ORF in a gateway‐compatible pCS2 + vector. Capped mRNA for microinjection was synthesized as described (Steinbach et al, 1997).
Morpholino Oligonucleotides
The pwMO morpholino oligonucleotide against the translational start site of pwwp2a mRNAs was purchased from GeneTools: 5′‐GCCGCCATTTTATCTTTCGCTTCTC‐3′. This oligonucleotide is fully complementary to the X. laevis pwwp2a mRNA (XeXenL6RMv10002517 m, Xenbase) and has a single mismatch to the X. tropicalis (XM_002940175) homolog. The unrelated standard control Morpholino (CoMO) served as control for specificity.
Embryo handling
Xenopus laevis and Xenopus tropicalis eggs were collected, in vitro fertilized, microinjected, and cultivated following standard procedures. Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop & Faber, 1994). The morpholino oligonucleotides were injected in one blastomere at the 2‐cell stage for morphological and molecular analyses. For CoMO and pwMO (20 ng/blastomere), Alexa Dextrans (Invitrogen) Fluor‐488 (green) or Alexa‐594 (red) were coinjected as lineage tracer. For rescue experiments, 250 pg of in vitro transcribed human GFP‐PWWP2A mRNAs (wild‐type and mutants) was co‐injected with Morpholinos and Alexa‐594 Dextran; synthesis of the PWWP2A proteins was verified by GFP fluorescence.
RNA in situ hybridization and cartilage staining
Whole‐mount RNA in situ hybridizations was performed as described (Sive et al, 2000). Cartilage was stained with Alcian Blue as described (Bellmeyer et al, 2003) and dissected from stained embryos for better visualization. Embryos were photographed under bright light with a Leica M205FA stereomicroscope.
Luciferase assay
The Morpholino oligo was injected into both blastomeres at the 2‐cell stage, followed by injection of Xl pwwp2a/luci mRNA at 4‐cell stage in all blastomeres. Luciferase chemiluminescence was measured at mid‐gastrula and normalized to CoMO conditions.
Ethics statements
Experiments on frogs were done in accordance with Deutsches Tierschutzgesetz; experimental use of Xenopus embryos has been licensed by the Government of Oberbayern (project/AK ROB: 55.2.1.54‐6.3‐11).
Primary accessions
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE78009 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78009).
Author contributions
SP, SLi, and SBH conceived of this study. SP performed all mononucleosome‐IP experiments for LFQ‐MS identification, nChIP‐seq (together with SLi) as well as all RNAi and cell cycle experiments. GW and EM conducted all Xenopus experiments with support of RAWR. ECK performed mass spectrometry with the support of MM and data analysis acquired by SP. NK, LMZ, and RMMS performed in vitro binding studies with the help of SLi. SLe, YM, KS, and SLi performed FRAP analysis and chromosome spreads with the support of HL. YM and SLi acquired time‐lapse microscopy pictures. RMMS carried out EMSA assays with advice of CR. TS undertook all nChIP‐seq and RNA‐seq bioinformatic analyses. CV generated SK‐mel147 cell lines stably expressing GFP‐tagged proteins with the support of EB. DT performed H2A.Z DKO IPs, MK constructed and analyzed DKO cells expressing Flag‐H2A.ZΔC with the help of MH. SP, SLi, and SBH designed the experiments and interpreted the results. SBH wrote the manuscript with support from all other coauthors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 7
Acknowledgements
We thank Andrea Schmid, Silvia Härtel, Martina Peritore, Takumi Narimiya, and Eva‐Maria Baur for technical help and Zuzana Storchova, Martha Smets, Ashish Singh, Sabrina Pfennig, and Irina Solovei for experimental advice. We are grateful to Christian J. Janzen for critically reading the manuscript. We thank Gregor Gilfillan (Oslo University Hospital) and Stefan Krebs from LAFUGA (Gene Center Munich) for help and advice in high‐throughput sequencing. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through the Collaborative Research Center SFB1064 (Project A10 to SBH, A12 to RAWR, A17 to HL, and Z04 to TS), by CIPSM to SBH, MM, and HL, the Weigand'sche Stiftung to SBH and Nanosystems Initiative Munich (NIM) to HL. EB was supported by Worldwide Cancer Research, Hirschl/Weill‐CaulierResearch Award, and NCI/NIH R01CA154683 and MH by JSPS KAKENHI grant numbers 25116009. SP and KS were fellows of the Munich International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS‐LS). LMZ, SLe, RMMS, and SLi are fellows of the Integrated Research Training Group (IRTG) of the SFB1064.
The EMBO Journal (2017) 36: 2263–2279
References
- Auckland P, McAinsh AD (2015) Building an integrated model of chromosome congression. J Cell Sci 128: 3363–3374 [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada‐Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C (2003) The protooncogene c‐myc is an essential regulator of neural crest formation in xenopus. Dev Cell 4: 827–839 [DOI] [PubMed] [Google Scholar]
- Bonisch C, Hake SB (2012) Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res 40: 10719–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch C, Schneider K, Punzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E, Leonhardt H, Mann M, Michaelis J, Schermelleh L, Hake SB (2012) H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res 40: 5951–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near‐optimal probabilistic RNA‐seq quantification. Nat Biotechnol 34: 525–527 [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW (2005) The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem 280: 13665–13670 [DOI] [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, Barsacchi G (1997) Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev 61: 187–198 [DOI] [PubMed] [Google Scholar]
- Chen T, Tsujimoto N, Li E (2004) The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 24: 9048–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Munoz‐Canoves P, Nebreda AR (2010) Essential role of p18Hamlet/SRCAP‐mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J 29: 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A, Elgin SC (1992) A histone variant, H2AvD, is essential in Drosophila melanogaster . Mol Biol Cell 3: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P (2012) A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet 8: e1003047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryhurst D, McMullen B, Fazli L, Rennie PS, Ausio J (2012) Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor‐mediated transcription and is upregulated in a model of prostate cancer progression. Cancer Lett 315: 38–47 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M (2013) Structural basis for high‐affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res 41: 3924–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin‐Lopez JM, Gonzalez‐Romero R, Dryhurst D, Ishibashi T, Ausio J (2009) The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z‐1 and H2A.Z‐2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol Biol 9: 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I (2001) Histone variant H2A.Z is required for early mammalian development. Curr Biol 11: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane‐Robinson C, Zhao K (2013) H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self‐renewal and differentiation. Cell Stem Cell 12: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouzalen N, Moreau J, Mechali M (1996) H2A.ZI, a new variant histone expressed during Xenopus early development exhibits several distinct features from the core histone H2A. Nucleic Acids Res 24: 3947–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet K, Cote J (2014) DNA repair: chromatin remodeling without H2A.Z? Cell Cycle 13: 1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P (2000) Immunofluorescence in cytogenetic analysis: method and applications. Genet Mol Biol 23: 1107–1114 [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S (2009) Chromosome‐wide Rad51 spreading and SUMO‐H2A.Z‐dependent chromosome fixation in response to a persistent DNA double‐strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Kim K, Punj V, Choi J, Heo K, Kim JM, Laird PW, An W (2013) Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenetics Chromatin 6: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe M, Oku H, Matsuda R, Hori T, Muto A, Igarashi K, Fukagawa T, Harata M (2016) Genetic complementation analysis showed distinct contributions of the N‐terminal tail of H2A.Z to epigenetic regulations. Genes Cells 21: 122–135 [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner‐Fraser M (1998) Neural crest induction in Xenopus: evidence for a two‐signal model. Development 125: 2403–2414 [DOI] [PubMed] [Google Scholar]
- Latrick CM, Marek M, Ouararhni K, Papin C, Stoll I, Ignatyeva M, Obri A, Ennifar E, Dimitrov S, Romier C, Hamiche A (2016) Molecular basis and specificity of H2A.Z‐H2B recognition and deposition by the histone chaperone YL1. Nat Struct Mol Biol 23: 309–316 [DOI] [PubMed] [Google Scholar]
- Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z (2014) Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res 24: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama EO, Hori T, Tanabe H, Kitamura H, Matsuda R, Tone S, Hozak P, Habermann FA, von Hase J, Cremer C, Fukagawa T, Harata M (2012) The actin family member Arp6 and the histone variant H2A.Z are required for spatial positioning of chromatin in chicken cell nuclei. J Cell Sci 125: 3739–3743 [DOI] [PubMed] [Google Scholar]
- Mayor R, Theveneau E (2013) The neural crest. Development 140: 2247–2251 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Mulholland CB, Smets M, Schmidtmann E, Leidescher S, Markaki Y, Hofweber M, Qin W, Manzo M, Kremmer E, Thanisch K, Bauer C, Rombaut P, Herzog F, Leonhardt H, Bultmann S (2015) A modular open platform for systematic functional studies under physiological conditions. Nucleic Acids Res 43: e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J (1994) Normal Table of Xenopus laevis (Daudin). New York: Garland Publishing Inc. [Google Scholar]
- Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, Dimitrov S, Romier C, Hamiche A (2014) ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505: 648–653 [DOI] [PubMed] [Google Scholar]
- Pimentel HJ, Bray N, Puente S, Melsted P, Pachter L (2016) Differential analysis of RNA‐Seq incorporating quantification uncertainty. Biorxiv https://doi.org/10.1101/058164 [DOI] [PubMed] [Google Scholar]
- Qin S, Min J (2014) Structure and function of the nucleosome‐binding PWWP domain. Trends Biochem Sci 39: 536–547 [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Greaves I, Tremethick DJ (2004) RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol 11: 650–655 [DOI] [PubMed] [Google Scholar]
- Ridgway P, Brown KD, Rangasamy D, Svensson U, Tremethick DJ (2004) Unique residues on the H2A.Z containing nucleosome surface are important for Xenopus laevis development. J Biol Chem 279: 43815–43820 [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC (2006) Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45: 5671–5677 [DOI] [PubMed] [Google Scholar]
- Sansoni V, Casas‐Delucchi CS, Rajan M, Schmidt A, Bonisch C, Thomae AW, Staege MS, Hake SB, Cardoso MC, Imhof A (2014) The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res 42: 6405–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P (2007) Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol 27: 6457–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Fuchs C, Dobay A, Rottach A, Qin W, Wolf P, Alvarez‐Castro JM, Nalaskowski MM, Kremmer E, Schmid V, Leonhardt H, Schermelleh L (2013) Dissection of cell cycle‐dependent dynamics of Dnmt1 by FRAP and diffusion‐coupled modeling. Nucleic Acids Res 41: 4860–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM (2000) Early development of Xenopus laevis. A Laboratory manual. New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT (2000) The PWWP domain: a potential protein‐protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473: 1–5 [DOI] [PubMed] [Google Scholar]
- Steinbach OC, Wolffe AP, Rupp RA (1997) Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature 389: 395–399 [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 7: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Grondin G, Gaudreau L (2010) H2A.Z overexpression promotes cellular proliferation of breast cancer cells. Cell Cycle 9: 364–370 [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R (2012) Neural crest delamination and migration: from epithelium‐to‐mesenchyme transition to collective cell migration. Dev Biol 366: 34–54 [DOI] [PubMed] [Google Scholar]
- Valdes‐Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ (2012) Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res 22: 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin CY, Lin CJ, Swiderski DL, Vogel H, Bernstein JA, Attie‐Bitach T, Chang CP, Wysocka J, Martin DM, Attardi LD (2014) Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardabasso C, Gaspar‐Maia A, Hasson D, Punzeler S, Valle‐Garcia D, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, Chung CY, Yao JL, Singh R, Segura MF, Fontanals‐Cirera B, Verma A, Mann M, Hernando E, Hake SB, Bernstein E (2015) Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell 59: 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Henikoff JG, Henikoff S (2010) H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol 17: 1500–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, Schermelleh L, Hake SB (2010) Identification and characterization of two novel primate‐specific histone H3 variants, H3.X and H3.Y. J Cell Biol 190: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratting D, Thistlethwaite A, Harris M, Zeef LA, Millar CB (2012) A conserved function for the H2A.Z C terminus. J Biol Chem 287: 19148–19157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Everett AD (2007) Hepatoma‐derived growth factor binds DNA through the N‐terminal PWWP domain. BMC Mol Biol 8: 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Ighaniyan S, Stathopoulos L, Rollo B, Landman K, Hutson J, Newgreen D (2014) The neural crest: a versatile organ system. Birth Defects Res C Embryo Today 102: 275–298 [DOI] [PubMed] [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD (2014) Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515: 582–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 7


