Summary
The mutant form of the GTPase KRAS is a key driver of pancreatic cancer but remains a challenging therapeutic target. Exosomes, extracellular vesicles generated by all cells, are naturally present in the blood. Here we demonstrate that enhanced retention of exosomes in circulation, compared to liposomes, is due to CD47 mediated protection of exosomes from phagocytosis by monocytes and macrophages. Exosomes derived from normal fibroblast-like mesenchymal cells were engineered to carry siRNA or shRNA specific to oncogenic KRASG12D (iExosomes), a common mutation in pancreatic cancer. Compared to liposomes, iExosomes target oncogenic Kras with an enhanced efficacy that is dependent on CD47, and is facilitated by macropinocytosis. iExosomes treatment suppressed cancer in multiple mouse models of pancreatic cancer and significantly increased their overall survival. Our results inform on a novel approach for direct and specific targeting of oncogenic Kras in tumors using iExosomes.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is in urgent need of effective new therapies1. Mutations in the GTPase KRAS are commonly encountered in PDAC2 and these drive initiation, progression and metastasis3,4. Dampening oncogenic Kras using genetic manipulation in mice inhibits tumor progression despite the presence of other genetic defects5. A direct and specific targeting of Ras has however been elusive6.
RNA interference (RNAi)-based approach to target wild-type Kras or downstream effectors using nanoparticles showed impact on tumor burden in lung and colorectal cancer models7–9. Targeting oncogenic Kras has been limited to delivery via direct electroporation10 or biopolymeric implants11 in xenograft models of pancreas cancer, and effective delivery of RNAi to non-liver parenchymal organs, especially pancreas, remains a challenge. While liposomes and nanoparticles may offer advantages for RNAi delivery over viral-based delivery systems, they exhibit low efficiency and rapid clearance from the circulation12. Here we probed whether exosomes can function as efficient carriers of RNAi. Exosomes are nano-sized extracellular vesicles (40–150 nm) with a membrane lipid bilayer that are released by all cells and efficiently enter other cells13.
Unlike liposomes and other synthetic drug nanoparticle carriers, exosomes contain transmembrane and membrane anchored proteins that likely enhance endocytosis, thus promoting the delivery of their internal content14,15. Exosomal proteins include CD4716,17, a widely expressed integrin associated transmembrane protein that functions in part to protect cells from phagocytosis18,19. CD47 is the ligand for signal regulatory protein alpha (SIRPα), and CD47-SIRPα binding initiates the ‘don’t eat me’ signal that inhibits phagocytosis20. Oncogenic RAS was shown to endow pancreatic cancer cells with enhanced macropinocytosis that may facilitate cellular uptake of exosomes21. The use of exosomes might also minimize cytotoxic effects observed when synthetic nanoparticles were used in vivo22. We identified the functional contribution of CD47 and Ras-induced macropinocytosis in suppressing exosomes clearance from circulation and enhancing specificity to pancreatic cancer cells, respectively. Such properties of exosomes enhanced their ability to deliver RNAi to specifically target oncogenic Kras in pancreatic tumors, and exosomes as ‘single targeted agent’ significantly enhances overall survival of all mouse models of PDAC tested.
Results
CD47 on exosomes suppress their clearance by monocytes
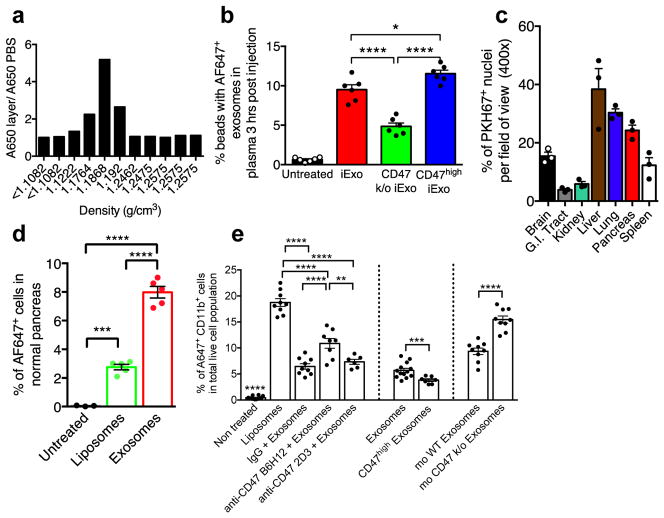
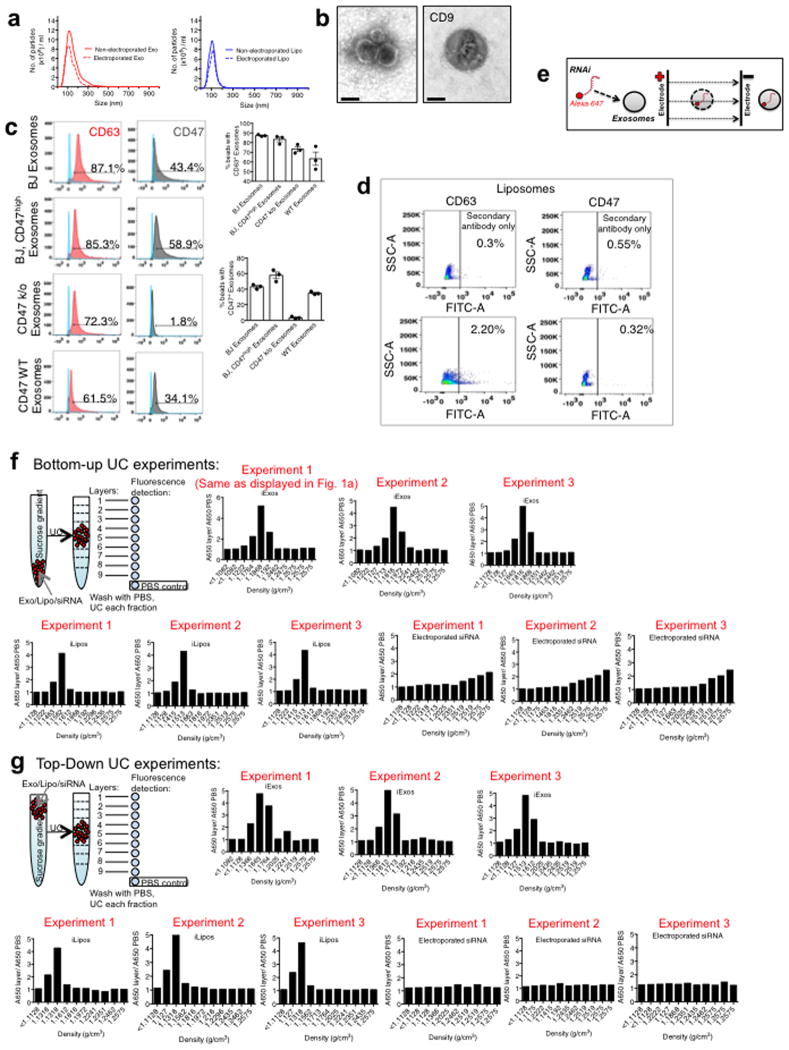
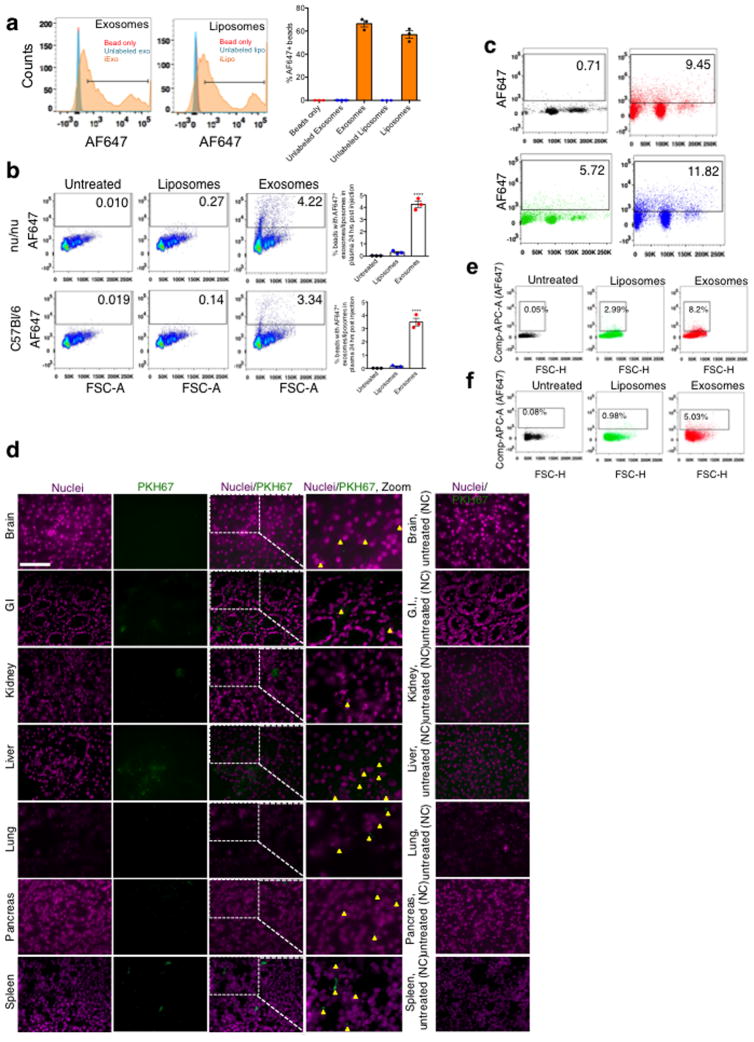
Exosomes were purified from the supernatant of normal human foreskin fibroblast (BJ) cultures (Extended Fig. 1A–B). CD47 detection on exosomes is noted in mass spectrometry analysis of exosomes (Supplementary Table 1, ExoCarta database, http://exocarta.org). In contrast with exosomes derived from CD47 knockout (k/o) mouse ear fibroblasts (CD47 k/o exosomes) and liposomes, BJ fibroblasts exosomes were positive for CD63 and CD47 (Extended Fig. 1C–D). We optimized the electroporation of Alexa-Fluor 647 (AF647)-tagged siRNA into exosomes (iExosomes) and liposomes (iLiposomes) without compromising their structural integrity (Extended Fig. 1A, E). Fractionation using sucrose gradient revealed detection of AF647, indicative of siRNA, in fractions characteristic of exosomes and liposomes, whereas electroporated siRNA alone (siRNA was mixed with electroporation buffer and subjected to electroporation without exosomes or liposomes) did not accumulate in these fractions (Fig. 1A, Extended Fig. 1F–G). Following intraperitoneal (i.p.) injection (24 hours), iExosomes but not iLiposomes were detected in the circulation of either C57Bl/6 or nude mice (Extended Fig. 2A–B). Three hours post i.p. injection, exosomes are also readily detected in the circulation, and while CD47 k/o exosomes showed diminished retention, exosomes with high levels of CD47 expression (CD47high exosomes, see methods) showed higher retention in the circulation (Fig. 1B, Extended Fig. 2C). Exosomes accumulated in the liver, lung and pancreas (Fig. 1C, Extended Fig. 2D), and a greater number of pancreas cells showed AF647-siRNA signal from iExosomes compared to iLiposomes (Fig. 1D, Extended Fig. 2E–F).
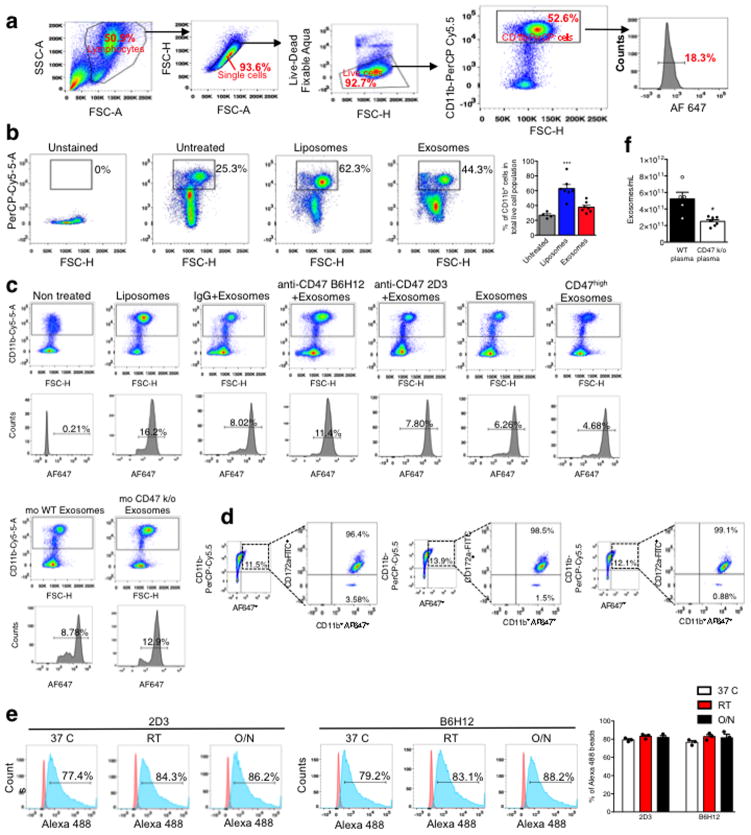
Figure 1. CD47 on exosomes limits their clearance by circulating monocytes.
(A) Fluorescence intensity in sucrose gradient layers from “Bottom-Up” method (see Supplementary Fig. 1E–F), (B) FC analysis of exosomes with AF647 tagged siRNA in the circulation (n=6 mice per group). (C) Quantification of PKH67 labeled exosomes in the indicated organs (n=3 mice) (D) FC analyses of pancreas cells 6 hours following injection of siKrasG12D Exos (n=5 mice), siKrasG12D Lipos (n=5 mice), and PBS (untreated, n=3 mice). (E) Quantification of Alexa 647+/CD11b+ monocytes in the blood 3 hours post i.p. injection. One way ANOVA: (Non treated, n=12 mice) vs (Liposomes, n=9 mice), IgG+siKrasG12D iExo (n=9 mice), anti-CD47 (B6H12)+siKrasG12D iExo (n=8 mice) and anti-CD47 (2D3)+siKrasG12D iExo (n=6 mice). Unpaired two-tailed t test: siKrasG12D iExo (n=13 mice) and (CD47high iExo, n=7 mice). Unpaired two-tailed t test: (mouse WT Exosomes, n=9 mice) and (mouse CD47 k/o exosomes, n=9 mice). The mean +/− SEM is depicted. Unless otherwise stated, one-way ANOVA was used. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001. FC: Flow cytometry; mo: mouse. See accompanying source data.
Generally, efficient phagocytosis by circulating monocytes and other cells removes dying/dead cells, cell debris and foreign particles. iLiposomes, compared to iExosomes, enhanced the mobilization of CD11b+ monocytes in circulation (Extended Fig. 3A–B). While control mouse blood (untreated) showed background levels of AF647 positivity in CD11b+ monocytes, an increase in the number of circulating AF647+ monocytes (indicative of phagocytosis) was noted when mice were treated with liposomes compared to exosomes (Fig. 1E, Extended Fig. 3C). Furthermore, the levels of CD47 on exosomes inversely correlated with circulating AF647+ monocytes in the blood (Fig. 1E, Extended Fig. 3C), supporting that the presence of CD47 on exosomes limits their clearance. Additionally, following iExosomes injection, the AF647+ monocytes in the circulation were positive for SIRP-α (Extended Fig. 3D). Using anti-CD47 B6H12 blocking antibody, which prevents CD47/SIRP inhibition of phagocytosis in contrast with nonblocking anti-CD47 2D3 antibody23, resulted in a specific and significant increase in AF647+CD11b+ circulating monocytes (Fig. 1E, Extended Fig. 3C). Notably, the binding of anti-CD47 2D3 and anti-CD47 B6H12 antibodies to exosomes was similar (Extended Fig. 3E). In contrast, decreased AF647+CD11b+ circulating monocytes were noted when using CD47High iExosomes (Fig. 1E, Extended Fig. 3C). Interestingly, CD47 k/o mice showed lower levels of circulating exosomes compared to age-matched controls (Extended Fig. 3F). Our results indicated a superior escape from phagocytic clearance of exosomes compared to liposomes, in part mediated by exosomal CD47-SIRPα ‘don’t eat me’ signal.
Specific targeting of KrasG12D in pancreatic cancer cells using iExosomes
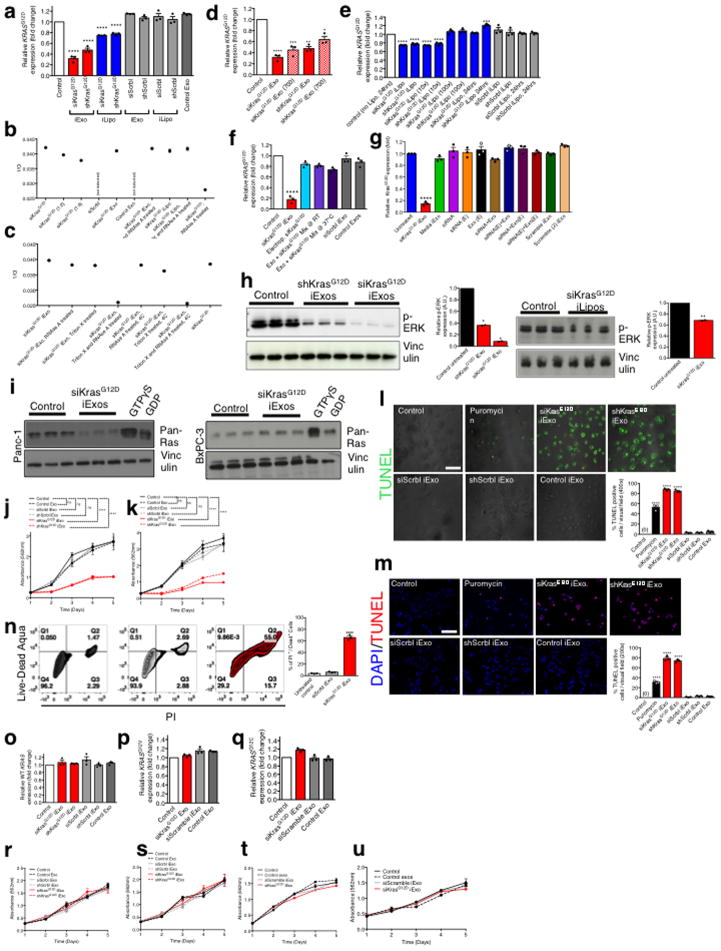
iExosomes (with siRNA or shRNA targeting KrasG12D) significantly reduced KrasG12D mRNA levels and phosphorylated-ERK protein levels in Panc-1 cells, with superior efficacy compared to iLiposomes despite a similar siRNA loading efficiency in both nanoparticles (Extended Fig. 4A–H, Supplementary text, Supplementary Fig. 1). iExosomes also suppressed Ras activity specifically in Panc-1 cells compared to BxPC-3 cells (KRASWT) (Extended Fig. 4I, Supplementary Figure 2), and impaired proliferation (Extended Fig. 4J–K) and enhanced apoptosis (Extended Fig. 4L–N) in Panc-1 cells, while leaving BxPC-3 (KRASWT), Capan-1 (KRASG12V), and MIA PaCa-2 (KRASG12C) cancer cells unaffected (Extended Fig. 4O–U).
iExosomes suppress KrasG12D-expressing human pancreatic orthotopic tumors
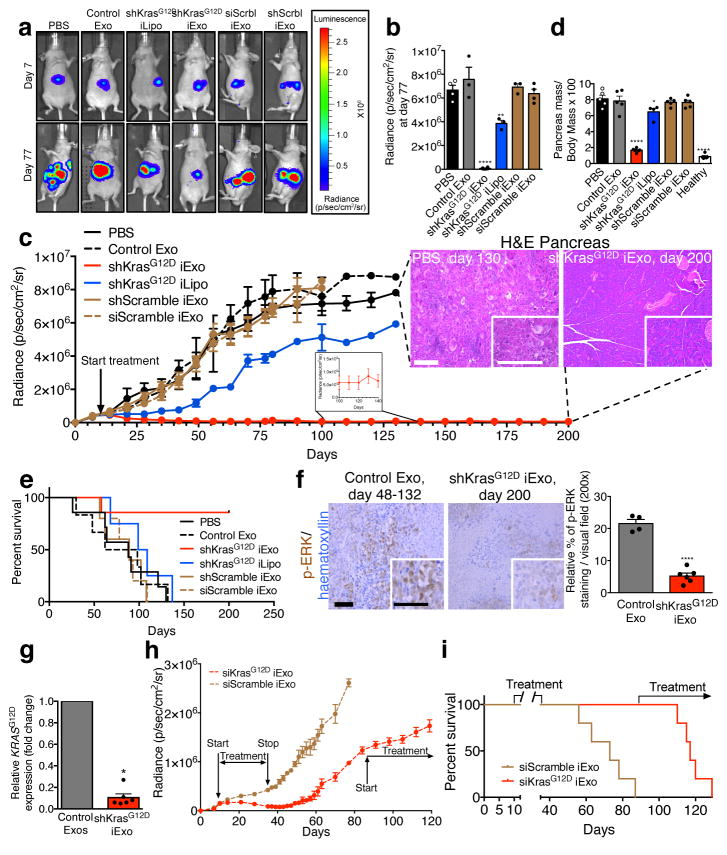
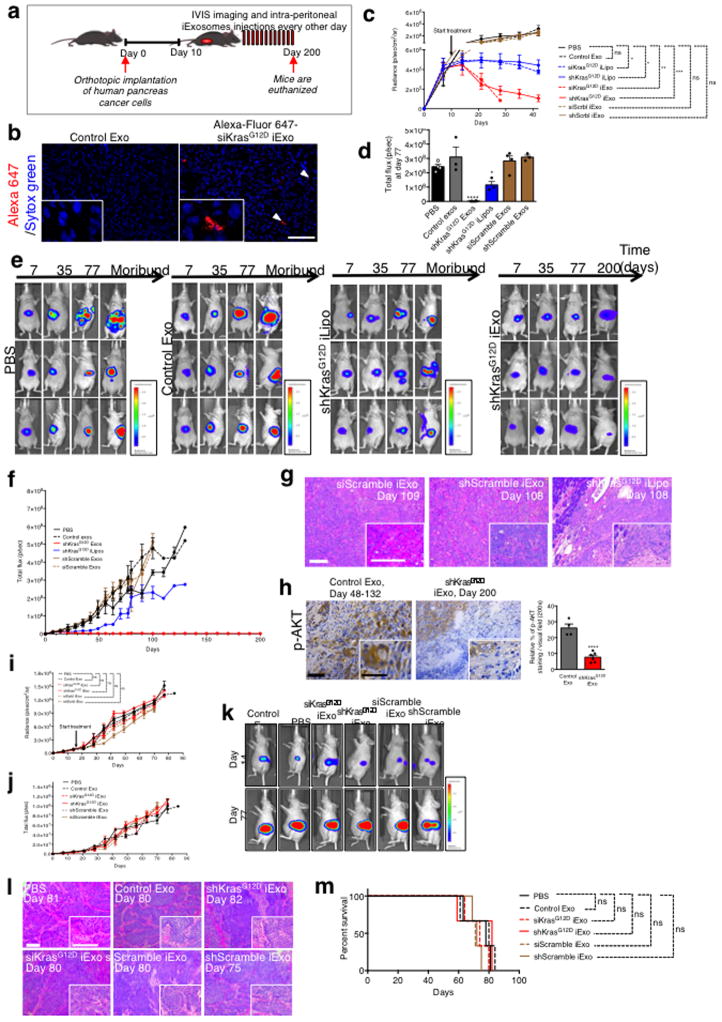
Mice with luciferase expressing orthotopic Panc-1 tumors were treated with repeated i.p. injection of ~108 iExosomes (every other day, 0.15 to 0.20 μg of exosomal protein per injection) or iLiposomes (Extended Fig. 5A). Accumulation of iExosomes payload (AF647-siRNA) was readily detected in the pancreas (Extended Fig. 5B). While the tumors of control mice (PBS vehicle, non-electroporated exosomes (control Exo) or exosomes and liposomes with scrambled RNAi) grew at an exponential rate, the tumors treated with iExosomes were significantly reduced after 30 days of treatment (Extended Fig. 5C). Tumor growth was blunted with iLiposomes, however to a much lesser extent than with iExosomes (Extended Fig. 5C).
When control mice revealed extensive tumor burden, tumors in iExosomes-treated mice were in contrast reduced to nearly undetectable level (Fig. 2A–B, Extended Fig. 5D–E), and this persisted after 200 days of treatment (Fig. 2C, Extended Fig. 5F). Histopathological analyses (endpoint), relative pancreas mass, and overall survival indicated robust improvement in iExosomes treated mice (Fig. 2C–E, Extended Fig. 5G). iExosomes suppressed downstream Kras signaling and KRASG12D expression in tumors/pancreas (Fig. 2F–G, Extended Fig. 5H). Suspending iExosomes treatment after initial tumor reduction showed sustained tumor suppression effects that lasted over 10 days following the last treatment with iExosomes (Fig. 2H). Control mice succumbed to pancreatic cancer, whereas the siKrasG12D iExosomes treated mice were all alive (day 87). Resuming iExosomes treatment at this point in time controlled the growth of these advanced tumors (Fig. 2H–I). Despite continuous treatment at an advanced disease state, the mice responded with partial tumor growth control but ultimately succumbed (Fig. 2H–I). iExosomes did not however impact orthotopic BxPC-3 (KRASWT) tumor growth or survival (Extended Fig. 5I–M). Loss of surface proteins on exosomes with proteinase K (PK) treatment, validated by flow cytometry (Extended Fig. 6A), significantly suppressed the anti-tumor efficacy of iExosomes, whereas RNAse A treatment alone did not have any impact on the anti-tumor efficacy of iExosomes (Extended Fig. 6B–J, Supplementary text).
Figure 2. iExosomes restrains Panc-1 tumor growth.
(A) Luciferase activity at day 7 and day 77-post cancer cell injection. (B) Tumor bioluminescence at day 77, PBS: n=4 mice, Control Exo: n=3 mice, shKrasG12D iExo: n=6 mice, shKrasG12D iLipo: n=4 mice, shScramble (Scrbl) iExo: n=5 mice, siScramble (Scrbl) iExo: n=5 mice. (C) Panc-1 orthotopic tumor growth (bioluminescence). PBS: n=7 mice, Control Exo: n=6 mice, shKrasG12D iExo: n=7 mice, shKrasG12D iLipo: n=4 mice, shScramble iExo: n=5 mice, siScramble iExo: n=5 mice. Representative H&E of the pancreas (scale bar: 100μm) is shown. (D) Relative pancreas mass (PBS, n=5 mice, Control exos, n=5 mice, shKrasG12D iExo, n=6 mice, shKrasG12D iLipo, n=4 mice, shScramble iExo, n=5, mice siScramble iExo, n=5, and normal healthy mice, n=5) mice. (E) Kaplan-Meier curve of Panc-1 tumor bearing mice, Log-rank Mantel-Cox test, PBS: n=7 mice, Control Exo: n=6 mice, shKrasG12D iExo: n=7 mice, shKrasG12D iLipo: n=4 mice, shScramble iExo: n=5 mice, siScramble iExo: n=5 mice. (F) p-ERK immunolabeling (scale bar: 100μm). Unpaired two-tailed t test, Control Exo: n=4 mice, shKrasG12D iExo: n=6 mice. (G) KrasG12D transcript levels in tumors, Control Exo (n=5 mice), or shKrasG12D iExo (n=6 mice), unpaired two-tailed t test. (H) Panc-1 orthotopic tumor growth, siScramble iExo (n=5) or siKrasG12D iExo (n=5). (I) Kaplan-Meier curve of Panc-1 tumor bearing mice. Log-rank Mantel-Cox test, n=5 mice per group. The mean +/− SEM is depicted. Unless stated otherwise, one-way ANOVA comparing experimental groups to control group (PBS) was used to determine statistical significance. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001, ns: not significant. See accompanying source data.
CD47 and Ras-dependent macropinocytosis facilitates enhanced efficacy of iExosomes
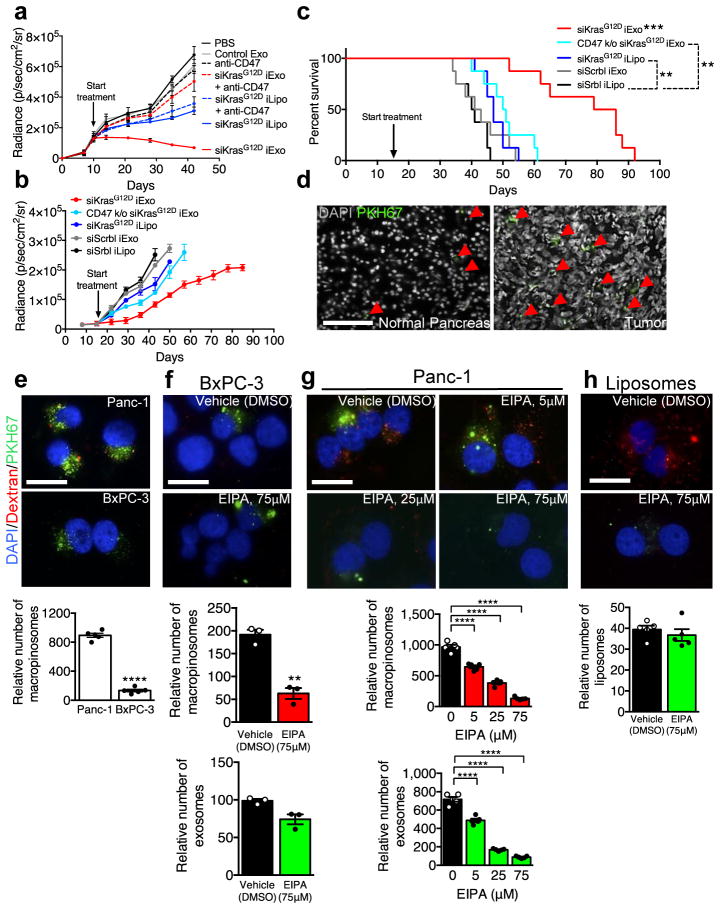
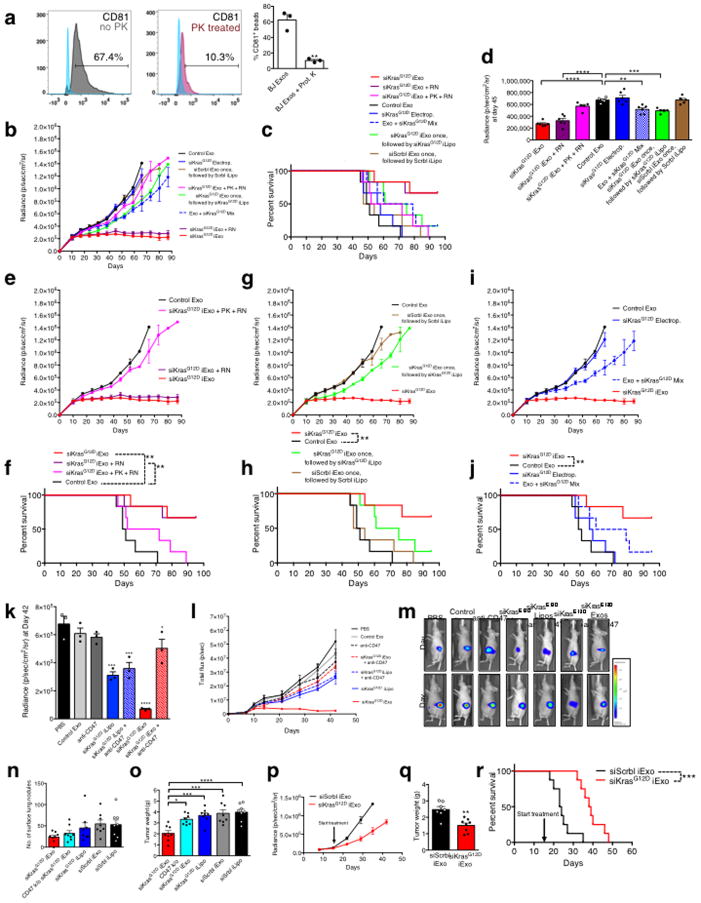
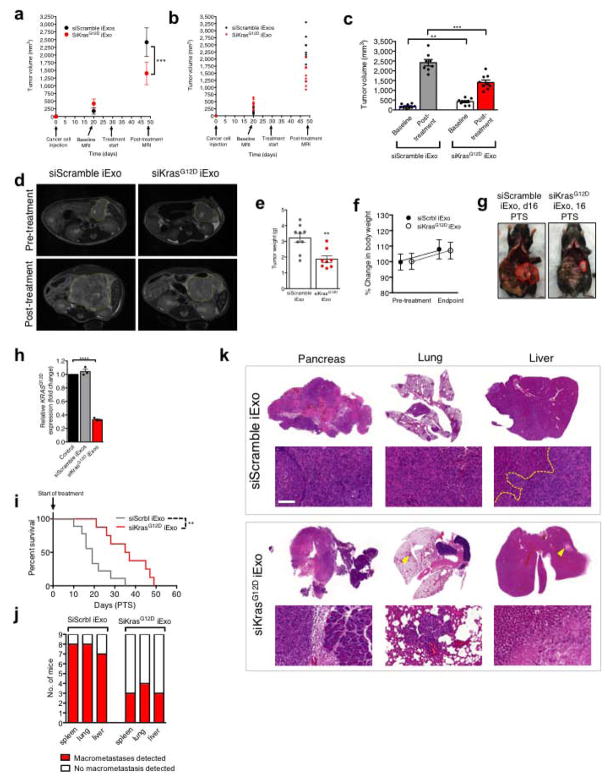
In mice with orthotopic Panc-1 tumors, iExosomes and iLiposomes were administered with and without incubation with anti-CD47 neutralizing antibodies. Unlike with iLiposomes or anti-CD47 antibodies alone, the efficacy of iExosomes was significantly inhibited with neutralization of CD47-SIRPα ‘don’t eat me’ signal (Fig. 3A, Extended Fig. 6K–M, Supplementary Fig. 4C). In immunocompetent mice with KPC689 orthotopic tumors, CD47 k/o iExosomes failed to robustly suppress tumor growth and improve survival compared to iExosomes (Fig. 3B–C, Extended Fig. 6N–O). In this model (treatment start 16 days post cancer cell injection), iLiposomes and control treatment were ineffective (Fig. 3B–C, Extended Fig. 6N–O). Similar results were obtained when performing the experiments in Nude mice, despite a more aggressive cancer progression in the immunocompromised background (Extended Fig. 6P–R).
Figure 3. CD47 and macropinocytosis enhance iExosomes uptake and therapeutic efficacy.
(A) Panc-1 orthotopic tumor growth, n=3 mice per group. (B) KPC689 orthotopic tumor growth, n=8 mice per group. (C) Kaplan-Meier curve, KPC689 orthotopic tumor bearing mice, Log-rank Mantel-Cox test, n=8 mice per group. (D) Confocal micrographs (scale bar: 100μm) of increased (preferential) entry of labeled exosomes into tumor tissue. (E) Macropinocytic uptake in Panc-1 or BxPC-3 cells, unpaired two-tailed t test. (F–G) Macropinocytic and exosomes uptake in BxPC-3 (unpaired two-tailed t test, F) or Panc-1 (one-way ANOVA comparing treated groups to non-treated group (0 μM EIPA, G) cells treated with vehicle (DMSO) or EIPA at the indicated concentrations (H) Macropinocytic and liposomes uptake, unpaired two-tailed t test. E, G, H: 5 distinct wells, F: 3 distinct wells. In E–H, scale bar: 50μm. The data is presented as the mean ± SEM. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001, ns: not significant. See accompanying source data.
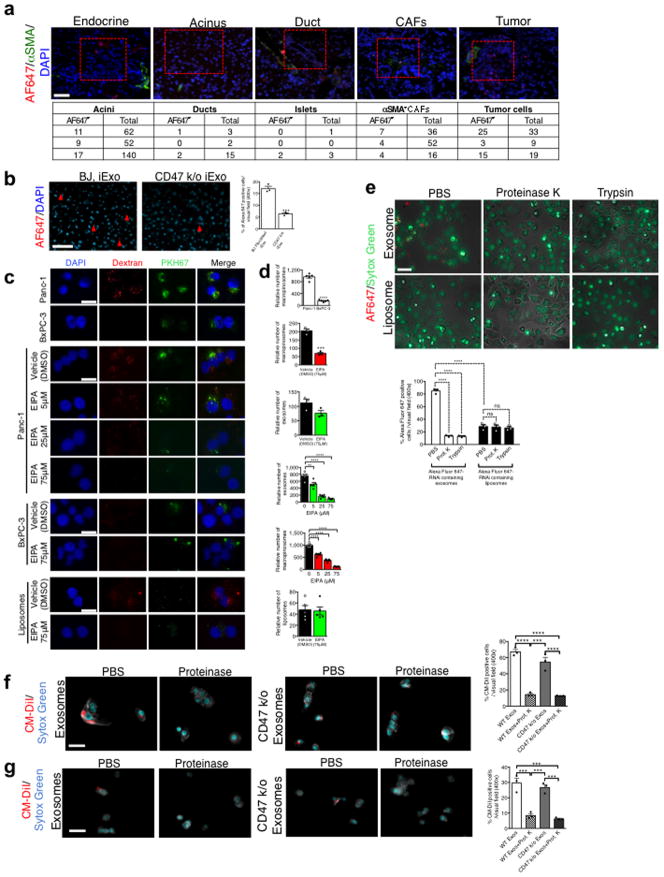
Accumulation of PKH67 labeled exosomes signal (fluorescent lipophilic dye) was predominantly detected in established Panc-1 tumors 3 hours after the injection, with less efficient accumulation noted in adjacent normal pancreas (Fig. 3D), and accumulation of AF647+ foci (iExosomes) in KTC mice were observed predominantly in tumors cells, as well as in normal acini, ducts, endocrine islet and αSMA+ CAFs (Extended Fig. 7A). Notably, decreased AF647+ foci in pancreas tumors was noted when using CD47 k/o iExosomes compared to iExosomes (Extended Fig. 7B). Collectively these data suggested that along with enhanced retention of iExosomes in systemic circulation, increased accumulation of iExosomes in tumors could reflect enhanced uptake of iExosomes in cancer cells when compared to normal pancreatic cells. This observation further complements the in vitro studies which demonstrate that exosomes more efficiently deliver RNAi molecules to the pancreatic cancer cells when compared to liposomes (Extended Fig. 4A–H). In this regard, oncogenic Ras has been implicated in intensifying macropinocytosis21,24. Our results confirmed increased macropinocytosis in Panc-1 cells compared to BxPC-3 cells (Fig. 3E, Extended Fig. 7C–D). Exosomes uptake mirrored the macropinocytosis frequency, and inhibition of macropinocytosis with EIPA reduced exosomes uptake but not liposomes (Fig. 3F–H, Extended Fig. 7C–D). Treatment of exosomes with PK or trypsin significantly reduced their entry into Panc-1 cells, yet this was independent of CD47 (Extended Fig. 7E–G). Three distinct mechanisms thus likely contribute to the enhanced anti-tumor response to iExosomes: CD47 presence on exosomes contributes to evasion from the host immune clearance in the circulation, Ras mediated enhanced macropinocytosis, and presence of proteins on the surface of exosomes that may increase pancreatic cancer cells uptake of iExosomes.
iExosomes inhibit advanced metastatic disease and increase overall survival
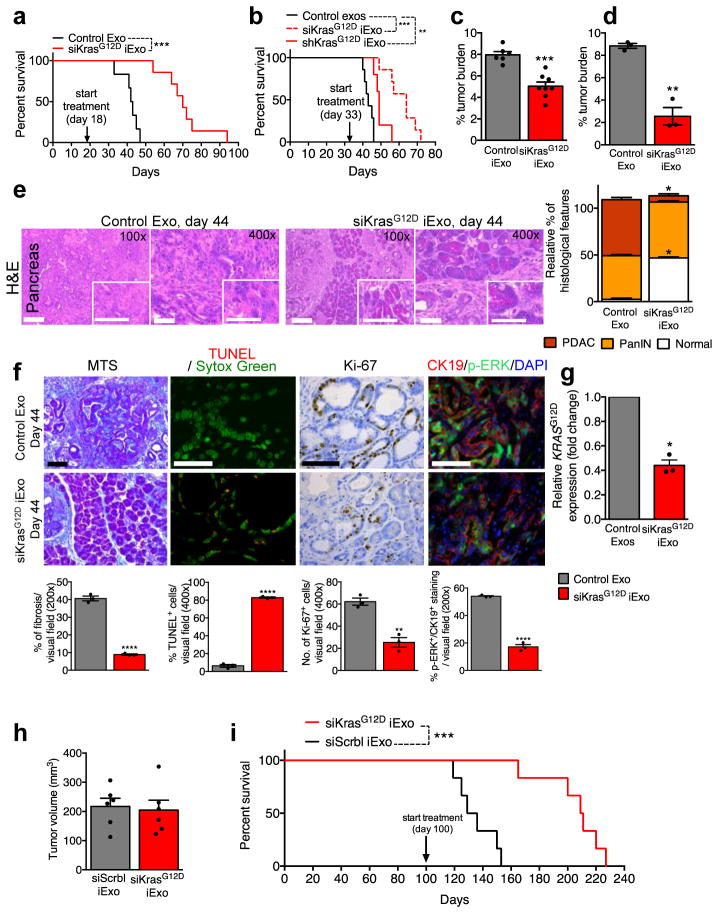
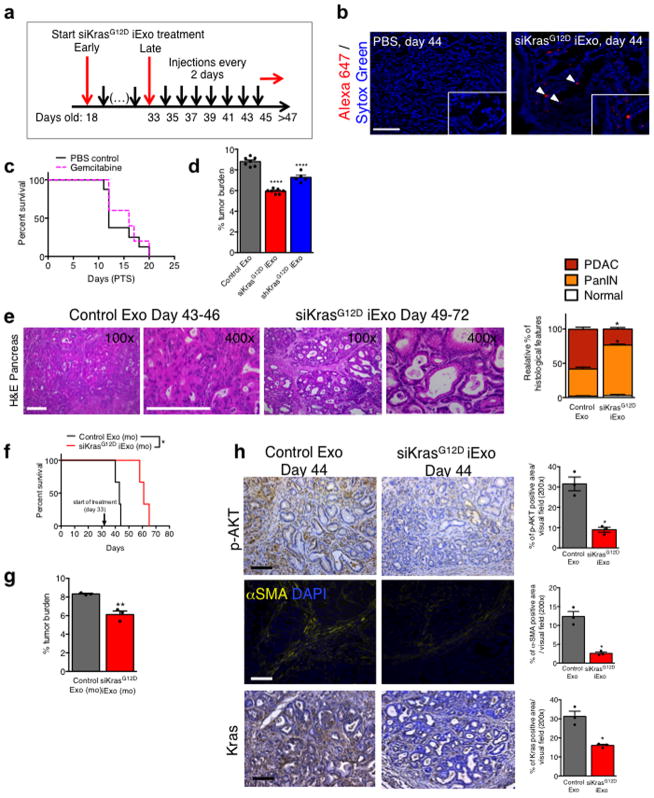
We treated KTC mice25,26 with iExosomes on day 18 (early treatment start) or day 33 (late treatment start) when mice present with PDAC (Extended Fig. 8A). Notably, accumulation of AF647-signal was detected in KTC tumors (Extended Fig. 8B). In both experiments, iExosomes increased lifespan compared to control exosomes (Fig. 4A–B), in contrast with a lack of response observed when mice were treated with gemcitabine (Extended Fig. 8C and shown by others25,27–29). iExosomes reduced the tumor burden (Fig. 4C–D, Extended Fig. 8D) and improved histopathology (Fig. 4E, Extended Fig. 8E). iExosomes from human or murine sources showed similar response (Extended Fig. 8F–G). Diminished pancreas desmoplasia, enhanced cancer cell apoptosis, suppressed cancer cell proliferation, reduced phospho-ERK, phospho-AKT and Kras levels are noted in KTC tumors, as well as diminished oncogenic KrasG12D expression with iExosomes treatment (Fig. 4F–G, Extended Fig. 8H). Prior to treatment start, KPC mice showed comparable tumor burden (Fig. 4H), and iExosomes in this model also showed anti-tumor efficacy with increased survival (Fig. 4I). Further, no obvert cytotoxicity was observed with iExosomes therapy (Extended Fig. 9A–C). iExosomes also suppressed PDAC progression in the advanced, highly metastatic KPC689 orthotopic tumor setting (Extended Fig. 10A–G), reducing KrasG12D expression (Extended Fig. 10H), increasing survival (Extended Fig. 10I) and limiting metastasis (Extended Fig. 10J–K).
Figure 4. iExosomes suppress pancreas cancer progression in KTC GEMM.
(A) Kaplan-Meier curve of KTC (early treatment) mice, Log-rank Mantel-Cox test, siKrasG12D iExo: n=8 mice, Control exos: n=6 mice. (B) Kaplan-Meier curve of KTC mice, Log-rank Mantel-Cox test, siKrasG12D iExo: n=7 mice, shKrasG12D iExo: n=5 mice, Control exos: n=7 mice. (C) Tumor burden (early treatment) at end point. siKrasG12D iExo: n=8 mice, Control exos: n=6 mice. (D) Tumor burden at 44 days of age, n=3 mice per group. (E) H&E stained tumors (scale bar: 100μm, inset scale bar: 50 μm) from 44 days-old KTC mice and relative percentages in histological phenotypes, n=3 mice per group. (F) Masson Trichrome staining (MTS, scale bar: 100μm), TUNEL, Ki-67, and phosphorylated-ERK/CK-19 immunolabeling of 44 days-old KTC mice, n=3 mice per group. (G) KrasG12D transcript levels in tumors of age-matched (44 days old) KTC mice, n=3 mice per group. (H) Tumor volume at baseline (MRI), siKrasG12D iExo: n=6 mice, siScrbl iExo: n=6 mice. (I) Kaplan-Meier curve of KPC mice, Log-rank Mantel-Cox test, n=6 mice in each group. The mean ± SEM is depicted. Unless stated otherwise, unpaired two-tailed t test was used to determine statistical significance. * p<0.05, ** p< 0.01, *** p< 0.001, **** p<0.0001. See accompanying source data.
Discussion
Mutations in KRAS are associated with cancer of the pancreas, lung and colon, among others30,31, and oncogenic KRAS mutations and activation of downstream effectors such as MEK, Akt and Erk, among others, are sufficient drivers of pancreas cancer3–5,30,32–35. A sound rationale for targeting Ras emerged for the treatment of cancer11,36,37, but Ras has remained largely undruggable6. Some efficacy were reported with methodologies developed to target oncogenic Kras using siRNA molecules7,8,10,11, but these approaches may have been limited by lack of specificity and inefficient delivery. Nonetheless, a recent clinical study demonstrated that siG12D-LODERTM was well-tolerated and showed potential efficacy in patients with locally advanced pancreas cancer38. We report that engineered iExosomes can control advanced PDAC in mice and this approach is clinically feasible39.
Our studies suggest that exosomes exhibit a superior ability to deliver RNAi and suppress tumor growth when compared to liposomes. Unlike liposomes, the plasma membrane-like phospholipids and membrane-anchored proteins of exosomes may contribute to their diminished clearance from the circulation12,40,41. Our results support that the presence of CD47 on exosomes allows for evasion from phagocytosis by the circulating monocytes and increases exosomes half-life in the circulation. While CD47 does not play a significant role in the entry of exosomes into pancreatic cells, enhanced macropinocytosis in Kras mutant cancer cells22,31 favored their exosomes uptake. Our results also support an efficient uptake of iExosomes despite the stroma dense features of pancreas tumors. Whether exosomes entering cells via this mechanism (macropinocytosis) protect themselves from lysosome dependent degradation of their content needs further exploration. Collectively, our study offers insight into the therapeutic potential of exosomes in specific targeting of oncogenic Kras in pancreatic cancer.
Methods
Cell culture
Human foreskin fibroblast (BJ), Capan-1, MIA PaCa-2 cells were cultured in DMEM supplemented with 10% exosomes depleted FBS and 1% penicillin-streptomycin. Panc-1, BxPC-3, and T3M4 cells were cultured in RPMI 10% FBS and 1% penicillin-streptomycin. All cells lines were from American Type Culture Collection (ATCC) except for T3M4 (Cell Bank, RIKEN BioResource Center); there were no additional validation of the cell lines performed. Luciferase expressing Panc-1 and BxPC-3 cells (expressing a CMV promoter 5′ to the firefly luciferase protein) were gifts from Dr. Thiruvengadam Arumugam, UT MDACC. KPC689 cancer cell line was established from the pancreas tumors of Pdx1cre/+;LSL-KRasG12D/+;LSL-Trp53R172H/+ mice (KPC) mice42. KPC689 cells were engineered to stably express GFP and luciferase following infection with F-Luc-GFP lentivirus (Capital Biosciences). C57BL/6 wild type (WT) fibroblasts were isolated from the ears of C57BL/6 mice by mincing the isolated ears in DMEM supplemented with collagenase type 4 (400 units/ml) and incubating overnight. The next day the cells and tissue pieces were washed with DMEM supplemented with 20% FBS and 1% penicillin-streptomycin and expanded in this media. For exosomes collection, the cells were cultured using exosomes-depleted FBS. The same procedure was performed on the CD47 knockout mice (B6.129S7-Cd47tm1Fpl/J) mice and C57BL/6 mice to generate ear and tail fibroblast lines. For overexpression of CD47 on BJ fibroblasts, transfections were performed using Lipofectamine 2000 reagent (Invitrogen) with pCMV6-AC-GFP CD47 plasmid (Origene, MG204706), after which exosomes were isolated using the standard protocol described below.
Isolation and purification of exosomes
Exosomes were purified by differential centrifugation processes, as described previously43–46. Exosomes-depleted FBS was prepared as follows: the FBS is filtered using a 100nm filter, then ultracentrifuged for 16 hours, and then filtered again using a 100nm filter. Supernatant was collected from cells that were cultured in media containing exosomes-depleted FBS for 48 hours, and was subsequently subjected to sequential centrifugation steps for 800g for 5 minutes, and 2,000g for 10 minutes. This resulting supernatant was then filtered using 0.2 μm filters, and a pellet was recovered at 100,000g in a SW 32 Ti rotor after 2 hours of ultracentrifugation (Beckman). The supernatant was aspirated and the pellet was resuspended in PBS and subsequently ultra-centrifuged at 100,000g for another 2 hours. The purified exosomes were then analyzed and used for experimental procedures. For treatment of exosomes with proteinase K, purified exosomes were incubated (37°C, 30 minutes) with 5mg/mL of proteinase K (Sigma-Aldrich, dissolved in RNase-free water) followed by heat inactivation (60°C, 20 minutes). For RNase treatment, purified exosomes were incubated (37°C, 30 minutes) with 2 mg/mL of protease-free RNase A (Thermo Scientific) followed by addition of 10X concentrated RNase inhibitor (Ambion). These exosomes were then subsequently used for FACS analysis, in vitro assays and treatment of tumor bearing mice, as described below.
Sucrose gradient47
Sucrose density gradients were performed to characterize the exosomes. For the “Bottom-Up” sucrose gradient separation (Extended Fig. 1F), the exosomes, resuspended in 2 mL of HEPES/sucrose solution (2.5M sucrose, 20mM HEPES/NaOH solution, pH 7.4), were loaded first in the bottom of the tube and overlaid with a 9mL linear sucrose gradient (2.0–0.25M sucrose, 20mM HEPES/NaOH, pH 7.4) in a SW41 tube (Beckman, 11mL). For the “Top-Down” sucrose gradient separation (Extended Fig. 1G), exosomes were resuspended in 1mL of HEPES/sucrose solution (0.25M sucrose, 20mM HEPES/NaOH, pH 7.4). A 10mL linear sucrose gradient (2.0–0.25M sucrose, 20mM HEPES/NaOH, pH 7.4) was built into a SW41 ultracentrifuge tube, and the exosomes suspension (1mL, 0.25M sucrose, 20mM HEPES/NaOH, pH 7.4) was deposited on top of this linear sucrose gradient. In both types of sucrose gradient experiments (Bottom-Up and Top-Down), the gradients were ultracentrifuged for 16 hours at 210,000g at 4°C. Gradient fractions of 1 mL were collected from the top to the bottom of the tube, and the densities of each fractions were evaluated using a refractometer. Each layer was placed into a separate centrifuge tube, and PBS was added to a final volume of 11 mL, then this was ultracentrifuged at 150,000g at 4°C for 2 hours. The pellets for each layer were resuspended in 200 μl of PBS and loaded onto a black 96-welled microplate to detect the Alexa fluor 647 (AF647) fluorophore-tagged siRNA contained in them using a fluorescence detection plate reader. A microplate containing 200 μl of PBS was used for background readings. The detection of the fluorescence of Alexa-Fluor 647 fluorophore is depicted as the ratio of fluorescent intensity of the sucrose gradient layer wells over the fluorescent intensity of PBS (negative control) containing wells. The sucrose gradient experiments (both Bottom-Up and Top-Down experiments) were also performed with the siKrasG12D iLiposomes and electroporated siKrasG12D siRNA. A total of three independent experiments were performed and the results from each of these experiments are shown in Extended Fig. 1F–G.
Electroporation of exosomes and liposomes
109 number of total exosomes (measured by Nanosight™ analysis) and 1 μg of siRNA or plasmid (for shRNA silencing) were mixed in 400 μl of electroporation buffer (1.15mM potassium phosphate pH 7.2, 25mM potassium chloride, 21% Optiprep). These exosomes were electroporated using a single 4 mm cuvette using a Gene Pulser Xcell Electroporation System (BioRad, catalog number 165–2081), as previously described45,46. The cuvette electrode plates are made of aluminum that allows for a uniform pulse delivery to the entire system. Briefly, after adding 400 μl of the RNAi-exosomes mixture to the cuvette, it was electroporated at 400V, 125μF and ∞ ohms, and the cuvette was immediately transferred to ice. Of note, when injecting multiple mice or using more than 109 exosomes and 1 μg of siRNA, a master mix of exosomes and siRNA is prepared in the electroporation buffer, and 400 μl of the mixture is aliquoted into each cuvette prior to electroporation. A similar procedure was performed using liposomes (100 nm, purchased from Encapsula Nano Sciences). After electroporation, exosomes were washed with PBS, as described above. After the wash, the exosomes are resuspended in PBS, and kept on ice and injected into the mice immediately. Following this wash step, the mice were dosed with, conservatively, 108 iExosomes per injection in 100 μl PBS volume. This dosage represents approximately 0.15 to 0.20 μg of exosomes protein, and mice thus received approximately 0.15–0.20 μg of exosomes protein every 48 hours. For in vitro transfection, exosomes and liposomes were electroporated and washed with PBS as described above, and 200,000 cells in a 6-well plate were treated with exosomes and liposomes for the indicated time as described for each assay and subsequently washed with PBS and used for further analysis. The siRNA sequence (sense strand 5′-GUUGGAGCUGAUGGCGUAGTT-3′, anti-sense 5′-CUACGCCAUCAGCUCCAACTT-3′) reflects a G to A nucleotide deviation from the wild-type Kras gene sequence (bold) to specifically target the Glycine to Aspartate amino acid substitution (KrasG12D) and include a TT nucleotide overhang to promote silencing efficiency, as described previously10,48,49. The central position of the nucleotide deviant in this KrasG12D siRNA enhances its specificity against the wild-type mRNA sequence. This was also labeled with an Alexa fluor 647 (AF647) fluorophore at the 3′ end on the sense strand to track its delivery. The siRNA was obtained from Qiagen (Cat. No.1027424). All Stars Negative siRNA (Scrambled siRNA) (1027287) was obtained from Qiagen. The siRNA sequences were also tagged with Alexa fluor 647. A second scrambled siRNA control was used (sense strand 5′-UUCUCCGAACGUGUCACGUTT-3′, anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′, Extended Fig. 4G) and the results were consistent with the scrambled siRNA from Qiagen. The KrasG12D shRNA sequence used was 5′CCGGGTTGGAGCTGATGGCGTAGTTCTCGAGCTACGCCATCAGCTCCAACTTTTTT-3′, and was flanked with Age1 and EcoR1 sequences to allow for cloning into the pLKO.1 vector, according to manufacturers protocol (Addgene). The shRNA sequence reflects a G to A nucleotide deviation from the wild type Kras gene sequence (bold) so that to specifically target the Glycine to Aspartate amino acid substitution in the KrasG12D mutation. Scrambled pLKO.1 shRNA was obtained from Addgene. For experiments performed on siRNA electroporated without exosomes, 1 μg of siRNA was added to 400 μl of electroporation buffer, and electroporated as described above. This mixture was then ultracentrifuged for 2 hours at 100,000g either by itself, or after it was mixed with 109 exosomes (RT and 37°C, in PBS for 30 minutes), and then used for further downstream assays. Notably, freshly prepared exosomes were used for every single assay reported in this manuscript, in both in vivo and in vitro experiments.
Immunogold Labeling and Electron Microscopy
Fixed specimens at an optimal concentration were placed onto a 300 mesh carbon/formvar coated grids and allowed to absorb to the formvar for a minimum of 1 minute. For immunogold staining the grids were placed into a blocking buffer for a block/permeabilization step for 1 hour. Without rinsing, the grids were immediately placed into the primary antibody at the appropriate dilution overnight at 4°C (monoclonal anti-CD9 1:10, Abcam). As controls, some grids were not exposed to the primary antibody. The next day, all of the grids were rinsed with PBS then floated on drops of the appropriate secondary antibody attached with 10nm gold particles (AURION, Hatfield, PA) for 2 hours at room temperature. Grids were rinsed with PBS and were placed in 2.5% Glutaraldehyde in 0.1M phosphate buffer for 15 minutes. After rinsing in PBS and distilled water the grids were allowed to dry and stained for contrast using uranyl acetate. The samples were viewed with a Tecnai Bio Twin transmission electron microscope (FEI, Hillsboro, OR) and images were taken with an AMT CCD Camera (Advanced Microscopy Techniques, Danvers, MA).
Flow cytometry analyses of exosomes
Exosomes from BJ Fibroblasts, BJ Fibroblasts over-expressing CD47, CD47 knockout mouse ear fibroblasts and WT-C57BL/6 mouse ear fibroblasts were isolated as described above and resuspended in 200 μL of PBS. Aldehyde/sulfate beads (10 μL, Life Technologies) were added to the solution and beads and exosomes mixture allowed to mix using a benchtop rotator for 15 minutes at room temperature. PBS (600 μL) was then added to the solution and mixing was continued overnight at 4°C. 1M Glycine (400 μL) was added and mixing was continued for 1 hour at room temperature. The mixture was then spun down at 12,000rpm at RT for 1 minute. The precipitate was then resuspended in 100 μL of 10% BSA in PBS, and mixed for 45 minutes at room temperature. The mixture was spun down at 12,000rpm for 1 minute at RT and the supernatant aspirated. The beads with the exosomes attached (pellet) were then resuspended in 40 μL of 2% BSA in PBS, and split equally into two tubes: one for staining for CD47, CD63 or CD81, the other for control (secondary antibody only). The exosomes bound to beads were then incubated with 1μL of anti-CD47 antibody (for mouse: BD biosciences, catalog no. 556045; for human: eBiosciences, catalog no. 14–0479) or 3μL of anti-CD63 (for mouse: Santa Cruz Biotech catalog no. SC-31211; for human: BD biosciences, catalog no. 556019) or anti CD-81 antibody (for human: BD Biosciences, catalog no. 555675) in 20 μL volume, and mixed at RT for 30 minutes. The mixture was then centrifuged at 12,000 rpm for 1 minute at RT, the supernatant aspirated, and the pellet resuspended in 20 μL of 2% BSA in PBS. Secondary antibodies were then added and the samples were mixed at RT for 1 hour. The samples were then centrifuged at 12,000 rpm for 1 minute at RT, supernatant aspirated, and pellet resuspended in 200 μL of 2% BSA in PBS. The exosomes bound to the beads were washed three times with 2% BSA in PBS. CD47, CD63 and CD81 detection on the beads was analyzed using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples.
Flow cytometry analysis for exosomes and liposomes biodistribution
Exosomes and liposomes were labeled with PKH67 (Sigma Aldrich) according to the manufacturer’s protocol. Alternatively, the exosomes and liposomes were electroporated with AF-647 tagged RNAi prior to injection in mice. These were then injected i.p. into C57BL/6 or Nude mice. Plasma was then obtained from these mice at the listed time points following injection of either exosomes or liposomes. The plasma was then diluted in 11 mL PBS and filtered through a 0.2 μm pore filter. Subsequently, the samples were then ultracentrifuged overnight at 150,000g at 4°C. The pellet was then washed with PBS, and followed by a second step of ultracentrifugation at 150,000g for 2 hours at 4°C. The samples were then resuspended in 200 μL of PBS. Aldehyde/sulfate beads (10 μL, Life Technologies) were added to the solution and the beads and exosomes/liposomes mixture was allowed to mix using a benchtop rotator for 15 minutes at room temperature. PBS (600 μL) was then added to the solution and mixing was continued overnight at 4°C. 1M Glycine (400 μL) was added and mixing was continued for 1 hour at room temperature. The mixture was then spun down at 12,000rpm at RT for 1 minute, supernatant aspirated, and pellet resuspended in 200 μL of 2% BSA in PBS. The exosomes/liposomes bound to the beads were washed three times with 2% BSA in PBS. FITC or APC+ beads were analyzed using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples.
Flow cytometry analysis for binding efficiency of CD47 neutralizing antibody on exosomes
Exosomes from BJ fibroblasts were isolated as described above, and then incubated with 10μg/mL of anti-CD47 neutralizing monoclonal antibody (Bio-Xcell, B6H12 or 2D3 antibodies, as specified) for 1 hour at either room temperature or 37°C, or overnight at 4°C, and then bound to aldehyde sulfate beads as described above. 1M Glycine (400 μL) was added and mixing was continued for 1 hour at room temperature. The mixture was then spun down at 12,000rpm at RT for 1 minute. The samples were washed with 2% BSA, and secondary antibody (Alexa 488) was then added and the samples were mixed at RT for 1 hour. The samples were then centrifuged at 12,000 rpm for 1 minute at RT, supernatant aspirated, and pellet resuspended in 200 μL of 2% BSA in PBS. The exosomes bound to the beads were washed three times with 2% BSA in PBS. Alexa 488 positive beads were then analyzed using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples.
Flow cytometry analysis for comparison of binding efficiency of exosomes and liposomes to aldehyde sulfate beads (Extended Fig. 2A)
Exosomes and liposomes were electroporated with A647 siRNA as described above. The samples were then resuspended in 200 μL of PBS. Aldehyde/sulfate beads (10 μL, Life Technologies) were added to the solution and the beads and exosomes/liposomes mixture was allowed to mix using a benchtop rotator for 15 minutes at room temperature. PBS (600 μL) was then added to the solution and mixing was continued overnight at 4°C. 1M Glycine (400 μL) was added and mixing was continued for 1 hour at room temperature. The mixture was then spun down at 12,000rpm at RT for 1 minute, supernatant aspirated, and pellet resuspended in 200 μL of 2% BSA in PBS. The exosomes/liposomes bound to the beads were washed three times with 2% BSA in PBS. A647+ beads were analyzed using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples.
Visualization of exosomes biodistribution in the tissue
Exosomes were labeled with PKH67 (Sigma Aldrich) according to the manufacturer’s protocol. Alternatively, the exosomes were electroporated with AF647 tagged RNAi prior to injection in mice. These were then injected i.p. into C57BL/6 mice. The specific organs were obtained from these mice at the listed times post injection and then were frozen. Sectioned tissue was stained with DAPI nuclear stain, and images were then captured using Zeiss Observer Z1 inverted microscope. Images were quantified by counting the number of nuclei that had PKH67 labeled/AF647 labeled exosomes surrounding it (PKH67/AF647 positive cells) and divided by the total number of nuclei, in five random visual fields per organ (400x). For evaluation of the entry of exosomes in the various pancreas structures, exosomes electroporated with AF647 tagged siRNA were injected i.p into 26-day old KTC mice. The pancreas of these mice was then harvested 24 hours later, mounted in O.C.T. compound and frozen. Sectioned tissue was stained with DAPI nuclear stain, and the images were then captured using Zeiss Observer Z1 inverted microscope. Images were quantified by counting the number of nuclei within a particular structure (Islet, Acinus, Duct, CAF, Tumor) that had AF647 labeled exosomes surrounding it (AF647 positive cells) and dividing by the total number of nuclei within that structure (400x). All control samples were run side by side with experimental samples.
Real-time PCR Analyses
Cells were incubated with iExosomes for 3 hours, after which RNA was retro-transcribed with MultiScribe Reverse Transcriptase (Applied Biosystems) and oligo-d(T) primers following total RNA purification with Trizol (Invitrogen), according to the manufacturer’s directions. Real-time PCR analyses were performed on an ABI PRISM 7300HT Sequence Detection System Instrument using SYBR Green Master Mix (Applied Biosystems). The transcripts of interest were normalized to 18S transcript levels. Primers for KrasG12D were designed as described in50, KrasG12C/V were designed as described in43, and Kras WT primers were designed as described in51. Each reaction included three technical replicates, which were averaged to define one biological replicate. The experiments were repeated three times on distinct days and each experiment defined a biological replicate. Statistical analyses were performed on dCt of biological replicates (mice or independent experiments) and the results expressed as relative fold change. Forward primer sequence for KRASG12D(hu): F-5′-ACTTGTGGTAGTTGGAGCAGA-3′. Reverse primer sequence for KRASG12D(hu): R-5′-TTGGATCATATTCGTCCACAA-3′. Forward primer sequence for KRASG12D(Mo): F-5′-ACTTGTGGTGGTTGGAGCAGC-3′. Reverse primer sequence for KRASG12D(Mo): R-5′-TAGGGTCATACTCATCCACAA-3′. Forward primer sequence for KRASWT(hu): F-5′-ATTGTGAATGTTGGTGT-3′. Reverse primer sequence for KRASWT(hu): R-5′-GAAGGTCTCAACTGAAATT-3′. Forward primer sequence for 18S: F-5′-GTAACCCGTTGAACCCCATT-3′. Reverse primer sequence for 18S: R-5′-CCATCCAATCGGTAGTAGCG-3′. Forward primer sequence for KRASG12V(Hu): F-5′-ACTTGTGGTAGTTGGAGCAGT-3′. Reverse primer sequence for KRASG12V(Hu): R-5′-TTGGATCATATTCGTCCACAA-3′. Forward primer sequence for KRASG12C(Hu): F-5′-AACTTGTGGTAGTTGGAGATT-3′. Reverse primer sequence for KRASG12C(Hu): R-5′-TTGGATCATATTCGTCCACAA-3′.
In some experiments, the exosomes were subjected to a variety of treatments as described below, prior to treatment of Panc-1 cells:
siKrasG12D iExo: Panc-1 cells treated with siKrasG12D iExo (BJ derived exosomes).
Media Exo: FBS-depleted culture medium was incubated without cells for 48hrs at 37°C, and then processed as to collect exosomes. The ultracentrifuged pellet was electroporated with siKrasG12D iExo as performed in the siKrasG12D iExo group.
siRNA: Panc-1 cells treated with siKrasG12D siRNA (no exosomes, no electroporation).
siRNA (E): Panc-1 cells treated with siKrasG12D siRNA that was electroporated (‘E’).
Exo (E): Panc-1 cells treated with just BJ derived exosomes were electroporated (‘E’) without siKrasG12D.
siRNA + Exo: Panc-1 cells treated with BJ derived exosomes and siKrasG12D added to the wells of cells concurrently.
siRNA (E) + Exo: Panc-1 cells treated with BJ derived exosomes that were mixed with electroporated siKrasG12D.
siRNA + Exo (E): Panc-1 cells treated with electroporated BJ derived exosomes that were mixed with siKrasG12D.
siRNA (E) + Exo (E): Panc-1 cells treated with electroporated BJ derived exosomes that were mixed with electroporated siKrasG12D.
Scramble iExo: Panc-1 cells treated with BJ derived exosomes that were electroporated with siScramble siRNA (from Qiagen, as described above).
Scramble (2) iExo: Panc-1 cells treated with BJ derived exosomes that were electroporated with a distinct siScramble siRNA (target sequence: AATTCTCCGAACGTGTCACGT).
All control samples were run side by side with experimental samples.
MTT, TUNEL and flow cytometry apoptosis assay
Panc-1, BxPC-3, Capan-1 and MIA PaCa-2 cells were seeded in a 96-well plate (1,000 cells/well) and allowed to seed for 24 hours, after which they were treated with exosomes electroporated with KrasG12D siRNA, KrasG12D shRNA, scrambled siRNA, scrambled shRNA, PBS or non-electroporated control exosomes. Treatment was given only once at the beginning, post seeding of cells. Subsequently, every 24 hours, MTT reagent (tetrazole, Sigma Aldrich) was added to the cell culture media for 3 hours at 37°C. The supernatant was then discarded, cells washed with PBS, and lysed with dimethyl sulfoxide to dissolve the formazan product. Absorbance was measured at an optical density of 562 nm in a spectrophotometric plate reader. In these MTT experiments, each treatment (e.g. iExosomes) were aliquoted into 5 partitions, and each partition was used to treat 3 wells of cells. The triplicate wells were averaged to define n=1 partition and each treatment thus totaled n=5 partitions. The MTT assay for Panc-1 and BXPC-3 was repeated again under the exact same conditions, as independent experiments (Extended Fig. 4). For TUNEL assay, cells were treated with iExosomes for 24 hours, and apoptosis measurement by TUNEL was assessed using the In Situ Cell Death Kit, TMR red (Roche), according to the manufacturer’s directions. The cells were fixed with 4% PFA at room temperature for 20 minutes, and SYTOX green nucleic acid stain (Invitrogen, 1:10,000 in PBS for 10 minutes at room temperature) or DAPI were used to delineate the nuclei. Images were taken by Zeiss LSM 510 confocal microscope, quantified by counting the number of cells with TUNEL positivity per visual field (400x), and the results were expressed as the percentage of cells with positive label out of the total number of cells counted per visual field. The TUNEL assay was repeated again under the exact same conditions, as independent experiments (Extended Fig. 4). For flow cytometry analysis of apoptosis in Panc-1 cells, Panc-1 cells were treated with iExosomes or scramble iExosomes for 24 hours, and apoptosis and dead cells were measured by LIVE/DEAD fixable aqua (ThermoFisher, L34957) and propidium idodide (5 μL of a 50μg/ml stock solution per reaction (from BD Biosciences, 556547), according to the manufacturers instructions. This was then analyzed by using the LSR Fortessa X-20 cell analyzer. All control samples were run side by side with experimental samples.
Visualization and quantification of Alexa Fluor 647/CM-DiI in cells treated with exosomes or liposomes
Exosomes isolated from BJ fibroblasts, CD47 knockout fibroblasts, and WT C57BL/6 fibroblasts were electroporated with Alexa fluor 647 tagged siRNA and treated with PBS, proteinase K, or trypsin (Life Technologies, 10X, 15 minutes at room temperature and ultracentrifuged with PBS for 2 hours at 4°C), were washed with PBS for 2 hours, and then added to Panc-1 cells cultures on glass coverslips for 3 hours. For staining of exosomes with CM-DiI dye (ThermoFisher), isolated exosomes were resuspended in 1 mL PBS, and 2 μL (1:500) of CM-DiI dye was added, after which the mixture was incubated at 37°C for 5 minutes, and then at 4°C for 10 minutes. This was then ultracentrifuged with PBS for 2 hours, and then added to Panc-1 and BxPC-3 cells on glass coverslips for 3 hours. The cells were then fixed by washing with cold PBS and incubating with 4% PFA at room temperature for 20 minutes. The cells were then washed with PBS, incubated with 0.05% Triton X for 10 minutes, washed with PBS and stained with Sytox green nuclear stain (Invitrogen) or DAPI. The coverslips were then mounted on to glass slides with mounting media. Accumulation of Alexa Fluor 647/CM-DiI was visualized using Zeiss Observer Z1 inverted microscope. The number of cells with Alexa Fluor 647/CM-DiI labels was counted per visual field (x400) and the results were expressed as the percentage of cells with positive label out of the total number of cells counted per visual field. All control samples were run side by side with experimental samples.
Quantification of Loading Efficiency within Exosomes/Liposomes by RT-PCR
109 exosomes or liposomes were electroporated with 1 μg siRNA as described above. When stated, the electroporated exosomes/liposomes were proteinase K treated and RNAse A treated (as described above). Specifically, when both treatments were required, they were performed sequentially. The samples were first proteinase K (PK) treated, the PK was inactivated, then the samples were washed with PBS and spun down using ultracentrifugation. The resuspended, PK-treated exosomes were then RNAse A treated, then the RNAse was inactivated, and the exosomes were washed with PBS and spun down using ultracentrifugation. We also treated exosomes with 1% Triton X-100 prior to RNAse A treatment. Briefly, exosomes were subjected to treatment with 1% Triton X-100 for 30 minutes at 37°C, after which RNAse A was added. One microgram of siRNA was used as input, and 1 μg siRNA was also used for RNAse A treatment following an identical procedure as listed above. Control exosomes consisted in non-electroporated exosomes. All samples were mixed with 500 μl of TRIzol reagent, and 200 μl of chloroform was added to the mixtures. The aqueous phase was recovered following 15 minutes of centrifugation at 10,000g at 4°C. The aqueous phase, 200 μl for each sample, was then mixed with 250 μl of 100% ethanol and bound to filters provided in the Total Exosomes RNA and Protein Isolation Kit (Invitrogen, catalog number 4478545). The protocol to purify the RNA was then followed according to the manufacturer’s directions. A total of 100 μl of eluted RNA for each sample was obtained. The Custom TaqMan® Small RNA Assay kit was purchased (Applied Biosystems) to specifically detect the sense strand of the KrasG12D siRNA and the manufacturer’s protocol was followed, using 5 μl of RNA template for the reverse transcription (RT) reaction, and 1.33 μl of 1:1000 diluted RT reaction product for the qPCR. The reactions were also performed by diluting the electroporated exosomes 1:1000 prior to proceeding with the described treatments, in which case the RT reaction product was not diluted. The RNA was extracted as described above, RT reaction performed as described above, and 1.33 μl of the RT reaction product was used for the pPCR. qPCRs were run with technical duplicates. The RT reaction product of the siRNA input sample was also further diluted 1:2 and 1:4 fold to establish a standard curve. No template control were included in the qPCR reaction and showed no detectable signal. Each exosomes and liposomes samples was prepared in triplicates, consisting in 3 independent preparations of exosomes/liposomes electroporation. The average 1/Ct and standard deviation of the 3 independent experiments is presented. All control samples were run side by side with experimental samples.
Protein identification by nano Liquid Chromatography coupled to tandem Mass Spectrometry (LC-MS/MS) analysis
Exosomes extraction was performed by ultracentrifugation (Beckman Optima XE 100) 100,000g, overnight, 4ºC, followed by two washing steps with NaCl 0.9% (saline). Protein extraction was done using a solution of 8M/2.5% SDS-Urea (Sigma), cComplete (Roche) and PMSF (Sigma), for 30 minutes on ice followed by centrifugation 17,000g for 30 minutes. Proteins were present in the supernatant. T3M4 CD24+CD44+ derived exosomes protein was precipitated using methanol/chloroform methodology and quantified with PIERCE 660nm. A total of 40 μg of protein was used for the analysis. The sample digestion was performed overnight using trypsin solution. The digestion product was purified with SEP-PAK C18 cartridge. For the analysis, 1 μg of peptides were subjected to nano liquid chromatography (Eksigent Technologies nanoLC Ultra 1D plus, AB SCIEX, Foster City, CA) coupled to high-speed Triple TOF 5600 mass spectrometer (AB SCIEX, Foster City, CA) with a Nanospray III source (1 technical replicate). The mass spectrometry data obtained was analyzed using Mascot Server v. 2.5.0 (Matrix Science, London, UK) as search engine against Homo sapiens database (including also the decoy database). The confidence interval for protein identification was set to ≥ 95% (p<0.05) and only peptides with an individual ion score above the 1% False Discovery Rates (FDR) threshold were considered correctly identified.
Western Blot
To deduce the protein levels in cell lysates after 24 hours of treatment with exosomes, Panc-1 cells were homogenized in RIPA lysis buffer and protein lysates were normalized using Bicinchoninic Acid (BCA) protein assay kit (Pierce, Thermo Fisher Scientific). Twenty micrograms of protein lysates were loaded onto acrylamide gels for electrophoretic separation of proteins under denaturing conditions and transferred onto PVDF membranes (ImmobilonP) by wet electrophoretic transfer. The membranes were then blocked for 1 hour at room temperature with 5% non-fat dry milk in PBS with 0.05% Tween-20, and incubated overnight at 4°C with the following primary antibodies: anti-rabbit p-Erk-p44/p42 MAPK (Erk1/2) (Thr202/Tyr 204) (Cell Signaling, 4376, 1:1,000), anti-rabbit Vinculin (Abcam, 129002, 1:10,000). Secondary antibodies were incubated for 1 hour at room temperature. Washes after antibody incubations were done with an orbital shaker, three times at 15 min intervals, with PBS containing with 0.05% Tween-20. Membranes were developed with chemiluminescent reagents from Pierce, according to the manufacturer’s directions. Supplementary Fig. 1 shows uncropped western blots from data presented in Extended Fig. 4H. The quantifications were performed on two independent experiments (n=2) with uncropped western blots shown in Supplementary Fig. 1. Western blots were quantified by ImageJ software, wherein the p-ERK peak intensity values (selecting both bands that represent p-Erk1 and p-Erk2) were normalized to those of vinculin (selecting the ~124 kDa band), in each blot. All control samples were run side by side with experimental samples.
RAS Binding Assay
Lysates from cells treated with iExosomes and controls were isolated according to the manufacturers instructions (Cytoskeleton, BK008), and subsequently the GTP bound vs. GDP bound RAS activity in Panc-1 and BxPC-3 cells was measured using the Ras pulldown activation assay kit (Cytoskeleton, BK008), and using western blotting as the final readout. The scanned film saturation was uniformly set to 0 using the format picture tool in PowerPoint and the cropped blots are shown in Extended Fig. 4I; and the uncropped, unmodified western blots are shown Supplementary Fig. 1. All control samples were run side by side with experimental samples.
Mice
Female athymic nu/nu mice (Charles Rivers) between 4 to 6 weeks of age were housed in individually ventilated cages on a 12 hours light-dark cycle at 21 to 23ºC and 40% to 60% humidity. Mice were allowed free access to an irradiated diet and sterilized water. Under general anesthesia, Panc-1, BxPC-3 (106 cells in 10μl PBS) or KPC689 cells (5×105 cells in PBS) were injected into the tail of the pancreas using a 27-gauge syringe. For detection of luciferase expression, the mice were injected i.p. with 100mg/kg of body weight of luciferin (200 μl of 10mg/ml luciferin in PBS) 12–15 minutes before imaging, anesthetized with isoflurane, and imaged using IVIS (Xenogen Spectrum). For tumor burden analyses, Living Image version 4.4 (Caliper Life Sciences) was used to quantify all tumors. A circular region of interest (ROI) around the pancreas and tumor was set within the same experimental groups. In addition, exposure conditions (time, aperture, stage position, binning) were kept identical for all measurements within each experiment. Tumor measurements for average radiance (p/sec/cm2/sr) or total flux (wherever mentioned, p/sec) were obtained under the same conditions for all experimental groups. All IVIS imaging analyses were ascertained by two independent experimentalists, one of which was blinded to the treatment groups. The mice were imaged regularly and randomly divided into groups for treatments. The mice were monitored for sign of distress daily, and two of the 3 experimentalists monitoring health status were blinded to the treatment groups. Mice received 108 exosomes or liposomes i.p. in 100 μl volume of PBS every other day. Exosomes or liposomes were electroporated with 1 μg of siRNA (Alexa 647 tagged siRNA), or shRNA, or were pre-treated with proteinase K and/or RNase A as described above, or were mixed with electroporated siRNA (RT and 37°C), and washed with PBS prior to injection. When using KTC (Ptf1acre/+;LSL-KrasG12D/+;Tgfbr2Lox/Lox)25 genetically engineered mice, exosomes treatment was initiated at 18 (early) or 33 (late) days of age. For exosomes biodistribution studies, adult C57BL/6 mice were injected i.p. with exosomes labeled with PKH67 (Sigma) or exosomes electroporated with AF647. For the KPC689 orthotopic study, 5×105 of KPC689 cells were injected orthotopically in the tail of the pancreas of adult female C57BL/6 mice (Jackson Laboratory). These mice were then imaged by IVIS or by Magnetic Resonance Imagining (MRI) 20 days post tumor cell induction. Treatment with exosomes (siKrasG12D iExo and siScramble iExo) was started on day 16 (early) and day 32 (late)-post tumor cell induction, and continued every other day. Another MRI was additionally performed on 48 days post tumor cell induction. MRI was performed and analyzed as previously described42. For experiments aimed to neutralize CD47, 10μg/mL of anti-CD47 neutralizing monoclonal antibody (Bio-Xcell, B6H12 or 2D3 antibodies, as specified) was incubated with either exosomes or liposomes for 1 hour at room temperature, washed with PBS by ultracentrifugation as described above, and injected into the mice. Treatment with both CD47 monoclonal antibody and iExosomes/iLiposomes along with controls was performed every other day. Treatment of KPC (Pdx1cre/+;LSL-KRasG12D/+;LSL-Trp53R172H/+) was started when the mice reached 100 days of age. Due to the variability of the model, the mice were subjected to MRI prior to treatment start, to determine baseline tumor size, and subsequently grouped into siKrasG12D iExo or siScrbl iExo groups. For gemcitabine studies, a 50mg/kg dosage, administered every 48 hours intraperitoneally was used. At time of necropsy or euthanasia, gross observation of the metastatic burden and measure of primary tumor burden was performed in a blinded fashion: the experimentalist performing the tissue collection, recording of disease burden and metastasis, was blinded to the treatment group. Blood urea nitrogen (BUN), aspartate transaminase (AST) and alanine transaminase (ALT) analyses were performed using plasma (collected using heparin) and BioAssay Systems blood chemistry assay kits (catalog DIUR-100, EASTR-100 EALT-100 respectively), following the manufacturer’s specifications. In all orthotopic mouse models (Panc-1, BxPC-3, KPC689), all control groups were treated side by side with the experimental groups. For KTC and KPC GEMM, mice were enrolled randomly into control or experimental treatment groups when they became available and reached the stated age for treatment start, however, care was taken to ensure, whenever possible, that mice were enrolled into both control and experimental groups side by side. For KPC mice, baseline MRI confirmed the presence of tumor, following which mice were enrolled into either control or experimental groups at 100 days of age, and control and experimental treatment were administered side by side when the treatment windows overlapped during the experiment. All animal procedures were reviewed and approved by the Institute for Animal Care and Use Committee at UT MDACC.
Histological analyses
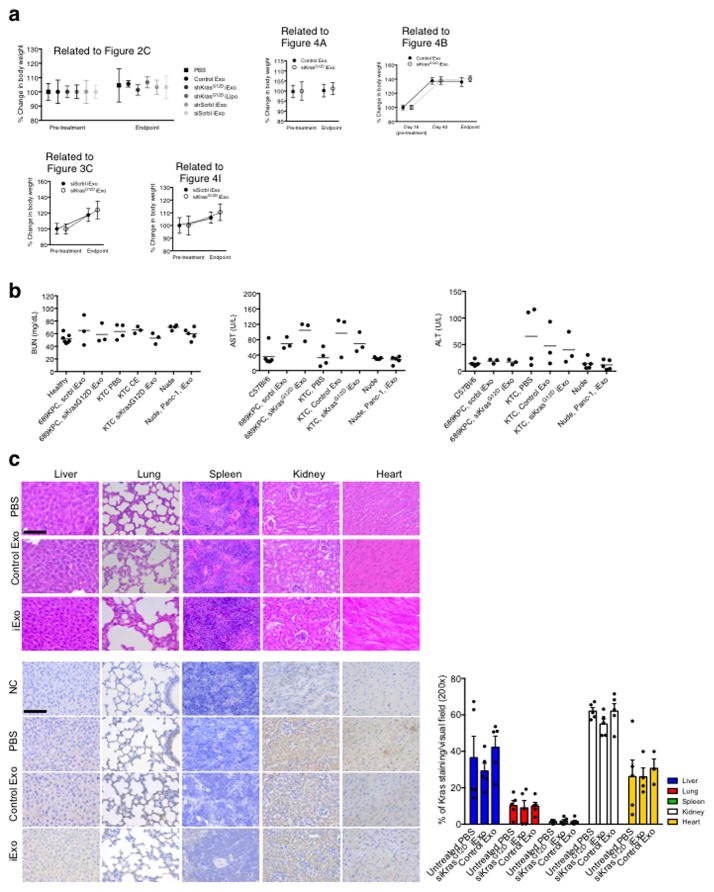
Tissues were fixed in formalin and processed for paraffin embedding. Tissue sections of 5 μm thickness were cut and stained for haematoxylin and eosin (H&E) and Masson’s trichrome (MTS) (Leica). For histopathological scoring, H&E stained slides were scored based on the morphological stages of pancreas cancer: normal, pancreatic intraepithelial neoplasia (PanIN) and pancreatic ductal adenocarcinoma (PDAC). For each tissue section, a percentage score for each of the three stages (Normal, PanIN, PDAC) were performed in a blinded fashion. Specifically, at least two (three in some experiments) experimentalists evaluated the slides. They each performed their analyses independently from one another, and one of the two, or two of the three, experimentalists were blinded to the treatment groups. All three experimentalists returned identical conclusions and the scores were averaged for each stages for each mouse. Note that while small foci of cancer cells can be seen in the shKrasG12D iExo treated pancreas, the vast majority of the pancreas was histologically unremarkable. For the analysis of fibrosis in mice, six 200× visual fields were randomly selected for each MTS stained pancreas section and fibrosis was quantified using a grid intersection analysis with Adobe Photoshop. For each image evaluated, a grid of a 100 squares was overlapped on each picture, and each intersection was counted for blue (fibrotic area) and purple/red (non-fibrotic area). A percentage score was then obtained for each tissue section. Tissue sections were also subjected to antigen retrieval (15 minutes in 10nM citrate buffer at pH6 and 98°C) prior to immunostaining. The tissue sections were incubated with 4% CWFS gelatin (Aurion) in either TBS or PBS, 1 hour prior to overnight incubation with the primary antibodies. The following primary antibodies were used for staining: anti-rabbit p-Erk-p44/p42 MAPK (Erk1/2) (Thr202/Tyr 204) (IHC, Cell Signaling, 4376, 1:400), anti-rabbit p-AKT-Anti AKT1 (phospho S473) (IHC, Abcam, ab81283, 1:100), anti-rabbit Ki-67 (IHC, Thermo Scientific, RM-9106-S, 1:400) and conjugated anti-actin α-SMA-Cy3 (IF, Sigma, C6198, 1:100). For CK19 p-ERK co-staining, the primary antibodies used were anti-rabbit CK19 (IF, Abcam, 52625) and anti-rabbit p-ERK p44/p42 MAPK (Erk1/2) (IF, Cell Signaling, 4370). For IHC, the sections were incubated with biotinylated goat anti-rabbit and streptavidin HRP (Biocare Medical), each for 10 minutes, and counterstained with haematoxylin. DAB positivity was analyzed. Note that the quantification was performed on measurably smaller tumor areas in the siKrasG12D iExo treated group compared to large tumor area in control group. This was performed on at least five, and up to eight 200× pictures per tissue section, and an average of relative percent positive score was obtained for each tissue section. Ki-67 staining was quantified by counting the number of positively stained nuclei, per visual field (400x), whereas p-Erk, p-AKT, and α-SMA staining was quantified with ImageJ to define a positively stained area, which was then expressed as a percentage of positively stained area to the total image area. Quantification of CK19 and p-ERK co-immunolabels was performed on 8 random 400× images per tissue section, and inserted into FIJI by Image J co-localization software. TUNEL assay was performed using the In situ cell death detection kit, TMR Red (Roche), according to the manufacturer’s directions. Alexa fluor 647 was detected on frozen tissue sections by staining the nuclei of the tissue with SYTOX green (1:10,000 in PBS for 10 minutes). Images were taken by Zeiss LSM 510 confocal microscope, and quantified by counting the number of cells with TUNEL positivity per visual field (400x) and the results were expressed as the percentage of cells with positive label out of the total number of cells counted per visual field. PKH67 labeled exosomes or AF647 electroporated exosomes were also injected into mice 3 hours prior to euthanasia, and the pancreas were fixed, processed and sectioned as described above. Sections were mounted on slides, the nuclei stained with DAPI, and the pancreas sections imaged using the Zeiss Observer Z1 inverted microscope. PKH67/AF647 positive cells were counted in each 400× visual field and differentiated according to tumor or normal peritumoral cells based on nuclear staining characteristics. For the exosomes biodistribution studies, the number of PKH67 positive cells was counted in 5 random 400× visual fields in the brain, G.I. Tract, kidney, liver, lung, pancreas and spleen from 3 mice. The results were expressed as the percentage of cells with positive label out of the total number of cells counted. For pancreas structure quantification, the number of AF647 positive cells was counted in each 400× visual field in the pancreas, and the results were expressed as the percentage of cells with positive label out of the total number of cells counted per visual field. Representative pictures of the structures were taken accordingly. For Kras staining, pancreas tumor, liver, lung, kidney, spleen and heart, 5 μm thick sections from formalin fixed, paraffin embedded tissues were processed for antigen retrieval (2 repeats of 15 minutes microwave based antigen retrieval using sodium citrate buffer (10mM sodium citrate, 0.05% Tween 20, pH 6.0)), then incubated at room temperature for 15 minutes with 3% H2O2 in methanol. The sections were washed in TBS, blocked with Rodent Block M solution (Biocare) for 30–45 minutes at room temperature, then incubated with 1:10 dilution of Kras antibody (ThermoFisher, 414700, clone 9.13) in 3% BSA containing PBS diluent, overnight at 4°C or at room temperature for 4 hours. The slides were then processed for secondary antibody application and DAB based development using Biocare’s MACH 4 universal HRP-polymer reagents, according to the manufacturer’s recommendation. Analyses for comparative DAB positivity was performed using ImageJ software by designing a macros to define a positively stained area, which was then expressed as a percentage of positively stained area to the total image area. Each organ had a unique macros programmed for quantification. All control samples were run side by side with experimental samples.
Quantification of the number of exosomes from the plasma of mice
CD47 knockout mice (CD47 k/o) vs. WT C57BL/6 mice were retro-orbitally bled using heparin and the plasma was isolated. 300 μl of plasma per mouse was then diluted in 11 mL PBS and filtered through a 0.2 μm pore filter. Subsequently, the samples were then ultracentrifuged overnight at 150,000g at 4°C. The pellet was then washed with PBS, and followed by a second step of ultracentrifugation at 150,000g for 2 hours at 4°C, after which the total number of exosomes in the plasma of the mice was measured by NanoSight™ analysis. All control samples were run side by side with experimental samples.
Macrophage clearance
Immunocompetent C57BL/6 mice between the ages of 10 and 14 weeks were injected i.p. with either exosomes or liposomes containing Alexa fluor 647 tagged siRNA. The blood of these mice was collected 3 hours post injection and processed for flow cytometry analyses. Red blood cells were depleted using ACK lysis buffer (Invitrogen), and the peripheral cells were blocked with FC block (1:1000, BD Pharmingen), stained with Live/Dead Aqua dye (1:200, Life technologies, 405nm) anti-CD11b (1:200, BD Pharmingen, PerCP/Cye 5.5) and anti CD172a (1:200, BD Pharmingen, FITC) antibodies for 30 minutes, washed with PBS, and analyzed using the LSR Fortessa X-20 cell analyzer (UT MDACC flow cytometry core facility). Immunocompetent C57BL/6 mice were also i.p. injected with exosomes that were electroporated with Alexa fluor 647-tagged siRNA and incubated with 10μg/mL of CD47 neutralizing monoclonal antibody (Bio-Xcell, B6H12 or 2D3 antibodies) for 1 hour at RT. The blood of these mice was collected 3 hours post injection and processed for flow cytometry analyses as described above. All control samples were run side by side with experimental samples.
Macropinosome visualization and quantification21
Fifty thousand cells (Panc-1 and BxPC-3) were seeded onto glass coverslips, and 24 hours after seeding the cells, they were serum starved for 18 hours. For 5-(N-ethyl-N-isopropyl) amiloride (EIPA, Sigma Aldrich) treatment, cells were pre-treated with 5μM, 25μM or 75μM EIPA for 30 minutes at 37°C. DMSO was used as a vehicle. Cells were then incubated with exosomes or liposomes, labeled with PKH67 (Sigma Aldrich) for 3 hours at 37°C. Macropinosomes were detected using a high molecular mass TMR-dextran (Invitrogen), wherein TMR dextran is added to the serum free media at a concentration of 1mg/mL for 30 minutes at 37°C. At the end of the incubation period, cells were rinsed 5 times with cold PBS and fixed with 4% PFA. Cells were then stained with DAPI nuclear stain, and then coverslips were mounted onto the slides. Images were then captured using Zeiss Observer Z1 inverted microscope, and at least three fields from at least three to five separate wells were randomly selected across each sample, and analyzed using the ‘Analyze Particles’ feature on Image J, according to Commisso C. and colleagues21. The particle density was then expressed as the relative number of macropinosomes. Briefly, the ‘macropinocytic index’ was computed by determining the total macropinosome area in relation to the total cell area for each field, and then determining the average across all fields. A detailed protocol to analyze and calculate the amount of macropinocytosis within a sample is listed in21. A similar quantification was performed using PKH67 label and the result was expressed as the relative number of exosomes or liposomes. The macropinocytosis assay was repeated again as an independent experiment in Extended Fig. 7. All control samples were run side by side with experimental samples.
Statistical analyses
Statistical analyses used are detailed in the figure legends. One-way ANOVA or unpaired two tailed student’s t test were used to establish statistical significance using GraphPad Prism (GraphPad Software). For survival analyses, Kaplan-Meier plots were drawn and statistical differences evaluated using the Log-rank (Mantel-Cox) test. A P value < 0.05 was considered statistically significant.
Data availability
Source data for all figures are provided with the paper and reagents will be provided upon availability and reasonable request.
Extended Data
Extended Figure 1. Exosomes purification and siRNA loading.
(A) Exosomes and liposomes numbers and size distribution-using NanoSight™. (B) Transmission electron micrograph of exosomes and stained for CD9 by immunogold (left panel: 2ary antibody only), scale bar: 100nm. (C) FC analyses for CD63 and CD47 on exosomes (n=3 distinct exosomes isolations). (D) FC analyses and quantification of exosomal proteins CD63 and CD47 in liposomes. (E) Schematic representation of electroporation of RNAi into exosomes. (F) Schematic and fluorescence intensity plot of sucrose gradient layers (from the “Bottom-Up” method, UC: ultracentrifuge). Results from three independent experiments are shown. (G) Schematic and fluorescence intensity plot of sucrose gradient layers (from the “Top-Down” method, UC: ultracentrifuge). Results from three independent experiments are shown. The data is presented as the mean ± SEM. FC: Flow cytometry. See accompanying source data.
Extended Figure 2. Tissue distribution and clearance of iExosomes.
(A) FC analyses and quantification of the comparison of the binding efficiency to aldehyde sulfate beads, n=3 distinct batches of exosomes and liposomes (B) FC analyses and quantification of AF647-tagged RNAi containing exosomes and liposomes isolated from the plasma of C57BL/6 (n=3 mice) and Nude (Nu/nu) mice (n=3 mice), 24 hours post injection. (C) FC analysis plots (from data shown in Fig. 1B) of exosomes with AF647 tagged siRNA in the circulation of mice. (D) Representative micrographs of the indicated organs of mice injected i.p. with PKH67 labeled BJ fibroblast exosomes (n=3 mice). Quantification is shown in Fig. 1C. (E–F) FC analyses of pancreas cells 6 hours (E) following injection of siKrasG12D Exos (quantification shown in Fig. 1D) and 24 hours (F) following injection of siKrasG12D Exos. The data is presented as the mean ± SEM. One-way ANOVA was used to determine statistical significance. **** p<0.0001. FC: Flow cytometry. See accompanying source data.
Extended Figure 3. CD47 induced monocyte clearance and iExosomes characterization.
(A) Schematic representation of gating strategy for data shown in Fig 1E. (B) FC analysis of CD11b+ cells in the circulation, liposomes (n=7 mice), exosomes (n=7 mice), Untreated mice (n=4). (C) Representative dot plots from Fig. 1E. (D) FC analyses of SIRP-α (CD172a) expression from Alexa 647+/CD11b+ monocytes. (E) FC analyses of the binding efficiency of CD47 neutralizing antibodies to exosomes (n=3 distinct batches of exosomes). (F) Quantification of the number of exosomes/mL in the plasma of WT C57BL/6 mice (n=5) vs. CD47 knockout mice (n=7), unpaired two-tailed t test. The data is presented as the mean ± SEM. Unless otherwise stated, one-way ANOVA was used to determine statistical significance. * p<0.05, *** p<0.001. FC: Flow cytometry. See accompanying source data.
Extended Figure 4. iExosomes specifically target KrasG12D expression.
(A) KRASG12D transcript levels in Panc-1 cells (n=3 independent experiments). (B–C) 1/Ct values from RT-PCR analysis under the listed conditions, to determine the loading efficiency of siRNA. Standards (siKrasG12D, 1:2 and 1:4 dilution): n=1, experimental groups: n=3 independent experiments. (D) KRASG12D transcript levels in Panc-1 cells, n=3 independent experiments. The experiments with 400 exos per cell is the same data that is also presented in panel A. (E–G) KRASG12D transcript levels in Panc-1 cells under the listed conditions. In all groups, n=3 independent experiments. (H) Western blotting (Panc-1 cells) for phosphorylated ERK (p-ERK) and Vinculin. si and sh KrasG12D iExo: One way ANOVA, iLipo: two-tailed t-test, n=2 independent experiments. (I) RAS pull-down assay. (J–K) Panc-1 cells MTT assay (n=5 partitions of indicated treatments with 3 or 6 wells for each partition of treatment) (J) and separate independent experiment (K). (L–M) TUNEL assay (n=3 distinct wells of Panc-1 cells) (L) and separate independent experiment (M). (N) FC analysis of apoptosis in Panc-1 cells. Three different treatments were used to treat n=3 distinct wells of cells. (O) Wild-type KRAS transcript levels in BxPC-3 cells (n=3 independent experiments). (P) KRASG12V transcript levels in Capan-1 cells (n=3 independent experiments) (Q) KRASG12C transcript levels in MIA PaCa-2 cells (n=3 independent experiments). (R–U) MTT assay: n=5 partitions of treatment given to 3 wells each, BxPC-3 cells (R) and separate independent experiment (S), n=3 partitions of treatment given to 10 wells each, Capan1 cells (T), n=3 partitions of treatment given to 10 wells each, MIA PaCa-2 cells, (U). The data is presented as the mean ± SEM. Unless otherwise stated, one-way ANOVA was used to determine statistical significance. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001. FC: Flow cytometry. See accompanying source data. For uncropped blots for H and I, refer to Supplementary Fig. 1.
Extended Figure 5. KrasG12D RNAi containing exosomes suppress Panc-1 orthotopic tumor growth but not BxPC-3 orthotopic tumor growth.
(A) Experimental scheme. (B) Representative micrographs (scale bar: 100μm,) depicting accumulation of internalized AF647-tagged siRNA from exosomes. (C) Panc-1 orthotopic tumor growth. PBS: n=6 mice, Control exos: n=6 mice, siKrasG12D iLipo: n=3 mice, shKrasG12D iLipo: n=3 mice, siKrasG12D iExo: n=7 mice, shKrasG12D iExo: n=7 mice, siScrbl iExo: n=5 mice, shScramble iExo: n=5 mice. Statistical test compares treatment groups to PBS control group at day 42-post cancer cell injection, or day 28 for siKrasG12D exos group. Unpaired two-tailed t test. This graph is an inset from the graph shown in Fig. 2C. (D) Tumor bioluminescence at day 77 (total flux), PBS: n=4 mice, Control Exo: n=3 mice, shKrasG12D iExo: n=6 mice, shKrasG12D iLipo: n=3 mice, shScramble iExo: n=3 mice, siScramble iExo: n=4 mice. (E) Luciferase activity at day 7, 35, 77 and moribund stage or day 200 (shKrasG12D iExo)-post cancer cell injection. Some of these panels are also shown in Fig. 2a. (F) Bioluminescence from Panc-1 orthotopic tumors over time (total flux). PBS: n=7 mice, Control Exo: n=6 mice, shKrasG12D iExo: n=7 mice, shKrasG12D iLipo: n=4 mice, shScramble iExo: n=5 mice, siScramble iExo: n=5 mice (G) Representative H&E of the Panc-1 orthotopic pancreas (scale bar: 100μm). (H) Representative micrographs (scale bar: 100μm) of tumors immunolabeled for phosphorylated AKT (p-AKT) and quantification. Control Exo, n=4 mice; shKrasG12D iExo, n=6 mice. Unpaired two-tailed t test. (I–J) BxPC-3 orthotopic tumor growth, n=3 mice per group. (K) Luciferase activity at day 14 and day 77-post cancer cell (BxPC-3) injection. (L) Representative H&E of the BxPC-3 orthotopic pancreas at the indicated experimental endpoints (scale bar: 100μm). (M) Kaplan-Meier curve of BxPC-3 tumor bearing mice, Log-rank Mantel-Cox, n=3 mice per group. The data is presented as the mean ± SEM. Unless otherwise stated, one-way ANOVA was used to determine statistical significance. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001. See accompanying source data.
Extended Figure 6. Anti-tumor response of iExosomes in orthotopic models.
(A) FC analyses and quantification of CD81 on exosomes under listed conditions, n=3 independent experiments. (B) Bioluminescence from Panc-1 orthotopic tumors over time, n=6 mice per group. (C) Kaplan-Meier curve of Panc-1 tumor bearing mice. Log-rank Mantel-Cox test, n=6 mice in each group. (D) Tumor bioluminescence at day 45, n=6 mice per group. One-way ANOVA. (E–J) Bioluminescence from Panc-1 orthotopic tumors over time depicting separate groups, from panel B, and Kaplan-Meier curve depicting the separate groups, from panel C, Log-rank Mantel-Cox test. n=6 mice per group. (K) Tumor bioluminescence at day 42, n=3 mice per group. Experimental groups compared to the PBS control group, one-way ANOVA. (L) Bioluminescence from Panc-1 orthotopic tumors over time (total flux), n=3 mice per group. (M) Luciferase activity at day 10 and day 42-post cancer cell (Panc-1) injection. (N) Surface lung nodules of KPC689 mice, n=8 mice per group. (O) Tumor weights (g: grams), n=8 mice per group, siKrasG12D iExo group is compared to other treatment groups, one-way ANOVA. (P) Bioluminescent KPC689 orthotopic tumors in nu/nu mice, n=8 mice per group. (Q) Tumor weights (g: grams), n=8 mice per group. (R) Kaplan-Meier curve of KPC689 nu/nu mice. Log-rank Mantel-Cox test, n=8 in each group. The data is presented as the mean ± SEM. Unless otherwise stated, unpaired two-tailed t test was used to determine statistical significance. ** p< 0.01, *** p<0.001, **** p<0.0001. See accompanying source data.
Extended Figure 7. Pancreas localization and macropinocytosis promotes iExosomes uptake into tumor cells.
(A) Quantification and representative pictures (scale bar: 100μm) of pancreas structure in KTC mice injected with exosomes with AF647 tagged siRNA, n=3 mice. (B) Quantification and representative images (scale bar: 100μm) of pancreas of mice injected with the indicated conditions, n=3 mice, unpaired two-tailed t test. (C) Representative images (scale bar: 50μm) for data presented in Fig. 3E–H. (D) Quantification of macropinocytic and exosomes uptake (independent experiment, identical statistical analyses). (E) AF647 RNAi-tagged exosomes/liposomes uptake in Panc-1 cells (scale bar: 100μm). n=3 independent experiments. (F) CM-DiI tagged CD47 k/o vs. WT exosomes uptake in Panc-1 cells (scale bar: 100μm). n=3 independent experiments. (G) CM-DiI tagged CD47 k/o vs. WT exosomes uptake in BxPC-3 cells (scale bar: 100μm). n=3 independent experiments. The data is presented as the mean ± SEM. Unless otherwise stated, one-way ANOVA was used to determine statistical significance. ns: not significant. * p<0.05, ** p< 0.01, *** p<0.001, **** p<0.0001. See accompanying source data.
Extended Figure 8. Treatment of KTC GEMM with iExosomes.
(A) Experimental scheme. (B) Accumulation of internalized AF647-tagged siRNA from exosomes (scale bar: 100μm). (C) Kaplan-Meier curve of KTC mice. PBS: n=8 mice, gemcitabine: n=5 mice. (D) Tumor burden at experimental end point (Control Exo: (n=7 mice), siKrasG12D iExo: (n=7 mice), shKrasG12D iExo: (n=5 mice)). One-way ANOVA. (E) H&E stained tumors (scale bar: 100μm) from KTC mice and relative percentages in histological phenotypes, n=4 mice per group. (F–G) Kaplan-Meier curve of KTC mice, n=3 mice per group, Log-rank Mantel-Cox test (F) and percent tumor burden (G). (H) Representative micrographs (scale bar: 100μm) of tumors immunolabeled for phosphorylated AKT, αSMA and Kras from 44 days old KTC mice in the indicated experimental groups (n=3 mice). Unless stated otherwise, unpaired two-tailed t test was used to determine statistical significance. * p<0.05, ** p< 0.01, **** p<0.0001. See accompanying source data.
Extended Figure 9. Cytotoxicity and off target effect of iExosomes.
(A) Change in the percentage of mouse body weights, pre- and post-treatment, in the listed groups and cohorts. (B) Mouse toxicity tests, consisting of BUN, AST and ALT in the listed groups. (C) Hematoxylin and eosin, and Kras immunostaining of the listed organs in KTC (early) mice. Three to five mice evaluated per organ, one-way ANOVA. The data is presented as the mean ± SEM. See accompanying source data.
Extended Figure 10. iExosomes suppress pancreas cancer progression in KPC orthotopic mouse model.
(A–B) Magnetic Resonance Imaging (MRI) of KPC orthotopic tumors n=9 mice per group (A) and each individual tumor (B). (C) Tumor volume as measured by MRI n=9 mice per group. One-way ANOVA. (D) Representative axial images. (E) Tumor weight (g: grams) at the experimental end point. siScrbl iExo: n=9 mice, siKrasG12D iExo: n=8 mice. (F) Change in the percentage of mouse body weights, pre and post treatment (endpoint), n=9 mice per group. (G) Representative gross images of two KPC orthotopic mice that died on day 16 PTS (siScrbl iExo) or was euthanized on day 16 PTS (siKrasG12D iExo). (H) KrasG12D transcript levels in KPC689 cells (n=3 independent experiments). One-way ANOVA. (I) Kaplan-Meier curve of KPC orthotopic tumor bearing mice, Log-rank Mantel-Cox test. siScrbl iExo group: n=9 mice, siKrasG12D iExo group: n=8 mice. (J) Macroscopic metastatic nodules, n=9 mice per group. (K) H&E stained tissues. Unless stated otherwise, the data is presented as the mean ± SEM and unpaired two-tailed t test was used to determine statistical significance. * p<0.05, ** p< 0.01, *** p< 0.001, **** p<0.0001. See accompanying source data.
Supplementary Material
Acknowledgments
This work was primarily supported by the Cancer Prevention and Research Institute of Texas and the Knowledge Gap funding of MD Anderson Cancer Center. Other support include: VSL laboratory: UT MDACC Khalifa Bin Zayed Al Nahya Foundation; High Resolution Electron Microscopy Facility: Institutional Core Grant CA16672; MDACC Flow Cytometry core facility & JJL: NIH P30CA16672; MDACC Small Animal Imaging Facility: NIH P30-CA016672 and 5U24-CA126577. The Mass Spectrometry related analysis (SAM) was supported by the NORTE-01-0145-FEDER-000029 (NORTE 2020; ERDF) and FEDER funds(POCI-01/0145-FEDER-016618) and FCT – PTDC/BIM-ONC/2754/2014. We thank Kenneth Dunner Jr. for the help with the transmission electron microscopy and immunogold techniques, Edward Chang for slide scanning, D. Lundy for tissue processing, J. Carstens and C. Kahlert for independent analyses of tissue histopathology, J. Kaye and L. Gibson for mouse husbandry support, C. Kingsley and D. Lundy for MRI support, E. Lawson for IVIS imaging help, and P. Correa-de-Sampaio for microscopy imaging help.
Footnotes
Author contributions
RK conceptually designed the strategy for this study, provided intellectual input and contributed to the writing of the manuscript. HS and VSL injected mice orthotopically with tumor cells. HS performed necropsy analyses and quantitation of the exosomes in pancreas tissue sections. VSL provided intellectual input, extracted the RNA and quantified the siRNA loading in exosomes by qPCR analyses, stained tissues for Kras, helped with the monitoring of in vivo experiments, designed the experimental strategy, prepared figures and wrote the manuscript. SK and SY prepared exosomes (cultured cells for exosomes collection and preparation by ultracentrifugation), generated iExosomes (performed electroporation and wash steps involved in the generation of iExosomes and Nanosight™ measurements), and treated mice with iExosomes. SY also performed an independent analysis of the in vivo imaging data, generated and genotyped KTC and KPC mice for iExosomes treatment, and participated in the preparation of the figures. SK provided intellectual input, helped design the experimental strategy, and performed all of the other experiments presented in the manuscript, including treatment of mice with iExosomes and preparation of the exosomes for sucrose gradient analyses and qPCR analyses. Unless otherwise noted above and in the methods section, SK performed the independent replication of the experiments. SK performed the sucrose gradient analyses, performed Nanosight™ measurements and flow cytometry experiments and analyses, processed tissues for immunostaining and analyzed the staining, extracted RNA and performed qPCR for gene expression analyses of treated cells, performed the MMT assays and TUNEL assays, performed and quantified the macropinocytosis assays and the western blot analyses, analyzed data, prepared figures, participated in the preparation of the methods section and wrote the manuscript. SAM advised and provided intellectual input to the experiments related to siRNA identification, MTT assay, KRASG12D qPCR, western blots for pERK, and one Panc-1 tumor study and one KTC in vivo experiment. CFR provided the mass spectrometry data and methods sections for the mass spectrometry analyses. JJL reviewed the source data, advised on experimental design, and supervised statistical analyses.
Conflict of interest
MD Anderson Cancer Center and RK hold patents in the area of exosome biology and are licensed to Codiak Biosciences Inc. MD Anderson Cancer Center and RK are stock equity holders in Codiak Biosciences Inc. RK receives research support from Codiak Biosciences Inc. and serves as a member of the board of directors. VSL served once as a paid consultant for Codiak Biosciences Inc.
References
- 1.Hidalgo M, Von Hoff DD. Translational therapeutic opportunities in ductal adenocarcinoma of the pancreas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:4249–4256. doi: 10.1158/1078-0432.CCR-12-1327. [DOI] [PubMed] [Google Scholar]
- 2.Chang DK, Grimmond SM, Biankin AV. Pancreatic cancer genomics. Current opinion in genetics & development. 2014;24:74–81. doi: 10.1016/j.gde.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Collins MA, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. The Journal of clinical investigation. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins MA, et al. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PloS one. 2012;7:e49707. doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes & cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecot CV, et al. Therapeutic Silencing of KRAS using Systemically Delivered siRNAs. Molecular cancer therapeutics. 2014 doi: 10.1158/1535-7163.MCT-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan TL, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer discovery. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue W, et al. Small RNA combination therapy for lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3553–3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rejiba S, Wack S, Aprahamian M, Hajri A. K-ras oncogene silencing strategy reduces tumor growth and enhances gemcitabine chemotherapy efficacy for pancreatic cancer treatment. Cancer science. 2007;98:1128–1136. doi: 10.1111/j.1349-7006.2007.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zorde Khvalevsky E, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Meel R, et al. Extracellular vesicles as drug delivery systems: Lessons from the liposome field. Journal of controlled release: official journal of the Controlled Release Society. 2014 doi: 10.1016/j.jconrel.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 13.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Current opinion in cell biology. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Johnsen KB, et al. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochimica et biophysica acta. 2014;1846:75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Advanced drug delivery reviews. 2016;106:148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Kaur S, et al. CD47-dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells. Matrix biology: journal of the International Society for Matrix Biology. 2014;37:49–59. doi: 10.1016/j.matbio.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kibria G, et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Scientific reports. 2016;6:36502. doi: 10.1038/srep36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends in cell biology. 2001;11:130–135. doi: 10.1016/s0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 19.Jaiswal S, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao MP, Weissman IL, Majeti R. The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Current opinion in immunology. 2012;24:225–232. doi: 10.1016/j.coi.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoes S, et al. Cationic liposomes for gene delivery. Expert opinion on drug delivery. 2005;2:237–254. doi: 10.1517/17425247.2.2.237. [DOI] [PubMed] [Google Scholar]
- 23.Willingham SB, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Scientific reports. 2015;5:10300. doi: 10.1038/srep10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ijichi H, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. The Journal of clinical investigation. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman MH, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Provenzano PP, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Molecular oncology. 2013;7:232–247. doi: 10.1016/j.molonc.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying H, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes & development. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. British journal of cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh H, Longo DL, Chabner BA. Improving Prospects for Targeting RAS. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 37.Zeitouni D, Pylayeva-Gupta Y, Der CJ, Bryant KL. KRAS Mutant Pancreatic Cancer: No Lone Path to an Effective Treatment. Cancers. 2016;8 doi: 10.3390/cancers8040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golan T, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6:24560–24570. doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kordelas L, et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973. doi: 10.1038/leu.2014.41. [DOI] [PubMed] [Google Scholar]
- 40.Clayton A, Harris CL, Court J, Mason MD, Morgan BP. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. European journal of immunology. 2003;33:522–531. doi: 10.1002/immu.200310028. [DOI] [PubMed] [Google Scholar]
- 41.Gomes-da-Silva LC, et al. Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Accounts of chemical research. 2012;45:1163–1171. doi: 10.1021/ar300048p. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015 doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015 doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melo SA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 46.El-Andaloussi S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nature protocols. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 47.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology/editorial board, Juan S. Bonifacino … [et al.] 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 48.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic acids research. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rachagani S, et al. Activated KrasG(1)(2)D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. British journal of cancer. 2011;104:1038–1048. doi: 10.1038/bjc.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for all figures are provided with the paper and reagents will be provided upon availability and reasonable request.