Abstract
iNKT cells are a unique lineage of T cells that recognize glycolipid presented by CD1d. In the thymus, they differentiate into iNKT1, iNKT2 and iNKT17 effector subsets, characterized by preferential expression of Tbet, Gata3 and ROR-γt and production of IFN-γ, IL-4 and IL-17, respectively. We demonstrate that the transcriptional regulator Runx1 is essential for the generation of ROR-γt expressing iNKT17 cells. PLZF-cre Runx1 cKO mice lack iNKT17 cells in the thymus, spleen and liver. Runx1-deficient iNKT cells have altered expression of several genes important for iNKT17 differentiation, including decreased expression of IL-7Rα, BATF and c-Maf and increased expression of Bcl11b and Lef1. However, reduction of Lef1 expression or introduction of an IL-7Rα transgene is not sufficient to correct the defect in iNKT17 differentiation, demonstrating that Runx1 is a key regulator of several genes required for iNKT17 differentiation. Loss of Runx1 leads to a severe decrease in iNKT cell numbers in the thymus, spleen and liver. The decrease in cell number is due to a combined decrease in proliferation at Stage 1 during thymic development and increased apoptosis. Thus, we describe a novel role of Runx1 in iNKT cell development and differentiation, particularly in orchestrating iNKT17 differentiation.
Introduction
Invariant natural Killer T (iNKT) cells are innate lymphocytes that express a semi-invariant TCR with an invariant TCRα-chain, Vα14-Jα18, paired with limited TCRβ-chains, Vβ7, Vβ8, or Vβ2. iNKT cells recognizes glycolipids presented on an MHC-like molecule CD1d1–4. They share a common developmental precursor with conventional T cells at the double positive (DP) thymocyte stage5, 6. Upon positive selection into the iNKT cell lineage at DP stage, iNKT cells go through four sequential developmental stages (Stage 0–3), where Stage 0 is the earliest stage, characterized by high CD24 expression7. Unlike conventional T cells that do not proliferate after selection into CD4 or CD8 single positive T cell lineages, iNKT cells undergo a post-selection expansion at Stage 1 where they down-regulate CD24 expression. The intra-thymic proliferation of iNKT cell is highly regulated by molecular networks that involve the transcription factor c-Myc and the other metabolic pathways8–10. After proliferating, iNKT cells express an effector/memory phenotype and upregulate the expression of CD44 at Stage 2. Expression of NK receptors such as NK1.1 is turned on at Stage 3, where IL-15 is required for their homeostasis and survival by regulating the expression of Bcl-xL in Stage 3 iNKT cells11–13.
Although the traditional linear developmental pathway is used often to examine iNKT cells, iNKT cells differentiate into effector subsets in the thymus within Stages 1 through 314, 15. Their ‘master’ transcription factor PLZF is important for iNKT cell development and effector functions16, 17. In the thymus, three subsets that develop are iNKT1, iNKT2 and iNKT17, although there is evidence of other functional subsets in peripheral tissues14, 18, 19. iNKT subsets are distinguished by the signature transcription factors they express and the predominant production of cytokines they produce. iNKT1 cells are Tbet+ PLZFlo, produce IFNγ and are found within NK1.1+ Stage 3. iNKT2 cells are PLZFhi Gata3hi, produce IL-4 and are found in both Stage 1 and Stage 2. iNKT17 cells are ROR-γt+ PLZFmed, produce IL-17 and are found exclusively in Stage 214, 20.
Various transcriptional regulators and signaling programs have been identified to play a role in regulating iNKT subset differentiation. The mammalian target of rapamycin (mTOR) signaling pathway is crucial for iNKT cell development and differentiation21. mTORC1 is essential for differentiation of Tbet expressing iNKT1 while mTORC2 is important for iNKT2 and iNKT17 differentiation10, 22, 23. iNKT cells also require autophagy for their survival and the differentiation of iNKT1 cells24, 25. The transcriptional repressor NKAP is also required for iNKT cell proliferation and differentiation of ROR-γt expressing iNKT17 cells26. The transcription factor Bcl11b is important for restraining the NKT17 differentiation program to allow for differentiation of iNKT2 and iNKT1 cells27. The transcription factor Lef1 is also important for iNKT cell proliferation and is crucial for differentiation of iNKT2 cells28, 29. Loss of Lef1 leads to an increased proportion and function of iNKT17 cells suggesting Lef1 may restrain iNKT17 differentiation to promote iNKT2 differentiation. The transcription factor BATF is also required for the development of IL-17 producing iNKT cells30, 31. Although there is increasing evidence of molecular mechanisms governing iNKT development and differentiation, the interplay of transcription regulators that build molecular networks critical for specific iNKT cell differentiation is not fully understood. Here we show the role of Runx1 in regulating the transcriptional network that drives iNKT17 differentiation.
The Runt related transcription factor Runx1 belongs to the Runx family of transcription factors, and is also known as CBF-α, AML1 or PEBP2alphaB32. Runx proteins associate with core binding factor-beta (CBF-β) to bind DNA. The DNA binding domain, Runt, binds to a specific core sequence TGTGGNNN (where NNN can either be TTT or TCA)33. Runx1 can mediate either gene activation or repression as determined by other transcriptional coactivators or corepressors it binds to, such as HDACs34. Runx1 has essential roles in the development and differentiation of several hematopoietic lineages, including being a central player for hematopoiesis, conventional T cell development, and Th17 differentiation35–39. Although Runx1 is required for the positive selection of iNKT cells (as demonstrated by Egawa and colleagues’ CD4-cre Runx1 cKO model)5, its role in later iNKT cell development and differentiation beyond positive selection is unknown. In this study, we show that Runx1 is absolutely required for iNKT17 differentiation. In addition, we also demonstrate that Runx1 is required for intra-thymic proliferation and survival of iNKT cells.
Results
Runx1 is required for iNKT cell development
Loss of Runx1 in T cells using the CD4-cre Runx1 cKO mice leads to a complete block in the generation of iNKT cells while development of conventional T cell was normal5. Thus, Runx1 is required for the positive selection of DP thymocytes into the iNKT cell lineage. To investigate Runx1’s role in later iNKT cell development and differentiation after positive selection, we generated PLZF-cre Runx1 cKO mice. Expression of PLZF starts at Stage 0 and is required for iNKT cell development. Using a cre-dependent lineage marker, several groups have reported that PLZF-cre is transiently expressed in a precursor to hematopoietic stem cell (HSCs) during embryogenesis, marking ~30–50% of the HSC pool18, 40. However, as Runx1 is required for the generation of HSCs, only Runx1-sufficient HSC will develop in PLZF-cre Runx1 cKO mice41, 42. This model allows us to study the role of Runx1 specifically in iNKT cells, as conventional T cells do not express PLZF. We confirmed that there was no change in Runx1 expression in DP thymocytes and that conventional T cell development is unaltered in PLZF-cre Runx1 cKO mice (Fig. 1a).
Figure 1.
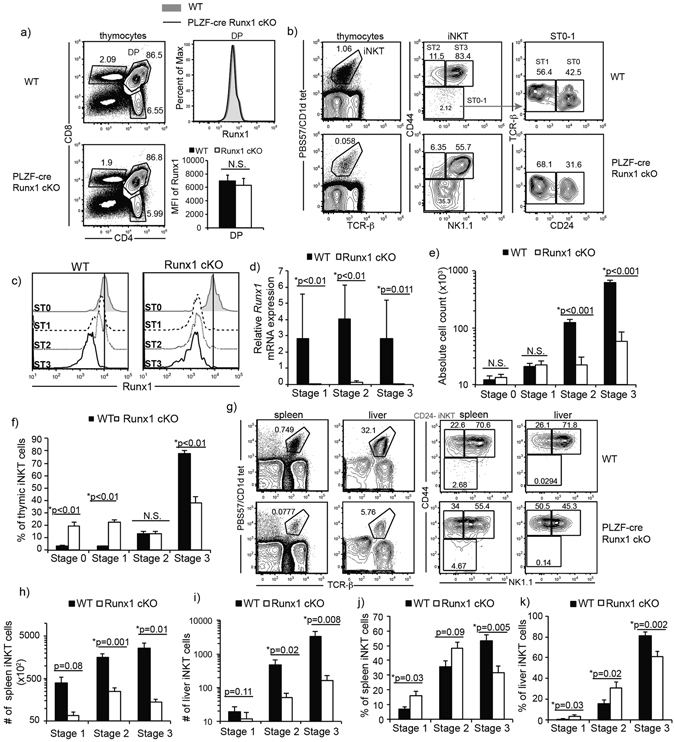
Runx1 is required for iNKT cell development. (a) Thymic CD4/CD8 profile in WT and PLZF-cre Runx1 cKO mice. Expression and quantification of Runx1 in DP thymocytes from WT (grey filled) and PLZF-cre Runx1 cKO (black line) mice. Data is representative and calculated from 11 mice per group. (b) Examination of thymic iNKT cells in their developmental stages in WT (top) and PLZF-cre Runx1 cKO mice (bottom). iNKT cells were identified using expression of TCR-β and CD1d/PBS57 loaded tetramers. Gated iNKT cells were further characterized into stages using NK1.1, CD44 and CD24 markers: Stage 0 (CD24+ CD44− NK1.1−) Stage 1 (CD24− CD44+ NK1.1−) Stage 2 (CD44hi NK1.1−) and Stage 3 (CD44+ NK1.1+). Data is representative of at least 30 mice per genotype from at least 20 independent experiments. (c) Expression of Runx1 in Stage 0–3 iNKT cells from WT (grey filled) and PLZF-cre Runx1 cKO (black line) mice. Data is representative of at least 7 mice/genotype from 5 independent experiments. (d) Relative mRNA expression of Runx1 in sorted Stage 1–3 iNKT cells of WT (black bars) and PLZF-cre Runx1 cKO (white bars) mice. Data is normalized to expression of Runx1 in WT Stage 3 iNKT cells = 1. Data is calculated from 3 WT and 3 PLZF-cre Runx1 cKO mice from 3 independent sorts. (e,f) Absolute cell count and percent of thymic iNKT cells at Stage 1, 2 and 3 in WT (black bars) and PLZF-cre Runx1 cKO mice (white bars). Data is calculated from at least 30 mice per group in the age range of 5–12 weeks old. (g) Representative frequencies of iNKT cells in spleen and liver of WT and PLZF-cre Runx1 cKO mice. Data is representative of at least 12 mice/genotype from 8 independent experiments in spleen and 9 mice/genotype from 7 independent experiments in liver. (h-i) Total Stage 1–3 iNKT cell count from spleen and liver. (j-k) Frequency of Stage 1–3 iNKT cells in the spleen and liver. (h-k) Data is calculated from at least 12 mice per group from 8 independent experiments in spleen and 9 mice/genotype from 7 independent experiments in liver. All statistical analysis was done using Student’s t-test. Means ± S.E.M.
In PLZF-cre Runx1 cKO mice, there is a dramatic alteration in iNKT cell development. The few iNKT cells present were skewed towards earlier stages (Stage 0–1) in development (Fig. 1b). Thus, Runx1 has additional critical roles in iNKT cell development beyond the DP to Stage 0 transition5. In PLZF-cre Runx1 cKO mice, Runx1 protein expression does not decrease until Stage 1 in iNKT cells (Fig. 1c). Thus, further analysis will focus on Stage 1 and subsequent stages of iNKT cell development. Deletion of Runx1 was also examined by q-PCR, where one primer was located within a floxed exon. Runx1 was efficiently deleted in Stage 1, Stage 2 and Stage 3 iNKT cells from PLZF-cre Runx1 cKO mice (Fig. 1d). Interestingly, WT Stage 3 iNKT cells downregulate Runx1 protein expression although mRNA levels were sustained at levels similar to earlier iNKT cell stages. Examination of the absolute cell count of iNKT cells in thymus of PLZF-cre Runx1 cKO mice revealed a significant reduction (p < 0.001) in cell number of Stage 2 (8.5 fold) and Stage 3 (19.5 fold) iNKT cells (Fig. 1e). The frequency of iNKT cells in Stage 0, Stage 1, Stage 2, and Stage 3 from WT and PLZF-cre Runx1 cKO mice was also quantified (Fig. 1f). There was a 6-fold and 7-fold increase in Stage 0 and Stage 1 iNKT cell frequency respectively, while there was a 2-fold decrease in Stage 3 iNKT cell frequency and no significant change in proportion of Stage 2 iNKT cells. In the periphery, there was also a dramatic decrease of 8-fold and 15-fold in iNKT cell numbers in the spleen and liver, respectively (Fig. 1g). The total cell count and frequency of Stage 1–3 iNKT cells in the spleen and liver was also quantified (Fig. 1h–k). In the spleen, there was a significant decrease in Stage 2 (6.5 fold) and Stage 3 (19 fold) iNKT cell numbers (Fig. 1h). In the liver, there was approximately 9.7-fold and 20-fold decrease in Stage 2 and Stage 3 iNKT cells, respectively (Fig. 1j). This shows that the block in thymic iNKT cell development also reflected in the numbers and proportions of splenic and liver iNKT cells. Hence, Runx1 is important for iNKT cell development after positive selection.
Runx1 regulates iNKT cell proliferation
After positive selection into lineage, unlike conventional T cells, iNKT cells undergo a burst of proliferation. To determine if Runx1-deficient iNKT cells have a defect in proliferation leading to a significant reduction in absolute cell number, we generated Rag1-GFP/PLZF-cre Runx1 cKO mice. The use of Rag1-GFP reporter mice to examine the rate of proliferation in iNKT cell has been described previously26. Briefly, GFP is knocked into one allele of the Rag1 locus43. Transcription of Rag1 stops at the DP thymocyte stage after successful rearrangement of TCR-α chain. However, GFP protein is stable with a long half-life44. In the periphery, Rag1-GFP persists in conventional T cells (which do not proliferate after DP thymocyte stage) for up to 3 weeks, and is used to mark recent thymic emigrants (RTE)44. However, iNKT cells undergo a burst of proliferation at Stage 1, leading to the dilution of GFP, allowing us to directly measure proliferation in vivo. Prior to the proliferative burst, the expression of Rag1-GFP reporter in Stage 0 iNKT cells of Rag1-GFP/PLZF-cre Runx1 cKO mice was equivalent to Stage 0 iNKT cells of Rag1-GFP (WT) mice. However, at Stage 1 and Stage 2, the Rag1-GFP reporter expression was significantly higher in iNKT cells of Rag1-GFP/PLZF-cre Runx1 cKO mice compared to Rag1-GFP (WT) mice (Fig. 2a), indicating a reduced rate of proliferation in the absence of Runx1. The expression of Rag1-GFP was 2.7 fold and 4 fold increased in Stage 1 and Stage 2 iNKT cells of Rag1-GFP/PLZF-cre Runx1 cKO, respectively, compared to Rag1-GFP WT mice. The expression of Ki-67, a proliferative capacity marker, was also significantly reduced by 3 fold in Runx1-deficient Stage 1 iNKT cells but not Stage 0 and Stage 2 iNKT cells (Fig. 2b). Similarly, the transcription factor c-Myc is required for iNKT cell proliferation and was reduced by 30% (p < 0.005) in Runx1-deficient Stage 1 iNKT cells (Fig. 2c). In HSCs, loss of both Runx1 and Runx3 lead to a defect in DNA repair resulting in increased DNA damage42. Thus, if Runx3 was also reduced in Runx1-deficient iNKT cells, there may be increased DNA damage which could contribute to the block in proliferation. The expression of Runx3 was significantly reduced by ~5 fold in Runx1-deficient Stage 1 and Stage 2 iNKT cells (Fig. 2d). To determine if loss of Runx1 with reduced expression of Runx3 led to increased DNA damage during proliferation of iNKT cells, we examined the expression of γH2AX by flow cytometry. Runx1-deficient Stage 1 iNKT cells had a statistically significant increase (~1.5 fold) in γH2AX expression compared to WT, demonstrating increased DNA damage (Fig. 2e). Hence, Runx1 is required for iNKT cell proliferation during thymic development.
Figure 2.
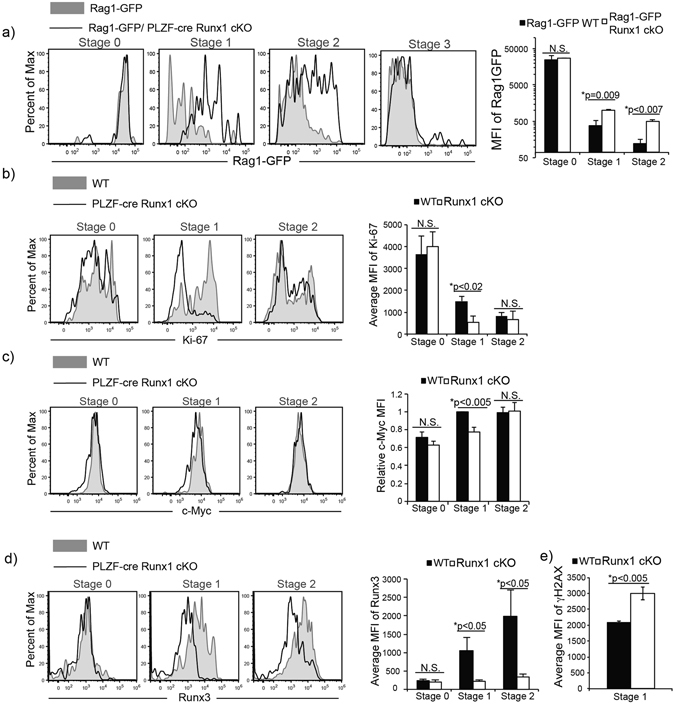
Loss of Runx1 leads to a defect in proliferation and increased DNA damage. (a) Expression of GFP in Stage 0–3 iNKT cells of Rag1-GFP (WT) (grey filled) and Rag1-GFP/PLZF-cre Runx1 cKO mice (black line). iNKT cells were gated and staged as described in Fig. 1. Data is representative of at least 3 mice/genotype from 3 independent experiments. Quantification of average mean fluorescent intensity (MFI) of GFP in Stage 0, 1 and 2 iNKT cells from Rag1-GFP (WT) (black bars) and Rag1-GFP/PLZF-cre Runx1 cKO mice (white bars). Data is calculated from at least 3 mice/genotype. (b) Intracellular expression of Ki-67 in Stage 0, Stage 1 and Stage 2 iNKT cells of WT (grey filled) and PLZF-cre Runx1 cKO mice (black line). Data is representative of at least 3 mice/genotype from 3 independent experiments. Quantification MFI of Ki-67 in Stage 0–2 in WT (black bar) and PLZF-cre Runx1 cKO (white bar). Data is calculated from 5 WT and 3 PLZF-cre Runx1 cKO mice from 3 independent experiments. (c) Intracellular expression of c-Myc in Stage 0–2 iNKT cells of WT (grey filled) and PLZF-cre Runx1 cKO mice (black line). Data is representative of at least 4 mice/genotype from 4 independent experiments. Relative MFI of c-Myc in Stage 0–2 iNKT cells of WT (black bar) and PLZF-cre Runx1 cKO mice (Runx1 cKO) (white bar). Data is normalized to MFI of c-Myc in WT Stage 1 iNKT cells = 1. Data is calculated from 4 mice/genotype from 4 independent experiments. (d) Expression of Runx3 in Stage 0–2 of WT (grey filled) and PLZF-cre Runx1 cKO (black line). Data is representative of at least 3 mice/genotype from 3 independent experiments. Quantification of MFI of Runx3 in Stage 0–2 in WT (black bar) and PLZF-cre Runx1 cKO (white bar). Data is calculated from 3 WT and 3 PLZF-cre Runx1 cKO mice from 3 independent experiments. (e) Average MFI of γH2AX was calculated for Stage 1 iNKT cells in WT (black bars) and PLZF-cre Runx1 cKO mice (white bars). Data is calculated from 3 mice/genotype from 3 independent experiments. All statistical analysis was done using Student’s t-test. Means ± S.E.M.
Runx1 is required for iNKT cell survival
To address whether increased cell death may also contribute to decreased iNKT numbers in PLZF-cre Runx1 cKO mice, we examined apoptosis by Annexin V and a fixable viability dye (FVD). Stage 1, Stage 2 and Stage 3 Runx1-deficient iNKT cells had an increased proportion of apoptotic or dead cells compared WT iNKT cells (Fig. 3a). The frequency of apoptotic Stage 1–3 iNKT cells in PLZF-cre Runx1 cKO mice was significantly increased by 1.6 fold in Stage1, 2.4 fold in Stage 2 and 3.7 fold in Stage 3 as compared to WT (Fig. 3b). However, no differences were observed in expression of pro-survival molecules Bcl-2 and Mcl-1 in Runx1-deficient iNKT cells as compared to WT iNKT cells (Fig. 3c,d). Thus loss of Runx1 results in increased cell death, which is not due to decreased expression of the Bcl-2 family molecules. Stage 3 iNKT cells require IL-15 signaling for their homeostasis and survival12, 13. To determine if loss of Runx1 leads to dysregulated homeostasis, we examined the expression of IL-15 receptor subunits (IL-15Rα, IL-2Rβ, γc) and their target, pro-survival molecule Bcl-xL. The expression of the IL-15 receptor subunits and Bcl-xL was not altered in Stage 3 iNKT cells of PLZF-cre Runx1 cKO mice compared to WT (Supplemental Fig. 1). This suggests that the 9-fold decrease in Stage 3 iNKT cell numbers in PLZF-cre Runx1 cKO mice (Fig. 1e) is not due to a defect in IL-15 mediated homeostasis. Together, decreased proliferation and increased apoptosis in the absence of Runx1 contribute to the severe decrease in iNKT cell numbers in PLZF-cre Runx1 cKO mice.
Figure 3.
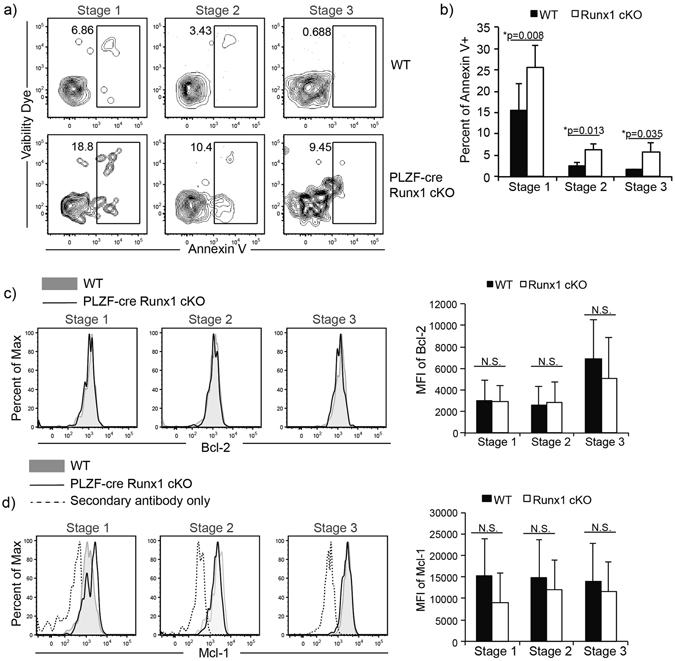
Runx1-deficient iNKT cells exhibit increased apoptosis but normal expression of pro-survival genes Bcl-2 and Mcl-1. (a) Representative frequency of apoptotic (Annexin V+) Stage 1–3 iNKT cells in WT (top) and PLZF-cre Runx1 cKO mice (bottom). Gating for iNKT cells and their developmental stages were defined as described in Fig. 1. Data is representative of at least 7 mice/genotype from 5 different experiments. (b) Quantification of apoptotic Stage 1–3 iNKT cells in WT (black bars) and PLZF-cre Runx1 cKO mice (white bars). Data was calculated from at least 7 mice/genotype from 5 different experiments. (c) Intracellular expression of pro-survival gene Bcl-2 in Stage 1–3 iNKT cells from WT (grey filled) and PLZF-cre Hdac3 cKO (black line). Data is representative of at least 3 mice/genotype from 3 different experiments. Quantification of MFI of Bcl-2 in Stage 1–3 iNKT cells in WT (black bars) and PLZF-cre Runx1 cKO mice (white bars). Data was calculated from 3 mice/genotype. (d) Intracellular expression of pro-survival gene Mcl-1 in Stage 1-3 iNKT cells from WT (grey filled) and PLZF-cre Hdac3 cKO (black line). Secondary only control for Mcl-1 is shown as dashed line. Data is representative of at least 3 mice/genotype from 3 different experiments. Quantification of MFI of Mcl-1 in Stage 1-3 iNKT cells in WT (black bars) and PLZF-cre Runx1 cKO mice (white bars). Data was calculated from 5 WT and 3 PLZF-cre Runx1 cKO mice. All statistical analysis was done using Student’s t-test. Means ± S.E.M.
Runx1 is required for the differentiation of ROR-γt expressing NKT17 cells
Although thymic iNKT cells were traditionally characterized using a linear developmental pathway, it is now established that iNKT cells also differentiate into effector subsets after Stage 0 in the thymus. Thymic iNKT cells differentiate into iNKT1, iNKT2 and iNKT17, although differentiation into other subsets such as iNKTFH and iNKT10 have been described in the periphery18, 19, 45. iNKT1, iNKT2 and iNKT17 cells are characterized by the preferential expression of transcription factors. iNKT1 cells are Tbet+ PLZFlo ROR-γt−, iNKT2 cells are PLZFhi ROR-γt− Tbet- and iNKT17 cells are ROR-γt+ PLZFmed Tbet−. iNKT1, iNKT2 and iNKT17 preferentially produce IFN-γ, IL-4 and IL-17A, respectively14, 46. In PLZF-cre Runx1 cKO mice, the proportion of iNKT1 (Tbet+ PLZFlo) cells was reduced (1.6 fold), while the proportion of Tbet− PLZF+ (containing iNKT2 and iNKT17) cells was increased by 3 fold. Examination of ROR-γt expression in the PLZF+ Tbet− gate to delineate iNKT17 from iNKT2 revealed the absence of ROR-γt expressing iNKT17 cells and increased proportion of PLZFhi ROR-γt− iNKT2 cells (Fig. 4a). Although the frequency of iNKT2 cells was increased, while the frequency of iNKT1 and iNKT17 cells was decreased in the PLZF-cre Runx1 cKO mice, the absolute count of thymic iNKT cell subsets revealed a statistically significant decrease in numbers in all three subsets. In the thymus, iNKT1 were decreased 16 fold, iNKT2 were decreased 3 fold and iNKT17 were decreased 60 fold in the PLZF-cre Runx1 cKO mice as compared to WT (Fig. 4b,c). The alteration in the frequency of thymic iNKT cell subsets (increased iNKT2 cells and decreased iNKT1 cells, absence of iNKT17 cells) in the PLZF-cre Runx1 cKO mice was also mirrored in the spleen and liver. Moreover, similar to thymic iNKT cell counts, there was a severe reduction in the number of iNKT1 (decreased 25 fold in the spleen and 12 fold in the liver) and iNKT2 cells (decreased 8 fold in the spleen and 29 fold in the liver), while there was a complete absence of iNKT17 cells in the periphery, spleen and liver (Supplemental Fig. 2). Thus, the severe defects in iNKT1, iNKT2 and iNKT17 cell generation in the thymus lead to similar reductions in the periphery. The expression of Runx1 also varied by effector subset. In WT mice, expression was highest in iNKT17 cells, while iNKT2 cells had intermediate Runx1 expression, and iNKT1 cells expressed the least Runx1 (Fig. 4d). Similar to our earlier analysis of Runx1 protein expression in PLZF-cre cKO mice, there was a large reduction in Runx1 protein expression in iNKT1 and iNKT2 cells (while iNKT17 cells, which are not generated in these mice, could not be examined) (Fig. 4d). Loss of Runx1 did not alter or significantly change the cytokine production of IFN-γ and IL-4 by iNKT1 and iNKT2, respectively (Fig. 4e,f). Quantification of either the percent positive cells for IFN-γ and IL-4 cytokine production and quantification of MFI of IFN-γ and IL-4 by stimulated iNKT1 and iNKT2 cells, confirmed that there was not a statistically significant change in cytokine production by Runx1-deficient iNKT1 and iNKT2 cells (Fig. 4g,h). Thus, Runx1 is essential for the differentiation of ROR-γt expressing iNKT17 cells. While there are fewer iNKT1 and iNKT2 cells present in PLZF-cre Runx1 cKO mice, Runx1 deficiency did not alter cytokine production by iNKT1 or iNKT2 cells.
Figure 4.
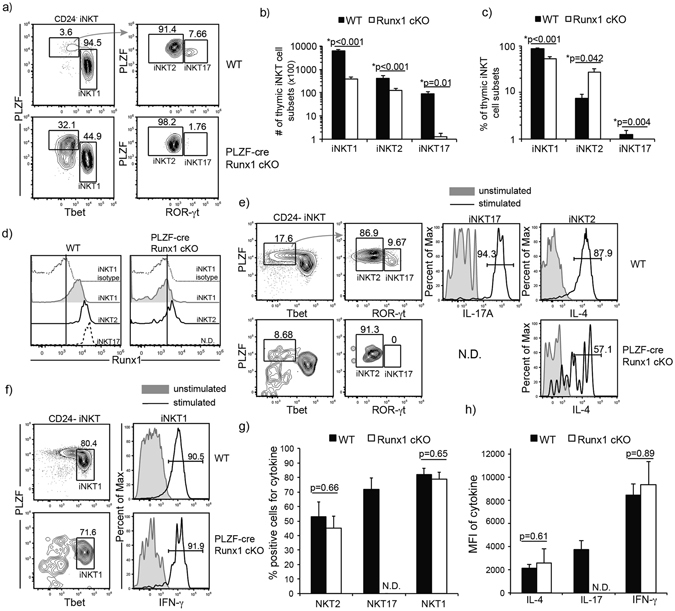
Runx1 is required for differentiation of ROR-γt expressing iNKT17 cells. (a) Representative frequency of iNKT subsets, iNKT1, iNKT2 and iNKT17 in WT (top) and PLZF-cre Runx1 cKO (bottom) mice. Functional subsets were defined by expression of PLZF, Tbet, ROR-γt. iNKT1 (Tbet+ PLZFlo), iNKT2, (PLZFhi ROR-γt− Tbet−) and iNKT17 (ROR-γt+ PLZFmed Tbet−). (b) Absolute cell number of iNKT1, iNKT2 and iNKT17 cells in WT (black bars) and PLZF-cre Runx1 cKO (white bars) mice. (c) Percent of thymic iNKT1, iNKT2 and iNKT17 cells in WT (black bars) and PLZF-cre Runx1 cKO (white bars) mice. (a-c) Data is representative and calculated from 22 WT and 12 PLZF-cre Runx1 cKO mice. (d) Expression of Runx1 in thymic iNKT1, iNKT2 and iNKT17 in WT and PLZF-cre Runx1 cKO mice with isotype control for WT iNKT1. Data is representative of 7mice/genotype. (e) Production of IL-4 and IL-17A by iNKT2 and iNKT17 cells in WT and IL-4 in PLZF-cre Runx1 cKO mice. Production of IL-17A by Runx1-deficient iNKT cells is not determined (N.D.) due to absence of ROR-γt expressing iNKT17 cells. Thymocytes were stimulated for 6hr with PMA/ionomycin, and then examined for cytokine productions using flow cytometry. (f) Production of IFN-γ by iNKT1 cells in WT (top) and PLZF-cre Runx1 cKO (bottom) mice. Thymocytes were stimulated for 6hr with PMA/ionomycin, and then examined for cytokine production using flow cytometry. (g) Frequency of cells positive for cytokine production (IL-4, IL-17A and IFN-γ) in stimulated iNKT2, iNKT17, and iNKT1 respectively in WT (black bars) and PLZF-cre Runx1 cKO (white bars) mice. Production of IL-17A by Runx1-deficient iNKT cells is not determined (N.D.) due to absence of ROR-γt expressing iNKT17 cells. (h) Quantification of MFI of cytokine (IL-4, IL-17A and IFN-γ) produced by stimulated iNKT2, iNKT17, and iNKT1, respectively in WT (black bars) and PLZF-cre Runx1 cKO (white bars) mice. Production of IL-17A by Runx1-deficient iNKT cells is not determined (N.D.) due to absence of ROR-γt expressing iNKT17 cells. (e-h) Data is representative and calculated from 8 WT and 4 PLZF-cre Runx1 cKO mice. All statistical analysis was done using Student’s t-test. Means ± S.E.M.
Runx1 regulates the transcription program that governs iNKT17 differentiation
The linear developmental pathway and differentiation pathway of iNKT cells can be aligned together to understand the dynamics of the development and differentiation occurring synchronously. iNKT1 cells are NK1.1+ Stage 3 iNKT cells; iNKT2 cells are found in both Stage 1 and Stage 2 iNKT cells where PLZF expression is the highest, whereas iNKT17 are found exclusively in Stage 2. As the loss of Runx1 leads to a defect in generating ROR-γt expressing iNKT17 cells, we utilized the linear developmental gating scheme (Stage 2) to examine Runx1 regulation of other genes important for iNKT17 differentiation. Although IL-15 is important for iNKT1/Stage 3 cell homeostasis, IL-7 is exclusively required for the differentiation of ROR-γt expressing iNKT17 cells47. The expression of IL-7Rα was reduced in Stage 2 Runx1-deficient iNKT cells (Fig. 5a). To determine whether lower expression of IL-7Rα could be responsible for the defect in iNKT cell generation in PLZF-cre Runx1 cKO mice, an IL-7Rα transgene was crossed into PLZF-cre Runx1 cKO mice to improve the expression of IL-7Rα in Stage 2 Runx1-deficient iNKT cells (Fig. 5a). However, the defect in the generation of iNKT17 cells was not rescued in IL-7Rα Tg/PLZF-cre Runx1 cKO mice (Fig. 5b). Interestingly, IL-7’s regulation on iNKT17 cells was independent of STAT signaling but was dependent on the PI3K/AKT/mTOR pathway47. To test this, we examined whether there were differences in expression of mTOR, phosphoAKT (Ser374) and phosphoS6 kinase in Runx1-deficient Stage 2 iNKT cells. However, this analysis did not reveal any alteration in the expression of these molecules in Runx1-deficient Stage 2 iNKT cells as compared to WT Stage 2 iNKT cells (Supplemental Fig. 2). Therefore, the defect in iNKT17 generation in the absence of Runx1 is not due to lower levels of IL-7Rα expression. The transcriptional repressor NKAP is required for the differentiation of ROR-γt expressing iNKT17 cells26. However, the expression of nkap was not altered in Runx1-deficient Stage 2 iNKT cells compared to WT iNKT cells (Fig. 6a), demonstrating that Runx1-mediated regulation of iNKT17 differentiation is independent of NKAP. The ectopic expression of PLZF in conventional T cells lead to an increase in IL-17A producing T cells48, suggesting that PLZF may regulate ROR-γt expressing cells. Previously, we showed that transgenic expression of PLZF in NKAP-deficient iNKT cells rescued the generation and function of ROR-γt expressing iNKT17 cells26. To determine if ectopic expression of PLZF in Runx1-deficient iNKT cells could also restore generation of iNKT17 cells, we generated Lck-PLZF Tg/PLZF-cre Runx1 cKO mice. However, the transgenic expression of PLZF also did not rescue the defect in differentiation of iNKT17 cells (Supplemental Fig. 3). To understand the molecular mechanism of how Runx1 regulates iNKT17 differentiation, we examined several transcription regulators known to alter either the differentiation of iNKT17 cells or rorc expression in Th17 differentiation. The transcription factor c-Maf is important for rorc expression in Th17 differentiation49 and is highly expressed in iNKT17 cells20. The transcription factor, BATF is also required for the differentiation of iNKT17 cells31. Runx1-deficient Stage 2 iNKT cells had decreased expression of both BATF and c-Maf (Fig. 6b) and quantification of the MFI revealed a statistically significant decrease in the expression of BATF (1.5 fold) and c-Maf (2 fold) (Fig. 6b). The transcription factors Bcl11b and Lef1 restrain the differentiation of iNKT17 cells as iNKT cells deficient for either transcription factor had increased frequencies of ROR-γt expressing iNKT17 cells27, 29. Interestingly, in PLZF-cre Runx1 cKO mice, there was a statistically significant increased expression of both Bcl11b (2 fold) and Lef1 (2 fold) in Stage 2 iNKT cells (Fig. 6c). Taken together, Runx1 plays an essential role in regulating the several genes previously implicated in iNKT17 differentiation. To understand if Runx1 mediates Lef1 expression to direct proper iNKT17 differentiation, we decreased Lef1 expression in Runx1-deficient iNKT cells by generating PLZF-cre Lef1fl/wt Runx1fl/fl mice (Fig. 6d). However, reduction of Lef1 expression in Runx1-deficient iNKT cells did not rescue the block in iNKT cell development (Fig. 6e) or the defect in iNKT17 differentiation (Fig. 6f). Therefore, Runx1 controls iNKT17 differentiation by regulating several genes involved in the differentiation of iNKT17 cells, and alterations in the expression of a single factor (Lef1 or IL-7Rα) was insufficient to restore iNKT17 generation.
Figure 5.
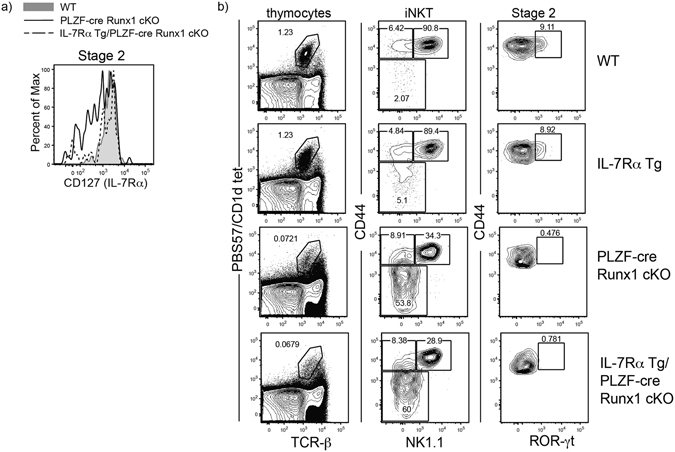
Transgenic expression of IL-7Rα in PLZF-cre Runx1 cKO mice does not rescue block in iNKT17 differentiation. (a) Expression of IL-7Rα in Stage 2 iNKT cells from WT (grey tint) and PLZF-cre Runx1 cKO (black line) and IL-7Rα Tg/PLZF-cre Runx1 cKO (dashed line) mice. Data is representative of 3 mice/genotype from 2 independent experiments. (b) FACS analysis of iNKT cell development in WT, IL-7Rα Tg, and IL-7Rα Tg/PLZF-cre Runx1 cKO mice. Analysis of iNKT cells was performed as described in Fig. 1. Data is representative of 3 mice/genotype from 2 independent experiments.
Figure 6.
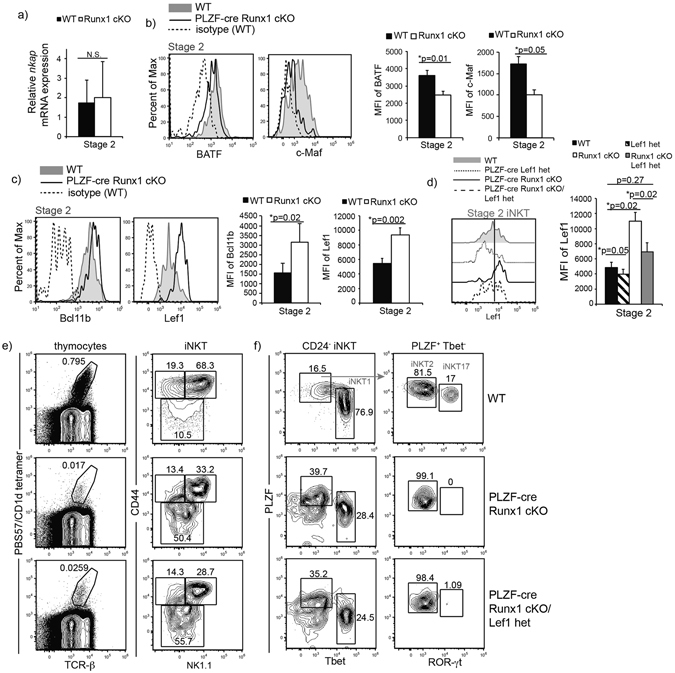
Runx1-deficient iNKT cells exhibit a dysregulated transcriptional program required for iNKT17 cell differentiation. (a) Relative mRNA expression of nkap in sorted Stage 2 iNKT cells of WT (black bar) and PLZF-cre Runx1 cKO mice (white bar). Data is calculated from 3 WT and 3 PLZF-cre Runx1 cKO mice. (b) Intracellular expression of BATF and c-Maf in Stage 2 iNKT cells of WT (grey filled), PLZF-cre Runx1 cKO mice (black line) and isotype control (dashed line). Quantification of MFI for BATF and c-Maf in Stage 2 iNKT cells of WT (black bar) and PLZF-cre Runx1 cKO mice (white bar). Data is representative (histogram overlays) and calculated from at least 11 mice/genotype from 5 independent experiments. (c) Intracellular expression of Bcl11b and Lef1 in Stage 2 iNKT cells in WT (grey filled) and PLZF-cre Runx1 cKO mice (black line). Quantification of MFI for Bcl11b and Lef1 in Stage 2 iNKT cells of WT (black bar) and PLZF-cre Runx1 cKO mice (white bar). Data is representative (histogram overlays) and calculated from at least 6 mice/genotype from 4 independent experiments for Bcl11b and 11 mice/genotype from 6 independent experiments for Lef1. (d) Lef1 expression in Stage 2 iNKT cells from WT (grey filled), PLZF-cre Lef1 het (dotted line), PLZF-cre Runx1 cKO (black line) and PLZF-cre Runx1 cKO/Lef1 het (dashed line) mice. Quantification of MFI of Lef1 in Stage 2 iNKT cells from WT (black bar), PLZF-cre Runx1 cKO (white bar) and PLZF-cre Runx1 cKO/Lef1 het (grey bar) mice and PLZF-cre Lef1 het (striped bar) mice. Data is representative (histogram overlay) and calculated from 4 mice/genotype from 4 independent experiments. (e) Development of iNKT cells in WT, PLZF-cre Runx1 cKO and PLZF-cre Runx1 cKO/Lef1 het mice. Gating was performed as described in Fig. 1. Data is representative of 4 mice/genotype from 4 independent experiments. (f) Representative frequency of iNKT subsets: iNKT1, iNKT2 and iNKT17 in WT, PLZF-cre Runx1 cKO and PLZF-cre Runx1 cKO/Lef1 het mice. Subsets gating were performed as described in Fig. 5. Data is representative of 4 mice/genotype from 4 independent experiments. All statistical analysis was done using Student’s t-test. Means ± S.E.M.
Discussion
Runx1 is required for the positive selection of DP thymocytes into the iNKT cell lineage, as demonstrated using CD4-cre Runx1 cKO mice5. However, it was unknown whether Runx1 plays additional roles in subsequent development and differentiation of iNKT cells. In this study, we show that Runx1 is critical in regulating proliferation and survival of iNKT cells, as well as the differentiation of ROR-γt expressing iNKT17 cells. In PLZF-cre Runx1 cKO mice, there is a dramatic decrease in Stage 2 (8.5 fold) and Stage 3 (19.5 fold) iNKT cell numbers. Loss of Runx1 leads to a defect in proliferation, with reductions in Ki-67 (3 fold) and c-Myc (30%) and an increase in γH2AX (1.5 fold) expression. Furthermore, Runx1-deficient iNKT cells exhibit increased cell death that is not due to altered expression of the pro-survival molecules Bcl-2 and Mcl-1. Loss of Runx1 in iNKT cells leads to a defect in the differentiation and function of ROR-γt expressing iNKT17 cells. Runx1-deficient iNKT cells exhibit changes in the expression of several genes required for the proper differentiation of iNKT17 cells. There is a decreased expression of c-Maf (2 fold) and BATF (1.5 fold), while there is an increased expression of Bcl11b (2 fold) and Lef1 (2 fold). Thus, Runx1 is a key regulator required for differentiation of iNKT17 cells.
Proliferation generates DNA damage which is immediately repaired upon detection at defined cell cycle checkpoints50. Runx1 and Runx3 mediate DNA damage repair in hematopoietic stem cells, and loss of both Runx1 and Runx3 lead to increased genetic instability with increased levels of γH2AX42. Runx1-deficient iNKT cells have reduced expression of Runx3 (5 fold) and increased γH2AX (1.5 fold) staining. Furthermore, in the PLZF-cre Runx1 cKO/Rag1-GFP mice, there is a defect in the rate of proliferation. This suggests that during the burst of proliferation at Stage 1, loss of Runx1 in iNKT cells coupled with decreased Runx3 expression leads to increased DNA damage. The increase in DNA damage may also contribute to the increased cell death observed in PLZF-cre Runx1 cKO iNKT cells (Fig. 3a,b). However, Runx1-deficient Stage 1 iNKT cells also exhibit reduced expression of c-Myc and the proliferative capacity marker Ki-67, demonstrating that it is not simply increased DNA damage which is responsible for the decreased proliferation. It is unclear whether Runx1 directly regulates c-Myc expression in iNKT cells. It has been reported that Runx1 can directly bind and repress c-Myc promoter in Jurkat T cells and murine primary hematopoietic cells51. However, as Runx proteins can function as either activators or repressors depending on its binding partners and cellular contexts, it does not exclude the possibility that Runx1 is a positive regulator of c-Myc in iNKT cells.
The Runx family of transcription factors consists of three members, Runx1, Runx2 and Runx3. In CD4-cre Runx3 cKO mice, there is minimal effect on the development of iNKT cells and iNKT differentiation52. Whereas in the CD4-cre Runx1 cKO mice, there is a complete block in iNKT cell development, suggesting that other Runx transcription factors cannot compensate for loss of Runx15. However, examination of any functional redundancy between Runx3 and Runx1 during later iNKT cell development and differentiation will need to await generation of PLZF-cre Runx1/Runx3 dKO mice.
Previously, we demonstrated that the transcriptional repressor NKAP is required for the positive selection of DP thymocytes into the iNKT cell lineage53. NKAP is also required for iNKT cell proliferation and the differentiation of iNKT17 cells26. Transgenic expression of PLZF (Lck-PLZF) in PLZF-cre NKAP cKO, restored iNKT17 differentiation defect but not proliferation26. Runx1 is also required for positive selection of DP thymocytes5 and in this study we have demonstrated additional roles for Runx1 in iNKT cell proliferation, survival and iNKT17 differentiation. However, Runx1 and NKAP have different roles in mediating iNKT17 differentiation, as nkap mRNA expression is normal and transgenic expression of PLZF in PLZF-cre Runx1 cKO mice did not rescue the defect in iNKT17 differentiation as it did in PLZF-cre NKAP cKO mice. Furthermore, expression of Runx1 is unaltered in the PLZF-cre NKAP cKO mice (P.T. and V.S.S., unpublished data). Therefore, Runx1 and NKAP regulate iNKT17 differentiation independently of each other.
Runx1 can induce rorc expression and associate with ROR-γt during conventional Th17 differentiation39. However, Runx1’s role in iNKT17 differentiation has not been defined. The transcriptional program governing the differentiation of ROR-γt expressing iNKT17 cells involve the interplay of various transcriptional regulators acting as activators and repressors. IL-7 signaling is crucial exclusively for iNKT17 homeostasis and survival. IL-7-mediated regulation of iNKT17 homeostasis was independent of STAT signaling but was dependent on the PI3K/AKT/mTOR pathway47. BATF is important for IL-17A producing CD4 T cells (Th17) and iNKT17 cells31, 54. The transcription factor c-Maf also regulates the differentiation of Th17 by associating with Sox5 to directly bind and activate the rorc promoter in conventional CD4 T cells49, 55. However, the functional role of c-Maf is not understood in iNKT17 differentiation, although a recent study reported high mRNA expression of maf in iNKT17 cells by RNA-seq20. The transcriptional repressor NKAP is also critical for iNKT cell proliferation and differentiation of iNKT17 cells26. While BATF, c-Maf and NKAP positively regulate iNKT17 differentiation; the transcription factors Bcl11b and Lef1 restrain iNKT17 differentiation. Deficiency in either factor in iNKT cells led to an increased frequency of iNKT17 cells27, 29. Although, Runx1 can induce rorc expression and associate with ROR-γt during Th17 differentiation39, its role in iNKT17 differentiation has not been defined. It is unlikely that the only role for Runx1 in iNKT cell differentiation is by directly regulating rorc expression, as we have shown that the loss of Runx1 leads to statistically significant dysregulation of the expression of transcription factors (BATF, c-Maf, Bcl11b and Lef1) involved in the iNKT17 differentiation program. Although the expression of NKAP was not altered, Runx1-deficient Stage 2 iNKT cells have decreased expression of the positive regulators, BATF and c-Maf, while the expression of negative regulators, Bcl11b and Lef1, is increased. Furthermore, decreasing Lef1 expression in Runx1-deficient iNKT cells did not correct the iNKT17 differentiation and IL-17A production defect, suggesting that restoration of a part of the dysregulated transcriptional network required for iNKT17 differentiation is not sufficient. This further supports Runx1’s role as a ‘key’ regulator for iNKT17 differentiation instead of simply a direct regulation of rorc expression during iNKT17 differentiation. However, it is not known whether Runx1 directly regulates the expression of BATF, c-Maf, Bcl11b and Lef1, or whether these are the only genes dysregulated during iNKT development in the absence of Runx1. To do this, future work will examine global changes in gene expression during iNKT cell development and differentiation in the absence of Runx1.
Methods
Mice
PLZF-cre18, Runx1fl/fl 56, Rag1-GFP43, IL-7Rα Tg57, Lck-PLZF Tg48, and Lef1fl/fl 58 mice has been previously described. All mice were on C57Bl/6 genetic background. Mice were housed in barrier facilities and all experiments were performed at the Mayo Clinic with the approval of the Institutional Animal Care and Use Committee. Mice of age 5–16 weeks were examined with either littermate or age-matched controls.
Ethics approval
All animal experiments in this study were approved by the the Institutional Animal Care and Use Committee of Mayo Clinic and were performed in accrodance with relevant guidelines and regulations for animal experimentation at the Mayo Clinic.
Flow Cytometry
All flow cytometry were performed on the Attune NxT (Life Technologies) or LSRII (BD Bioscience). Experiments were analysed using FlowJo (Tree Star). Each analysis had doublet cells excluded and live/dead gating using fixable viability dye (Tonbo) done before analysis, unless otherwise stated. Fluorophore conjugated or unconjugated antibodies were purchased from Tonbo, BD Bioscience, eBioscience, Biolegend, Cell Signaling Technology, AbCam or R&D Systems. The list of antibodies purchased is as follows: Tonbo: TCR-β (H57.597), CD4 (RM-5, GK1.5), NK1.1 (PK136), IL-4 (11311), IL-17A (TC11-18H10-1), IFN-γ (XMG1.2). eBioscience: Ki-67 (SolAl5), CD132 (TUGm2), IL-15R (DNT15Ra), Annexin V, CD24 (M1/69), NK1.1 (PK136), PLZF (Mags.21F7), Tbet (4B10), Bcl-2 (10C4), Runx1(RXDMC), Rat IgG2a,κ, (eBR2a), c-Maf (SYM0F1), mouse IgG2b,κ (eBMG2b). Biolegend: CD44 (IM7), CD8α (53-6.7), CD122 (TM-B1), CD24 (M1/69), IL-4 (11311), IL-17A (TC11-18H10-1), IFN-γ (XMG1.2), PLZF (Mags.21F7, 9E12), Tbet (4B10), Bcl-2 (10C4), CD4 (RM-5, GK1.5), CD127(A7R34). Cell Signaling Technologies: Mcl-1 (D2W9E), c-Myc (D84C12), pAKT(ser374) (193H12), BATF (D7C5), Lef1 (C12A5), Anti-Rabbit IgG (AF647), mTOR (7C10), pS6(D68F8). R&D: NK1.1 (KLRG-1C). AbCam: Bcl-xL (7B2.5), Bcl11b (25B6), Rat IgG2a,κ, (ab18446), BD Bioscience: ROR-γt (B2D), Runx3 (R3-5G4). CD1d tetramers loaded with PBS57 or empty were obtained from the NIH Tetramer Core Facility. Cells were stained with fixable viablity dye (FVD) in 1x PBS before staining for surface antigens in FACS buffer. For intracelluar stains, cells were stained with FVD and surface antigens before fixation and permeablization with Foxp3 fix:perm buffer kit (eBioscience or Tonbo) and then stained for intracellular antigens.
Isolation of iNKT cells
Sorts were performed using a FACSAria (Becton Dickinson) in the Mayo Clinic Flow Cytometry Core Facility. Thymocytes were first stained with CD1d-PBS57 loaded tetramer (PE conjugated), then incubated with anti-PE coated magnetic beads (Miltenyi Biotec) to positively select iNKT cells. iNKT cells were then separated using (LS) MACS separation column. Isolated cells were then stained with FVD before surface stains with TCR-β, BV421-CD1d/PBS57 tetramer, CD24, CD44 and NK1.1 to distinguish iNKT cell developmental stages. Gated iNKT cells (PBS57-CD1d+ TCR-β+) were distinguished in stages: Stage 1 (CD24− CD44lo NK1.1−) Stage 2 (CD24− CD44hi NK1.1−) Stage 3 (CD24− CD44hi NK1.1+). iNKT cells were sorted directly into lysis buffer from RNeasy kit (Qiagen).
Quantitative PCR
Using the RNeasy kit (Qiagen), mRNA from sorted iNKT cells were isolated. cDNA was generated and amplified using an Ovation PicoSL WTA V3 kit (NuGen) from mRNA isolated from sorted iNKT cells. Runx1, nkap and Gapdh Taqman assays used for q-PCR were purchased from Applied Biosystems. Assay used for Runx1 spanned the deleted exon in the Runx1 gene. Q-PCR was performed in ABI RT-PCR StepOne Plus System (Applied Biosystems) to calculate relative quantity (using 2−ΔΔCT method, ref. 59) of Runx1. Relative Runx1 expression in iNKT cells from PLZF-cre Runx1 cKO was calculated with WT Stage 3 (=1).
Stimulation of iNKT cells
Stimulated iNKT cells were examined for cytokine production. Thymocytes were stimulated or unstimulated for total of 6 hrs in complete culture media (RPMI) with 200 nM PMA and 1uM Ionomycin. BFA/Monensin from eBioscience was added an hour after stimulation. Cell were harvested after additional 5 hours and washed before FACS staining. Cells were stained for FVD and surface antigens before fixed and permed to stain for cytokines and intracellular antigens.
Apoptosis
Apoptotic iNKT cells were identified using FVD (Tonbo) and Annexin V (Biolegend) staining. Thymocytes were first stained as usual for FVD before surface stain. After surface stain, cells were washed and resuspended in 1x binding buffer then stained for Annexin V for 30 min on ice. Cell were washed and immediately analyzed by flow cytometry.
Statistical analysis
Two-tailed Student’s t test was used for statistical analysis with p value < 0.05 considered significant.
Electronic supplementary material
Acknowledgements
We thank the National Institute of Health (NIH) Tetramer Core Facility for generously providing us with PE and BV421 conjugated CD1d tetramers. We thank Dr. Nobuo Sakaguchi (Kumamoto University, Kumamoto, Japan) for Rag1-GFP knock in mice. We thank Dr. Al Singer (NCI, Bethesda, MD) for the IL-7Rα Tg mice. We thank all laboratory members of V.S.S. and Dr. Kay Medina (Mayo Clinic) for helpful discussions. This work was supported by NIH grants AI1069031 and AI083279 to V.S.S. and AI121080 to H-H.X.
Author Contributions
P.T. designed, performed research, analyzed data and wrote the paper; B.M. designed, performed research, and analyzed data; J.Y.C. and S.R.A. managed mouse colony; H.-H.X. contributed reagents and insights; D.B.S. contributed reagents and insights; V.S.S. designed research, analyzed data and edited the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07365-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bix M, Coles M, Raulet D. Positive selection of V beta 8+ CD4-8- thymocytes by class I molecules expressed by hematopoietic cells. The Journal of Experimental Medicine. 1993;178:901–908. doi: 10.1084/jem.178.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. The Journal of Experimental Medicine. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. The Journal of Experimental Medicine. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendelac A, et al. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 5.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Dao T, et al. Development of CD1d-restricted NKT cells in the mouse thymus. European journal of Immunology. 2004;34:3542–3552. doi: 10.1002/eji.200425546. [DOI] [PubMed] [Google Scholar]
- 7.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. The Journal of Experimental Medicine. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dose M, et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mycko MP, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. Journal of Immunology. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 10.Prevot N, et al. Mammalian target of rapamycin complex 2 regulates invariant NKT cell development and function independent of promyelocytic leukemia zinc-finger. Journal of Immunology. 2015;194:223–230. doi: 10.4049/jimmunol.1401985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodolce JP, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/S1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda JL, et al. Homeostasis of V alpha 14i NKT cells. Nature Immunology. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 13.Gordy LE, et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. Journal of Immunology. 2011;187:6335–6345. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature Immunology. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Current Opinion in Immunology. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature Immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch L, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nature Immunology. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. The Journal of Clinicalinvestigation. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel I, et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nature Immunology. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Gorentla B, Zhong XP, Shin J. mTOR and its tight regulation for iNKT cell development and effector function. Molecular Immunology. 2015;68:536–545. doi: 10.1016/j.molimm.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. The Journal of Clinical Investigation. 2014;124:1685–1698. doi: 10.1172/JCI69780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. Journal of Immunology. 2014;193:1759–1765. doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 24.Salio M, et al. Essential role for autophagy during invariant NKT cell development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5678–5687. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei B, et al. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. Journal of Immunology. 2015;194:5872–5884. doi: 10.4049/jimmunol.1402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thapa P, et al. NKAP Regulates Invariant NKT Cell Proliferation and Differentiation into ROR-gammat-Expressing NKT17 Cells. Journal of Immunology. 2016;196:4987–4998. doi: 10.4049/jimmunol.1501653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin MN, et al. Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7608–7613. doi: 10.1073/pnas.1521846113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berga-Bolanos R, Zhu WS, Steinke FC, Xue HH, Sen JM. Cell-autonomous requirement for TCF1 and LEF1 in the development of Natural Killer T cells. Molecular Immunology. 2015;68:484–489. doi: 10.1016/j.molimm.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr T, et al. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. The Journal of Experimental Medicine. 2015;212:793–807. doi: 10.1084/jem.20141849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao T, et al. Early growth response gene-2 controls IL-17 expression and Th17 differentiation by negatively regulating Batf. Journal of Immunology. 2013;190:58–65. doi: 10.4049/jimmunol.1200868. [DOI] [PubMed] [Google Scholar]
- 31.Jordan-Williams KL, Poston S, Taparowsky EJ. BATF regulates the development and function of IL-17 producing iNKT cells. BMC Immunology. 2013;14:16. doi: 10.1186/1471-2172-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Wijnen AJ, et al. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23:4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- 33.Bowers SR, Calero-Nieto FJ, Valeaux S, Fernandez-Fuentes N, Cockerill PN. Runx1 binds as a dimeric complex to overlapping Runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids research. 2010;38:6124–6134. doi: 10.1093/nar/gkq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H, Friedman AD. Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. The Journal of Biological Chemistry. 2011;286:208–215. doi: 10.1074/jbc.M110.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuda T, Nishimura M, Nakao M, Fujita Y. RUNX1/AML1: a central player in hematopoiesis. International Journal of Hematology. 2001;74:252–257. doi: 10.1007/BF02982057. [DOI] [PubMed] [Google Scholar]
- 36.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/S0092-8674(02)01111-X. [DOI] [PubMed] [Google Scholar]
- 37.Motoda L, et al. Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells. 2007;25:2976–2986. doi: 10.1634/stemcells.2007-0061. [DOI] [PubMed] [Google Scholar]
- 38.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. The Journal of Experimental Medicine. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nature Immunology. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichikawa M, Asai T, Chiba S, Kurokawa M, Ogawa S. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle. 2004;3:722–724. doi: 10.4161/cc.3.6.951. [DOI] [PubMed] [Google Scholar]
- 42.Wang CQ, et al. Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Reports. 2014;8:767–782. doi: 10.1016/j.celrep.2014.06.046. [DOI] [PubMed] [Google Scholar]
- 43.Kuwata N, Igarashi H, Ohmura T, Aizawa S, Sakaguchi N. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. Journal of Immunology. 1999;163:6355–6359. [PubMed] [Google Scholar]
- 44.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nature Immunology. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 45.Chang PP, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nature Immunology. 2011;13:35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 46.Lee YJ, et al. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity. 2015;43:566–578. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster KE, et al. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunology. 2014;7:1058–1067. doi: 10.1038/mi.2013.122. [DOI] [PubMed] [Google Scholar]
- 48.Kovalovsky D, et al. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. Journal of Immunology. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka S, et al. Sox5 and c-Maf cooperatively induce Th17 cell differentiation via RORgammat induction as downstream targets of Stat3. The Journal of Experimental Medicine. 2014;211:1857–1874. doi: 10.1084/jem.20130791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature Reviews. Molecular Cell Biology. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 51.Jacobs PT, et al. Runx transcription factors repress human and murine c-Myc expression in a DNA-binding and C-terminally dependent manner. PloS one. 2013;8:e69083. doi: 10.1371/journal.pone.0069083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, et al. Runt-related transcription factor 3 is involved in the altered phenotype and function in ThPok-deficient invariant natural killer T cells. Cellular & Molecular Immunology. 2014;11:232–244. doi: 10.1038/cmi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thapa P, et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nature Communications. 2013;4:1582. doi: 10.1038/ncomms2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato K, et al. Marked induction of c-Maf protein during Th17 cell differentiation and its implication in memory Th cell development. The Journal of Biological Chemistry. 2011;286:14963–14971. doi: 10.1074/jbc.M111.218867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Growney JD, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JH, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 58.Yu S, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T-cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


