Abstract
Eukaryotes have evolved elaborate splicing mechanisms to remove introns that would otherwise destroy the protein-coding capacity of genes. Nuclear premRNA splicing requires sequence motifs in the intron and is mediated by a ribonucleoprotein complex, the spliceosome. Here we demonstrate the presence of a splicing apparatus in the protist Trichomonas vaginalis and show that RNA motifs found in yeast and metazoan introns are required for splicing. We also describe the first introns in this deep-branching lineage. The positions of these introns are often conserved in orthologous genes, indicating they were present in a common ancestor of trichomonads, yeast, and metazoa. All examined T. vaginalis introns have a highly conserved 12-nt 3′ splice-site motif that encompasses the branch point and is necessary for splicing. This motif is also found in the only described intron in a gene from another deep-branching eukaryote, Giardia intestinalis. These studies demonstrate the conservation of intron splicing signals across large evolutionary distances, reveal unexpected motif conservation in deep-branching lineages that suggest a simplified mechanism of splicing in primitive unicellular eukaryotes, and support the presence of introns in the earliest eukaryote.
Keywords: evolution, gene expression, old introns, splicing
Splicing of nuclear premRNA plays a central role in eukaryotic gene expression and contributes to gene regulation and protein diversity (1, 2). Splice-site (SS) motifs, recognized by small nuclear RNAs and protein components of the spliceosome, coordinate multiple steps in splicing (1). Precision is required to generate mRNAs that encode functional proteins, yet the SS motifs that define exon/intron junctions are short and often weakly conserved. Two types of spliceosomal introns, U2- and U12-dependent, have been described in eukaryotes (reviewed in ref. 3). Groups I and II introns, capable of self splicing in vitro and present in some eubacterial genomes, are also found in organellar genomes of unicellular eukaryotes and plants (reviewed in refs. 4 and 5). Most spliceosomal introns are U2-dependent and are bordered at the 5′ end by a consensus of 6 nt, encompassing the invariant 5′ GT. The 5′ splice-site consensus in yeast (Saccharomyces cerevisiae) introns is generally highly conserved, whereas the consensus in mammalian introns is often limited to the 5′ GT. The 3′ SS motif of mammalian and yeast U2-dependent introns is shorter and typically limited to the last 3nt(T/CAG3′) (1, 3, 6, 7). In addition to the 5′ and 3′ SS motifs, yeast introns also require a conserved branch-point sequence (TACTAAC), which is only loosely conserved in mammalian U2-dependent introns.
Spliceosomal introns are common in animal and plant genes, absent in eubacterial and archaeal genes, and found in several unicellular protists. The prevalence of splicing in protists is unknown, and the properties of protist introns vary considerably. In euglenoids and trypanosomes, both transsplicing and cis-splicing U2-dependent introns are present (8). Although virtually all trypanosome premRNAs undergo transsplicing, only one cis-splicing intron has been identified (9). In euglenoids, U2-dependent cis-splicing introns are common; however, a subset of nuclear genes are also present that contain nonconventional introns thought to be excised by a spliceosome-independent mechanism (10, 11). In contrast, apicomplexan parasites Plasmodium and Toxoplasma and entamoebids (12) appear to have only conventional cis-splicing introns and no transsplicing.
The two deepest-branching protist lineages are the diplomonad and parabasalid lineages (13–15). Only three diplomonad genes are known to contain introns (16, 17), and no intron-containing genes have been reported in the parabasalid lineage. However, the presence of a highly conserved essential spliceosomal protein, PRP8, in the parabasalid, Trichomonas vaginalis (18), provides evidence that splicing is likely to occur. The spliceosomal machinery and the RNA motifs required for splicing have not been examined for either of these lineages. Such analyses are critical, because the only known intron from the diplomonad, Giardia, lacks the canonical 5′ GT essential for splicing in yeast and metazoa (16), suggesting that splicing mechanisms in deep-branching protists may differ (19).
Here we examine the motifs required for the parabasalid T. vaginalis to splice heterologous introns and characterize the first introns found in this lineage. These introns share an extremely conserved extended 3′ SS motif that encompasses the branch point and is necessary for splicing with the only known intron in Giardia. Their positions within respective genes are also often conserved in orthologous genes in distal lineages, indicating the presence of introns in this deep-branching lineage before its divergence from its common ancestor with plants and animals.
Materials and Methods
Cultures. T. vaginalis strain T1 was maintained in Diamond's complete media supplemented with iron (20).
Generation of Chloramphenicol Acetyltransferase (CAT) Constructs Containing Introns. The CAT gene was inserted into the Asp-718 and BamHI restriction sites of a pBluescript vector that had previously been subjected to site-specific mutagenesis to disrupt the two PvuII restriction sites. Forward and reverse oligomers corresponding to the sequence of each specific intron tested were annealed and blunt-end ligated into the PvuII site of the CAT gene within this construct. The CAT genes containing these introns were subsequently inserted at the Asp-718 and BamHI restriction sites to the T. vaginalis expression vector Master-Neo (21) that contains a neomycin phosphotransferase selectable marker. The sequence of each construct was verified by sequencing.
Transfections. Transfections were conducted as described (20), and cultures were selected with the drug Gentamicin (GIBCO) (G418) for 5 days before harvesting to assess CAT activity.
CAT Assay. CAT assays were performed on cell lysates by using radioactively labeled chloramphenicol C14, as described by Promega.
DNA and RNA Isolation. Genomic DNA from T. vaginalis was isolated by using the guanidium hydrochloride method as described (22). Total RNA was isolated by using a lithium chloride procedure and enriched in mRNA by using a Promega mRNA isolation kit.
RT-PCR Analyses. The first-strand synthesis for RT-PCR was generated with SuperScript RTIII (Invitrogen) on RNaseH-treated mRNA with a CAT-specific reverse primer, AAGGCCGGATAAAACTTGTGCT. Four microliters of the resulting cDNA was then used for PCR with a combination of CAT-specific primers (CATFor2, TCAGTTGCTCAATGTACCTATA ACCAG; CATRT2, TCATCAGGCGGGCA AGAATGT). The final PCR products were resolved on a 2% agarose gel.
Confirmation of Trichomonas Endogenous Introns. RNA was isolated by TRIzol preparation (Invitrogen). The first-strand cDNA was generated on total RNA with the oligo dT primer by using Superscript RT III reverse transcriptase. A list of oligonucleotides can be found in Supporting Text, which is published as supporting information on the PNAS web site.
RNA Blot Analysis. Total RNA or mRNA was separated on a 1.2% agarose gel and blotted to nitrocellulose by using standard procedures. Hybridization probes were labeled by nick translation by using DNA polymerase (Boehringer), and hybridizations were conducted at 65°C in 3× SSC, followed by washing at the same temperature, reducing the salt concentration to 0.1 × SSC.
DNA Sequencing. All RT-PCR products were subcloned into the TOPO vector (Invitrogen). DNA sequencing was performed with M13 forward and reverse primers by using the Big Dye version 3 labeling kit (Applied Biosystems). The sequence was obtained by using an Applied Biosystems 3700 Capillary DNA Analyzer.
Algorithm for the Intron Search. A perl script was written to search the preliminary T. vaginalis sequence data, obtained from The Institute for Genomic Research through the web site (www.tigr.org/tdb/e2k1/tvg), for the presence of the following motif: GYNNGYN(n)ACTAACACACAG, where Y, T or C and n, any nucleotide. As indicated by N(n), no size limit was set for the distance between the putative 5′ SS motif (GYNNGY) and the putative 3′ SS motif (ACTAACACACAG).
Results
Trichomonads Are Capable of Splicing Short Introns and Require Conserved Splicing Motifs. To address whether the primitive eukaryote T. vaginalis contains the spliceosomal machinery required for intron removal, we developed a functional assay to test its ability to splice heterologous introns. Introns were inserted into the PvuII site in the CAT reporter gene contained in a T. vaginalis expression vector (21), creating either an in-frame stop codon or shifting the ORF. This would require accurate splicing of the intron from the premRNA for the production of CAT activity in cell transfected with these constructs.
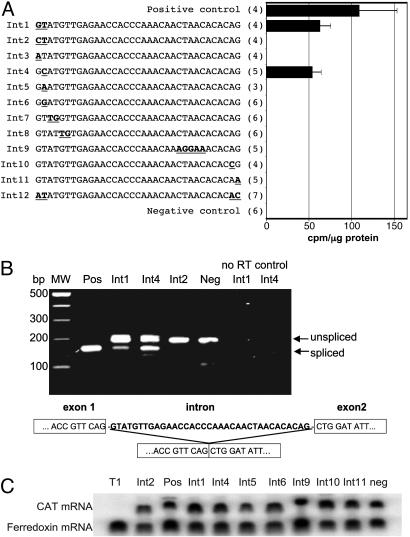
Initially, we tested whether T. vaginalis could remove introns from two related anaerobic parasitic protists, a 35-nt intron from the Giardia intestinalis ferredoxin gene (16) and a 46-nt intron from an Entamoeba histolytica serine/threonine kinase gene (23), as well as a 302-nt intron from the Trypanosoma cruzi poly(A) polymerase gene (9) and a 46-nt modified version of an adenovirus intron (24). Although the three introns from E. histolytica, T. cruzi, and adenovirus-2 contained canonical 5′ GT-AG3′ SS motif and putative branch site sequences, none of the introns were removed from the CAT gene (data not shown). In contrast, analysis of T. vaginalis transfectants harboring a CAT gene containing the G. intestinalis intron (16) in which the 5′ CT was modified to a 5′ GT revealed the presence of CAT activity (Fig. 1A, Int1). Transfectants possessing the CAT gene with the wild-type Giardia intron (e.g., 5′CT-AG3′ SS) do not produce CAT activity (Fig. 1 A, Int2). These data demonstrate that T. vaginalis contains a functional premRNA splicing machinery and suggest it requires a 5′GT SS.
Fig. 1.
T. vaginalis can splice a 35-nt intron. (A) CAT activity in T. vaginalis cells transfected with plasmid constructs of a CAT gene containing the wild-type Giardia intron (Int2) and mutations of this 35-nt sequence (Int1 and Int3–12). Mutations are indicated in bold and underlined. The positive control (at the top) contains a construct with an intron-less CAT gene and the negative control (at the bottom) has a CAT gene with the Int1 intron inserted in the opposite orientation. CAT activity is expressed as cpm of n-butyrylated chloramphenicol per microgram of protein. The number of independent transfections tested are designated in brackets, and standard error bars are shown. (B) RT-PCR products separated on a 2% agarose gel show two populations of CAT cDNAs corresponding to spliced and unspliced mRNA in T. vaginalis cells transfected with the Int1 (GT) and the Int4 (GC) constructs. No spliced CAT cDNA is detected in cells transfected with the Int2 construct containing wild-type Giardia intron (CT) or the Int1 construct with the intron inserted in the reverse orientation (neg). Pos, RT-PCR reaction by using cells transfected with the intronless CAT construct. (C) RNA blot analysis of total RNA from transfected cells. The blot was hybridization with a CAT gene probe showing that cells without CAT activity do transcribe the CAT RNA. A ferredoxin probe was used as an internal standard for RNA loading.
Additional mutational analyses of the motifs necessary for splicing in T. vaginalis transfectants established the requirement of standard eukaryotic canonical 5′ and 3′ SS for splicing. Alterations at any of these positions in the Giardia intron abolished CAT activity (Fig. 1 A, Ints 2, 3, 5, 6, and 10–12), with the exception of an intron containing 5′ GC-AG3′ sites (Int4). These findings are consistent with reports demonstrating that a minority of introns contain a 5′ GC SS (25). We also observed that changing the third and fourth or the fifth and sixth dinucleotides at the 5′ SS to TG abolished CAT activity (Fig. 1 A; Int7 and Int8), indicating a critical role for the first 6 nt at the 5′ SS in premRNA splicing in T. vaginalis. To test whether a sequence identical to the consensus branch site (ACTAAC) of yeast introns (26) is required for splicing in T. vaginalis, we mutated this sequence in the 35-nt intron (Fig. 1 A, Int9) and found that CAT activity was abolished. Combined, these results show that the splicing machinery necessary to remove U2 spliceosomal introns from premRNAs is present in trichomonads, and that the same motifs used by yeast and metazoa are required for splicing in this deep-branching eukaryote.
To confirm that CAT activity reflects splicing and to demonstrate precise intron removal, we used RT-PCR to clone and sequence CAT mRNA. As shown in Fig. 1B, two products of the predicted size for unspliced and spliced mRNAs are found in cells transfected with the CAT gene introns bordered by either a GT (Int1) or a GC (Int4) 5′ SS. In contrast, transfectants that did not express detectable CAT activity (e.g., CT/Int2 and negative control cells/Int13) produce only a single RT-PCR product, corresponding in length to unspliced RNA (Fig. 1B). These data confirm that transfectants without activity produce CAT premRNA but are incapable of splicing it. Sequencing of the RT-PCR products showed accurate splicing by T. vaginalis of the canonical introns in Int1 and Int4 transfectants (Fig. 1B) and demonstrated that longer RT-PCR products contain the intron. The presence of unspliced RNA in Int1 and Int4 transfectants indicates inefficiency of splicing that might be attributed to the short length of the intron or the artificial exons that flank it. To demonstrate that the lack of gene transcription does not account for the absence of CAT activity, Northern blot analysis of total RNA was also performed, and all examined T. vaginalis transfectants harboring intron-containing CAT genes were shown to contain CAT premRNA (Fig. 1C).
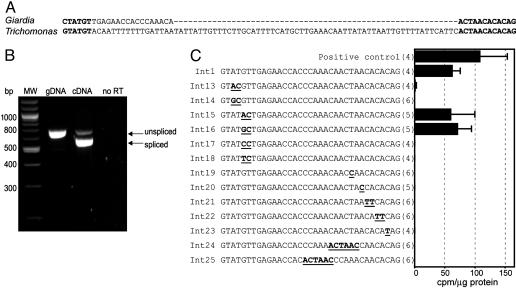
A T. vaginalis Gene Contains an Intron with Extended Conserved Motifs Found in the Only Known Giardia Intron. Having demonstrated the presence of splicing machinery in T. vaginalis, we searched for intron-containing genes on release of sequence data from the T. vaginalis Genome Sequencing Project (www.tigr.org/tdb/e2k1/tvg) by examining homologues of intron-containing genes in other protists. An ORF encoding a putative poly(A) polymerase (PAP1) (9) that is interrupted by a 94-nt sequence flanked by canonical 5′ GT-AG3′ SS was identified. Inspection of this putative intron exposed an unexpected identity of the last 12 nt, ACTAACACACAG, with those of the 35-nt Giardia intron (Fig. 2A). Moreover, 5 of the 6 nt at the putative 5′ SS of the two sequences are also identical (Fig. 2 A and C) and similar to that found in yeast introns. RT-PCR with PAP1-specific primers was used to show that the 94-nt sequence is an intron, because it is removed from the premRNA precisely at the predicted positions (Fig. 2B). The conservation at the 5′ SS and the atypical extended strict conservation at the 3′ SSs strongly suggest a functional role for these motifs. To test this, these sequences were mutated in the 35-nt CAT intron (Int1) and assayed for CAT activity. Single- or double-nucleotide mutations of 6 nt at the 5′ SS revealed that several positions could vary and still support splicing (T to C at position 3 and GT to AC or GC at positions 5 and 6; see Fig. 2C, Int13, Int15, and Int16). However, splicing activity was lost when the C at the fourth position was preceded by a G (GTGCGT) (Fig. 2C; Int14). Similarly, CAT activity was observed only when the C at position 6 was preceded by an A or a G (Fig. 2C; Int15 and Int16), because placing a T or C at position 5 abolished splicing (Fig. 2C; Int17 and Int18). Strict sequence requirements for the 12 nt at the 3′ end were found to be necessary for splicing (Fig. 2C, Int19 to Int25). Two single-nucleotide mutations in the putative branch site, the conserved A and T (Int 19, ACCAAC and Int 20, ACTACC) also abolished splicing, consistent with the role for this sequence as a branch site motif. Similarly, we observed that splicing is inactivated when the branch motif ACTAAC is moved 2 or 7 nt upstream (Int24 and Int25), indicating a spatial requirement for the motif immediately upstream of the 3′ SS. Finally, three additional mutants provide evidence that the entire 12-nt motif is required for splicing, because alterations in all positions 3–7 nt upstream of the 3′ AG disrupted splicing (Fig. 2C; Int 21, Int 22, and Int 23). These results led to the prediction that T. vaginalis introns are typically bordered by 5′-GYAYGY and ACTAACACACAG-3′ conserved motifs.
Fig. 2.
T. vaginalis poly(A) polymerase gene contains an intron with extended conserved 5′ and 3′ SS that are necessary for splicing. (A) Alignment of the only known Giardia intron and the T. vaginalis poly(A) polymerase (PAP) intron reveals identity at 5 of 6 nt at the 5′ SS and 12 or 12 at the 3′ SSs. (B) RT-PCR products generated by using T. vaginalis genomic DNA (gDNA) or mRNA (cDNA) as the template and primers flanking the putative intron in the PAP gene were separated on a 2% agarose gel. (C) CAT activity in T. vaginalis cells transfected with plasmid constructs of a CAT gene containing the Giardia 35-nt intron with mutations (Int13–25), as indicated in bold and underlined. The positive control is described in Fig. 1. CAT activity is expressed as cpm of n-butyrylated chloramphenicol per microgram of protein. The number of independent transfections assayed is shown in brackets, and bars designate standard error. A derived consensus sequence required at the 5′ and 3′ SS for CAT activity is listed in bold.
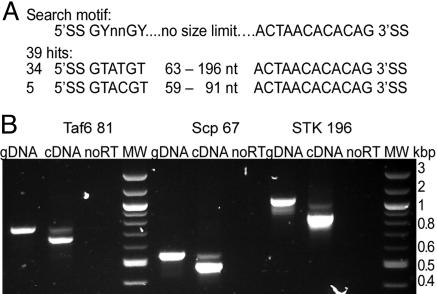
Large-Scale Search for Intron-Containing Genes in the T. vaginalis Genome. Strict conservation of 5′ and 3′ SS and placement of the putative branch site adjacent to the 3′ SS motif in introns from Giardia and T. vaginalis, as well as the observed requirement of these sequences for splicing, led us to search for other T. vaginalis intron-containing genes by using in silico methods. A database containing 2.7-fold coverage of the T. vaginalis genome (www.tigr.org/tdb/e2k1/tvg) was searched with the motif 5′-GYnnGY... no size limit... ACTAACACACAG-3′, and 39 putative introns were identified. These were all short and varied in length from 59 to 196 nt. Although our search motif allowed any nucleotide to be in positions 3 and 4 at the 5′ end search motif, 34 of the 39 hits contain the same 6 nt at the putative 5′ SS, GTATGT, and the remaining five differ by only 1 nt (GTACGT) (Fig. 3A). Further analyses of the 5′ end of these sequences revealed a strong bias for an A at position 7 (32 of 39) and a T at position 8 (33 of 39). Positions 9–13 likewise showed a relatively strong preference for a T (21 of 39 and 25 of 39, respectively) (Fig. 4, which is published as supporting information on the PNAS web site). Although a consensus sequence is common for the first 6 nt of U2 introns (3), extension of this conservation to 13 nt was unexpected.
Fig. 3.
In silico analysis reveals the presence of additional intron-containing genes. (A) The sequence used to search the T. vaginalis genome database (www.tigr.org/tdb/e2k1/tvg) for introns is shown (at the top), and the 5′ and 3′ motifs and length of the 39 sequences identified are listed (at the bottom). (B) RT-PCR products generated by using T. vaginalis genomic DNA (gDNA) or mRNA (cDNA) as the template and primers flanking the putative introns in genes encoding a TATA-binding protein-associated factor (Taf6–81), a small C-terminal domain phosphatase (Scp 67), and a STK (STK 196).
A search of the regions surrounding these putative introns by using artemis (27) was then performed, revealing the presence of interrupted ORFs in 26 introns, and blast analyses showed that 20 of these introns are orthologues of known genes in other organisms, whereas the other 8 encode unique proteins (Tables 2–4, which are published as supporting information on the PNAS web site). To directly test whether these sequences are introns, specific primers flanking three genes encoding a putative C-terminal domain phosphatase (scp67), a serine threonine kinase (STK) gene (stk196), and the transcription factor TAF6 (taf6–81) were used in RT-PCR. The sequences interrupting these ORFs were found to be precisely removed from their mRNAs, demonstrating they are introns (Fig. 3B).
Analyses of T. vaginalis Introns. Classification of the types of genes containing introns shows that 10 of 18 ORFs are homologous to different types of STKs. The remaining eight encode three C-terminal domain phosphatases (Scp), two centaurins, two TATA-binding protein-associated factors (Taf6), and one poly(A) polymerase (PAP1) (Table 2 and Fig. 4). To determine whether paralogous copies of these genes might also contain introns with an imperfect match to the search motif, blast analyses were performed, and two additional introns, one in a putative centaurin gene (cent115) and another in a STK gene (stk70), were found. These putative introns have a precisely conserved 5′-GTATGT motif but differ in 1 of 12 nt at the 3′ SS (Table 2 and Fig. 4).
We found that the position of introns in T. vaginalis genes is strongly biased toward the first third of the gene. Also slightly over half (15 of 28) of the introns are inserted between two codons [e.g., phase zero introns (28, 29)]. Eight interrupt codons after the first nucleotide (phase 1 introns) and five after the second nucleotide (phase 2 introns), resulting in a phase distribution of 5:2.7:1.7 (phases 0, 1, and 2, respectively; Table 5, which is published as supporting information on the PNAS web site). No strong nucleotide preference was observed in the exon immediately adjacent to the SS; however, an A or a T is often found in the last exon position bordering the 5′ SS. This is in contrast to that observed for U2 exon/intron borders of other eukaryotic introns where this position is G > 80% of the time (3).
T. vaginalis introns found in orthologous genes are of similar length and structure and are often, but not always, located in roughly the same position within the gene (Tables 3–5). This positional conservation indicates that the intron was present before gene multiplication and divergence. Not all orthologous genes have positionally conserved introns; for example, only one of the four PAP genes in the T. vaginalis genome database contains an intron (data not shown).
Intron Positions in Orthologous Genes from Organisms Representing Other Eukaryotic Lineages. Introns found in the same position in homologous genes of phylogenetically diverse species are interpreted to be “old” introns, whereas those in a unique position are referred to as “new” introns (30). To determine whether T. vaginalis introns are old or new, we compared their position to that of introns in orthologous genes from a protist and various yeast and metazoa. Sixty percent of the T. vaginalis introns were found to interrupt the gene in the exact position of an intron in an orthologous gene from at least one other eukaryotic lineage (Tables 1 and 5). Moreover, an intron was found in another eukaryotic orthologue within four amino acids of the position observed for all T. vaginalis introns. Given the extreme divergence of T. vaginalis proteins, this likely reflects amino acid divergence around an old intron as opposed to the addition of a new intron. These data indicate that some T. vaginalis genes contain old introns, present before the divergence of parabasalids from other eukaryotic lineages. Moreover, they reinforce the conclusion that introns were present when unicellular eukaryotes first evolved, and that their paucity in most extant protists and yeasts is the result of subsequent loss (31, 32).
Table 1. Conservation of intron position in T. vaginalis genes and in orthologous genes from other eukaryotic lineages.
| Gene/organism | H.s. | D.m. | C.e. | A.t. | S.p. | S.c. | P.f. |
|---|---|---|---|---|---|---|---|
| PAP1 94 | 0 | — | — | — | — | — | — |
| Cent 116, 122, 127 | 0 | 0 | — | — | — | — | — |
| SCP 67, 70, 76 | — | — | 2 | — | — | — | — |
| TAF6 81, 91 | 3 | — | 1 | 4 | — | — | — |
| STK 93 | 3 | 3 | 0 | 2 | 4 | — | — |
| STK 59, 68, 105 | — | — | — | 0 | — | — | 4 |
| STK 70, 78 | — | 2 | 3 | 4 | 1 | — | — |
| STK 110, 114, 134 | 0 | 1 | 0 | 3 | 3 | — | — |
| STK 99 | 0 | — | 2 | 3 | — | — | — |
| STK 196 | 2 | 3 | 2 | 2 | — | — | 4 |
Genes are abbreviated as follows: PAP, poly(A) polymerase; SCP, small C-terminal domain phosphatase; and TAF6, TBP-associated factor TAF6, where numbers correspond to intron length. Organisms are abbreviated as follows: H.s., Homo sapiens; D.m., Drosophila melanogaster; C.e., Caenorhabditis elegans; A.t., Arabidopsis thaliana; S.c., Saccharomyces cerevisiae; S.p., Schizosaccharomyces pombe; and P.f., Plasmodium falciparum. 0 indicates an intron at the same position, and 1, 2, 3, or 4 indicates the presence of an intron shifted in position by one to four amino acids relative to the T. vaginalis intron. — indicates the absence of an intron in a similar position in the orthologous gene. See Table 5 and Supporting Text for detailed analyses.
Discussion
We have shown that T. vaginalis, a protist of the parabasalid deep-branching eukaryotic lineage, possesses splicing machinery that functionally depends on standard conserved motifs and have identified the first intron-containing genes in this lineage. Our studies illustrate the conservation of functional splicing signals across large evolutionary expanses, show that T. vaginalis contain old introns, and imply that spliceosomal introns were present in genes of the common ancestor of all extant eukaryotes.
In addition to the conservation of typical U2 introns, extended 5′ and 3′ splicing motifs were found in all examined T. vaginalis introns. These extended motifs, which are also found in the only reported intron in Giardia (16, 33), a protist belonging to the sister diplomonad lineage, appear to be required for splicing, because their mutation abolishes splicing. Strikingly, 100% nucleotide identity was observed in five of six positions at the 5′ SS with 92% identity at the remaining position in 41 putative T. vaginalis introns and the Giardia intron. Likewise, 11 or 12 nt at the 3′ SS of these introns are identical, with the remaining position exhibiting 95% identity. Although the first 6 nt at the 5′ SS are often conserved in U2 introns, particularly in yeast introns (3), the extraordinarily high level of conservation of 12 nt at the 3′ SS in T. vaginalis and Giardia introns is unprecedented. Moreover, the remarkable conservation of this trait between two lineages and their requirement for splicing imply a difference in SS recognition and/or intron excision/ligation by spliceosomes in these unicellular organisms. On the other hand, endogenous genes with introns of variable lengths and exon/intron compositions that allow for a degree of nucleotide flexibility incompatible with our assay may be present that do not require strict conservation of SS sequences.
Remarkably, the putative branch site motif (ACTAAC) required for splicing in T. vaginalis is contained within the highly conserved 12-nt 3′ motif, from –7 to –12 nt. Attempts to exchange this sequence with upstream sequences eliminated splicing, indicating that the distance between the 3′ SS and this element is critical. This spacing requirement is also consistent with the ability of T. vaginalis to splice the 5′ GT-Giardia intron (Int1) and not three others, all of which contain standard 5′ and 3′ SSs but do not have their branch site motifs adjacent to the 3′ SS. This immediate proximity of the branch site and the 3′ end in T. vaginalis introns, an extremely rare trait of yeast and metazoan introns, promotes speculation that their spliceosomes may combine the steps of branch site and 3′ SS recognition. Interestingly, an essential spliceosomal protein, Prp8, that interacts with both 5′ and 3′ canonical motifs and the branch site (34), is the only reported spliceosomal protein in T. vaginalis (18). Thus, the close juxtaposition of the branch site and the 3′ SS may reflect a simplified mechanism for spliceosomal assembly in T. vaginalis, where Prp8 plays an indispensable role. In silico analyses to determine whether genes encoding other critical spliceosomal proteins are present in trichomonads await a complete coverage of the genome sequence. The structural similarity between T. vaginalis and Giardia spliceosomal introns and group II self-splicing introns, a class of introns thought to be evolutionarily related to spliceosomal small nuclear RNA (snRNA) machinery (5, 35), is noteworthy. The distance (7 nt) between the branch site and the 3′ SS of group II introns is the same as that observed for the putative branch site contained with the conserved 12 nt at the 3′ SS of Trichomonas and Giardia introns. The use of analogous splicing pathways by group II introns and nuclear premRNA has been taken to support a common evolutionary origin (35). The possibility that T. vaginalis and Giardia introns represent evolutionary intermediates possessing features of both systems is an exciting one, with important mechanistic implications. Despite strong conservation of canonical 5′ GT-AG3′ and branch site motifs in T. vaginalis introns implying that trichomonad snRNAs would likewise be conserved, these RNAs have yet to be identified.
We have shown that Trichomonas is capable of splicing a 35-nt intron, whereas most eukaryotic introns have a minimal length of ≈50 nt (36). This minimal length has been interpreted to ref lect physical constraints imposed by the spliceosome. Whether the ability to splice such short introns by T. vaginalis and Giardia reflects significant differences in their splicing components and/or mechanisms awaits further analyses.
The complete sequence and annotation of genomes of numerous eukaryotes have allowed large-scale comparisons of intron position(s) in phylogenetically diverse organisms (31). With rare exceptions, intron-poor unicellular eukaryotes have been found to have a significant 5′ bias in intron position, a property not observed in metazoan genes (31, 37). Our analyses of T. vaginalis introns are consistent with these findings, because introns are typically found within the first one-third of the gene. These data support the argument that the paucity of introns in unicellular eukaryotes results from a secondary loss, driven by homologous recombination between the gene and an intronless cDNA derived by reverse transcription of the corresponding mRNA (31, 37, 38), as opposed to introns simply being less abundant during early eukaryotic evolution.
Two theories have dominated the “origin of introns” debate. The first posited that introns were present in the common ancestor of all cells and were subsequently lost in extant eubacteria, archaea, and certain unicellular eukaryotes (the “intronearly” hypothesis or the exon theory of genes) (28, 39–42), and the second postulated that introns were added to genes in eukaryotes and never existed in orthologous genes in introndeficient organisms (43, 44) (the “intron-late” hypothesis). At the crux of this debate is the issue of whether introns were present in the last common ancestor of archaea, eubacteria, and eukaryotes. The data presented here support the intron-early hypothesis.
A mixed model of intron evolution incorporating features of both the intron-early and intron-late hypotheses has recently been embraced (27–30, 37, 39, 45–48). Comparative genomics have revealed that numerous introns present in a common ancestor of fungi, nematodes, and arthropods were lost in these lineages but retained in vertebrates and plants (29, 39). When present, such introns are located in the same position in divergent orthologues and are referred to as old introns. Unique positions for introns in the genes of specific groups of vertebrates and plants have also provided evidence that introns can be added to genes (49), so-called new introns (39, 48, 50). Consistent with the hypothesis that the 5′ positional bias of introns in T. vaginalis genes is indicative of secondary loss of introns, we have shown that many of the retained introns in T. vaginalis genes are old. The position of ≈60% of examined T. vaginalis introns are conserved in orthologous genes from at least one other eukaryotic lineage. This observation and the requirement of canonical motifs for splicing in this deep-branching eukaryote indicate that introns and the spliceosomal machinery necessary for their excision were present in the ancestral eukaryote. This places spliceosomes on the roster of essential defining eukaryotic features and establishes the presence of introns in all examined eukaryotes.
Supplementary Material
Acknowledgments
We thank Drs. Doug Black, Richard Stefl, and Nancy Sturm, as well as our colleagues in the lab, for helpful discussions and critical comments on the manuscript, and we thank Reviewer no. 1 for insightful comments. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant R01A I30537, and a Co-Operative Agreement Award (U01AI050913) to The Institute for Genomic Research to sequence the genome of T. vaginalis. P.J.J. is a Burroughs Wellcome Scholar in Molecular Parasitology.
Author contributions: S.V., W.Y., and P.J.J. designed research; S.V. and W.Y. performed research; J.M.C. contributed new reagents/analytic tools; S.V. and P.J.J. analyzed data; and S.V. and P.J.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SS, splice site; CAT, chloramphenicol acetyltransferase; STK, serine-threonine protein kinase.
References
- 1.Reed, R. (2000) Curr. Opin. Cell. Biol. 12, 340–345. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. (2000) Cell 103, 367–370. [DOI] [PubMed] [Google Scholar]
- 3.Sharp, P. A. & Burge, C. B. (1997) Cell 91, 875–879. [DOI] [PubMed] [Google Scholar]
- 4.Belfort, M., Derbyshire, V., Parker, M. M., Cousineau, B. & Lambowitz, A. M. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 761–783.
- 5.Bonen, L. & Vogel, J. (2001) Trends Genet. 17, 322–331. [DOI] [PubMed] [Google Scholar]
- 6.Tarn, W. Y. & Steitz, J. A. (1997) Trends Biochem. Sci. 22, 132–137. [DOI] [PubMed] [Google Scholar]
- 7.Moore, M. J. (2000) Nat. Struct. Biol. 7, 14–16. [DOI] [PubMed] [Google Scholar]
- 8.Ebel, C., Frantz, C., Paulus, F. & Imbault, P. (1999) Curr. Genet. 35, 542–550. [DOI] [PubMed] [Google Scholar]
- 9.Mair, G., Shi, H., Li, H., Djikeng, A., Aviles, H. O., Bishop, J. R., Falcone, F. H., Gavrilescu, C., Montgomery, J. L., Santori, M. I., et al. (2000) RNA 6, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchhal, U. S. & Schwartzbach, S. D. (1994) Nucleic Acids Res. 22, 5737–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessier, L. H., Paulus, F., Keller, M., Vial, C. & Imbault, P. (1995) J. Mol. Biol. 245, 22–33. [DOI] [PubMed] [Google Scholar]
- 12.Wilihoeft, U., Campos-Gongora, E., Touzni, S., Bruchhaus, I. & Tannich, E. (2001) Protist 152, 149–156. [DOI] [PubMed] [Google Scholar]
- 13.Baldauf, S. L., Roger, A. J., Wenk-Siefert, I. & Doolittle, W. F. (2000) Science 290, 972–977. [DOI] [PubMed] [Google Scholar]
- 14.Horner, D. S. & Embley, T. M. (2001) Mol. Biol. Evol. 18, 1970–1975. [DOI] [PubMed] [Google Scholar]
- 15.Simpson, A. G., Roger, A. J., Silberman, J. D., Leipe, D. D., Edgcomb, V. P., Jermiin, L. S., Patterson, D. J. & Sogin, M. L. (2002) Mol. Biol. Evol. 19, 1782–1791. [DOI] [PubMed] [Google Scholar]
- 16.Nixon, J. E., Wang, A., Morrison, H. G., McArthur, A. G., Sogin, M. L., Loftus, B. J. & Samuelson, J. (2002) Proc. Natl. Acad. Sci. USA 99, 3701–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson, A. G., MacQuarrie, E. K. & Roger, A. J. (2002) Nature 419, 270. [DOI] [PubMed] [Google Scholar]
- 18.Fast, N. M. & Doolittle, W. F. (1999) Mol. Biochem. Parasitol. 99, 275–278. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, P. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond, L. S. (1957) J. Parasitol. 43, 488–490. [PubMed] [Google Scholar]
- 21.Ortiz, D. & Johnson, P. J. (2003) Mol. Biochem. Parasitol. 128, 43–49. [DOI] [PubMed] [Google Scholar]
- 22.Delgadillo, M. G., Liston, D. R., Niazi, K. & Johnson, P. J. (1997) Proc. Natl. Acad. Sci. USA 94, 4716–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urban, B., Blasig, C., Forster, B., Hamelmann, C. & Horstmann, R. D. (1996) Mol. Biochem. Parasitol. 80, 171–178. [DOI] [PubMed] [Google Scholar]
- 24.Konarska, M. M., Grabowski, P. J., Padgett, R. A. & Sharp, P. A. (1985) Nature 313, 552–557. [DOI] [PubMed] [Google Scholar]
- 25.Burset, M., Seledtsov, I. A. & Solovyev, V. V. (2000) Nucleic Acids Res. 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp, P. A. (1994) Cell 77, 805–815. [DOI] [PubMed] [Google Scholar]
- 27.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 28.Roy, S. W. (2003) Genetica 118, 251–266. [PubMed] [Google Scholar]
- 29.Zhaxybayeva, O. & Gogarten, J. P. (2003) Curr. Biol. 13, 764–766. [DOI] [PubMed] [Google Scholar]
- 30.Fedorov, A., Merican, A. F. & Gilbert, W. (2002) Proc. Natl. Acad. Sci. USA 99, 16128–16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mourier, T. & Jeffares, D. C. (2003) Science 300, 1393. [DOI] [PubMed] [Google Scholar]
- 32.Roy, S. W., Fedorov, A. & Gilbert, W. (2003) Proc. Natl. Acad. Sci. USA 100, 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McArthur, A. G., Morrison, H. G., Nixon, J. E., Passamaneck, N. Q., Kim, U., Hinkle, G., Crocker, M. K., Holder, M. E., Farr, R., Reich, C. I., et al. (2000) FEMS Microbiol. Lett. 189, 271–273. [DOI] [PubMed] [Google Scholar]
- 34.Siatecka, M., Reyes, J. L. & Konarska, M. M. (1999) Genes Dev. 13, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacquier, A. (1996) Biochimie 78, 474–487. [DOI] [PubMed] [Google Scholar]
- 36.Deutsch, M. & Long, M. (1999) Nucleic Acids Res. 27, 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakurai, A., Fujimori, S., Kochiwa, H., Kitamura-Abe, S., Washio, T., Saito, R., Carninci, P., Hayashizaki, Y. & Tomita, M. (2002) Gene 300, 89–95. [DOI] [PubMed] [Google Scholar]
- 38.Fink, G. R. (1987) Cell 49, 5–6. [DOI] [PubMed] [Google Scholar]
- 39.Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G. & Koonin, E. V. (2003) Curr. Biol. 13, 1512–1517. [DOI] [PubMed] [Google Scholar]
- 40.de Souza, S. J., Long, M., Schoenbach, L., Roy, S. W. & Gilbert, W. (1996) Proc. Natl. Acad. Sci. USA 93, 14632–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell, J. E. & Doolittle, W. F. (1986) Proc. Natl. Acad. Sci. USA 83, 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert, W., Marchionni, M. & McKnight, G. (1986) Cell 46, 151–153. [DOI] [PubMed] [Google Scholar]
- 43.Palmer, J. D. & Logsdon, J. M., Jr. (1991) Curr. Opin. Genet. Dev. 1, 470–477. [DOI] [PubMed] [Google Scholar]
- 44.Cavalier-Smith, T. (1991) Trends Genet. 7, 145–148. [PubMed] [Google Scholar]
- 45.Fedorov, A., Roy, S., Fedorova, L. & Gilbert, W. (2003) Genome. Res. 13, 2236–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fedorov, A., Roy, S., Cao, X. & Gilbert, W. (2003) Genome. Res. 13, 1155–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoltzfus, A. (2004) Curr. Biol. 14, 351–352. [DOI] [PubMed] [Google Scholar]
- 48.Qiu, W. G., Schisler, N. & Stoltzfus, A. (2004) Mol. Biol. Evol. 21, 1252–1263. [DOI] [PubMed] [Google Scholar]
- 49.Logsdon, J. M., Jr., Stoltzfus, A. & Doolittle, W. F. (1998) Curr. Biol. 8, 560–563. [DOI] [PubMed] [Google Scholar]
- 50.Sverdlov, A. V., Rogozin, I. B., Babenko, V. N. & Koonin, E. V. (2003) Curr. Biol. 13, 2170–2174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.