ABSTRACT
Interleukin-15 (IL-15) is a critical regulator of immune responses, especially at mucosal interfaces within the gastro-intestinal tract. Here, we describe the discovery and characterization of a humanized antibody to IL-15. Data from its epitope and mode of action, cell biology and primate pharmacology, as well as translational studies in human samples and in vivo proof-of-concept experiments in mouse models demonstrate the therapeutic potential of this new antibody targeting IL-15 for refractory celiac disease and eosinophilic esophagitis.
KEYWORDS: Antibody, celiac disease, cis, eosinophilic esophagitis, humanized, interleukin-15, IL-15, trans
Introduction
Interleukin-15 (IL-15) is member of the 4 α-helix bundle family of cytokines (for reviews, see refs.1,2,3). It uses the common gamma chain (IL-15Rγ) shared with IL-2, IL-4, IL-7, IL-9 and IL-21 as part of its receptor complex to trigger signaling pathways leading to cellular activation, proliferation and survival. It also shares the receptor chain IL-2/IL-15Rβ with IL-2, but uniquely binds to the IL-15Rα receptor chain with very high affinity. Cells of the innate immune system such as macrophages and dendritic cells, but also a variety of structural cells including fibroblasts and epithelial cells, can produce IL-15. Production of mature IL-15 occurs after stimulation with a variety of immune stimuli including Toll-like receptor ligands, microbes and cytokines. The IL-15Rα receptor chain is rather widely expressed by haematopoietic and non-haematopoietic cells, while IL-15Rβγ expression is more tightly regulated. IL-15 can signal in a cell contact-dependent manner through the trans presentation of IL-15/IL-15Rα complexes to a different cell that expresses the IL-15Rβγ complex.4 It can also act as a free cytokine on cells that express IL-15Rαβγ or IL-15Rβγ, including through recycling of intracellular IL-15/IL-15Rα complexes, a signaling mode herein referred as cis presentation. It is believed that the equilibrium between cis and trans presentation plays an important role in regulating IL-15 functions.5 IL-15 receptor signaling induces Janus kinase 1 (JAK1) – signal transducer and activation of transcription 3 (STAT-3) and JAK3–STAT5 activation through the β and γ chains, respectively.1,3
While it was originally described mostly as a T and natural killer (NK) cell growth factor, in a way not that different from IL-2,1 IL-15 has emerged through recent years as a more complex regulatory cytokine, at the interface between innate and adaptive immunity and playing a major role in the control of homeostasis and immune responses within specific environments (for reviews, see refs.2,3,6). Hence it appears that dysregulation of responses controlled by IL-15 could be an important component of certain human immune pathologies. A causative role for IL-15 was first evoked for diseases such as rheumatoid arthritis and psoriasis, but more recently IL-15 has been most convincingly implicated in gastro-intestinal pathologies.2,7,8,9 This is likely owing to the very special role of IL-15 in shaping and controlling the mucosal interface within the gastro-intestinal tract.2
In inflammatory bowel diseases (IBD), both IL-15 and IL-15Rα expression are increased compared with healthy controls.10,11 Mice rendered genetically deficient for IL-15 are protected in both the acute and chronic dextran sulfate sodium-induced models of colitis.12 Celiac disease (CeD) is another gastro-intestinal immune pathology that has frequently been associated with IL-15 (for reviews, see refs.13 and14). Early on, it was recognized that IL-15 could drive the immune pathological features of CeD by acting both on intestinal epithelial cells and on discrete population of mucosa-associated lymphocytes.15 IL-15 can be expressed by the intestinal epithelium or cells within the lamina propria, but dual expression in both compartments seems to be necessary to trigger villous atrophy in CeD.14 Current knowledge suggests that IL-15 triggers CeD through multiple mechanisms including: 1) loss of tolerance to oral antigens (gluten) through stimulation of mucosal dendritic cells that acquire an inflammatory phenotype; 2) loss of immune regulation since IL-15 can render effector T cells resistant to regulatory T cell (Treg) suppression; 3) increased epithelial destruction through activation of lymphokine killer activity; and 4) increased survival of deleterious populations of intra-epithelial lymphocytes that can even develop in a small number of patients into a cryptic form of lymphoma known as type II refractory celiac disease (RCD).13,14,16,17,18,19 It has been proposed that IL-15 plays such pleiotropic effects in CeD because its physiologic role is to serve as a danger signal to regulate tissue-resident T cells and tissue destruction in the gut.6
Owing to this newly recognized function as a master immune checkpoint in gut immunology, it is likely that the incrimination of IL-15 will further expand into other pathologies of the gastrointestinal tract. Indeed, a deleterious role for IL-15 has been hypothesized for pathologies that are currently attracting substantial attention due to their increasing impact on human health. Those include eosinophilic esophagitis (EoE), an immune-mediated chronic inflammation of the esophagus triggered by food antigens and characterized by a local influx of eosinophils20 (for reviews, see refs.21,22,23), type I diabetes6,24 and non-alcoholic fatty liver disease.25
Thus, IL-15 represents an attractive target for several human pathologies with high medical need. There is, however, no approved treatment that specifically neutralizes IL-15, despite some early encouraging studies of a monoclonal antibody in rheumatoid arthritis patients.8,9 This might be related to the fact that IL-15 has been an elusive and difficult target for the development of potent and fully in vivo active neutralizing antibodies.26,27 We describe herein the discovery and characterization of CALY-002, a best-in-class neutralizing antibody against IL-15, and provide data supporting its relevance for the treatment of RCD and EoE.
Results
Characterization of CALY-002 epitope and mechanism of action
Epitope mapping of CALY-002 was performed by analyzing its binding to libraries of structured peptides designed to represent linear but also discontinuous epitopes of IL-15 by using the Chemically Linked Peptides on Scaffolds (CLIPS).28 The CLIPS technology allows peptides to be structured into single loops, double loops, triple loops, sheet-like folds, helix-like folds, and combinations thereof. It was found using this technique that CALY-002 preferentially binds to linear and constrained peptides containing the peptide sequence 109DTVENLIILAN119. This observation is similar to that reported for the originator antibody, B-E29.27 Using single amino-acid substitution, residues found to be most essential for the binding of CALY-002 to IL-15 were D109, E112, I116 and N119. However, in contrast with what was proposed for the originator B-E29 antibody,27,26 we found that CALY-002 did not prevent binding of IL-15 to IL-15Rα in an in vitro binding assay using biotinylated IL-15 as described previously (Fig. 1A).26 Indeed, since the quaternary structure of IL-15 in complex with IL-15Rαβγ was resolved, it was possible to visualize the epitope recognized by CALY-002 within this complex (Fig. 1B).29 The 109DTVENLIILAN119 motif is part of the IL-15 α-helix C, located within the vicinity of the IL-15Rβ chain. Furthermore, the D109 residue found by us to be critical for CALY-002 epitope was described as one of the key residues for the direct interaction between IL-15 and IL-15Rβ (Fig. 1B).29 We therefore propose that the mechanism of action of CALY-002 is to prevent IL-15 from interacting with IL-15Rβ, hereby preventing signaling through the IL-15Rβγ complex.
Figure 1.
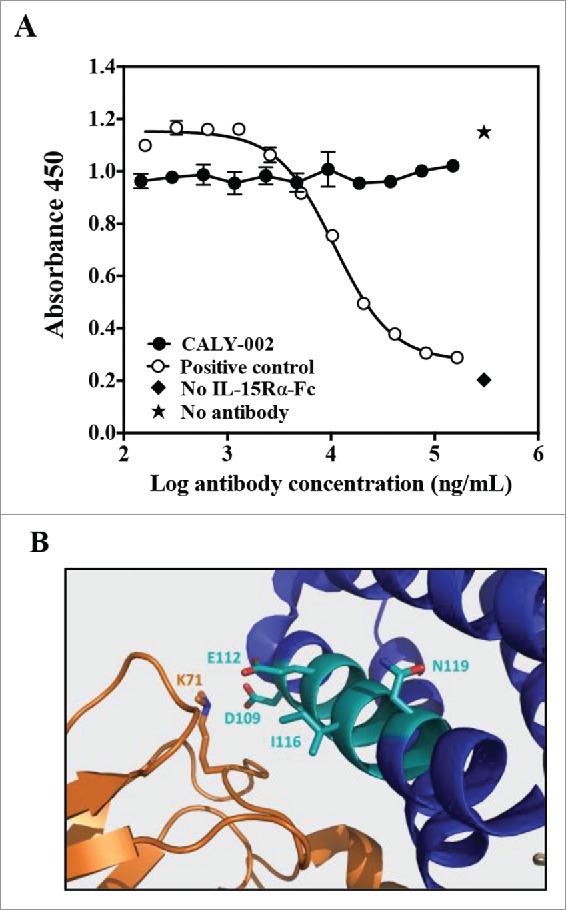
CALY-002 epitope and mode of action. (A) CALY-002 does not prevent IL-15 binding to IL-15Rα. Biotinylated hIL-15 at a single concentration was incubated with immobilized IL-15Rα-Fc in the presence of a titration of antibodies, as described. Data represent mean ± SD of IL-15 binding signal expressed as 450 nM absorbance from one representative experiment. (B) The published structure of the IL-15/IL-15Rαβγ quaternary complex was used to visualize the IL-15 binding epitope of CALY-002. Interleukin-15 is shown in blue, the IL-15 binding motif of CALY-002 in cyan, and IL-15Rβ in orange. The side chain of the critical residues of the binding motif of CALY-002 is highlighted: D109, E112, I116, and N119. K71 of IL-15Rβ directly interacts with D109 of IL-15.
Characterization of in vitro CALY-002 effects in IL-15-dependent assays
Alongside and following the humanization and sequence optimization of the anti-IL-15 antibody CALY-002, its activity was tested in a variety of specific in vitro assays. First, kinetic interaction of CALY-002 with immobilized IL-15 was measured by surface plasmon resonance (SPR), showing that CALY-002 binds to IL-15 with a tight 9.4 pM affinity (Table 1). Furthermore, the CALY-002/IL-15 complex was found to be very stable with a measured off-rate (koff) close to the limit of the measurement range (Table 1). In an ELISA-type assay, CALY-002 was shown to bind human and cynomolgus monkey IL-15 in a dose-dependent manner with similar potency, as measured by a subnanomolar half-maximum inhibitory binding capacity (BC50) (Table 1). This was improved compared with a chimeric antibody comprising the variable domain of the original B-E29 antibody grafted onto a human IgG1 framework similar to that of CALY-002, showing an average BC50 of 4.23 nM on human IL-15. On the other hand, CALY-002 did not bind to either mouse IL-15, rat IL-15 or human IL-2 (Table 1). Of note, CALY-002 binds equally well to recombinant human IL-15 from E. coli or mammalian cell origin (data not shown).
Table 1.
Characterization of the binding affinity, species specificity and selectivity of CALY-002. Kinetic interaction of CALY-002 with recombinant IL-15 was measured by surface plasmon resonance. Dissociation (koff) and association (kon) rate constants were obtained by nonlinear regression analysis. KD = koff /kon. Selectivity and selectivity of CALY-002 toward recombinant human, monkey, mouse and rat IL-15 as well as human IL-2 was tested by ELISA. Whenever appropriate, the half-maximum binding concentration of CALY-002 (BC50) was calculated. Results are expressed as the mean of 3 independent experiments ± SD.
| Surface plasmon resonance |
ELISA binding assays |
|||||||
|---|---|---|---|---|---|---|---|---|
| Assays | kon | Koff | KD | BC50 | BC50 | Binding | Binding | Binding |
| (1/Ms) | (1/s) | (pM) | Human | Monkey | rat | mouse | human | |
| |
|
|
|
IL-15 (nM) |
IL-15 (nM) |
IL-15 |
IL-15 |
IL-2 |
| CALY-002 characteristics | 1.15 × 106 | 1.07 × 10−5 | 9.4 | 0.55 ± 0.46 | 0.43 ± 0.11 | No Binding | No Binding | No Binding |
Since IL-15 enhances proliferation and survival of a variety of cell lines,26,27 effects of various concentrations of CALY-002 were tested on cell survival/proliferation induced by recombinant IL-15, and CALY-002 potency measured as its half-maximum inhibitory concentration (IC50). Two different cell lines were tested. The M-07e cell line only expresses IL-15Rβ and IL-15Rγ.26 This system allows for testing the effect of antibodies on IL-15 cis presentation when IL-15 is given as a soluble cytokine. In addition, trans presentation can also be assessed using M-07e cells by first immobilizing recombinant IL-15 onto recombinant IL-15Rα –Fc and then adding M-07e cells, as previously shown.26 A second cell line was used: Kit 225 cells that express IL-15Rα, IL-15Rβ and IL-15Rγ. In this test system, a mixture of cis and trans presentation of IL-15 can co-exist, since IL-15 can possibly be presented from one cell to the other following binding to IL-15Rα; it might therefore represent a more physiologic situation. In all systems tested, CALY-002 showed potent inhibition of human IL-15-induced proliferation (Table 2), demonstrating that CALY-002 is a strong neutralizer of IL-15 cis and trans presentation. Furthermore, whenever tested, CALY-002 inhibited equally well human and cynomolgus monkey recombinant IL-15 (Table 2).
Table 2.
CALY-002 inhibits IL-15-mediated cell proliferation. The effect of CALY-002 on the proliferation and survival of M-07e and Kit 225 cells induced by recombinant soluble human or monkey IL-15, or human IL-15 immobilized onto recombinant human IL-15Rα-Fc, was measured as described in Materials and Methods. The half-maximum inhibitory concentration (IC50) of CALY-002 in each assay was determined and results are expressed as mean IC50 ± SD for at least 3 experiments for each condition tested.
| Soluble IL-15 |
hIL-15 immobilized on hIL-15Rα-Fc | ||||
|---|---|---|---|---|---|
| Stimulus |
Human |
Human |
Monkey |
Monkey |
Human |
| Cell line | M-07e | Kit 225 | M-07e | Kit 225 | M-07e |
| IC50 on cell proliferation (nM) | 0.26 ± 0.38 | 0.05 ± 0.03 | 0.33 ± 0.15 | 0.19 ± 0.15 | 0.19 ± 0.15 |
Characterization of in vivo CALY-002 effects in IL-15-dependent assays and pharmacokinetics profile in cynomolgus monkeys
It has been shown that pre-formed IL-15/IL-15Rα complexes have potent agonistic effect in vivo, stronger than IL-15 alone when compared on a molar basis.26 This model was used to test the in vivo inhibitory capacity of CALY-002 for human IL-15. Administration of freshly prepared human IL-15/IL-15Rα-Fc complexes on Day 1, Day 2 and Day 3 induced strong NK cell expansion in mouse spleen at Day 4 that was fully inhibited by a single 100 μg injection of CALY-002 at Day 1, just prior injection of IL-15/IL-15Rα-Fc (Fig. 2A).
Figure 2.
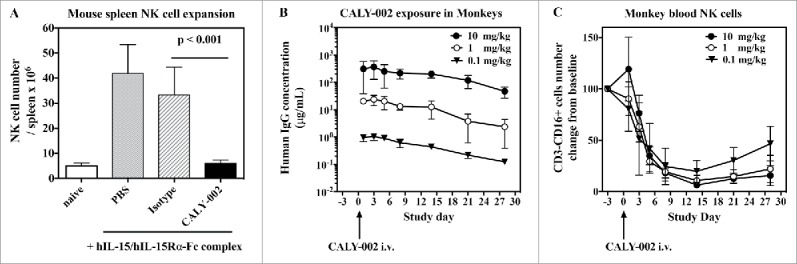
Pharmacodynamic inhibition of IL-15 by in vivo in mice and monkeys. (A) NK cell expansion was induced in groups of 5 mice by injections of hIL-15 mixed with hIL-15Rα-Fc at Day 1, 2 and 3. PBS (gray bar), CALY-002 (black bar) or control isotype antibody (dashed bar) were injected 20 minutes before the first injection of hIL-15/hIL-15Rα-Fc. Animals were killed at Day 4 and number of NK cells present in individual spleens were determined. Naive mice were injected with saline alone (no hIL-15/hIL-15Rα-Fc, white bar). Data are plotted as mean ± SD. Statistical analysis: Student's t test. (B) Serum levels of human IgG in cynomolgus monkeys receiving a single i.v. dose of 10 mg/kg (closed triangles), 1 mg/kg (open circles) or 0.1 mg/kg (closed circles) CALY-002. Results are expressed as mean ± SD for 3 to 4 animals per group. (C) CALY-002 reduces circulating NK cell numbers in cynomolgus monkeys. Following a single dose injection of 10 mg/kg (closed triangles), 1 mg/kg (open circles) or 0.1 mg/kg (closed circles) CALY-002, animals underwent a 4-wk observation period and NK cell counts were determined by flow cytometry at Day −3 (pre-dose), and 1, 3, 5, 8, 14, 21 and 28 d after treatment. Data are plotted as mean absolute number of blood CD3−CD16+ cells NK cells ± SD for 3 to 4 animals per group.
Administration of anti-IL-15 antibodies to cynomolgus or rhesus monkeys was shown to induce a decrease of circulating NK cell numbers within a 2-week period.39,40 Since CALY-002 is able to bind and neutralize cynomolgus monkey IL-15 (Tables 1 and 2), we intravenously (i.v.) administered a single injection of CALY-002 at 3 different doses (0.1 mg/kg, 1 mg/kg and 10 mg/kg) to groups of cynomolgus monkeys. We first measured along time serum levels of the administered antibody (Fig. 2B). The pharmacokinetics parameters of CALY-002 after i.v. injection to cynomolgus monkeys were calculated and found to be similar to that reported for other IgG1 therapeutic antibodies, with a terminal half-life of 6 to 9 d (Table S1).41 Furthermore, a dose-related and gradual decrease in the number of peripheral NK cells was observed from Day 3 following administration of CALY-002 (Fig. 2C). The lowest mean NK cell counts were reached on Day 14, and represented respectively 19.8, 10.8 and 6.4% of the low, mid and high dose groups corresponding baseline value. No similarly sustained changes were observed in response to CALY-002 with the different T cell subsets analyzed using the panel of markers described in Materials and Methods: Total T cells, helper T cells, cytotoxic T cells, naïve T cells, central memory T cells (TCM) and effector memory T cells (TEM) generally remained comparable to the baseline levels. This experiment shows that CALY-002 is a very potent inhibitor of IL-15 activity in primates, with almost maximal effect observed after a single injected dose of 0.1 mg/kg.
Effects of CALY-002 in in vitro and in vivo models of type II refractory celiac disease
type II RCD is a form of cryptic intra-epithelial lymphoma that very seldom develops in CeD patients, leading to severe epithelial lesions and malnutrition. IL-15 is thought to play an important role in the pathogenesis of CeD, and especially in that of type II RCD, notably by promoting enteropathy and favoring the onset and expansion of intra-epithelial lymphocyte subset(s) that can transform into lymphoma.14,16,18,19 In vitro, recombinant IL-15 was shown to enhance proliferation and survival of lymphocyte cell lines isolated from type II RCD patients.16 We show, in 3 different type II RCD patient cell lines, that CALY-002 is able to fully revert the protection from apoptosis provided by IL-15, and to fully block IL-15-induced STAT5 phosphorylation (Fig. 3). In one of these cell lines, a full dose-response was performed and we obtained an IC50 of 2.4 nM for reversion of IL-15-induced prevention of apoptosis by CALY-002 (data not shown).
Figure 3.
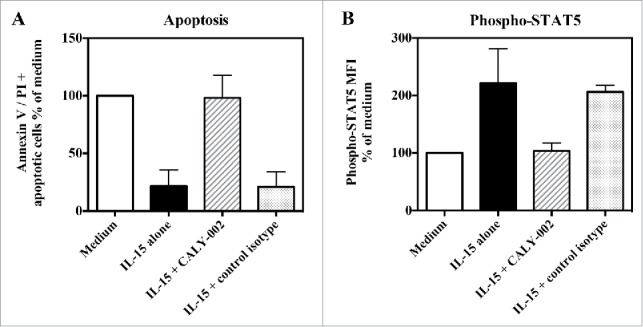
CALY-002 inhibits IL-15–induced survival and STAT5 phosphorylation of type II RCD patient IEL cell lines. Type II RCD IEL cell lines were cultured with medium (white bars), IL-15 (black bars), IL-15 with CALY-002 (hatched bars) or IL-15 with control isotype (gray bars) for 48h. (A) Percentages of apoptotic cells in cultured IELs were determined by labeling using an AnnexinV-APC/PtdIns Kit and (B) phosphorylated STAT5 was stained with PEcy7-labeled anti-p-STAT5 antibody. Results were transformed as percentage of control (medium) for the percentage of apoptotic cells or the MFI of phosphorylated STAT5 intracellular expression induced by IL-15, and expressed as mean ± SD of results obtained with 3 different type II RCD IEL cell lines.
type II RCD can partly be modeled in vivo in mice overexpressing human IL-15 in the gut epithelium (IL-15TgE mice). Those mice show massive accumulation of gut intra-epithelial lymphocyte populations, as well as cell expansion in other lymphoid compartments.16,42 Compared with normal B6 mice, IL-15TgE mice were characterized by enlarged spleens with massive accumulation of CD8 T cells (CD3+CD8+), NK cells (CD45+CD3−NK1.1+) and NKT cells (TCRαβ+NK1.1+), whereas CD4 T cells were only slightly increased and B cell levels were normal (Fig. 4). In the gut, intra-epithelial populations of CD4 and CD8 T cells as well as NK and NKT cells, but not B cells, were increased. Treatment with CALY-002 for 4 weeks significantly reduced, when compared with treatment with a control isotype antibody, all subsets of spleen lymphocytes described above except B and CD4 T cells, and all subsets of IELs except B cells (Fig. 4). In addition, CALY-002 treatment also normalized populations of lamina propria lymphocytes (LPL) (data not shown).
Figure 4.
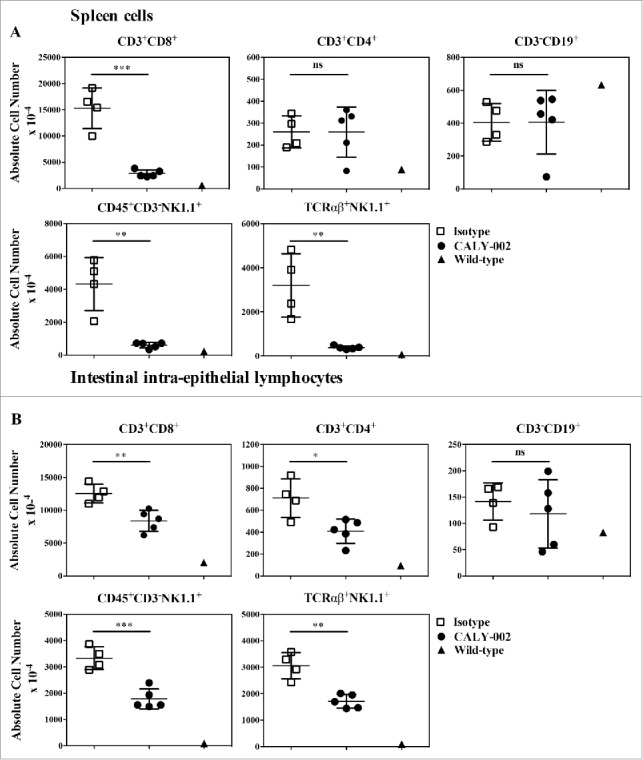
CALY-002 effect on spleen and intestinal intra-epithelial lymphocyte subsets in IL-15 transgenic mice. Total number of (A) spleen and intestinal intra-epithelial (B) CD3+CD8+, CD3+CD4+, CD3−CD19+, CD45+CD3−NK1.1 and TCRαβ+NK1.1+ lymphocytes at Day 28. Symbols represent individual IL-15 transgenic mice treated with either CALY-002 (closed circles) or control isotype (open squares) antibodies, or control C57BL/6 mice (closed triangles). Results are expressed as mean ± SD of absolute cell number/μL for a 1 mL suspension per animal. Statistical analysis: Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001; ns: not significant.
Thus, as described previously, anti-IL-15 antibody therapy is a promising treatment of type II RCD,16 and CALY-002 proved to be efficient in both in vitro and in vivo models of type II RCD.
IL-15 expression is increased in patients with EoE and in the experimental Aspergillus fumigatus mouse model of EoE
It was previously reported that IL-15 as well as IL-15Rα mRNA expression were increased in esophageal biopsies from active EoE patients compared with healthy individuals.20,43 We confirmed this finding using quantitative PCR in a well-characterized cohort of adult EoE patients (Fig. 5A, B). Furthermore, we demonstrate that IL-15 and IL-15Rα mRNA expression is significantly higher in patients who do not clinically and histologically respond to corticoid treatment compared with corticoid responders (Fig. 5A, B), while previous data showing a similar trend were collected only from a small number of patients that did not allow proper statistical analysis.20 As reported before,20 IL-15 mRNA expression correlated well with eosinophil infiltration in simultaneously collected esophageal biopsies (data not shown).
Figure 5.
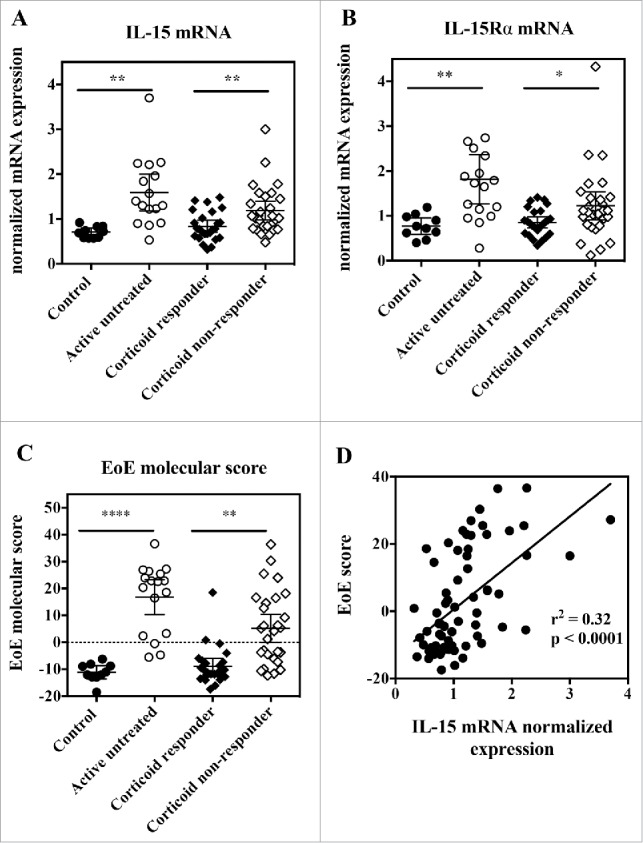
Esophageal biopsies from active EoE patients show increased IL-15 and IL-15Rα gene expression, and IL-15 expression correlates with EoE molecular score. RNA was extracted from esophageal biopsies, and subjected to quantitative RT-PCR gene expression analysis. (A) IL-15 mRNA expression, (B) IL-15Rα mRNA expression and (C) EoE molecular score from control healthy subjects (closed circles), active EoE non-treated patients (open circles), inactive EoE corticoid-responder patients (closed diamonds), and active EoE corticoid non-responder patients (open diamonds). Each symbol represents a single biopsy and mean ± 95% CI is indicated for each group. Statistical analysis: Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001. (D) Correlation between EoE molecular score and IL-15 mRNA expression for all subjects. Each symbol represents a single biopsy.
It has been shown that a molecular diagnostic test consisting of the quantitative analysis of the expression of 96 genes could identify patients with active EoE with high sensitivity and specificity.36 We selected a list of representative genes covering the major gene categories previously reported, and included another gene, CAPN14, the expression of which has been more recently linked to EoE through genome-wide association analysis (Table S2), performed quantitative PCR in our series of EoE esophageal biopsies, and calculated an EoE molecular score as described previously.36,37 We observed a significant increase in the EoE molecular score in active EoE patients, either treatment-naïve or unresponsive to corticoids, compared with healthy subjects or corticoid non-responsive EoE patients (Fig. 5C). Furthermore, we observed a clear and significant correlation between this EoE molecular score and IL-15 mRNA expression (Fig. 5D).
We then used immuno-fluorescence (IF) and immunohistochemistry (IHC) methods to further analyze IL-15 protein expression in esophageal biopsies. Detection of IL-15 in tissues is notoriously elusive, but can reliably be performed using well-validated techniques.44 To validate our method, we used as positive controls not only human tissues with presumably high expression of IL-15 (tonsil), but also duodenal biopsies from the above-mentioned IL-15TgE mice that overexpress human IL-15 in the gut (data not shown). Esophageal tissues of active EoE patients strongly express IL-15, particularly in the basal layers of the epithelium (Fig. 6). In contrast, epithelial cells from normal esophagus expressed no detectable level of IL-15 (Fig. 6). In corticosteroid-responsive patients, IL-15 expression was similar to that found in control subjects, while in corticosteroid-unresponsive patients, IL-15 expression was only slightly reduced (Fig. 6A, B). Normal esophageal epithelium was infiltrated with small numbers of T cells, while almost no eosinophils were detectable (Fig. 6). Patients with active EoE or corticosteroid-unresponsive patients exhibited strong T cell and eosinophil infiltration of which a subpopulation of both stained for IL-15 (Fig. 6). Since, to our knowledge, T cells and eosinophils do not express IL-15, it is possible that IL-15 staining reflects exogenous IL-15 bound to those T cells and eosinophils. The number of T cells was reduced with corticoid treatment, with no sensible difference between corticoid non-responders and corticoid-responders, except that only the former showed co-staining with IL-15 (Fig. 6). Eosinophils were decreased in patients treated with corticoids, and twice as much in corticoid-responders versus corticoid non-responders (Fig. 6). As for T cells, IL-15 staining of eosinophils was mostly observed in corticoid non-responders (Fig. 6).
Figure 6.
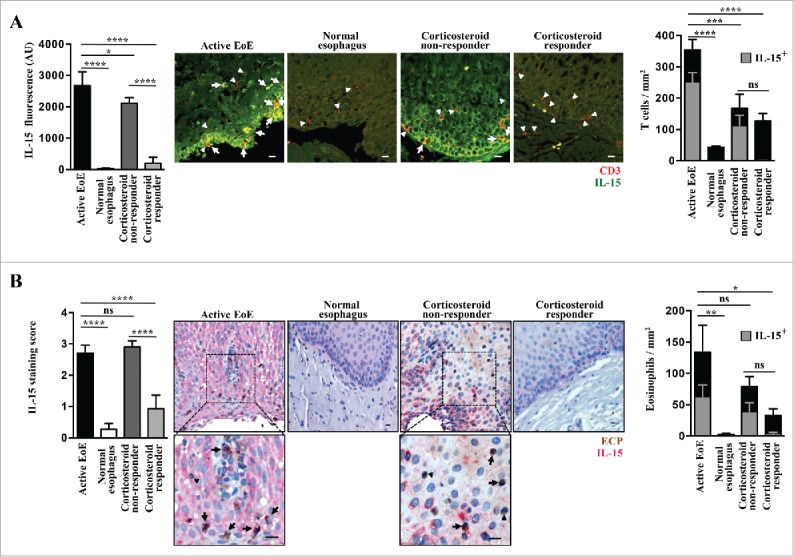
Increased IL-15 protein expression in esophageal biopsies from active EoE patients. (A) Immunofluorescence. Staining for IL-15 (green) and CD3 (red) was performed in 4 active EoE patients [4 x proximal and 4 x distal tissue samples], 2 normal control individuals [2 x proximal and 2 x distal tissue samples], 3 corticosteroid responders [3 x proximal and 3 x distal tissue samples] and 2 corticosteroid non-responders [2 x proximal and 2 x distal tissue samples]. Representative images are shown for each group. White arrows indicate examples of double stained cells, and filled white triangles indicate T cells without detectable IL-15 staining. Scale bars, 10 μm. Quantification of IL- 15 expression in epithelial cells is shown in the left panel. The right panel shows the quantitative analysis of infiltrating T cells that were either IL-15 negative (black bars) or positive (gray bars). (B) Immunohistochemistry. Staining for IL-15 (red) and ECP (brown) in active EoE, normal control individuals, corticosteroid responders, and corticosteroid non-responders (same esophageal tissues as described in panel A). Representative images are shown for each group. Insets demonstrate infiltrating eosinophils that are positive for IL-15 (black arrows) or IL-15 negative (filled black triangles). Scale bars, 10 μm. The right panel shows the quantitative analysis of infiltrating eosinophils that were either IL-15 negative (black bar) or positive (gray bar). Statistical analysis: one-way ANOVA followed by Tukey's multiple comparisons. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
We also attempted to measure IL-15 circulating levels in serum samples from the same patient cohorts using either Mesoscale or ELISA techniques. However, unlike what was previously reported in EoE patients,20,45 we did not observe any significant increase of circulating IL-15 levels in our cohort of EoE patients, and indeed in all subjects IL-15 levels were very low or undetectable (data not shown). Our data are not surprising given the fact that several other inflammatory molecules known to be locally associated with EoE could not be detected in serum,46 yet we do not know the source of discrepancy with the previously reported results.
There have been several experimental mouse models of EoE described, among which the aero-allergen Aspergillus fumigatus-induced model seems to be the one best reflecting the human pathology (for review see ref.47). It was previously reported that both IL-15 and IL-15Rα mRNA expression were increased in the esophagus of mice challenged with Aspergillus.20 We measured circulating levels of free IL-15 as well as IL-15/IL-15Rα complexes in the serum of mice before (Day 0) and at several time points (Day 7, Day 14 and Day 19) after a first challenge with Aspergillus, with or without treatment with dexamethasone. Mice remained regularly challenged with Aspergillus until Day 18. In mice administered with Aspergillus alone, circulating levels of free IL-15 as well as of IL-15/IL-15Rα complexes steadily increased throughout the study (Fig. 7A and B). In mice treated with dexamethasone, circulating levels of free IL-15 as well as IL-15/IL-15Rα increased in a much lower proportion, and especially were around basal levels at Day 14 (Fig. 7A and B). Dexamethasone is able to control esophageal eosinophilia in this model (Fig. 7C). Interestingly, at Day 19, both IL-15 and IL-15/IL-15Rα were at their highest levels, significantly different from baseline levels. Thus, the Aspergillus experimental mouse model is characterized by an increase in both free IL-15 and IL-15/IL-15Rα circulating levels, which at first are well controlled by corticoid treatment, but that have a tendency to escape corticoid control with chronic antigen challenge.
Figure 7.
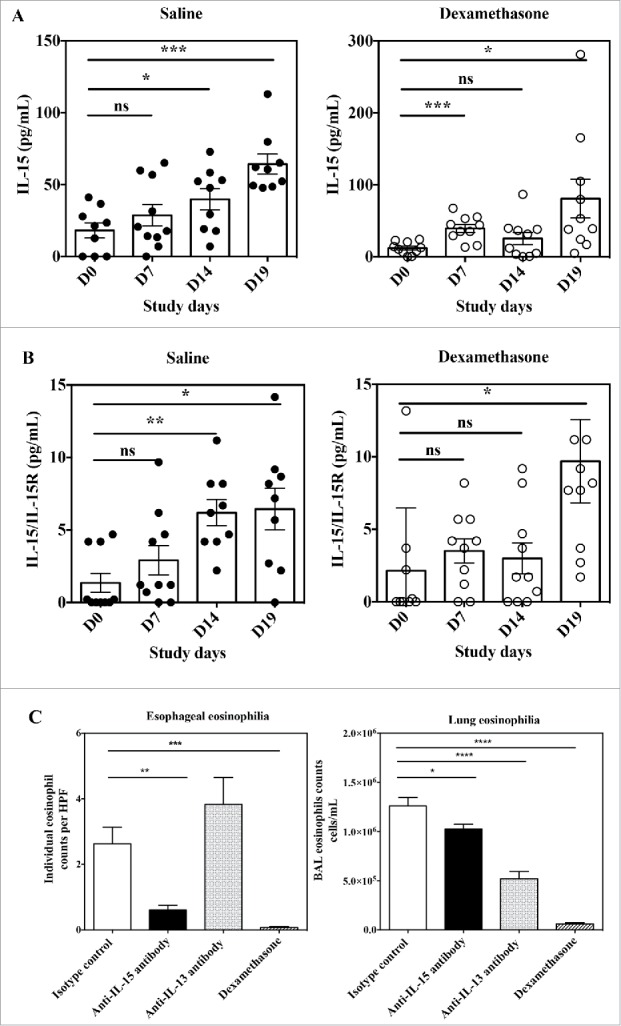
Effects of anti-IL-15 monoclonal antibody in the Aspergillus fumigatus mouse model of EoE. Serum concentration of free IL-15 (A) and IL-15/IL-15Rα heterodimer (B) in mice at Day 0 (before antigen challenge) and study Days 7, 14, and 19 of repeated intranasal challenge with Aspergillus fumigatus. Groups of 10 mice were treated either with saline control (closed circles) or dexamethasone (open circles). Data show individual and group mean ± SEM data for each group of animals at each time point. Statistical analysis: Student's t test. (C) Individual high power field (HPF) esophageal eosinophil counts (left panel) and total numbers of eosinophils in BAL fluid (right panel) of mice at Day 19 of repeated intranasal challenge with Aspergillus fumigatus and treated with control isotype antibody (white bar), anti-IL-15 antibody (black bar), anti-IL-13 antibody (dashed bar) or dexamethasone (hatched bar). Data are expressed as mean ± SEM for each group of animals. Statistical analysis: one-way ANOVA followed by Tukey's multiple comparisons. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
Anti-IL-15 treatment reduces esophageal eosinophilia in the experimental Aspergillus mouse model of EoE
It was previously shown that mice genetically deficient in IL-15Rα are protected from the development of esophageal eosinophilia in the Aspergillus experimental model.20 On the other hand, IL-15Rα-deficient mice poorly controlled lung eosinophilia in the same model.20 In contrast, mice genetically deficient in IL-13 challenged with Aspergillus still developed esophageal eosinophilia, but had much reduced eosinophils in their bronchoalveolar lung fluid (BAL).48 Because animals deficient in a given gene since birth are fundamentally different from those subjected to temporary blockade of a target by pharmacological means, we investigated the effect of neutralizing anti-IL-15 vs. anti-IL-13 antibodies in the Aspergillus mouse experimental model of EoE. We could not use CALY-002 in this model because this antibody does not recognize mouse IL-15 (Table 1). We obtained results remarkably similar to those reported with genetically deficient animals. When compared with control isotype antibody, monoclonal anti-IL-15 antibody abrogated esophageal eosinophilia, but only had a modest effect on BAL eosinophil numbers (Fig. 7C). Conversely, the anti-IL-13 monoclonal antibody did not decreased esophageal eosinophilic infiltration, but induced a major reduction in BAL eosinophil numbers (Fig. 7C). Taken together, these results confirm IL-15 as a valid target for antibody neutralization in EoE, with a biologic role clearly differentiated from that of IL-13.
Discussion
Intercepting IL-15 is a promising therapeutic avenue for several inflammatory or autoimmune diseases, especially those affecting the gastrointestinal tract. There are, however, no approved therapeutic monoclonal antibodies against IL-15. We have discovered CALY-002, a new potent and selective anti-IL-15 antibody resulting from the humanization and sequence optimization of the described previously B-E29 mouse anti-IL-15 antibody.27 The binding capacity of CALY-002 for human IL-15 was slightly increased compared with the original B-E29 antibody, but unlike what was shown for B-E29,26,27 CALY-002 does not interfere with IL-15 recognition of IL-15Rα. Our findings further demonstrate that subtle modifications in the epitope recognized by anti-IL-15 antibodies can have important consequences for their mode of action, as previously shown for DISC0280 and B-E29 anti-IL-15 antibodies that have very close, overlapping epitopes.26 The DISC0280 antibody was shown to fully block IL-15 binding to IL-15Rα as well as, potentially, IL-15Rβγ,26 whereas the original B-E29 antibody was shown to only partly interfere with IL-15 binding to IL-15Rα.26 In mice injected with IL-15, B-E29 was shown to be an antagonist, but DISC0280 was an agonist.26 Because CALY-002 acts as a full antagonist in the same in vivo assay but does not interfere with IL-15 binding to IL-15Rα, we propose that inhibition of IL-15 binding to IL-15Rα is fully dispensable for proper in vivo neutralizing activity. On the other hand, since one of the amino acid residues critical for binding of CALY-002 to IL-15 (D109) was shown to be in direct contact with IL-15Rβ in the quaternary structure of IL-15 in complex with its receptors,29 we hypothesize that CALY-002 exerts potent neutralization of IL-15 effects via blockade of IL-15 binding to IL-15Rβ.
We found, based on in vitro effects on cell lines, that CALY-002 was a potent inhibitor of cis as well as trans presentation of IL-15. The respective importance of cis vs. trans presentation in the biology of IL-15 is still debated. Trans presentation has been proposed to play an important role in IL-15 functions, especially during homeostasis.4 However, certain IL-15-driven outcomes are not dependent upon IL-15Rα, and therefore upon trans presentation.5,49,50 Indeed, as speculated earlier,4 it is possible that in inflammatory conditions an increase in IL-15 production could override the mechanism of trans presentation.51,52 Moreover, the presumably prominent role of trans presentation is supported by the hypothesis that IL-15 in the circulation predominantly, if not only, exists as a heterodimer with IL-15Rα.38 This is, however, not always the case, as we demonstrate here that intranasal Aspergillus administration to mice triggers an increase in both free and IL-15Rα-bound IL-15 circulating levels. It was also recently demonstrated that free IL-15 and IL-15/IL-15Rα complexes co-exist in mouse serum, even in steady-state conditions.53 It is therefore plausible that, especially in the case of dysregulated IL-15 production linked to diseased conditions, optimal neutralization of IL-15 deleterious effects will require potent blockade of both cis and trans presentation, such as that provided for by CALY-002.
When administered to cynomolgus monkeys, CALY-002 decreased peripheral NK cell numbers, but had no apparent effect on other populations of T cells. This is in contrast with a recent report showing that a rhesusized anti–IL-15 monoclonal antibody could also decrease circulating CD4 and CD8 TEM when administered to rhesus macaques.40 It is possible that this discrepancy is related to a different choice of markers to identify TEM cells between the 2 reports. Nevertheless, CALY-002 was very potent in cynomolgus monkeys, with almost maximal effect at the 0.1 mg/kg dose. For comparison purpose, mepolizumab, a humanized anti-IL-5 antibody that has been tested for EoE in humans and is approved for the treatment of severe eosinophilic asthma administered at 100 mg every 4 weeks, showed maximal pharmacodynamic activity in an allergic asthma model in cynomolgus monkeys at doses equal or above 5 mg/kg.54,55
We provide here further evidence that neutralizing IL-15 is an interesting approach for some forms of complicated CeD as well as EoE. The fact that IL-15 could be involved in these seemingly quite distinct diet-related diseases remains intriguing, but recent evidence supports that there could indeed be common underlying, IL-15-driven mechanisms in CeD and EoE. The association between CeD and EoE has been the focus of multiple studies with variable results, and the latest published study and meta-analysis of the literature suggests that there is no increased of EoE in children diagnosed with CeD.56 Conversely, genome-wide association studies of EoE patients have revealed a shared genetic etiology, at least in part, with atopy and other autoimmune conditions.37 Furthermore, study of a large Utah population database revealed that patients with EoE were at excess risk of auto-immune diseases, among which the most significant was CeD.57 Whereas it is still unclear why such variable results are observed, it could be due to the age of patients, with an association being more prominent in adults.58 Indeed, EoE has different features between children in adults, with higher rates of asthma and positive allergic reactivity to food antigens in children.59 There is also a clear, still unexplained, gender-bias for both diseases since EoE is more prominent in men, whereas CeD is more prominent in women. While a role for IL-15 in CeD has been proposed for more than a decade and supported by ample experimental data13,14 the implication of IL-15 in a Th2-driven disease such as EoE was less obvious. There is, however, a significant body of evidence showing that IL-15 is a potent driver of Th2 responses. Very early on, it was shown that IL-15 could induce IL-5 production by human Th2 clones, a property that was more recently extended to IL-13.60 IL-15, in synergy with tumor necrosis factor (TNF)-like cytokine 1A, is also able to stimulate strong IL-5 and IL-13 production by primary mucosal IL-18Rα+ CD4 T cells.61 Interestingly, IL-18 itself appears to be involved in EoE pathogenesis.62 In vivo, overexpression of IL-15 in the skin induces the expression of GATA3, which is a master transcriptional switch for Th2 as well as Th9 and ILC2 cells.63 Intriguingly, mice treated with the anti-IL-15 DISC0280 antibody together with IL-15, which results in enhancing IL-15 agonistic effects, showed a sharp increase in circulating levels of the Th2 cytokine IL-5.26 Furthermore, IL-15 is a potent vaccine adjuvant resulting in elevated levels of Th2-dependent IgG1 antibodies in a mouse model.64 Of note, IgG4 antiobdies play an important role in EoE pathogenesis and are the human counterpart of mouse IgG1.65 Last, recent evidence has linked IL-15 with the induction of cysteinyl leukotrienes, which are well-known mediators of Th2 and allergic responses, in the pathogenesis of CeD.66
Besides the amplification of Th2-driven immunity, IL-15 could play other roles in EoE pathogenesis. First, IL-15 was shown to directly prevent human eosinophil apoptosis and to stimulate primary mouse human esophageal cells to produce the eosinophil-selective chemokine eotaxin.20,67 Second, IL-15 is a well-known critical factor for the homeostasis and activation of various NK cell populations, and it was recently demonstrated that invariant NK T cells (iNKT) play a major role in experimental and human EoE pathogenesis.68 Third, IL-15 can broadly influence adaptive immunity. It was shown already a decade ago that adaptive T cell immunity is required for the control of experimental EoE.69 More recently, it was observed that esophageal eosinophilia is dispensable for the development of clinical and endoscopic features of EoE in siblings from EoE patients.70 These “EoE-like patients,” however, showed strong infiltration of T cells in their esophagus, albeit the frequency of CRTH2+ cells was reduced.70 Another team looked at the phenotype of T cells in the esophagus of EoE pediatric patients and its relationship with treatment responsiveness.71 They found that an increased proportion of CD8 T cells with the capacity to produce TNF and interferon-γ characterizes active EoE, with no difference observed regarding the capacity of CD4 T cells to produce IL-5 and IL-13.71 Thus, EoE appears to have much more diverse and complex underlying pathogenesis than originally suspected. Within this scope, IL-15, which can act as a regulator of Th2 cells, CD8 T cells, iNKT cells and directly affect epithelial cells and eosinophils, appears to be a very relevant target for therapeutic intervention. Neutralizing IL-15 could therefore have broader efficacy than neutralizing more downstream targets. Indeed, the fact that anti-IL-15 but not IL-13 blockade reduced esophageal eosinophilia in the Aspergillus model as we report herein, confirming previous results obtained in mice deficient for either IL-15Rα or IL-13,20,48 supports a superior efficacy profile of CALY-002 for the treatment of EoE.
We therefore propose that IL-15 is a master controller of immune and tissue responses in EoE, as it has been proposed for CeD and other gastrointestinal pathologies.2,6 Both diseases have seen sharp increase in incidence and prevalence in recent years, independently of better disease awareness or improved diagnostic modalities.72,73 An emerging environmental hypothesis to explain the recent increase in incidence of several inflammatory, allergic, auto-immune or metabolic diseases is a modification of the microbiome associated with specific organs, also called dysbiosis. Dysbiosis is now presented as one of the major factor that could promote CeD, although there is not yet a consensus regarding the special microbial species affecting positively or negatively the disease process.74 There have been recent reports showing that EoE is also associated with dysbiosis.75,76 Furthermore, it was demonstrated that IL-15 can favor gut inflammation by promoting intestinal dysbiosis.77 In mice, IL-15 overexpression in the intestinal epithelium restructures the composition of the microbiota with a decrease in butyrate-producing bacteria that is associated with a reduction in luminal butyrate levels across all intestinal compartments. Butyrate is an important dietary metabolite, part of the short chain fatty acids that are thought to play a role in the recent increase of ailments linked to Western-style diet.78 This reconfiguration of the microbiota by IL-15 in mice was associated with increased susceptibility to dextran sodium sulfate-induced colitis.77 While these data provide an interesting link between IL-15, dysbiosis and pathology, they do not clarify what triggers IL-15 production in human diseases. One critical avenue of research for our understanding of CeD and EoE pathogenesis will be to understand the interplay between the host and its environment for the initial induction and maintenance of sustained IL-15 levels in genetically susceptible individuals.
To conclude, we have discovered a new potent best-in-class anti-IL-15 humanized monoclonal antibody with a unique molecular mode of action, and provide additional rationale for its use to treat CeD, EoE, and potentially other diseases caused by IL-15 dysregulation. Its unique molecular mode of action indicates that this compound has the potential for therapeutic use in the treatment of RCD and EoE, and eventually other diseases caused by IL-15 dysregulation.
Materials and methods
Discovery of CALY-002 antibody
The anti-human IL-15 mouse monoclonal antibody B-E29 was produced following the immunization of BALB/c mice with recombinant human IL-15 as described elsewhere.27 The murine monoclonal antibody variable domains were sequenced and complementarity- determining regions (CDR) identified and humanized by Fusion Antibodies Ltd (Belfast, Ireland), using a combination of industry standard CDR-grafting technologies coupled with the latest research on antibody structure and up-to-date database of mature human IgG sequences. A number of human framework sequences were used as “acceptor” frameworks for the CDR sequences. These acceptor sequences have all come from mature human IgG from a human source. The variable sequence of various humanized candidates was further optimized to remove potentially problematic post-translational modifications sites and the final CALY-002 sequence was selected and synthetized with an IgG1 constant domain.
CALY-002 antibody production
The CALY-002 antibody was manufactured by transient recombinant DNA transfection into Chinese hamster ovary cells followed by purification on protein A Fast Protein Liquid Chromatography columns (performed by Evitria, Zurich, Switzerland) and re-suspended in phosphate-buffered saline (PBS, Lonza, ref. BE17512Q), pH 7.4. Protein concentration was measured by the manufacturer using the value of absorbance at 280 nm in a spectrophotometer with an extinction coefficient of 1.43.
Surface plasmon resonance kinetics analysis
Kinetic interaction of CALY-002 with IL-15 was measured at 25°C using a Biacore 3000 SPR biosensor (Biacore AB) by the SPR platform at the Integrated Structural Biology Institute, Grenoble, France. After immobilization of recombinant IL-15 (Prospec, ref. CYT-230) onto the carboxymethylated dextran surface of a CM5 sensor chip (GE Healthcare, ref. 29149604) at a level of about 101 response units, 120 µl of serially diluted CALY-002 antibody in a buffer containing Dulbecco's PBS (Gibco ref. 14040141) plus 0.005% surfactant P20 (GE Healthcare, ref. BR100054) was injected into the flow cell at a rate of 40 μl/min. Dissociation (koff) and association (kon) rate constants were obtained by nonlinear regression analysis of the primary sensorgram data according to a 1:1 (Langmuir) binding model after subtraction of reference signal and 0 nM antibody sample (double referencing). The dissociation constant KD was calculated using the formula KD = koff/kon.
Binding to recombinant cytokines by ELISA
Plastic 96-well Maxisorp plates (ThermoFisher ref. 442404) were coated with 100 ng/well of recombinant cytokine, washed and then incubated with various concentrations of CALY-002 or control antibodies, tested in duplicate wells. Recombinant cytokines used were: human IL-15 produced in E. Coli (Prospec, ref. CYT-230), rhesus/cynomolgus monkey IL-15 produced in yeast (myBioSource ref MBS948894), rat IL-15 produced in E. Coli (Sigma ref. SRP-4172), mouse IL-15 produced in E. Coli (R&D Systems ref. 447-ML), and human IL-2 produced in E. Coli (R&D Systems ref. 202-IL). Of note, reported rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) monkey IL-15 amino-acid sequences are identical (see Uniprot database entries G7P6C1 and G1K380). Control antibodies were: rabbit anti-mouse IL-15 (Acris ref. AP01124PU-S), rabbit anti-rat IL-15 (Biovision ref. 5172–100), and rabbit anti-human IL-2 (NovusBio ref. NBP1–4391). After washing off excess antibody from plates, a secondary anti-immunoglobulin goat antibody coupled to horseradish peroxidase (HRP; Sigma ref. A8275) was added and the enzymatic reaction revealed with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Interchim ref. UPS08171), then optical density signal read at 450 nM using a Fluostar Optima plate reader (BMG Labtech). Secondary peroxidase-labeled antibodies were: goat anti-human (Millipore ref. AP309P) or goat anti-rabbit (Sigma ref. A 8275) polyclonal antibodies. Dose-response curves were established and used to calculate the half maximal binding capacity (BC50) of each antibody to the cytokine tested, when appropriate, using Prism software (Graphpad) version 6.
Inhibition of biotinylated IL-15 binding to immobilized IL-15RαFc
Human IL-15Rα-Fc (R&D Systems ref. 7194-IR-050) in PBS at a concentration of 600 pM was coated onto MaxiSorp 96-well plates by incubation at 4°C overnight. The wells were washed twice with 150 μl PBS and blocked with 150 μl PBS containing 3% bovine serum albumin (BSA, Sigma) for 2 h at room temperature and washed again twice with 150 μl PBS. CALY-002 and controls were diluted in PBS with 0.1% BSA and added to the IL-15Rα-coated assay wells in a 50 μl volume. Serial dilutions were made in duplicate. Fifty μl of biotinylated human IL-15 (R&D Systems ref. NF150) at a final concentration of 100 pM was added and the assay plates incubated for 1 h at room temperature. The plates were then washed 6 times with PBS containing 0.1% Tween 20 followed by addition of 1/10,000 diluted high sensitivity streptavidin-peroxidase (Pierce ref. 21130). After 30 min incubation at room temperature, the plates were washed 6 times with PBS containing 0.1% Tween 20 followed by addition of 100 μl TMB peroxidase substrate. The intensity of the colorimetric reaction was measured by reading its 450 nm optical density (OD), and is proportional to binding.
Mapping of IL-15 epitope recognized by CALY-002
This study was conducted at Pepscan Presto BV (Lelystad, The Netherlands). To reconstruct discontinuous epitopes of human IL-15, a library of structured peptides was synthesized based on the published sequence of human IL-15 (Uniprot P40933). This was done using Pepscan's CLIPS technology.28 CLIPS templates are coupled to side-chain thiol groups of cysteine residues. A first set (Set 1) of peptides consisted of linear peptides of length 15 with an overlap of 14 residues, derived from the target IL-15 sequence. A second set of peptides (Set 2) was identical to Set 1, with alanine replacements on positions 10 and 11. A naturally occurring alanine was replaced by glycine. Peptides of Set 3 were identical to Set 1, with naturally occurring cysteines being replaced by serine, and cysteines being introduced at positions 1 and 4. These cysteines were linked by an mP2 CLIPS. This linkage staples peptides with a tendency to attain α helical conformations. Peptides of set 4 were peptides of Set 1 in which naturally occurring cysteines were replaced by serine, and cysteines were introduced at positions 1 and 8. These cysteines were linked by an mP2 CLIPS. Peptides of Set 5 were peptides of Set 1 in which naturally occurring cysteines were replaced by serine, and cysteines are introduced at positions 4 and 11. These cysteines were linked by an mP2 CLIPS. Peptide Set 6 were peptide variants on the base sequence ASIHDTVENLIILANNSLSS. In variant peptides, residues 5 through 16 were replaced by one of the 20 proteogenic amino acids. The peptide arrays were prepared as reported.28 The binding of CALY-002 to each of the synthesized peptides was tested in a PEPSCAN-based ELISA. In this assay, the peptide arrays were incubated with CALY-002 antibody solution (overnight at 4°C). After washing, the peptide arrays were incubated with a 1/1,000 dilution of an antibody peroxidase conjugate for one hour at 25°C. After washing, the peroxidase substrate 2,2′-azino-di-3-ethylbenzthiazoline sulfonate and 2 μl/ml of 3% H2O2 were added. After one hour, the color development was quantified with a charge coupled device camera and an image processing system. The published structure of the IL-15 quaternary complex (PDB ID: 4GS7) was used to visualize the IL-15 binding epitope of CALY-002, as well as critical residues using PyMOL software.29
Inhibition of IL-15-induced rescue from cytokine withdrawal-induced apoptosis in cell lines
The Kit 225 cell line was established from a patient with T cell chronic lymphocytic leukemia30 and was obtained from Merck-Serono, Geneva, Switzerland. The M-07e cell line was established from the peripheral blood of a 6-month-old girl with acute megakaryoblastic leukemia31 and was obtained from DSMZ, Leibniz, Germany. Kit 225 cells express the 3 chains of the IL-15 receptor (IL-15Rα, IL-15Rβ and IL-15Rγ), whereas M-07e cells express only the IL-15Rβ and IL-15Rγ chains.26 M-07e and Kit 225 cells were maintained in culture medium consisting of RPMI 1640 medium (InVitrogen ref. 52400–025) plus 10 mM Hepes (InVitrogen ref. 15630–056), 1 x penicillin/streptomycin (InVitrogen ref. 15070–063), 1 x L-Glutamine (InVitrogen 25030–024), 1 x sodium pyruvate (InVitrogen ref. 11360–039), 10% fetal bovine serum (FBS, PAA ref. A15–501) plus 200 U/ml human IL-2 (R&D Systems). One day before each assay, cells were starved of IL-2 by washing them twice in washing medium (Hanks' Balanced Salt Solution, InVitrogen ref. 14025–050, plus 0.1% BSA, Sigma ref. A4503–50G and 10mM Hepes), then returning them to culture flasks in culture medium without IL-2. For the assay, various concentrations of CALY-002 antibody were mixed with a fixed 1 ng/mL concentration of soluble recombinant human (R&D Systems) or monkey (MyBiosource) IL-15 in culture medium in 96-well flat-bottomed plastic microwell plates (VWR ref. 734–1794). Then 5 × 104 M-07e or Kit 225 cells were added to each well for a total volume of 100 μl, and cell cultures incubated for 48 hours at 37°C in the presence of 5% CO2. Proliferation/survival was assessed using a luminescent cell viability assay measuring ATP concentration (Titerglo, Promega ref. G7572) following manufacturer's instructions. In some experiments, before the addition of IL-15 and CALY-002 antibody, plastic plates were coated with 100 μL per well of a 1 μg/ml solution of recombinant human IL-15Rα-Fc (R&D Systems ref. 7194-IR-050) in PBS and left overnight at 37°C in a CO2 incubator. This additional step allowed for measuring the effects of CALY-002 on IL-15 trans presentation. Dose-response curves were established to calculate the half maximal inhibitory capacity (IC50) of the CALY-002 antibody in this assay using Prism software version 6.
Blockade of IL-15/IL-15RαFc agonist activity in vivo
All animal care and experimental procedures involved in this study were performed by Fildeta (Zagreb, Croatia) and were approved by its internal Ethical Review Committee. Fidelta's animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Six-week-old male C57BL/6 mice were purchased from Charles River (Calco, Italy) and 5 mice were randomized per each experimental group. Mice were administered intraperitoneally on study Day 1 with 100 μg of either CALY-002 or human IgG1 control isotype antibody (anti-HIV-1 gp120 b12, Evitria) diluted in PBS (Lonza), or PBS alone. A stock solution of human IL-15 (Prospec) was prepared at a concentration of 20 µg/mL in PBS, as well a human IL-15Rα-Fc (R&D Systems) stock solution at a concentration of 72 µg/mL in PBS. An appropriate volume of IL-15 and IL-15Rα-Fc solutions was mixed at 1:1 ratio within 30 to 40 minutes prior injection. Mice were administered 20–30 minutes after antibody injection on study Day 1 with 100 μL of preformed IL-15/IL-15Rα-Fc solution, via the intraperitoneal route. Administration of freshly prepared IL-15/IL-15Rα-Fc solution was repeated at study Day 2 and Day 3. On study Day 4, all mice were killed by subtotal exsanguination under ketanest + rompun anesthesia. Whole spleens from each mouse were collected, washed with PBS and a single cell suspension prepared by gently pushing the spleens through 40 μm Falcon cell strainers (Becton Dickinson ref. 08–771-1) with a syringe plunger. Cell count was determined for each splenocyte sample using a hematological counter (Sysmex, Yverdon, Switzerland). Cells were further washed in staining buffer (PBS + 2% heat-inactivated FBS), followed by the immunophenotyping protocol. Staining for flow cytometry was performed on 1 × 106 cells for each individual suspension. All samples were first incubated with FACSLysing solution (Becton Dickinson re. 349202) to remove erythrocytes, then with the Fc blocking reagent (Anti-mouse CD16/CD32, eBioscienceref. 14–0161) to prevent the unspecific binding of antibodies. Samples were then stained with appropriate dilutions of CD45-eFluor450 (ref. 48–0451), CD3-APC (ref. 17–0032), and NK1.1-PE (ref. 12–5942), all from e-Bioscience, as well as with the Live/Dead Fixable Yellow Dead Cell Stain (Life Technologies ref. L34959). Samples were processed on a FACScan/Cytek DxP8 flow cytometer (Becton Dickinson) and analyzed with FlowJo software (Ashland, OR, USA). Mouse spleen NK cells were determined as live CD45+CD3-NK1.1+ lymphocytes. Further analyses were performed with Prism 6 software.
Pharmacokinetics and effect of CALY-002 administration on circulating NK and T cells in Cynomolgus Macaques
This study was performed at ITR Canada (Baie d'Urfé, QC, Canada) and approved by its internal Ethical Review Committee. All animals used on this study were cared for in accordance with the principles outlined in the current “Guide to the Care and Use of Experimental Animals” as published by the Canadian Council on Animal Care and the “Guide for the Care and Use of Laboratory Animals,” an NIH publication. Six male and 4 female cynomolgus macaques (Macaca Fascicularis) monkeys (ChinaWorldwide Primates, Inc., FL, USA) were transferred to the study from the ITR spare colony and were assigned to the appropriate dose group by a process of block randomization based on body weight. Body weights range from 2.8 to 6.3 kg for males and 3.0 to 4.9 kg for females. CALY-002 was administered once by i.v. injection (slow bolus over approximately 10 minutes) to 3 different dose groups. One group of 2 males and 1 female received 0.1 mg/kg antibody, another group of 2 males and 1 female received 1 mg/kg antibody, and the third group of 2 males and 2 females received 10 mg/kg antibody. The dose volume administered to each animal was 2 ml/kg. A series of 8 blood samples for pharmacokinetic evaluation were taken over a 28-day period (pre-dose, 24 hours post dose, Day 3, 5, 8, 14, 21 and 28). The serum concentrations of CALY-002 were analyzed using an ELISA method quantifying human IgG and qualified for use in cynomolgus monkey serum. Briefly, Maxisorp plates were coated with 50 μl anti-human IgG (ThermoFisher Scientific, ref. MA5–16929) at 500 ng/mL in PBS buffer and incubated overnight at 4°C. Residual binding sites were blocked by incubation with 200 μl blocking casein buffer (ThermoFisher Scientific ref. 37528) and incubated for 1 hour at room temperature. Appropriate dilutions of samples or standard (CALY-002) in PBS plus 0.05% Tween (Sigma) were added to each well and incubated for 1 hour at room temperature. After 4 washes in PBS-Tween, an appropriate dilution of biotinylated secondary antibody (same as capture antibody) in PBS-Tween was added and incubated for 1 hour at room temperature. Plates were washed and further incubated with streptavidin HRP (Pierce) diluted in PBS-Tween for 45 min at room temperature. After a last wash, plates were incubated with TMB substrate (Interchim) and reaction stopped with stop solution (Sigma). Optical absorbance was read on an ELISA reader at 450 nm and 620 nm within 30 minutes after brief agitation. Standard curves of OD 450 nm – OD 620 nm vs. IgG concentration were plotted and fitted using the Prism 6 software after log transformation and a 4 parameter logistic curve (4PL). Pharmacokinetics analysis was performed using the Summit PK Solution software (version 2.0) using a built-in 2 non-compartmental analysis of concentration-time data obtained following i.v. injection. Data were plotted and further analyzed using Graphpad Prism software version 6.
For pharmacodynamics evaluation (circulating NK and T cells numbers), a series of 9 blood samples were collected into tubes containing EDTA anticoagulant at pre-dose (in duplicate) and Days 1, 3, 5, 8, 14, 21 and 28. The white blood cell counts (total absolute and percent differential lymphocyte count) were measured from each blood sample using the ADVIA 120 hematology analyzer (Siemens Healthcare). The total absolute lymphocyte count was used to calculate the absolute value of the NK cells and T lymphocyte subsets. Each sample was analyzed by flow cytometry using 2 panels of makers, one for NK cells (CD3, CD16, CD45) and the other one for T cells (CD3, CD4, CD8, CD45, CD45RA, CCR7) using standard methods at ITR. Following staining, red blood cells were lysed by the addition of a FACS Lysing solution (Becton Dickinson). Data acquisition was then performed with a flow cytometer (BD FACSCanto™ II with FACSDiva™ software (version 6.1.3)). Data analysis was performed using FCS Express software (version 4). A gate was drawn around the lymphocytes, identified as CD45high events with low side scatter (SSC). Within the events comprised in the lymphocyte gate, the percentage of NK cells (CD3−CD16+) was determined from the NK cell panel, and the percentage of total T lymphocytes (CD3+), helper T lymphocytes (CD3+CD4+), cytotoxic T lymphocytes (CD3+CD8+), naïve cytotoxic lymphocytes (CD3+CD8+CD45RA+CCR7+), TCM lymphocytes (CD3+CD8+CD45RA+CCR7−) and TEM lymphocytes (CD3+CD8+CD45RA−CCR7−) were determined from the T cell panel.
Effect on primary intestinal intraepithelial lymphocyte (IEL) cell lines from patients with type II refractory celiac disease
Primary cell lines of CD103+sCD3− (surface CD3 negative) IEL were derived from duodenal biopsies from type II RCD patients as described previously.32 Three independent cell lines were tested, called HAM_RAC, BER_JEA, and CHE_GAB. The efficiency of the CALY-002 antibody to inhibit IL-15-driven STAT5 phosphorylation and to revert IL-15-induced prevention of apoptosis was tested in the 3 different CD103+sCD3− IEL lines at a 4 μg/ml antibody concentration previously estimated to give 80% apoptosis inhibition in HAM_RAC cells. Cell lines (0.25 × 106 cells) in culture medium (RPMI 1640 with 10% FBS and antibiotics) with or without 5 ng/ml recombinant human IL-15 (R&D Systems) were treated with CALY-002 or control isotype for 48 h, then fixed (BD Biosciences ref. 557870), permeabilized (BD Biosciences ref. 55850), stained with PEcy7-labeled anti-p-STAT5 antibody (BD Biosciences ref. 560177) and analyzed with a FACS CANTO II flow cytometer (BD Biosciences). Results were expressed as mean fluorescence intensity (MFI). Percentages of apoptotic cells in cultured primary IELs were determined using an Annexin V-APC/PI Kit according to the manufacturer's instructions (Sony biotechnology ref. 3804660) and analyzed with a FACS CANTO II flow cytometer.
In vivo effect in human IL-15 transgenic mice
All animal care and experimental procedures involved in this study were approved by Ile-de-France- René Descartes Ethics committee for animal experimentation. Groups of 12–14 week-old T3b-IL-15 Tg mice16 were treated on study days: D0, 1, 3, 7, 10, 14, 17, 21, 24, 27 with 100 μg CALY-002 or human IgG1 control isotype, injected intraperitoneally in 0.1 mL PBS. At D28, mice were killed and cells were isolated from the spleen and intraepithelial (IEL) intestinal lymphocytes, then specific lymphocyte subsets defined by surface expression of cellular markers were analyzed by flow cytometry as described previously.16 Two control non-transgenic C57BL/6 mice (wild-type control) were also used at D0 and D28, respectively. Data were plotted and further analyzed using Graphpad Prism software version 6.
Eosinophilic Esophagitis patient biopsies
The patients included in this study were diagnosed between August 2012 and February 2014 at a single gastroenterology clinic in Olten, Switzerland. The local Ethics Committee approved data collection into the Swiss EoE Cohort Study. All participants provided written informed consent. This study included 2 groups of subjects. First, 54 male or female patients of 20 y of age or older with EoE that was diagnosed according to recommended diagnostic criteria.22 Following these guidelines, all EoE patients had symptoms of esophageal dysfunction and eosinophilic infiltration with at least 15 eosinophils/high power field (hpf) (400x magnification) after an 8-week course with high-dose proton pump inhibitors. Histologic diagnosis of EoE was based on the analysis of 4 biopsies from the proximal and 4 biopsies from the distal esophagus. Other causes of esophageal eosinophilia such as concomitant gastro-esophageal reflux disease, eosinophilic gastroenteritis or parasitic infection were excluded. Taking into account EoE's patchy histologic distribution ,22,33,34 2 biopsy specimens per patient or control were analyzed, one from the upper half (proximal) and one from the lower half (distal) of the esophagus. EoE was defined as active at the time-point of biopsy sampling – independent on the current treatment status – when patients had symptoms of esophageal dysfunction and the peak eosinophil count was higher than 15 eosinophils/hpf. The second group consisted of 12 volunteers with healthy esophagus. Detailed patient characteristics, including medication, are provided in Table S3.
Targeted mRNA expression analysis study
Expression of selected genes was analyzed by Reverse-Transcriptase Quantitative Polymerase Chain reaction (RT-qPCR) at Biogazelle (Gent, Belgium). Briefly, RNA was isolated from 117 samples using the miRNeasy Mini kit (Qiagen ref. 217004) following the manufacturer's instructions. On-column DNase digestion was performed during the RNA extraction. The RNA concentration was determined using the Nanodrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific). RNA quality control was performed using the 2100 Bioanalyzer microfluidic gel electrophoresis system (Agilent). Contamination by genomic DNA was assessed on 25% of the samples by means of 2 DNA-specific qPCR assays using RNA as input, and confirmed the complete absence of genomic DNA in all tested samples. In conclusion, after RNA extraction and quality control, 96 samples were available for RT-qPCR analysis. Synthesis of cDNA was performed using the iScript Advanced cDNA Synthesis Kit (BioRad ref. 1725038) with 50 ng of total RNA as input. The cDNA quality of each sample was assessed by means of 2 universally expressed human genes. To ensure sufficient template for reliable quantification, before cDNA quantification a gene-specific PCR-based pre-amplification step consisting of 12 cycles was performed using the SsoAdvanced PreAmp Supermix (Bio-Rad ref. 172–5160). For gene expression profiling, we used validated approaches for qPCR amplification (BioRad, see Table S2). All measurements were performed in 384-well plates (Bio-Rad ref. CFX384) in a reaction volume of 5 μl, using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad ref. 172–5270). Each PCR reaction was done in duplicate using 5 ng cDNA equivalents of RNA as input in each reaction. A pilot experiment was performed to select proper, stably expressed reference genes for data normalization using a set of 9 candidate reference genes on all samples. Data analysis was done in Biogazelle's qbase+ software (version 3.0) using Biogazelle's qbase+ quantification model.35 Data were plotted and further analyzed using Graphpad Prism 6 software.
EoE activity molecular score
The EoE molecular scores have been calculated as described elsewhere using expression data on 24 genes known to be up- or downregulated in EoE, as a representative subset of 94 described previously genes36 as well as CAPN1437 (Table S2). In brief, the sums of the log10 of the calibrated normalized relative quantities (CNRQ) values were calculated separately for upregulated and downregulated gene groups. A negative weight was endowed to the upregulated gene sum before the addition of the 2 ∑(log10 CNRQ) values to establish the EoE activity score.
Immunofluorescence
Paraffin-embedded tissue sections were deparaffinized and rehydrated with graded ethanol dilutions. After antigen retrieval in EDTA-Tris buffer (10 mM, pH 9.0), immunofluorescence staining was performed by incubating the paraffin sections with goat anti-human IL-15 antibody (R&D Systems ref. AF315) or normal goat IgG control antibody (R&D Systems ref. AB-108-C) both used 1:100, and monoclonal mouse anti-CD3 antibody (Dako ref. M7254; clone F7.2.38) or mouse IgG1 isotype control antibody (R&D Systems ref. MAB002; clone # 11711) both used 1:50. Thereafter, secondary Alexa Fluor® 488 - conjugated donkey anti-goat (1:400; ThermoFisher ref. A-11055) and Cy3-conjugated donkey anti-mouse (1:100) antibodies (Jackson ImmunoResearch Laboratories, Inc., ref. 705–166–147) were applied and tissue samples incubated at room temperature for 1 h. Samples were washed and mounted in Prolong Gold mounting medium (ThermoFisher ref. P36930) and image acquisition was performed using confocal laser scanning microscopy LSM 700 (Carl Zeiss Micro Imaging, Jena, Germany) with a 40x/1.40 Oil DIC objective and analyzed with IMARIS software (Bitplane AG, Zurich, Switzerland). Five representative micrographs from each tissue were subjected to automated analysis to measure mean fluorescence intensity for IL-15 using automated isosurface module of IMARIS software. Numbers of T cells were counted in 5 high power fields (HPF) for each patient/control individual.
Immunohistochemistry
Paraffin-embedded tissue sections were deparaffinized and rehydrated with graded ethanol dilutions, after which antigen retrieval was performed in EDTA-Tris buffer (10 mM, pH 9.0). Double-staining IHC was performed using 2 different sets of IHC detection kits. Firstly, eosinophils were labeled with a monoclonal anti-ECP antibody (1:20; Pharmacia & Upjohn Diagnostics AB, clone EG1) or mouse IgG1 isotype control antibody (R&D Systems ref. MAB002; clone 11711), and staining was detected by Dako EnVision®+ Dual Link System-HRP (Dako ref. K4065) according to the manufacturer's instructions. Tissues were then subjected for a second staining procedure to detect IL-15 using the Dako REAL Detection System, alkaline phosphatase/RED kit (Dako ref. K5005), applying goat anti-human IL-15 antibody (R&D Systems ref. AF315) or normal goat IgG control antibody (R&D Systems ref. AB-108-C), both diluted 1:100, and subsequently polyclonal rabbit anti-goat biotinylated secondary antibody (Dako ref. E0466) according to instructions provided with the kit. Image acquisition was performed using a Zeiss Axiovert 200 inverted microscope (Carl Zeiss Micro Imaging) with a 40x objective and analyzed using the Zen Blue software (Carl Zeiss Micro Imaging). Numbers of eosinophils were evaluated in 5 HPF for each patient/control individual. Quantification of IL-15 expression in epithelial cells was done independently by 2 scientists and scored (0 = none; 1 = mild; 2 = medium; 3 = strong, 4 = very strong).
Mouse experimental eosinophilic esophagitis model
This study was performed at Preclin Biosystems AG animal facility (Epalinges, Switzerland). All animal care and experimental procedures involved in this study were approved by the Veterinary Office of Canton de Vaud. Female BALB/c mice (Charles River Laboratories) were acclimatized for 1 week before the study. The mice were 8 weeks of age on day 0 of the study. Mice were anesthetized by intraperitoneal injection of 9.75 mg xylasol and 48.75 mg ketasol per kg (Dr. E. Graeub AG, Bern, Switzerland) and administered with 15 μg of Aspergillus fumigatus (GREER Laboratories ref. XPM3D3A25, Lot N° 260980) in a volume of 50 μL PBS per nasal on study days: Day 0, 2, 4, 7, 9, 11, 14, 16, 18. All injections of test articles and controls were performed under a 200 μL volume, in PBS. Antibodies were i.v. administered at a 100 μg dose per mouse on study days: Day 0, 4, 7, 11, 14, 18. Control isotype antibody anti-trinitrophenol rat IgG2a, clone 2A3, was manufactured by Bioxcell (ref. BE0089). Rat anti-mouse IL-15 antibody clone AIO.3 (ref. 16–7154) and rat anti-mouse IL-13 antibody clone eBio1316H (ref. 16–7135) were manufactured by eBioscience. Freshly prepared dexamethasone (Sigma-Aldrich, ref. D1159) was administered intraperitoneally at 3 mg/kg dose on study days: Day 0, 2, 4, 7, 9, 11, 14, 16, 18. On study Day 0, administration of test articles and controls took place just prior challenge with Aspergillus fumigatus. On study Day 19, 18–20 hours after the last antigen challenge, all animals were killed by lethal i.p. injection with pentabarbitol (Streuli Pharma ref. 1170139A) immediately followed by bronchoalveolar lavage (BAL) in 500 μL of saline. Cells were isolated from the BAL fluid and differential cell counts were performed based upon standard morphological and cytochemical criteria. The complete length of the esophagus of each mouse was carefully isolated from behind the cricoid cartilage superiorly, to just above the diaphragm inferiorly. Following the isolation, the esophagus was put flat on a histology sample cassette, immersed in a beaker containing formalin and stored at 4°C until processing for histopathological analysis. Formalin-fixed tissues were embedded in paraffin and sectioned. Slides were de-paraffinized and thereafter stained for eosinophils using a commercially available staining kit (Abcam ref. ab150665), applying manufacturer's staining protocol designed for a selective staining of eosinophils. The subsequent quantification of eosinophilia was performed blinded as follow: 10 random fields of view per sample were quantified at 40x magnification with eosinophils counted based on their donut, ring or bi-lobed shaped and nuclei and pink-colored cytoplasm.
Measure of circulating IL-15 and IL-15/IL-15Rα levels in experimental EoE
Since IL-15 can exist in the circulation as a heterodimer with IL-15Rα,38 2 ELISA assays were used in the mouse experimental model of EoE. The first ELISA assay was a kit to specifically measure IL-15/IL-15Rα heterodimer levels (e-Bioscience ref. 88–7215) and used as per the manufacturer's instructions. The second ELISA assay was a kit to measure IL-15 purchased from Cusabio (ref. CSB-E04604m) and used as per the manufacturer's instructions. It was not known whether this kit would measure only free IL-15 or could detect both free IL-15 and IL-15/IL-15Rα heterodimers, and therefore controls were prepared to investigate the kit specificity: several dilutions of a fresh vial of the e-Bioscience IL-15/IL-15Rα standard in the Cusabio sample diluent buffer were tested alongside a standard curve of free IL-15 (Cusabio standard) in the Cusabio assay. None of the 3 tested concentrations of recombinant IL-15/IL-15Rα complex (250, 125 and 62.5 pg/mL) gave significant optical density reading above background (data not shown). Hence, the Cusabio assay measures only free IL-15 and not free IL-15 and IL-15/IL-15Rα heterodimer. Levels of IL-15 or IL-15/IL-15Rα complex were only measured in groups that received saline or dexamethasone, since it is unknown whether the anti-IL-15 antibody administered in the study would interfere with the ELISA assays, and attempts to investigate this were not conclusive. All serum samples were diluted 1:5 for the assay in the appropriate assay diluent (corresponding to each kit).
Supplementary Material
Disclosure of potential conflict of interest
Alain Vicari, Laurence Goffin, Yolande Chvatchko: this research is sponsored by Calypso Biotech SA and may lead to the development of products, in which we have a business and financial interest. We have disclosed those interests fully to Taylor & Francis, and have in place an approved plan for managing any potential conflicts arising from this arrangement.
Olivier Léger: in accordance with Taylor & Francis policy and my ethical obligation as a researcher, I am reporting that I am a consultant to Calypso Biotech SA, a company that may be affected by the research reported in the enclosed paper. I have disclosed those interests fully to Taylor & Francis, and I have in place an approved plan for managing any potential conflicts arising from that involvement.
Alain M. Schoepfer: in accordance with Taylor & Francis policy and my ethical obligation as a researcher, I am reporting that I received consulting fees, speaker fees or research grants from Aptalis Pharma, Inc., Dr. Falk Pharma, GmbH, Germany, Glaxo Smith Kline, AG, and Nestlé S. A., Switzerland.
Funding
This work was supported by grants from the Swiss National Science Foundation (32003B_135665 to AMS and AS and 32003B_160115/1 to AMS, and 310030_166473 to HUS).
References
- 1.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood 2001; 97:14-32; PMID:11133738; https://doi.org/ 10.1182/blood.V97.1.14 [DOI] [PubMed] [Google Scholar]
- 2.Pagliari D, Cianci R, Frosali S, Landolfi R, Cammarota G, Newton EE, Pandolfi F. The role of IL-15 in gastrointestinal diseases: a bridge between innate and adaptive immune response. Cytokine Growth Factor Rev 2013; 24:455-66; PMID:23791986; https://doi.org/ 10.1016/j.cytogfr.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 3.Waldmann TA. The Biology of IL-15: Implications for cancer therapy and the treatment of autoimmune disorders. J Investig Dermatol Symp Proc 2013; 16:S28-30; PMID:24326545; https://doi.org/ 10.1038/jidsymp.2013.8 [DOI] [PubMed] [Google Scholar]
- 4.Stonier SW, Schluns KS. Trans-presentation: A novel mechanism regulating IL-15 delivery and responses. Immunol Lett 2010; 127:85-92; PMID:19818367; https://doi.org/ 10.1016/j.imlet.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ota N, Takase M, Uchiyama H, Olsen SK, Kanagawa O. No requirement of trans presentations of IL-15 for human CD8 T cell proliferation. J Immunol 2010; 185:6041-8; PMID:20926799; https://doi.org/ 10.4049/jimmunol.0901834 [DOI] [PubMed] [Google Scholar]
- 6.Jabri B, Abadie V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat Rev Immunol 2015; 15:771-83; PMID:26567920; https://doi.org/ 10.1038/nri3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldmann TA. Targeting the interleukin-15 system in rheumatoid arthritis. Arthritis Rheum 2005; 52:2585-8; PMID:16142738; https://doi.org/ 10.1002/art.21363 [DOI] [PubMed] [Google Scholar]
- 8.Baslund B, Tvede N, Danneskiold-Samsoe B, Larsson P, Panayi G, Petersen J, Petersen LJ, Beurskens FJ, Schuurman J, van de Winkel JG, et al.. Targeting interleukin-15 in patients with rheumatoid arthritis: a proof-of-concept study. Arthritis Rheum 2005; 52:2686-92; PMID:16142748; https://doi.org/ 10.1002/art.21249 [DOI] [PubMed] [Google Scholar]
- 9.Villadsen LS, Schuurman J, Beurskens F, Dam TN, Dagnaes-Hansen F, Skov L, Rygaard J, Voorhorst-Ogink MM, Gerritsen AF, van Dijk MA, et al.. Resolution of psoriasis upon blockade of IL-15 biological activity in a xenograft mouse model. J Clin Invest 2003; 112:1571-80; PMID:14617758; https://doi.org/ 10.1172/JCI200318986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Geboes K, Colpaert S, D'Haens GR, Rutgeerts P, Ceuppens JL. IL-15 is highly expressed in inflammatory bowel disease and regulates local T cell-dependent cytokine production. J Immunol 2000; 164:3608-15; PMID:10725717; https://doi.org/ 10.4049/jimmunol.164.7.3608 [DOI] [PubMed] [Google Scholar]
- 11.Nishiwaki T, Ina K, Goto H, Watanabe O, Tsuzuki T, Furuta R, Ando T, Hibi K, Kusugami K. Possible involvement of the interleukin-15 and interleukin-15 receptor system in a heightened state of lamina propria B cell activation and differentiation in patients with inflammatory bowel disease. J Gastroenterol 2005; 40:128-36; PMID:15770395; https://doi.org/ 10.1007/s00535-004-1510-y [DOI] [PubMed] [Google Scholar]
- 12.Yoshihara K, Yajima T, Kubo C, Yoshikai Y. Role of interleukin 15 in colitis induced by dextran sulphate sodium in mice. Gut 2006; 55:334-41; PMID:16162679; https://doi.org/ 10.1136/gut.2005.076000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meresse B, Malamut G, Cerf-Bensussan N. Celiac disease: an immunological jigsaw. Immunity 2012; 36:907-19; PMID:22749351; https://doi.org/ 10.1016/j.immuni.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 14.Abadie V, Jabri B. IL-15: a central regulator of celiac disease immunopathology. Immunol Rev 2014; 260:221-34; PMID:24942692; https://doi.org/ 10.1111/imr.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiuri L, Ciacci C, Auricchio S, Brown V, Quaratino S, Londei M. Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology 2000; 119:996-1006; PMID:11040186; https://doi.org/ 10.1053/gast.2000.18149 [DOI] [PubMed] [Google Scholar]
- 16.Malamut G, El Machhour R, Montcuquet N, Martin-Lannerée S, Dusanter-Fourt I, Verkarre V, Mention JJ, Rahmi G, Kiyono H, Butz EA, et al.. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease-associated inflammation and lymphomagenesis. J Clin Invest 2010; 120:2131-43; PMID:20440074; https://doi.org/ 10.1172/JCI41344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korneychuk N, Ramiro-Puig E, Ettersperger J, Schulthess J, Montcuquet N, Kiyono H, Meresse B, Cerf-Bensussan N. Interleukin 15 and CD4+ T cells cooperate to promote small intestinal enteropathy in response to dietary antigen. Gastroenterology 2014; 146:1017-27; PMID:24361466; https://doi.org/ 10.1053/j.gastro.2013.12.023 [DOI] [PubMed] [Google Scholar]
- 18.Schulthess J, Meresse B, Ramiro-Puig E, Montcuquet N, Darche S, Bègue B, Ruemmele F, Combadière C, Di Santo JP, Buzoni-Gatel D, et al.. Interleukin-15-dependent NKp46+ innate lymphoid cells control intestinal inflammation by recruiting inflammatory monocytes. Immunity 2012; 37:108-21; PMID:22705105; https://doi.org/ 10.1016/j.immuni.2012.05.013 [DOI] [PubMed] [Google Scholar]
- 19.Ettersperger J, Montcuquet N, Malamut G, Guegan N, Lopez-Lastra S, Gayraud S, Reimann C, Vidal E, Cagnard N, Villarese P, et al.. Interleukin 15-Dependent T Cell-like Innate Intraepithelial Lymphocytes Develop in the Intestine and Transform into Lymphomas in Celiac Disease. Immunity 2016; 45:610-25; PMID:27612641; https://doi.org/ 10.1016/j.immuni.2016.07.018 [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Wang M, Mavi P, Rayapudi M, Pandey AK, Kaul A, Putnam PE, Rothenberg ME, Mishra A. Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice. Gastroenterology 2010; 139:182-93.e7; PMID:20381491; https://doi.org/ 10.1053/j.gastro.2010.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology 2009; 137:1238-49; PMID:19596009; https://doi.org/ 10.1053/j.gastro.2009.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG Clinical Guideline: Evidenced Based Approach to the Diagnosis and Management of Esophageal Eosinophilia and Eosinophilic Esophagitis (EoE). Am J Gastroenterol 2013; 108:679-92; PMID:23567357; https://doi.org/ 10.1038/ajg.2013.71 [DOI] [PubMed] [Google Scholar]
- 23.Dellon ES. Eosinophilic Esophagitis. Gastroenterol Clin North Am 2013; 42:133-53; PMID:23452635; https://doi.org/ 10.1016/j.gtc.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bobbala D, Chen XL, Leblanc C, Mayhue M, Stankova J, Tanaka T, Chen YG, Ilangumaran S, Ramanathan S. Interleukin-15 plays an essential role in the pathogenesis of autoimmune diabetes in the NOD mouse. Diabetologia 2012; 55:3010-20; PMID:22890824; https://doi.org/ 10.1007/s00125-012-2675-1 [DOI] [PubMed] [Google Scholar]
- 25.Cepero-Donates Y, Lacraz G, Ghobadi F, Rakotoarivelo V, Orkhis S, Mayhue M, Chen YG, Rola-Pleszczynski M, Menendez A, Ilangumaran S, et al.. Interleukin-15-mediated inflammation promotes non-alcoholic fatty liver disease. Cytokine 2016; 82:102-11; PMID:26868085; https://doi.org/ 10.1016/j.cyto.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 26.Finch DK, Midha A, Buchanan CL, Cochrane D, Craggs RI, Cruwys S, Grahames C, Kolbeck R, Lowe DC, Maltby J, et al.. Identification of a potent anti-IL-15 antibody with opposing mechanisms of action in vitro and in vivo. Br J Pharmacol 2011; 162:480-90; PMID:20942844; https://doi.org/ 10.1111/j.1476-5381.2010.01061.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard J, Harb C, Mortier E, Quéméner A, Meloen RH, Vermot-Desroches C, Wijdeness J, van Dijken P, Grötzinger J, Slootstra JW, et al.. Identification of an interleukin-15alpha receptor-binding site on human interleukin-15. J Biol Chem 2004; 279:24313-22; PMID:15039446; https://doi.org/ 10.1074/jbc.M312458200 [DOI] [PubMed] [Google Scholar]
- 28.Timmerman P, Puijk WC, Meloen RH. Functional reconstruction and synthetic mimicry of a conformational epitope using CLIPS™ technology. J Mol Recognit 2007; 20:283-99; PMID:18074397; https://doi.org/ 10.1002/jmr.846 [DOI] [PubMed] [Google Scholar]
- 29.Ring AM, Lin JX, Feng D, Mitra S, Rickert M, Bowman GR, Pande VS, Li P, Moraga I, Spolski R, et al.. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat Immunol 2012; 13:1187-95; PMID:23104097; https://doi.org/ 10.1038/ni.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori T, Uchiyama T, Tsudo M, Umadome H, Ohno H, Fukuhara S, Kita K, Uchino H. Establishment of an interleukin 2-dependent human T cell line from a patient with T cell chronic lymphocytic leukemia who is not infected with human T cell leukemia/lymphoma virus. Blood 1987; 70:1069-72; PMID:3115332 [PubMed] [Google Scholar]
- 31.Brizzi MF, Avanzi GC, Veglia F, Clark SC, Pegoraro L. Expression and modulation of IL-3 and GM-CSF receptors in human growth factor dependent leukaemic cells. Br J Haematol 1990; 76:203-9; PMID:2151248; https://doi.org/ 10.1111/j.1365-2141.1990.tb07872.x [DOI] [PubMed] [Google Scholar]
- 32.Mention JJ, Ben Ahmed M, Bègue B, Barbe U, Verkarre V, Asnafi V, Colombel JF, Cugnenc PH, Ruemmele FM, McIntyre E, et al.. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology 2003; 125:730-45; PMID:12949719; https://doi.org/ 10.1016/S0016-5085(03)01047-3 [DOI] [PubMed] [Google Scholar]
- 33.Gonsalves N, Policarpio-Nicolas M, Zhang Q, Rao MS, Hirano I. Histopathologic variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc 2006; 64:313-9; PMID:16923475; https://doi.org/ 10.1016/j.gie.2006.04.037 [DOI] [PubMed] [Google Scholar]
- 34.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, Burks AW, Chehade M, Collins MH, Dellon ES, et al.. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol 2011; 128:3-20.e6; quiz 21–2; PMID:21477849; https://doi.org/ 10.1016/j.jaci.2011.02.040 [DOI] [PubMed] [Google Scholar]
- 35.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007; 8:R19; PMID:17291332; https://doi.org/ 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, Franciosi JP, Garza JM, Kaul A, King EC, et al.. Molecular Diagnosis of Eosinophilic Esophagitis by Gene Expression Profiling. Gastroenterology 2013; 145:1289-99; PMID:23978633; https://doi.org/ 10.1053/j.gastro.2013.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, Weirauch MT, Vaughn S, Lazaro S, Rupert AM, et al.. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet 2014; 46:895-900; PMID:25017104; https://doi.org/ 10.1038/ng.3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergamaschi C, Bear J, Rosati M, Beach RK, Alicea C, Sowder R, Chertova E, Rosenberg SA, Felber BK, Pavlakis GN. Circulating IL-15 exists as heterodimeric complex with soluble IL-15Rα in human and mouse serum. Blood 2012; 120:e1-8; PMID:22496150; https://doi.org/ 10.1182/blood-2011-10-384362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebrec H, Horner MJ, Gorski KS, Tsuji W, Xia D, Pan WJ, Means G, Pietz G, Li N, Retter M, et al.. Homeostasis of human NK cells is not IL-15 dependent. J Immunol 2013; 191:5551-8; PMID:24184554; https://doi.org/ 10.4049/jimmunol.1301000 [DOI] [PubMed] [Google Scholar]
- 40.DeGottardi MQ, Okoye AA, Vaidya M, Talla A, Konfe AL, Reyes MD, Clock JA, Duell DM, Legasse AW, Sabnis A, et al.. Effect of Anti-IL-15 administration on T cell and NK cell homeostasis in Rhesus Macaques. J Immunol 2016; 197:1183-98; PMID:27430715; https://doi.org/ 10.4049/jimmunol.1600065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oitate M, Masubuchi N, Ito T, Yabe Y, Karibe T, Aoki T, Murayama N, Kurihara A, Okudaira N, Izumi T. Prediction of human pharmacokinetics of therapeutic monoclonal antibodies from simple allometry of monkey data. Drug Metab Pharmacokinet 2011; 26:423-30; PMID:21606605; https://doi.org/ 10.2133/dmpk.DMPK-11-RG-011 [DOI] [PubMed] [Google Scholar]
- 42.Yokoyama S, Takada K, Hirasawa M, Perera LP, Hiroi T. Transgenic mice that overexpress human IL-15 in enterocytes recapitulate both B and T cell-mediated pathologic manifestations of celiac disease. J Clin Immunol 2011; 31:1038-44; PMID:21938511; https://doi.org/ 10.1007/s10875-011-9586-7 [DOI] [PubMed] [Google Scholar]
- 43.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, et al.. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest 2006; 116:536-47; PMID:16453027; https://doi.org/ 10.1172/JCI26679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Setty M, Discepolo V, Abadie V, Kamhawi S, Mayassi T, Kent A, Ciszewski C, Maglio M, Kistner E, Bhagat G, et al.. Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology 2015; 149:681-91.e10; PMID:26001928; https://doi.org/ 10.1053/j.gastro.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita Y, Furuta K, Ishimura N, Ishihara S. Elevated plasma cytokines in Japanese patients with eosinophilic esophagitis and gastroenteritis. Digestion 2012; 86:238-43; PMID:22964662; https://doi.org/ 10.1159/000341421 [DOI] [PubMed] [Google Scholar]
- 46.Dellon ES, Rusin S, Gebhart JH, Covey S, Higgins LL, Beitia R, Speck O, Woodward K, Woosley JT, Shaheen NJ. Utility of a noninvasive serum biomarker panel for diagnosis and monitoring of eosinophilic esophagitis: A prospective study. Am J Gastroenterol 2015; 110:821-7; PMID:25781367; https://doi.org/ 10.1038/ajg.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra A. Significance of mouse models in dissecting the mechanism of human eosinophilic gastrointestinal diseases (EGID). J Gastroenterol Hepatol Res 2013; 2:845-53; PMID:25866707; https://doi.org/ 10.6051/j.issn2224-3992.2013.02.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niranjan R, Rayapudi M, Mishra A, Dutt P, Dynda S, Mishra A. Pathogenesis of allergen-induced eosinophilic esophagitis is independent of interleukin (IL)-13. Immunol Cell Biol 2013; 91:408-15; PMID:23689305; https://doi.org/ 10.1038/icb.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schluns KS, Williams K, Ma A, Zheng XX, Lefrançois L. Cutting edge: requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J Immunol 2002; 168:4827-31; PMID:11994430; https://doi.org/ 10.4049/jimmunol.168.10.4827 [DOI] [PubMed] [Google Scholar]
- 50.Burkett PR, Koka R, Chien M, Chai S, Chan F, Ma A, Boone DL. IL-15R alpha expression on CD8+ T cells is dispensable for T cell memory. Proc Natl Acad Sci U S A 2003; 100:4724-9; PMID:12671073; https://doi.org/ 10.1073/pnas.0737048100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanoni I, Spreafico R, Bodio C, Di Gioia M, Cigni C, Broggi A, Gorletta T, Caccia M, Chirico G, Sironi L, et al.. IL-15 cis presentation is required for optimal NK cell activation in lipopolysaccharide-mediated inflammatory conditions. Cell Rep 2013; 4:1235-49; PMID:24055061; https://doi.org/ 10.1016/j.celrep.2013.08.021 [DOI] [PubMed] [Google Scholar]
- 52.Polansky JK, Bahri R, Divivier M, Duitman EH, Vock C, Goyeneche-Patino DA, Orinska Z, Bulfone-Paus S. High dose CD11c-driven IL15 is sufficient to drive NK cell maturation and anti-tumor activity in a trans-presentation independent manner. Sci Rep 2016; 6:19699; PMID:26822794; https://doi.org/ 10.1038/srep19699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson BG, Quinn LS. Free IL-15 is more abundant than IL-15 complexed with soluble IL-15 receptor-alpha in murine serum: Implications for the mechanism of IL-15 secretion. Endocrinology 2016; 157:1315-20; PMID:26812159; https://doi.org/ 10.1210/en.2015-1746 [DOI] [PubMed] [Google Scholar]
- 54.Hart TK, Cook RM, Zia-Amirhosseini P, Minthorn E, Sellers TS, Maleeff BE, Eustis S, Schwartz LW, Tsui P, Appelbaum ER, et al.. Preclinical efficacy and safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in cynomolgus monkeys. J Allergy Clin Immunol 2001; 108:250-7; PMID:11496242; https://doi.org/ 10.1067/mai.2001.116576 [DOI] [PubMed] [Google Scholar]
- 55.Stein ML, Collins MH, Villanueva JM, Kushner JP, Putnam PE, Buckmeier BK, Filipovich AH, Assa'ad AH, Rothenberg ME. Anti-IL-5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol 2006; 118:1312-9; PMID:17157662; https://doi.org/ 10.1016/j.jaci.2006.09.007 [DOI] [PubMed] [Google Scholar]
- 56.Hommeida S, Alsawas M, Murad MH, Katzka DA, Grothe RM, Absah I. The Association Between Celiac Disease and Eosinophilic Esophagitis: Mayo Experience and Meta-analysis of the Literature. J Pediatr Gastroenterol Nutr 2016; [Epub ahead of print]; PMID:28045773; https://doi.org/ 10.1097/MPG.0000000000001499 [DOI] [PubMed] [Google Scholar]
- 57.Peterson K, Firszt R, Fang J, Wong J, Smith KR, Brady KA. Risk of autoimmunity in EoE and families: A population-based cohort study. Am J Gastroenterol 2016; 111:926-32; PMID:27215923; https://doi.org/ 10.1038/ajg.2016.185 [DOI] [PubMed] [Google Scholar]
- 58.Johnson JB, Boynton KK, Peterson KA. Co-occurrence of eosinophilic esophagitis and potential/probable celiac disease in an adult cohort: a possible association with implications for clinical practice: Gluten Spectrum in EoE. Dis Esophagus 2016; 29(8):977-82; PMID:26541352; https://doi.org/ 10.1111/dote.12419 [DOI] [PubMed] [Google Scholar]
- 59.Straumann A, Aceves SS, Blanchard C, Collins MH, Furuta GT, Hirano I, Schoepfer AM, Simon D, Simon HU. Pediatric and adult eosinophilic esophagitis: similarities and differences. Allergy 2012; 67:477-90; PMID:22313241; https://doi.org/ 10.1111/j.1398-9995.2012.02787.x [DOI] [PubMed] [Google Scholar]
- 60.Mori A, Suko M, Kaminuma O, Inoue S, Ohmura T, Nishizaki Y, Nagahori T, Asakura Y, Hoshino A, Okumura Y, et al.. IL-15 promotes cytokine production of human T helper cells. J Immunol 1996; 156:2400-5; PMID:8786297 [PubMed] [Google Scholar]
- 61.Holmkvist P, Roepstorff K, Uronen-Hansson H, Sandén C, Gudjonsson S, Patschan O, Grip O, Marsal J, Schmidtchen A, Hornum L, et al.. A major population of mucosal memory CD4+ T cells, coexpressing IL-18Rα and DR3, display innate lymphocyte functionality. Mucosal Immunol 2015; 8:545-58; PMID:25269704; https://doi.org/ 10.1038/mi.2014.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niranjan R, Rajavelu P, Ventateshaiah SU, Shukla JS, Zaidi A, Mariswamy SJ, Mattner J, Fortgang I, Kowalczyk M, Balart L, et al.. Involvement of interleukin-18 in the pathogenesis of human eosinophilic esophagitis. Clin Immunol 2015; 157:103-13; PMID:25638412; https://doi.org/ 10.1016/j.clim.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishra A, La Perle K, Kwiatkowski S, Sullivan LA, Sams GH, Johns J, Curphey DP, Wen J, McConnell K, Qi J, et al.. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov 2016; 6:986-1005; PMID:27422033; https://doi.org/ 10.1158/2159-8290.CD-15-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saikh KU, Kissner TL, Nystrom S, Ruthel G, Ulrich RG. Interleukin-15 increases vaccine efficacy through a mechanism linked to dendritic cell maturation and enhanced antibody titers. Clin Vaccine Immunol 2008; 15:131-7; PMID:18045883; https://doi.org/ 10.1128/CVI.00320-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, Lowichik A, Chen X, Emerson L, Cox K, et al.. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology 2014; 147:602-9; PMID:24907494; https://doi.org/ 10.1053/j.gastro.2014.05.036 [DOI] [PubMed] [Google Scholar]
- 66.Tang F, Sally B, Lesko K, Discepolo V, Abadie V, Ciszewski C, Semrad C, Guandalini S, Kupfer SS, Jabri B. Cysteinyl leukotrienes mediate lymphokine killer activity induced by NKG2D and IL-15 in cytotoxic T cells during celiac disease. J Exp Med [Internet] 2015; 212:1487-95; PMID:26304964; https://doi.org/ 10.1084/jem.20150303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoontrakoon R, Chu HW, Gardai SJ, Wenzel SE, McDonald P, Fadok VA, Henson PM, Bratton DL. Interleukin-15 inhibits spontaneous apoptosis in human eosinophils via autocrine production of granulocyte macrophage-colony stimulating factor and nuclear factor-kappaB activation. Am J Respir Cell Mol Biol 2002; 26:404-12; PMID:11919076; https://doi.org/ 10.1165/ajrcmb.26.4.4517 [DOI] [PubMed] [Google Scholar]
- 68.Rayapudi M, Rajavelu P, Zhu X, Kaul A, Niranjan R, Dynda S, Mishra A, Mattner J, Zaidi A, Dutt P, et al.. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin Transl Immunol 2014; 3:e9; PMID:25505954; https://doi.org/ 10.1038/cti.2013.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol 2007; 81:916-24; PMID:17194734; https://doi.org/ 10.1189/jlb.1106653 [DOI] [PubMed] [Google Scholar]
- 70.Straumann A, Blanchard C, Radonjic-Hoesli S, Bussmann C, Hruz P, Safroneeva E, Simon D, Schoepfer AM, Simon HU. A new eosinophilic esophagitis (EoE)-like disease without tissue eosinophilia found in EoE Families. Allergy 2016; 71:889-900; PMID:26970242; https://doi.org/ 10.1111/all.12879 [DOI] [PubMed] [Google Scholar]
- 71.Sayej WN, Ménoret A, Maharjan AS, Fernandez M, Wang Z, Balarezo F, Hyams JS, Sylvester FA, Vella AT. Characterizing the inflammatory response in esophageal mucosal biopsies in children with eosinophilic esophagitis. Clin Transl Immunol 2016; 5:e88; PMID:27525061; https://doi.org/ 10.1038/cti.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arias A., Pérez-Martínez I, Tenías JM, Lucendo AJ. Systematic review with meta-analysis: the incidence and prevalence of eosinophilic oesophagitis in children and adults in population-based studies. Aliment Pharmacol Ther 2015; 25:208-12; PMID:26510832; https://doi.org/ 10.1111/apt.13441 [DOI] [PubMed] [Google Scholar]
- 73.Lebwohl B, Ludvigsson JF, Green PHR. Celiac disease and non-celiac gluten sensitivity. BMJ 2015; 351:h4347; PMID:26438584; https://doi.org/ 10.1136/bmj.h4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nylund L, Kaukinen K, Lindfors K. The microbiota as a component of the celiac disease and non-celiac gluten sensitivity. Clin Nutr Exp 2016; 6:17-24; PMID:26990286; https://doi.org/ 10.1016/j.yclnex.2016.01.00226990286 [DOI] [Google Scholar]
- 75.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, Moore W, Stevens MJ, Yeckes A, Amsden K, et al.. Esophageal microbiome in eosinophilic esophagitis. PloS One 2015; 10:e0128346; PMID:26020633; https://doi.org/ 10.1371/journal.pone.0128346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, Wang ML. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome 2015; 3:2; PMID:25705378; https://doi.org/ 10.1186/s40168-015-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meisel M, Mayassi T, Fehlner-Peach H, Koval JC, O'Brien SL, Hinterleitner R, Lesko K, Kim S, Bouziat R, Chen L, et al.. Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J 2017; 11:15-30; PMID:27648810; https://doi.org/ 10.1038/ismej.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thorburn AN, Macia L, Mackay CR. Diet, Metabolites, and “Western-Lifestyle” inflammatory diseases. Immunity 2014; 40:833-42; PMID:24950203; https://doi.org/ 10.1016/j.immuni.2014.05.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


