Abstract
Protective high-affinity antibody responses depend on competitive selection of B cells carrying somatically mutated B-cell receptors by follicular helper T (TFH) cells in germinal centres. The rapid T-B-cell interactions that occur during this process are reminiscent of neural synaptic transmission pathways. Here we show that a proportion of human TFH cells contained dense-core granules marked by chromogranin B, which are normally found in neuronal presynaptic terminals storing catecholamines such as dopamine. TFH cells produce high amounts of dopamine and released it upon cognate interaction with B cells. Dopamine causes rapid translocation of intracellular ICOSL (inducible T-cell co-stimulator ligand, also known as ICOSLG) to the B-cell surface, which enhances accumulation of CD40L and chromogranin B granules at the human TFH cell synapse and increases the synapse area. Mathematical modelling suggests that faster dopamine-induced T-B-cell interactions increase total germinal centre output and accelerate it by days. Delivery of neurotransmitters across the T-B-cell synapse may be advantageous in the face of infection.
Nervous and immune systems enable higher organisms to monitor their environments. Afferent signals register cues that are usually processed by complex cell-cell interactions in the central nervous system or secondary lymphoid organs. Growing evidence suggests that the central nervous system and the immune system share signalling pathways previously considered system-specific. Lymphocytes co-opt elements of the molecular apparatus of neurons to form synapses that focus reception of antigen and costimulatory signals, and secretion of cytokines1. B cells can take up, release and/or respond to neurotransmitters such as catecholamines (CTs) (adrenaline, noradrenaline and dopamine)2–8, and human dendritic cells and T cells have been reported to produce dopamine9,10.
The generation of long-lived B cell responses takes place in germinal centres (GCs), where B cells and follicular helper T (TFH) cells form multiple short-lived interactions11 that ensure efficient selection of rapidly evolving B-cell clones competing for limiting T-cell help12,13. As such, signals that enhance T-B-cell interactions are likely to increase or accelerate the chances of selection and subsequent generation of long-lived B-cell responses. The speed and complexity of cellular interactions taking place in the germinal centre is analogous to the cellular connections within the nervous system. Therefore, we asked whether synaptic interactions involving secretion of neurotransmitters participate in germinal centre selection.
TFH cells contain chromogranin B+ granules
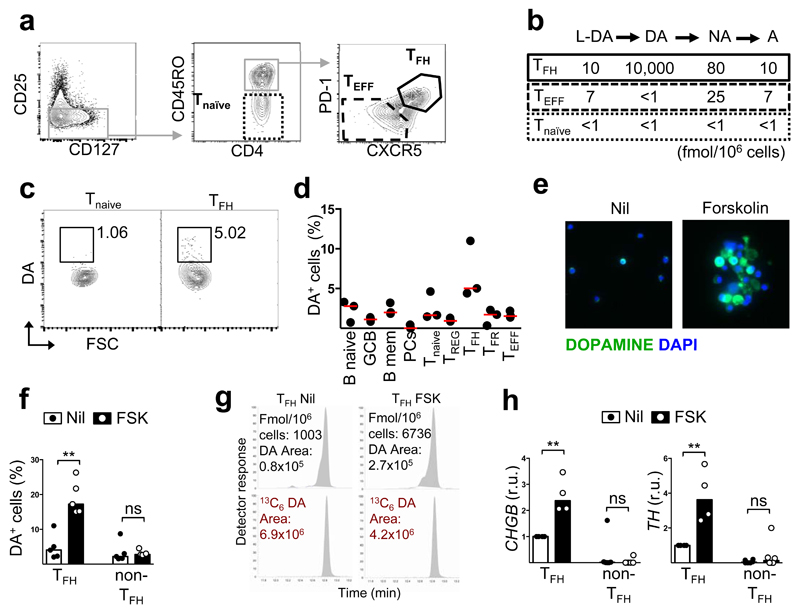
We stained human secondary lymphoid tissues with antibodies against molecules involved in synaptic transmission, whose transcripts were upregulated in human TFH cells14. Chromogranin B (CgB, encoded by CHGB), a protein that marks dense-core neuroendocrine secretory granules containing catecholamines15,16, showed selective reactivity within cells in human tonsil, lymph node and spleen germinal centres (Fig. 1a and Extended Data Fig. 1a, b), staining 3-5% of all germinal centre T cells (Extended Data Fig. 1c-e). CHGB RNA transcripts were also high in TFH cells (Fig. 1b, c). CgB+ cells expressed CD3 and the TFH markers PD-1, ICOS, CXCR5 and BCL-6 (Fig. 1d and Extended Data Fig. 1f). In mice, no CgB-expressing cells were detected in spleen or Peyer’s patches from immunised or lupus-prone mice despite CgB+ cells being visible in neuroendocrine tissues (Extended Data Fig. 2a-j). CHGB transcripts were not detected either in mouse T cells (Extended Data Fig. 2k). Analysis of CHGB transcripts using a live-cell RNA detection probe revealed high amounts of CHGB mRNA in the vast majority of human germinal centre TFH cells and intermediate amounts in pre-TFH and other effector T cells (Fig. 1e). This suggests that CgB protein can be rapidly regulated post-transcriptionally, rather than being confined to a subset of TFH cells.
Fig. 1. Human TFH cells express chromogranin B and contain dense-core vesicles.
a, Immunohistochemistry stain of human GCs: CgB (brown) (n=50). Scale bar 100μm. b, c CHGB mRNA by qPCR (b) (normalised to β2-microglobulin; r.u.: relative units) and RNA-sequencing (c) in lymphoid cell subsets, CPM: count per million. (n=3). d, Immunofluorescence on paraffin-embedded tonsil for CgB (red) and TFH markers (green). Magnification 400X (n=10). e, Flow cytometric plots showing CHGB mRNA in live CD3+ cells and fluorescence intensity within the indicated cell subsets (n=5). f, CgB stain in IgG4-related disease (n=5). g, CgB+ cells per mm2 tissue; bars represent medians; each dot is the average of 10 areas from each patient. ns, not significant, *p ≤ 0.05, **p ≤ 0.01; Mann-Whitney test. h, i, Ultrastructure of dense vesicles (arrows) within GC cells by electron-microscopy. ER=endoplasmic reticulum (n=3). j, k, Immunogold labelling for CgB in GC cells. ex.sp.: extracellular space; V: spaces generated during processing. Scale bar 2 μm (n=3). l, Immunofluorescence stain on sorted TFH cells; CgB+ (red) (n=3).
CgB+ cells were increased in IgG4-related disease (IgG4-RD, Fig. 1f, g) and three neoplasms of germinal centre origin: T-Cell-Rich B-Cell Lymphoma (T/HRBCL), nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL)17,18 and angioimmunoblastic T cell lymphoma (AITL) (Fig. 1g), consistent with increased TFH cells in these conditions (Extended Data Fig. 1d). CgB+ cells were also visible in the ectopic GCs of Hashimoto’s thyroiditis and were reduced in follicular lymphoma (FL) (Fig. 1g). Electron microscopy of human germinal centres confirmed the presence of typical neurosecretory dense-core granules (Fig. 1h, i) that stained positive for CgB (Fig. 1j, k), and cytoplasmic CgB+ granules were visualised in sorted TFH cells (Fig. 1l).
TFH cells produce and release dopamine
The above findings suggested that TFH cells may contain catecholamines. We used highly specific tandem gas chromatography-mass spectrometry (GC-MS/MS) to quantify dopamine (DA), adrenaline (A) and noradrenaline (NA) (the three most abundant catecholamines in dense-core granules from presynaptic neurons) in sorted tonsil T-cell subsets (Fig. 2a). The only abundant catecholamine in T cells was dopamine, which was found at high concentrations in TFH cells and was barely detectable in the other T-cell subsets (Fig. 2b). Flow cytometric staining using an anti-dopamine antibody10 also revealed ~5% of TFH cells contained dopamine ex vivo (Fig. 2c, d). Incubation of purified TFH cells for 24 hours with forskolin (FSK), the cAMP-inducing agent reported to enhance dopamine synthesis10, increased the proportion of dopamine-containing TFH cells 4-7 fold as determined by flow cytometry (Fig. 2e, f) and GC/MS/MS (Fig. 2g and Table 1). Dopamine did not increase in FSK-stimulated naïve T cells or non-TFH cells. FSK treatment also increased transcription of chromogranin B (CHGB) and tyrosine hydroxylase (TH) (Fig. 2h), the enzyme that converts tyrosine to the dopamine precursor L-DOPA (3,4(OH)2-phenylalanine)19, in TFH cells. Human TFH cells also expressed dopamine β-hydroxylase (DBH) RNA14 (Extended Data Fig. 3a, b), the enzyme that further metabolises dopamine. By contrast, in mice DBH expression appeared restricted to B cells (Extended Data Fig. 3c-f) and FSK induced minimal dopamine in TFH cells (Extended Data Fig. 4a, b).
Fig. 2. Human TFH cells produce dopamine.
a, Gating strategy for sorting T cell subsets from human tonsil. b, Quantification of catecholamines by GC/MS/MS (n=2). c, Representative dopamine FACS stain. d, Quantification of DA-expressing cells (n=3). e, Representative immunofluorescence dopamine stains of TFH cells untreated or treated with forskolin (FSK) (n=5). f, DA expression in untreated or FSK-treated TFH and non-TFH (effector and naïve T). (n=5). g, Representative GC/MS/MS peaks showing DA content in FSK-treated and untrated TFH cells compared to internal standard (13C6-labelled DA), which controls for losses during extraction. h, CHGB and TH mRNA expression by qPCR (normalised to RPL13A) in FSK-treated TFH and non-TFH cells. r.u., relative unit. (n=3). f,h, Bars represent median values; each dot represents one donor. ns, not significant and **p ≤ 0.01; Mann-Whitney test.
Table 1. Dopamine and DOPAC before and after FSK treatment.
| DOPAMINE (fmol/106 cells) | DOPAC (fmol/106 cells) | ||||
|---|---|---|---|---|---|
| conjugated | Fold change | conjugated | Fold change | ||
|
|
|||||
| TFH cells | Nil | 1003 | 55 | ||
| FSK | 6736 | 6.71 | 672 | 12.2 | |
|
| |||||
| naïve T cells | Nil | 116 | 20 | ||
| FSK | n.d. | - | 127 | 6.35 | |
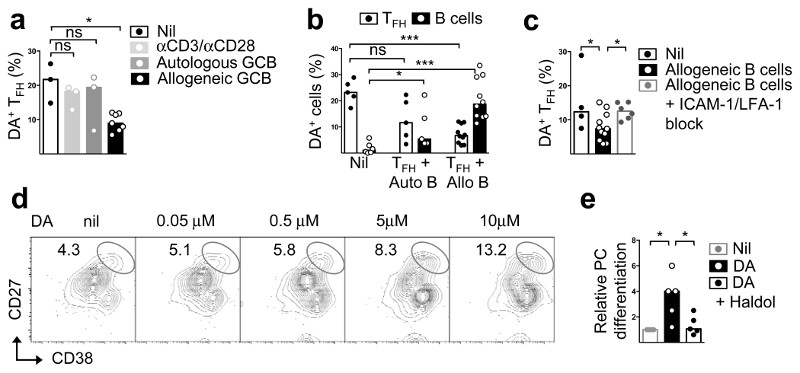
Having demonstrated that TFH cells can store and synthesise dopamine, we next investigated the conditions that could trigger dopamine release. Culture for 30 min with anti-CD3 plus anti-CD28 alone had no effect, and culture with autologous germinal centre B cells in the presence or absence of anti-CD3 led to dopamine release only in some donors (Fig. 3a, b, and Extended Data Fig. 5a). By contrast culture with allogeneic germinal centre B cells to facilitate antigen-specific TFH:B cell contacts caused over 50% reduction in dopamine in most donors (Fig. 3a-c), suggesting that cognate interactions with B cells are required. As TFH cells released dopamine, co-cultured B cells appeared to capture it (Fig. 3b). Inhibition of LFA1/ICAM1 interactions prevented dopamine release (Fig. 3c), suggesting T-cell antigen receptor signals and B-cell-derived integrin signals are both needed for dopamine transmission from TFH cells to germinal centre B cells.
Fig. 3. Dopamine is released from TFH cells upon cognate interactions.
a-c Flow cytometric quantification of dopamine content in FSK-stimulated TFH cells after 30 min incubation with anti-CD3/CD28 beads (1:1) or autologous or allogeneic GC B cells (1:2) (n=3) (b) also showing changes in DA content in GC B cells (autologous or allogeneic) cultured separately (“nil”), or together with TFH cells (n=5); and with or without ICAM-1 (5 μg/ml) and LFA-1 (10 μg/ml) block (c) (n=3); Mann-Whitney test. d, Flow cytometric plots showing plasma cells (PCs), identified as CD27hiCD38hi, induced in cultures of GC B cells stimulated for five days with anti-CD40 (2 μg/ml), IL-21 (20 ng/ml) and different concentrations of freshly-prepared DA (n=5). e, Fold changes in PC differentiation from GC B cells stimulated for 2h with or without DA (5μM) and Haloperidol (Haldol, 50nM), and cultured in the presence of anti-CD40 (2 μg/ml) and IL-21 (20 ng/ml) for 5d; two tailed student t-test. a-c, e: Bars represent median values and each dot represents a single experiment conducted in triplicates (n=5). Two tailed student t-test; ns, not significant, *p ≤ 0.05, ***p ≤ 0.001.
Dopamine translocates ICOSL to the surface
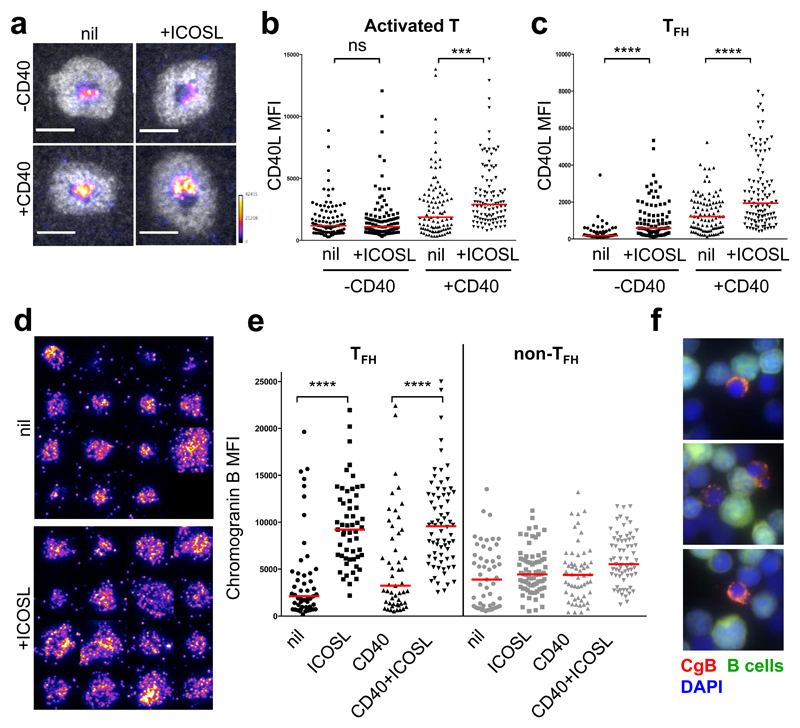
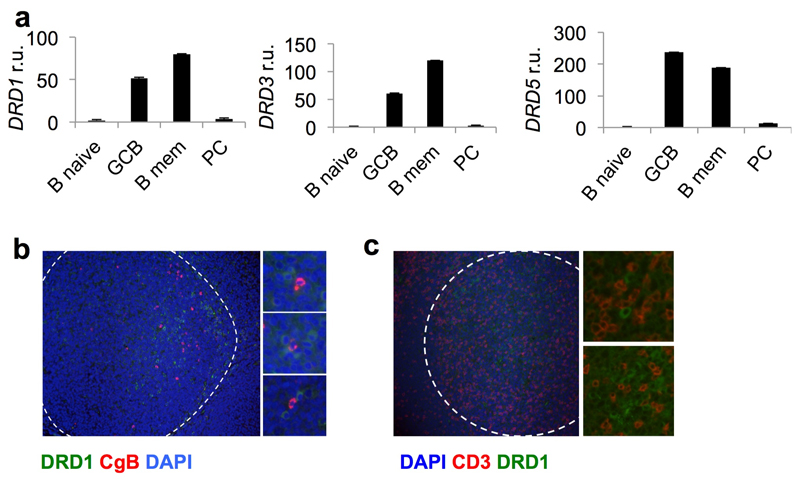
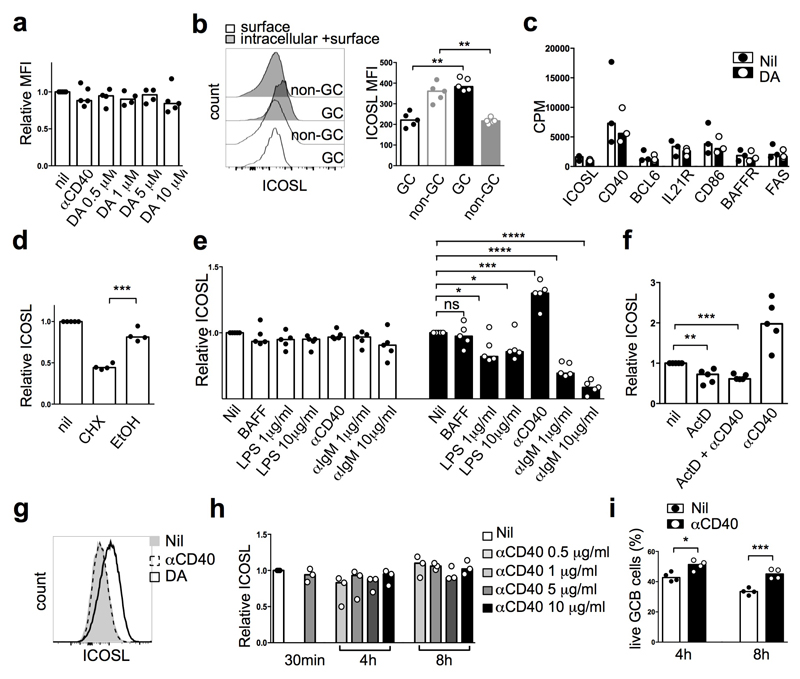
Analysis of expression of the dopamine receptors DRD1, 3 and 5 in human B cells20 revealed abundant transcripts in germinal centre and memory B cells (Extended Data Fig. 6a). DRD1+ cells were enriched in germinal centre light zones, some times in close proximity to CgB+ T cells (Extended Data Fig. 6b, c). Dopamine augmented germinal centre B-cell differentiation to plasma cells induced by interleukin (IL)-21 (Fig. 3d, e). Specificity was confirmed by inhibition with the dopamine receptor antagonist haloperidol21 (Fig. 3e). This effect was variable, with some donors exhibiting small or no effect (Fig. 3e). Dopamine did not influence germinal centre B-cell proliferation, cell cycle kinetics, or death (data not shown). Next, we investigated whether dopamine could regulate molecules important for germinal centre B-cell homeostasis or their ability to elicit T-cell help. Whereas no changes were observed in IL-21R, CD40, CD86, BAFFR, FAS or in intracellular BCL-6 (Fig. 4a), dopamine significantly upregulated surface ICOSL expression within 30 mins (Fig. 4a, b) without affecting germinal centre B-cell survival (Fig. 4c). The dopamine agonist SKF38393 also induced ICOSL upregulation in human germinal centre B cells, an effect that was blocked by haloperidol and the more selective dopamine receptor 1 (DRD1) antagonist SKF83566 (Fig. 4d). In contrast to the response observed in human germinal centre B cells, dopamine did not induce ICOSL upregulation in mouse germinal centre B cells (Extended Data Fig. 7a).
Fig. 4. Dopamine induces ICOSL upregulation on human GC B cells.
a, Gating of GC B cells and fluorescence intensity of specified proteins 30 minutes after stimulation with DA (10μM) (n=3). b, Fold changes of surface ICOSL expression with medium control set as unit 1 (n=8). c, Survival of GC B cells after DA stimulation (n=8). d, Fold changes of surface ICOSL expression on GC B cells stimulated with DA (10μM), DA agonist SKF38393 (10nM), Haloperidol (50nM) and DA antagonist SKF83566 (10nM) for 30 min, with medium control set as unit 1 (n=5). e, f, Representative histograms (e) and quantification (f) of surface and intracellular ICOSL on naïve, memory and GC B cells (n=4); Mann-Whitney test. g, RNA counts per million (CPM) of indicated transcripts in human GC B cells stimulated with or without DA (5μM) for 2h (n=3). h, Fold changes of surface ICOSL expression on human GC B cells treated with cycloheximide (CHX, 10 μg/ml) and stimulated with DA (10μM) for 30 min. i, j, Fold changes of surface ICOSL expression on human GC B cells stimulated with DA (10μM), anti-CD40 (1 μg/ml) or recombinant CD40L (10 μg/ml) (i), IL-21 (10, 50 or 100 ng/ml) or IL-4 (10 νγ/ml) (j) for 30 min (n=5). b, d, h-j, Bars represent medians and each dot represents a single experiment conducted in triplicate (n=10); two tailed student t-test. ns, not significant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001.
This rapid dopamine-induced upregulation of ICOSL suggested the possibility that preformed protein was translocating to the surface22. Comparison of surface and intracellular ICOSL expression revealed that mouse and human germinal centre B cells expressed high amounts of intracellular ICOSL (Fig. 4e, f and Extended Data Fig. 7b). RNA-sequencing did not reveal significant changes in ICOSL mRNA after incubation of germinal centre B cells with dopamine for 2h (Fig. 4g and Extended Data Fig. 7c) and dopamine-induced ICOSL upregulation still occurred in human germinal centre B cells treated with a selective blocker of protein synthesis (cycloheximide, CHX) (Fig. 4h). This treatment reduced ICOSL surface expression in mouse germinal centre B cells (Extended Data Fig. 7d). Together, these results suggest that dopamine triggers rapid export of ICOSL from pre-formed stores to the surface of human germinal centre B cells.
TFH-derived CD40L is the only signal known to-date to cause ICOSL upregulation in mouse germinal centre B cells23, an observation we replicated and found to occur at 4h but not within 30 min (Extended Data Fig. 7e); this upregulation was dependent on transcription24 (Extended Data Fig. 7f). Whereas BAFF caused no ICOSL modulation by 4h, lipopolysaccharides and anti-IgM stimulation induced ICOSL downregulation (Extended Data Fig. 7e); the latter was also seen in mice and is proposed to facilitate T-B-cell dissociation23. In contrast to mice, CD40 signalling triggered by either an agonistic anti-CD40 antibody or soluble CD40L did not induce ICOSL upregulation on human germinal centre B cells within 30 min, 4h or 8h stimulation (Fig. 4i and Extended Data Fig. 7g, h) across a broad dose-range. CD40 ligation did increase human germinal centre B cell survival in these experiments as expected25 (Extended Data Fig. 7i). IL-21 but not IL-4 also induced some ICOSL upregulation (Fig. 4j).
ICOSL augments CD40L at the synapse
In mice, ICOSL on germinal centre B cells engages ICOS on TFH cells leading to rapid CD40L externalization23. We used the in vitro supported lipid bilayer (SLB) to precisely quantify the effects of ICOS ligation on CD40L accumulation at the synaptic cleft of human T cells26. Addition of ICOSL to the SLB increased the amount of CD40L delivered to the synaptic cleft as detected by an anti-CD40L antibody (Fig. 5a-c and Extended Data Fig. 8a). This effect was dependent on (activated T cells) or enhanced by (TFH cells) the addition of CD40 to the SLB (Fig. 5b, c). Most TFH cells forming an immune synapse displayed chromogranin B signals within vesicle-like structures proximal to the synapse (Fig. 5d and Extended Data Fig. 8b). It is therefore likely that the small fraction of CgB+ TFH cells present within germinal centres are those engaged in synaptic interactions. In allogeneic co-cultures CgB granules within TFH cells also appeared polarised towards the B cell (Fig. 5f). As observed for CD40L, the abundance of CgB signals also increased upon ICOS ligation (Fig. 5d, e). It is therefore likely that cognate interactions with B cells and synapse formation rapidly promotes translation of the abundant CHGB mRNA present in most TFH cells and further dopamine release (Fig. 1b, c, e). Such a feed-forward loop is supported by the observed partial inhibition of dopamine release upon ICOSL blockade (Extended Data Fig. 5b).
Fig. 5. Effects of ICOS ligation at the immunological synapse.
a, Representative images of ICAM-1 ring (white) around CD40L (pseudocolor scale) in the presence or absence of CD40 and ICOSL at physiological densities on the supported lipid bilayer (SLB) containing ICAM-1 and UCHT1. Scale bar 5 µm. b, c, Plots represent CD40L MFI of individual activated human T (b) or TFH (c) cells forming synapses (n=3). d, Representative images of chromogranin B stain in the presence or absence of ICOSL at the immunological synapse. e, Plots represent CgB fluorescent intensity of individual activated TFH and non-TFH cells forming synapses (n=3). b,c,e, ns, not significant, (***p ≤ 0.001) and (****p ≤ 0.0001) nonparametric Mann-Whitney test (U test). f, Representative images of CgB+ TFH cells (red) forming synapses with allogeneic B cells (green).
Besides increasing CD40L at the synaptic cleft, addition of ICOSL to the SLB also increased the ICAM-1+ synapse area (Fig. 6a, b), which is likely to contribute to the increased T-B-cell entanglement reported in mice23. To investigate whether increasing dopamine content in TFH cells had a comparable effect in human TFH-germinal-centre B-cell interactions, we performed live imaging during allogeneic co-cultures. Indeed increasing dopamine content in TFH cells enhanced the TFH-germinal-centre B-cell contact interface without changing the duration of the interactions (Fig. 6c-e and Supplementary Video 1).
Fig. 6. Modelling of DA effect on TFH cell-GC B cell synapse and GC output.
a, Representative ICAM-1 area quantification. b, ICAM-1 area expressed as relative units (r.u.). c-e, Interactions (white) among untreated (green), or FSK-stimulated (blue) TFH cells and allogeneic GC B cells (red) cultured in the same well. For each interaction two frames are shown, numbers indicate time after starting imaging (see corresponding Supplementary Video 1). Plots represent quantification of T:B interaction duration (d) and contact area (e). b,d,e, ns, not significant, ***p ≤ 0.001; Mann-Whitney test. f, Impact of the speed of ICOSL upregulation (ΔtICOSL; fast, black and grey lines; and slow, colored lines) in GC B cells onto GC characteristics (mean affinity (left) and produced output (right)) estimated with computer simulations. Simulations were repeated with short (black, red, orange lines) and long (grey, magenta, cyan lines) periods of search for TFH cells. Slow ICOSL upregulation had the tendency to shrink GCs (see Ω in Table M2, red and magenta lines), therefore, the GC strength was restored by parameter adaptation (see Table M2, orange and cyan lines). Lines show mean of 100 simulations, grey shades show the standard deviations (details in supplementary methods). g, Graphic model of the proposed positive feedback between human TFH and GC B cells. Upon cognate interactions between TFH and GC B cells (1), dopamine (DA) is released from CgB+ granules (2). DA activates dopamine receptor 1 (DRD1) on GC B cells (3) and induces increase ICOSL surface expression (4), which in turn binds to ICOS on TFH cells, inducing CD40L membrane relocation (5) and CgB+ granule formation (6).
Dopamine accelerates germinal centre output
Our results demonstrated rapid dopamine-dependent ICOSL upregulation in human germinal centres but not in mice. As it is not possible to test the implications of fast versus slow ICOSL upregulation in experiments involving humans, we are forced to speculate about possible implications with the help of computer simulations. We started from the state-of-the-art mathematical model of germinal centre reactions, validated with many experimental data27–29, and explicitly included fast and slow up-regulation of ICOSL in germinal centre-B cells (Supplementary methods). Following the finding that TFH signalling to B cells involves a positive feedback loop between ICOS and CD40 signalling (Fig. 5c and 23), we assumed that the level of ICOSL in the B cell modulates the amount of signals received from the TFH cell. To our surprise, no impact on affinity maturation of germinal centre B cells was found using fast or slow ICOSL upregulation in these cells (Fig. 6f left). However, a significant reduction and retardation of output production was found when ICOSL was upregulated slower (Fig. 6f right). These results were robust against changes in the model assumptions and did not rely on the details of how B cells differentiated to output cells in the simulations (Extended Data Fig. 9a-c). In conclusion, the simulation results lead us to speculate that the dopamine-mediated improvement in the time required for B cells to elicit T-cell-help accelerates output of B cells from the germinal centres while keeping affinity maturation of germinal centre B cells unchanged.
Together, our data suggest a model in which human TFH cells engaging in synaptic interactions with germinal centre B cells release dopamine stored in CgB+ granules, which causes rapid externalization of ICOSL. In turn, ICOS ligation on TFH cells externalizes CD40L, increases the synapse area and enhances formation of dopamine-rich CgB granules (Fig. 6g). The resulting feed-forward loop allows maximal T-B-cell entanglement and the CD40L expression threshold required for germinal centre B-cell survival and/or selection. IL-21 was also able to cause rapid translocation of ICOSL in humans, albeit to a lesser extent than dopamine; it is possible that this cytokine is also stored in dense-core granules.
It is intriguing that the signals causing ICOSL translocation and the composition of dense-core granules in TFH cells are not conserved between mice and humans. Given the presence of intracellular ICOSL in mouse germinal centre B cells, other TFH-derived neurotransmitters and/or cytokines may also cause ICOSL translocation in mice. Our model predicts that the optimised selection conferred by dopamine signalling via DRD1 is translated into more rapid and enhanced GC output. This may provide a survival advantage in the face of infection by rapidly evolving viruses, toxins, and other infectious threats that are kept in check by high-affinity antibodies. Agonistic DRD1 signalling thus emerges as a novel strategy to boost vaccination whereas DRD1 antagonism may ameliorate pathogenic selection of self-reactive B cells into the long-lived memory or plasma cell pools that occurs in some autoimmune conditions.
Materials and Methods
Human tonsil and lymphoid tissues
Human tonsils were obtained from children undergoing routine tonsillectomy at The Canberra Hospital and Calvary John James Hospital. A single-cell suspension was obtained by mechanical disruption of the tissue followed by cell separation using Ficoll Hypaque (GE Healthcare Life Sciences) gradient. Informed consent was obtained from all patients. All experiments with human samples were approved by the Australian National University's Human Experimentation Ethics Committee and the University Hospitals Institutional Review Board. All tissue samples used for histology were retrieved from the paraffin and cryopreserved archives of the Pathology Unit of San Raffaele Scientific Institute and utilized following Institutional Review Board-approved institutional rules.
Mice, immunizations and spleen cell suspensions
C57BL/6 (B6), DBHGFP 30, Il21GFP 31 and Sanroque mice were bred and maintained in specific-pathogen-free conditions at the Australian Phenomics Facility, Australian National University. All procedures carried out were approved by the Australian National University's Animal and Human Experimentation Ethics Committees. To generate thymus-dependent germinal centre responses, 8-week-old female mice were immunized intravenously with 2 × 108 SRBCs (Applied Biological Products Management, Australia) and were taken down at day 7 post immunization. Single-splenocyte cell suspensions were prepared by mechanically disrupting the tissue through 70 μm nylon mesh filters (BD Bioscience) in complete RPMI 1640 media (Sigma). For RNA analysis T-cell subsets were isolated using FACS sorting following surface staining with CD4 FITC (RM4-5, BioLegend), CD44 Alexa Fluor 700 (IM7, BioLegend), CD25 APC (PC61, BD Pharmingen), CXCR5 biotin (2G8, BD Pharmingen), PD-1 Brilliant Violet 421 (29F.1A12, BioLegend), Streptavidin PE-Cy7 (BioLegend) and 7-AAD (Invitrogen). For ICOSL induction experiments, B cells were isolated using MACS columns (Miltenyi Biotec) according to the manufacturer’s instructions. 5 x 105 cells were stimulated with BAFF (100 ng/ml, R&D), lipopolysaccharides (LPS, 1 or 10 μg/ml, Sigma) from E. coli, anti-CD40 (10 μg/ml, BioXCell), anti-IgM (1 or 10 μg/ml, Jackson ImmunoResearch Laboratories) and indicated concentration of DA (Sigma) for 30 min or 4 h. Cells were stained for Zombie Aqua fixable viability kit (BioLegend), B220 (RA3-6B2, BD Pharmingen), GL-7 (GL7, BioLegend). Fas (Jo2, BD Pharmingen), CD3 (17A2, BD Pharmingen), ICOSL (HK5.3, BioLegend). Intracellular staining for ICOSL was performed using the FOXP3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions.
Immunohistochemistry
Using immunohistochemistry technique, the presence and location of CgB positive cells were evaluated in a series of randomly selected non-neoplastic lymphoid tissues, from different anatomical sites including tonsils (n=50), lymph-nodes (n=10) and spleens (n=10). Immunohistochemistry was also performed on pancreas and spleens of mice immunised with sheep red blood cells (n=3). Heat-induced antigen retrieval in Tris EDTA buffer (pH 9.0) for 30 min at 97 ºC was used, followed by blocking of endogenous peroxidase with 3% H2O2 and incubation with 3% normal bovine serum. Primary antibody against CgB (H300, Santa Cruz) was incubated for 1 h at room temperature, followed by detection with HRP conjugate-polymer (Thermo Scientific, Fremont, CA, USA) and developed with DAB chromogen. Selected samples underwent also double immunohistochemistry technique to evaluate simultaneous expression of CD3 (LN10, Dako) with CgB. Stained slides, underwent a second round of blocking in Tris EDTA buffer (pH 9.0), overnight incubation with the second primary antibody at 4°C and developed using AP conjugate-polymer and Fast Red chromogen (Thermo Scientific, Fremont, CA, USA). Tissue sections were counterstained with haematoxylin and scanned with Aperio ScanScope. Selected areas were analyzed with Aperio Color Deconvolution v9 algorithm and then, to produce multicolor composite images, digital snapshots were individually pseudocolored and overlaid in Adobe Photoshop CS3 (Adobe Systems, Inc., San Jose, CA)32.
To establish the number of CgB+ cells in different reactive, autoimmune and neoplastic conditions, immunohistochemistry CgB was performed on paraffin-embedded sections of randomly selected reactive lymph-node/tonsil, T cell rich/histiocytic B cell Lymphoma (THRLBCL), Nodular Lymphocyte Predominant Hodgkin Disease (NLPHL), Angioimmunoblastic-like T-cell lymphoma (AITL), Hashimoto's thyroiditis, IgG4-related disease (IgG4-RD) and Follicular Lymphoma (FL) (n=5). Slides were digitally scanned using Aperio ScanScope software and ten germinal centres for each sample were manually and randomly selected and analyzed for CgB expression in a blinded fashion. CgB+ cells were counted in ten areas for each sample. The results were expressed as the number of CgB+ cells per unit area.
Immunofluorescence
Frozen tonsil sections were fixed in cold 4% PFA for 20 min and blocked using 3% BSA. Heat-induced antigen retrieval in Tris EDTA buffer (pH 9.0) for 30 min at 97ºC was used to retrieve antigens on paraffin-embedded tonsil samples. Sections were then stained using CD3 (LN10 or polyclonal, Dako), PD-1 (NAT105, CNIO, Madrid), CXCR5 (51505, R&D), ICOS (AF169, R&D), BCL6 (LN22, Novocastra), CgB (H300, Santa Cruz), DRD1 (L205G1, BioLegend) for 1h at RT in the dark, followed by anti-mouse Alexa Fluor 488 (A-21202, Invitrogen) and anti-rabbit Alexa Fluor 594 (A-21207, Invitrogen) for 30 min at RT in the dark. Stained sections were mounted using Vectashield with DAPI mounting media (Vector Laboratories, #H-1200). Images were collected with an Olympus IX71 microscope with DP Controller software (Olympus) and compiled by using ImageJ software.
To visualise TFH-germina-centre B-cell synapses, isolated TFH cells were culture for 30 min with CFSE (Invitrogen)-labelled allogeneic germinal cell B cells. To stop the reaction, cells were pelleted and fixed with 4% PFA at 4 °C for 20 minutes without resuspending the pellet. Cells were than gently washed and transferred by cytospin to microscope slides. After blocking for 10 min with 3% BSA and permeabilizing for 10 min with 0.5% Triton, cells were incubated for 1 h at room temperature with primary antibody against CgB (H300, Santa Cruz), followed by anti-rabbit Alexa Fluor 594 (A-21207, Invitrogen) for 30 min at RT in the dark. Stained slides were mounted using Vectashield with DAPI mounting media (Vector Laboratories, #H-1200). Images were collected with a Zeiss Axio Observer microscope using ZEN software (Zeiss) and compiled by using ImageJ software.
Electron microscopy
Five samples randomly selected of human reactive tonsil were prepared and analyzed with an LEO 912AB (LEO Electron Microscopy Ltd. Cambridge, UK) electron microscope. To confirm the localization of CgB in dense-core vesicles, human reactive tonsils (n=3) were analyzed by immunoelectron microscopy33, using an anti-CgB polyclonal antibody (H300, Santa Cruz).
Gas Chromatography-Mass Spectrometry
T follicular helper cells, naïve and non-TFH effector cells were isolated and pooled from randomly selected human tonsils and were extracted using an ice-cold solution of 5M formic acid in n-butanol (1:4) and frozen till further analysis. Dopamine, noradrenaline, adrenaline and L-DOPA were measured by a highly specific Gas Chromatography Tandem Mass spectrometry (GC/MS/MS) assay in a blinded fashion. Catecholamine extraction and derivatization procedures previously described34 were slightly modified to determine Adrenaline, noradrenaline, dopamine and DOPA (no internal standards were used to ensure that no significant deuteron exchange contribution occurred during sample preparation considering expected low endogenous levels). Standards were prepared in 0.05M aqueous formic acid and calibrators were prepared within the 0.5-500 nM range. Standards and cell extracts were transferred to 13x100 mm glass culture tubes, dried and reconstituted in 2ml Tris buffer (pH 8.6) before adding a scoop (~200mg) of acid activated alumina and shaken gently for 15 min. After alumina settled, supernatant buffer was aspirated to waste and 5-6 water washes were made (aspirated and discarded) to reach neat water pH. The aqueous alumina slurry containing adsorbed catecholamines was transferred to glass Pasteur pipettes with silanized glass wool plugs. Packed alumina was rinsed with 1ml of water and then with 0.5 ml of 50% methanol. Catecholamines were carefully and slowly desorbed with 400 μL elution solvent (methanol:water:formic acid in the ratio of 6:1:0.25, pH3) into clean screw cap glass culture tubes. Alumina was flushed twice (2x400 μL) with eluting solvent to complete recovery. The contents were dried down under vacuum using a SpeedVac, (Thermo Scientific, Asheville, NC, USA). The dried samples were reconstituted using 80 μL of trifluoroethanol (TFE), 40 μL of trifluoroacetic anhydride (TFAA) and 30 μL of toluene. Tubes were capped and heated at 75°C for 30 min. Derivatives (Dopamine derivative, Extended Data Fig. 10) were transferred to glass vials and four μL were then injected into the GC. Selected reaction monitoring (SRM) transitions used in this method were evaluated for each individual trifluoroacetyl derivative and instrument conditions were optimized to ensure maximum sensitivity, collision energy was set at 20V for all transitions monitored. Analysis was performed using a Trace GC Ultra interfaced with a TSQ XLS Mass Spectrometry Detector and a Triplus autosampler (Thermo Scientific, San Jose, CA), operated in electron capture negative ionization (ECNI) mode. Methane and argon were used as the ECNI reagent and collision gas, respectively. Chromatographic separation was carried out using an Agilent 30 metre HP-5MS (0.25 mm ID × 0.25 μm film thickness) column. Helium was used as a carrier gas with flow rates of 1.2 mL/min. Argon flow in the collision cell was 1.0 mL/min. Four μL injection volumes were made at 250°C in splitless mode. The oven temperature program was as follows: 90°C for 0.5 min, 20°C/min to 160°C, held 2 min; then 5°C/min to 170°C. The temperature was again increased to 280°C at 20°C/min. The temperature of the CI source and of the quadrupoles (Q1and Q3) was 200°C and the auxiliary MS transfer line 275°C. The ECNI reagent gas flow was set at 2.0 mL/min SRM transitions for each analyte are listed in Supplementary Table 1. Data analysis was performed using Xcalibur™ software (Thermo Scientific, San Jose, CA). The quantification method detailed all compound information, which included expected peaks and their target signals as well as their expected retention times. Standard samples were tagged and the integration results (peak areas vs. concentration) were used to determine a linear regression curve for each analyte. These equations were used to calculate the concentration of endogenous catecholamines in samples. All integrated peaks were checked for accuracy.
To analyse conjugated dopamine naïve T cells or TFH cells before and after forskolin treatment were resuspended into 0.4N perchloric acid and frozen till further analysis. Hydrolysis of cell pellet with 500ul of 7% perchloric acid at 100 °C for 15 minutes allowed the release of protein bound (conjugated) dopamine. An aliquot of 13C6 labeled dopamine (Cambridge Isotope Laboratories, Andover, MA USA) was added as internal standard to correct for any losses occurred during acid hydrolysis. The supernatant was alkalinized and extracted with alumina as previously described. Similarly calibrators containing internal standard hydrolized in 7% perchloric acid, were prepared. Peak area ratios of dopamine to 13C6 labeled dopamine were plotted against concentration to construct the calibration curve used to estimate concentration of endogenous bound dopamine in the treated cells.
Human Flow cytometry
Tonsillar lymphocytes were stained with the following anti-human antibodies – CD4 APCCy7 (RPA-T4, BD Biosciences), CXCR5 Alexa 488 or Alexa 647 (RF8B2, BD Biosciences), PD-1 PE (MIH4, eBioscience) or BV605 or BV421 (EH12.2H7, BioLegend), CD127 FITC (11-1278, eBioscience) or BV 421 (A019D5, BioLegend), CD25 biotin (BC96, eBioscience or BioLegend) or PE-Cy7 (BC96, BD Biosciences or BioLegend), BCL6 Alexa 647 or PE-Cy7 (K112-91, BD Biosciences), CD3 APC (HIT3a, BD Biosciences) or Alexa 700 (UCHT1, BD Biosciences), CD27 FITC or APC (M-T271, BD Biosciences), CD38 FITC (HIT2, BD Biosciences) or PE (HB7, BD Biosciences), ICOSL APC (2D3, BioLegend), FAS PE-CF594 (DX2, BD Bioscience), CD40 APCCy7 (5C3, BioLegend), BAFFR PECy7 (11C1, BioLegend), CD19 PECy7 or BV605 (SJ25C1, BD Bioscience), IL21R BV421 (17A12, BioLegend), CD86 BV421 (2331/FUN-1, BD Bioscience). All surface stains were performed in the presence of Human TruStain FcX (cat. 422302, BD Bioscience). Intracellular staining was performed using the FOXP3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. Cells were stained with primary antibodies followed by secondary reagents for 30 min at 4 °C. Data were collected on a LSRII or Fortessa cytometer (BD) and analysed with FlowJo software (TreeStar). 7-AAD (Invitrogen) or Zombie Aqua (BioLegend) staining was used to exclude dead cells from analysis.
Immunofluorescent and flow cytometric detection of dopamine
Freshly isolated human TFH and non-TFH cells and mouse naïve and follicular T (IL-21+ or IL-21-) cells were stimulated with 10 μM forskolin (Sigma) for 24h in RPMI 1640 medium supplemented with 3% BSA, 50 μg/ml D-glucose (Sigma), 2 mM L-glutamine, 100 U penicillin-streptomycin, 0.1 mM non-essential amino acids and 100 mM Hepes (Gibco, Thermo Fisher Scientific). Cells were then collected, prefixed with 50 mM cacodylate and 1% sodium metabisulfite (MBS, Sigma) and fixed with 5% glutaraldehyde (GA) in 0.1 M cacodylate and 1% MBS for 15 min at RT. After washing with 1% MBS in 50 mM Tris (Tris–MBS) twice, the cells were incubated with or without rabbit anti-dopamine pAb (Millipore) in Tris–MBS containing 0.05% Triton-X for 1h at RT, followed by Alexa Fluor 488-conjugated anti-rabbit IgG (Invitrogen) for 30 min at RT. After washing with Tris–MBS, samples were mounted on cover glasses and visualized with a Zeiss Axio Observer microscope and then were analyzed with a LSR Fortessa (BD).
Dopamine release
Following 24 h stimulation with forskolin, as previously described, 1x105 freshly isolated human TFH cells were cultured with 1x105 anti-CD3/CD28 beads, 2x105 autologous or allogeneic CellTrace Violet (CTV, Thermo Fisher Scientific) labeled GC B cells for 30 min. In some of the culture with autologous B cells 1x105 anti-CD3/CD28 beads were added. To evaluate the effect of direct interaction 5 μg/ml of anti-ICAM-1 (clone HA58, BD) and 10 μg/ml of anti-LFA-1 (clone HI111, Biolegend) blocking antibodies were added to allogeneic co-culture. In experiments with ICOSL block allogeneic B cells pre-stimulated with 10 μM of freshly prepared dopamine (Sigma) before incubating with TFH cells in the presence or not of 10 μg/ml of anti-ICOSL blocking antibody (clone #873724, R&D). After culture cells were fixed and stained for dopamine following the protocol described previously and analyzed with a LSR Fortessa (BD).
Dopamine in vitro stimulation
Dopamine concentrations used in this study have ben calculated to mimic physiological concentration. Studies in neurons showed that a typical synaptic vesicle has a diameter in the range of 20–100 nm and a catecholamine concentration of 0.05–0.5 M. considering a 50 nm vesicle, this would contain ∼2000–20 000 molecules. Upon vesicle release this would result in a concentration of 0.3–3 mM at the synaptic cleft. Catecholamine concentration could be even higher when many vesicles are released simultaneously (i.e. 3–30 mM for 10 vesicles)35. For short term stimulation, 2 x 105 sorted human germinal centre B or 5 x 105 enriched B cells were stimulated for 30 min with 10 μM of freshly prepared DA (Sigma), 10 nM of dopamine receptor 1 agonist (SKF38393, Tocris), 10 nM of dopamine receptor 1 antagonist (SKF83566, Tocris) or 50nM of Haloperidol (Tocris) in RPMI 1640 medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U penicillin-streptomycin, 0.1 mM non-essential amino acids and 100 mM Hepes. Because Dopamine HCl (dopamine hydrochloride) is sensitive to alkalis, iron salts and oxidizing agents (i.e. light and air), solutions were always prepared from powder immediately before use. Cells were stained for ICOSL, CD40, CD86, FAS, IL21R, BAFFR, Zombie live-dead marker and intracellular Bcl6, and analysed on LSRII. For long term culture 2 x 105 sorted germinal centre B cells were stimulated with a range of DA dilutions for 5 days and stained for 7-AAD, CD27, CD4, CD19 and CD38 and analysed on LSRII. For the experiment with dopamine receptor block, 2 x 105 sorted germinal centre B cells were stimulated with 5 μM of freshly prepared DA with or without 50nM of Haloperidol (Tocris) for 2 h. Incubation media was then replaced with fresh media containing anti-CD40 (1 μg/ml, BioLegend) and IL-21 (10 ng/ml, Peprotech), cells were incubated for 5 days and plasma cell differentiation was assessed.
RT- PCR analysis
Total RNA was isolated from freshly isolated T and B cell subsets (n=5) using trizol (Invitrogen), then transcribed into cDNA by MLVRT synthesis (Invitrogen) and used as a template for qPCR to asses CHGB (F, 5’-TGC CAG TGG ATA ACA GGA AC-3’; R, 5’-TCT TCA GGA CTT GGC GGC A-3’)36, DRDs (DRD1 F, 5'-CAG TCC ACG CCA AGA ATT GCC-3'; DRD1 R, 5'-ATT GCA CTC CTT GGA GAT GGA GCC-3'; DRD3 F, 5'- TGG ATG TCA TGA TGT GTA CAG CC-3'; DRD3 R, 5'- TCC CCT GTG GTA TTA AAG CCA AAC-3'; DRD5 F, 5'-GTC GCC GAG GTG GCC GGT TAC-3'; DRD5 R, 5'-GCT GGA GTC AGA ATT CTC TGC AT-3’)20 and DBH (F, 5’- TCC AAG CTC CCA ATA TCC AG-3’; R, 5’- TCG GGT TTC ATC TTG GAG TC-3’) expression. To evaluate changes in the expression of CHGB and TH (F, 5’- TGT GAA GGT GTT TGA GAC GTT TG-3’; R, 5’-TCG AGG CGC ACG AAG TAC T-3’), total RNA was isolated from TFH and non-TFH cells after 24 h forskolin treatment. β2-microglobulin (F, 5’-TGC TGT CTC CAT GTT TGA TGT ATC T-3’; R, 5’-TCT CTG CTC CCC ACC TCT AAG T-3’), RPL13A (F, 5’-CCT GGA GGA GAA GAG GAA AGA GA-3’; R, 5’-TTG AGG ACC TCT GTG TAT TTG TCA A-3’) or GSα (F, 5’- GTG ATC AAG CAG GCT GAC TAT-3’; R, 5’- GCT GCT GGC CAC CAC GAA GAT GAT-3’) have been used as housekeeping gene. Mouse CHGB expression was evaluated using a TaqMan based assay (CHGB, Mm00483287_m1; GAPDH, Mm99999915_g1). The relative expression was calculated using the 2−ddct method37.
RNA sequencing
Human follicular T helper, follicular T regulatory, T naive cells, memory, and germinal centre B cells were FACS purified from three randomly selected fresh tonsils. Memory and germinal centre B cells were stimulated with 5 μM of freshly prepared DA in complete RPMI 1640 for 2 h. mRNA was then extracted and sent to the Australian Cancer Research Foundation Biomolecular Resource Facility, The John Curtin School of Medical Research, Australian National University for Library construction using the TruSeq Stranded mRNA LT Sample Prep Kit (Ilumina). Library samples were sequenced on a HiSeq2000 with a coverage of 25 million reads. The data was then sent to the Genome Discovery Unit (ANU Bioinformatics Consulting Unit, JCSMR, ANU) for analysis. There were a total of 621,384,768 raw 100bp paired-end reads from the sequencer across 18 samples and two lanes. Initial quality control checks were performed using FastQC and reads were subsequently trimmed using Trimmomatic38 version 0.32 with conservative settings (LEADING:15 TRAILING:15 SLIDINGWINDOW:4:20 MINLEN:60) which retained 506,390,951 high-quality read pairs (81.5%). All reads were aligned to the H. sapiens genome reference sequence using TopHat version 2.0.13 with default parameters. Read counts were then generated for each gene in each sample using featureCounts version 1.4.6-p1 by using annotated gene locations. Differential expression analysis was performed using the edgeR package version 3.10. Read counts per gene were normalized by trimmed mean of M-values (TMM). As edgeR uses the negative binomial distribution as its basic model for differential expression data, dispersion estimates are obtained using the quantile-adjusted conditional maximum likelihood (qCML) method for single factor experiments. Then, the qCML-based exact test for the negative binomial distribution was performed to test for differentially expressed genes in our groups of samples. We used a Benjamini-Hochberg adjusted p-value threshold of 0.05 to identify significantly differentially regulated genes.
Live RNA detection
Freshly isolated human tonsil cells were also used for transfection experiments with SmartFlare™ RNA detection probes (Millipore) specific for CHGB (5’-CCCAGCTTAGAGCTTGATAAGATGGCA-3’). 5 × 104 cells/well were treated with 4 μL diluted probe (1:20 in sterile PBS) and incubated overnight (16 h) at 37°C in an atmosphere of 5% CO2. In each experiment, two controls were included: a scramble SmartFlare™ Probe, which does not recognize any cellular sequence and served as a control to determine the background; an uptake SmartFlare™ Probe, which permanently fluoresces and provided the information that the SmartFlare™ particles were incorporated by the target cell type. Cells were stained for CD3, CD4, CXCR5, CD19, PD-1, CD45RO and DAPI (as live-dead marker). Fluorescence was evaluated using a LSR Fortessa flow cytometer.
Supported Lipid Bilayer (SLB) and TIRFM Imaging
SLB were formed as previously described26,39. Briefly, glass coverslips were cleaned with acid piranha solution, rinsed extensively, dried, and assembled into disposable six-channel chambers (Ibidi). SLB were formed by incubation of each channel with small unilammellar vesicles containing 12.5 mol% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] (nickel salt) and 0.05 mol% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt) in 1,2-dioleoyl-sn-glycero-3-phosphocholine at total phospholipid concentration 0.4 mM. Chambers were flooded with human serum albumin (HSA)-supplemented HEPES buffered saline (HBS), subsequently referred to as HBS/HSA. Following blocking with 5% casein in PBS containing 100 μM NiSO4, to saturate NTA sites, unlabelled streptavidin was then coupled to biotin head groups. Biotinylated UCHT1 (10 molecules/μm2) and His-tagged ICAM-1 (200 molecules/μm2), CD40 (300 moleculesa/μm2) and ICOSL (200 molecules//μm2) were then incubated with the bilayers at concentrations to achieve the indicated site densities. Anti-CD3/CD28 Dynabead stimulated CD4+ T cells (day 7) were suspended in HBS/HSA and labelled with 5 μg/mL Alexa Fluor 488 anti-CD40L antibody prior to incubation with the bilayers at 37°C for 15 minutes. Cells were then fixed at room temperature for 10 minutes with 2% formaldehyde in PHEM buffer. For experiments involving TFH cells frozen tonsil cell suspension was enriched for T cells (EasySep negative selection, Stemcell) and sorted on CXCR5hi and CXCR5low. Following synapse formation of 20 minutes the cells were fixed in 2% PFA in PHEM buffer for 10 minutes at RT and permeabilised with 0.1% Triton X-100 in HBS/HSA. Cells were then blocked with 5% Casein containing 5% Donkey serum for 1hour, washed with HBS/HSA and stained with anti-CgB in 5% Casein overnight. Followed by a secondary antibody in 5% Casein containing 5% Donkey serum for 60 minutes. Imaging was done on an Olympus IX83 inverted microscope equipped with a TIRF module. The instrument was fitted with an Olympus UApON 150x TIRF N.A 1.45 objective, 405 nm, 488 nm, 568 nm and 640 nm laser lines and Photomertrics Evolve delta EMCCD camera. Score for CD40L and CgB intensity and ICAM-1 positive area was conducted on blinded images.
Live cell in vitro TFH:GCB interaction imaging
Freshly isolated TFH cells have been stimulated for 24h with or without 10 μM forskolin. Treated TFH cells were labelled with CellTrace™ Violet (CTV, Thermo Fisher Scientific) and untreated TFH cells with CellTrace™ CFSE (CFSE, Thermo Fisher Scientific). Allogeneic GC B cells have been stimulated with 40 ng/ml of IL-4 (Peprotech) for 24h and labeled with CellTracker™ Red CMTPX (Thermo Fisher). Before imaging labeled cells were mixed together (1:1:4=TFH-FSK:TFH-nil:GCB), transferred to imaging chambers coated with 1.75 μg/cm2 of CellTak (VWR). Once stable the preparation was transferred to a Fluoview FVMPE-RS multiphoton microscope system (Olympus) equipped with a XLPLN25XWMP2 objective (25x; NA1.05; water immersion; 2mm working distance). Temperature of cells was maintained at 37°C via a heating mat underneath a thermoconductive polydimethylsiloxane (PDMS) platform, and monitored via a feedback probe within the platform. For analysis of the cells a 20 μm Z-stack (2 μm/ slice) was acquired using the resonant scanner with 5x line averaging for at least 30 minutes. Images of randomly selected areas were acquired using FV30 software (Olympus) and exported to Imaris (Bitplane) for downstream processing. Raw imaging data was analyzed in a blinded fashion with Imaris software (Bitplane). Z-stack was used to verify interactions and overlay between cells. Prior to cell tracking the Z-stack was resampled to a single slice for tracking. Tracking of individual cells was performed by using the ‘Surfaces’ function in Surpass mode, and detection of individual cells relied upon their relative fluorescent intensity and size (diameter ≥9µm). Masks of the tracked cells were then used to generate a clear final movie, on which cell interactions were defined by channel colocalisation from the Coloc mode. Interactions between cells were tracked by the ‘Surfaces’ function on regions of colocalisation. Duration of contact between cells was manual scored from the detected colocalisation, whilst the length of the contact area was determined by the longest ellipsoid axis from the colocalised region between cells. Cell counts were performed in ImageJ using the Cell Counter plugin.
Modelling the effect of Dopamine on GC reaction
In view of restrictions to test the impact of fast and slow signalling in GC-B cells in humans, we employed mathematical modelling to speculate on possible implications. Predictions from mathematical models bear the risk of relying on particular assumptions. We therefore repeated the analysis in different models and only accepted the results consistently reproduced in all models.
At first, we used the current state-of-the-art model of the GC reaction27–29 to investigate the impact of fast versus slow acquisition of help signals from TFH to B cells. The interaction time of B and T cells was set to 36 min and 4 h, to mimic the fast dopamine dependent ICOSL upregulation versus the slow dopamine-independent ICOSL upregulation. In order to make GC simulations comparable, the required duration of TFH signalling to B cells was adapted from 0.5 to 1.5 h. Longer T-B-interactions induced reduced output while keeping affinity maturation unchanged (Extended Data Fig. 9a, black curves for fast and red curves for slow ICOSL upregulation). Note that the GC strength (Ω), defined as the area under the curve of the GC B cell population kinetics27, was comparable in both simulations (Extended Data Fig. 9a, legend box), such that the effect on output production is not simply reflecting smaller GC sizes.
This result might rely on the secondary effect on the DZ to LZ ratio. To exclude this explanation for the observed reduced output, simulations with long T-B-interactions were retuned to match GC strength and DZ to LZ ratio between simulations with short and long T-B-interactions. Comparable GC volume and GC strength as well as the physiological DZ to LZ ratio were restored in silico by adapting three parameters: (i) the amount of collected antigen at which TFH induce half maximum numbers of divisions in selected B cells (KD), (ii) the duration of signalling by TFH required for B cell selection (ΔTmin), and (iii) the amount of antigen presented per follicular dendritic cells (AgFDC). The result that output production is reduced in simulations with longer T-B-interactions while keeping affinity maturation unchanged was confirmed in all settings (Extended Data Fig. 9a, black curves for fast and coloured curves for slow ICOSL upregulation, non-red curves with GC strength correction). By variation of the model parameters, we were not able to find a simulation in which longer T-B-interaction would not reduce output production.
In vivo, B cells integrate signals from many short contacts of 5 minutes to TFH cells40. The phenomenological representation of this signal-integration by a single interaction between TFH and GC-B cells in silico might be considered as a limitation of the previous approach. We therefore extended the mathematical model to better reflect the in vivo situation. Each instance of the B cell class was extended by a variable representing the amount of TFH signals received and by a variable representing the degree of ICOSL upregulation. B cells search for TFH help for a period derived from the amount of antigen they collected from FDCs. In this period (ΔtTfh), they interact with different TFH for 5 minutes each. As in the old model, TFH polarise towards the B cell which presents most pMHC in the case that more than one B cell attempts to get signals from the same TFH. The amount of signals received by a B cell in each interaction with a TFH is assumed to be proportional to the duration of TFH polarisation towards the B cell multiplied by the ICOSL level (fraction of max expression) of the B cell. This latter assumption reflects the positive feedback loop between ICOS and CD40 signalling in T-B-interactions23. The amount of integrated signal determines the number of divisions that is attributed to the B cell in the next round of recycling27,41. The number of divisions is calculated from the integrated signal with a Hill-function with Hill-coefficient 2 and characterised by the amount of signal required to induce two divisions (S(Np=2)). Note that the number of divisions was derived from the amount of collected antigen in the previous version of the model. In this new model, fast and slow ICOSL upregulation is directly represented by shifting the K-value (ΔtICOSL) of a Hill-function describing ICOSL upregulation over time. The level of ICOSL at any time impacts on the signalling strength in T-B-interactions (and by this on the number of B cell divisions) rather than on the duration of T-B-interactions (as in the previous model).
Despite a different selection model and a different impact point of the speed of ICOSL upregulation, the result that the production of output cells is retarded and reduced in simulations with slow ICOSL upregulation was confirmed (Extended Data Fig. 9b, black lines for fast versus green and red lines for slow ICOSL upregulation; grey lines for fast versus magenta lines for slow ICOSL upregulation). This also holds true, when the lower GC strength in simulations with slow ICOSL upregulation (Extended Data Fig. 9b, red lines) was compensated by a longer phase of search for TFH help leading to higher numbers of divisions attributed to B cells (Extended Data Fig. 9b, orange lines).
While there was no impact on affinity maturation in GC B cells (Extended Data Fig. 9b), the fewer output cells generated exhibited a consistently higher affinity (Extended Data Fig. 9b, all coloured lines show higher affinity than black or grey lines). Even though we consider the new model as more realistic, we excluded the impact on output affinity from the presentation in the main text, as it was not observed in all model variants.
The effect of slow ICOSL dynamics on speed and amount of output was reduced but persisted when a memory for ICOSL upregulation was assumed (Extended Data Fig. 9b, blue lines). In this setting, a selected B cell with upregulated ICOSL is thought to return to the DZ for further rounds of division. The dilution of ICOSL expression and of its mRNA onto the daughters is ignored (thus, overestimating memory) and all daughters, which restart search for TFH help in the next round of selection, are assumed to keep full ICOSL competence. Even under this extreme and unrealistic condition, the effect on output production was maintained, but less pronounced.
As the main impact point of the speed of ICOSL upregulation concerned output production we asked whether this result would depend on the choice of the output differentiation model. In the LEDA model, B cells that passed selection always return to the DZ and divide before they leave through the DZ. In the classical textbook GC, a subset of B cells that passed selection directly differentiates to output cells and leave the GC through the LZ. Both models lead to the same result (Extended Data Fig. 9c), such that we conclude that the particular choice of how B cells differentiate to output cells and leave the GC is not at the origin of the dependence of output production on the speed of ICOSL upregulation.
What is the reason for less and slower output in simulations with slow ICOSL upregulation? The number of generated output cells depends on the GC size which is reduced by slow ICOSL upregulation. But less output is also observed in simulations in which the GC size was compensated. Note that the number of generated output cells is proportional to the number of B cell selection events. By slow ICOSL upregulation, the average level of ICOSL is lower in B cells. As a consequence, the amount of TFH signals collected by B cells is reduced in silico which is supported by experimental data showing that ICOS signalling is part of a positive feedback loop in T-B-interactions23. This implies a stronger selection pressure and less selection events with slow ICOSL upregulation. In simulations with compensated GC size, the number of induced divisions in those B cells, which still passed selection, is higher (data not shown). The diversification of B cell receptors by mutation, which more likely induce bad than good mutations, compensates the gain in affinity maturation induced by the higher selection pressure. This induces three effects by slow ICOSL upregulation:
GC B cells have the same affinity maturation, which is the result of two concurring effects: higher selection pressure and higher B cell receptor diversification.
The number of selection events is reduced, which leads to less output cells being generated by the same total number of GC B cells.
The affinity of output cells is enhanced, because these are derived from a successful selection event with strong selection pressure before further diversification.
This explanation of the impact of the speed of ICOSL upregulation on output production is rather generic and involves the network of interactions active in GC reactions.
Each curve in Extended Data Fig. 9a-c shows the mean and standard deviation of 100 independent simulations. Simulations were programmed in C++ and performed under Linux Ubuntu on Xeon E5 2690 v3 CPUs. Visualisation was done with the GLE software package.
Statistical Analysis
No statistical methods were used to predetermine sample size. All samples were randomly selected and researchers were blinded whenever possible. All data were analysed with two tailed non-parametric Mann-Whitney test (U test) except for ICOSL induction and DA in vitro stimulation experiments, in which two tailed unpaired Student’s t test was used. Statistical tests with appropriate underlying assumptions on data distribution and variance characteristics were used. All statistical analysis was performed with Prism software (version 6, GraphPad Software). Statistically significant differences are indicated as *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001; ****p ≤ 0.0001 and ns = not significant.
Extended Data
Extended Data Fig. 1. CgB+ cells in human GC.
a,b, Representative immunohistochemistry for CgB (brown) of human lymph node (a) and spleen (b). (n=10). c, Quantification of CD3+CgB+ cells in human tonsils, lymph nodes (n=10) and spleens (n=5). d, Percentage of CgB+ T cells in human reactive and neoplastic conditions. c,d, ns, not significant, *p ≤ 0.05 and **p ≤ 0.01; nonparametric Mann-Whitney test (U test). e, Representative double immunohistochemistry for CgB (left) and CD3 (middle) after colour deconvolution. Pseudo-colour image (right) showing signal colocalisation. Original magnification 40X. Scale bar 100 μm (n=3). f, Representative immunofluorescence images for CD3 (green) and ICOS (red) in human GCs.
Extended Data Fig. 2. Mouse chromogranin B expression.
a-i, IHC staining shows no CgB reactivity in mouse GCs of immunised WT or Sanroque spleens and Peyer’s patches (n=3). j, IHC control staining for CgB in mouse pancreas islets. (a-j). Scale bar 100 μm. n=3 k, Relative mouse CHGB mRNA expression in different T cell subsets with adrenal gland as positive control. T cells were FACS sorted as follows: Tnaïve (CD4+ CD44lo CD25-); T effector memory (TEM, CD4+ CD44hi CD25- PD-1-/lo CXCR5-/lo); TFH (CD4+ CD44hi PD-1hi CXCR5hi); TREG (CD4+ CD25+ CD44int). GAPDH was used as housekeeping gene (n=3).
Extended Data Fig. 3. Dopamine β-hydroxylase expression in human and mouse lymphocytes.
a, Gel shows PCR products after amplification of human dopamine β-hydroxylase (DBH) mRNA in TFH cells, total tonsil and B cells. Gsα was used as housekeeping gene. For gel source data, see Supplementary Figure 1. b, RNA-sequencing showing expression of DBH mRNA in human Tnaive, TFH and TFR cells extracted from three tonsils, expressed as count per million (CPM). c, Immunofluorescence images showing GFP expression in adrenal medulla of DBHgfp/w mice. d, FACS plot showing GFP expression in splenocytes of DBHgfp/w mice. e, Quantification of DBH-GFP expression in mouse splenocytes. Bars represent median values and each dot represents a mouse (n=10). f, FACS plot showing DBH-GFP expression in B cells localising outside GCs of SRBC immunised mice (n=10).
Extended Data Fig. 4. Mouse endogenous and induced dopamine content.
a, b, Quantification and representative FACS plot of dopamine content in mouse naïve and follicular T cells (TFO) differentiated by the expression of IL-21. T cell subsets were FACS sorted into Tnaïve (CD4+ CD44lo), TFO IL-21+ (CD4+ CD44hi IL-21gfp/w) and TFO IL-21- (CD4+ CD44hi IL-21w/w) and dopamine content were analysed by flow cytometry before and after 24 h treatment with forskolin (FSK). Bars represent median values and each dot represents a mouse (n=5). *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001; nonparametric Mann-Whitney test (U test).
Extended Data Fig. 5. Dopamine release from human TFH cells.
a, Bar plot showing dopamine release from TFH cells after 30 min stimulation with autologous B cells (1:2) alone or with anti-CD3/CD28 beads (1:1). TFH cells were pre-stimulated with forskolin before inducing DA release. Bars represent median and each dot represent a single experiment conducted in triplicates (n=4). b, Bar plot showing dopamine release from TFH cells after 30 min stimulation with allogeneic GC B cells (1:2) alone or in the presence of ICOSL blocking antibody (10 μg/ml). TFH cells were pre-stimulated with forskolin before inducing DA release and B cells were pre-stimulated with 10 μM DA to increase ICOSL surface levels before incubation with TFH cells. Bar represent median of DA level in TFH cells (n=3) and each triangle represent allogeneic B cells from a single donor paired with its control (square, n=11). *p ≤ 0.05; paired t-test.
Extended Data Fig. 6. Dopamine receptors (DRDs) expression in human B cell subsets.
a, relative expression of DRDs mRNA in human B cell subsets normalised to naïve B cells. β2-microglobulin was used as housekeeping gene (n=3). Error bars show s.d. b, c, Representative images of dopamine receptor 1+ cells (green) localisation in human GC (dashed line), showing close proximity to CgB+ (b) or CD3+ (c) cells (red) (n=3).
Extended Data Fig. 7. Regulation of ICOSL upregulation in mouse and human B cells.
a, Fold changes of surface ICOSL expression on mouse GC B cells that were treated with anti-CD40 (10 μg/ml) and DA (0.5, 1, 5, 10μM) for 30 minutes, with medium control set as unit 1 (n=5). b, Representative histogram and quantification of surface and intracellular ICOSL on GC and non-GC B cells (n=5). **p ≤ 0.01; nonparametric Mann-Whitney test (U test). c, RNA counts per million of ICOSL, CD40, BCL6, IL21R, CD86, BAFFR and FAS mRNA in human memory B cells stimulated with or without DA (5μM) for 2h (n=3). d, Fold changes of surface ICOSL expression on mouse GC B cells that were treated with cycloheximide (CHX, 10 μg/ml) for 4h, with medium control set as unit 1. Bars represent median values and each dot represents a single mouse. e, Fold changes of surface ICOSL expression on mouse GC B cells that were stimulated with BAFF (100ng/ml), LPS (1 or 10 μg/ml), anti-CD40 (10 μg/ml) and anti-IgM (1 or 10 μg/ml) for 30 min and 4h. Unit 1 set on medium control. f, Fold changes of surface ICOSL expression on mouse GC B cells that were treated with actinomycin D (ActD, 5 μg/ml), anti-CD40 (10 μg/ml) for 4h, with medium control set as unit 1. Bars represent median and each dot represent a single mouse (n=5). d-f, ns, not significant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001; two tailed student t-test. g, Representative histogram of surface ICOSL expression on human GC B cells that were stimulated with DA (10μM) or anti-CD40 (1 μg/ml) for 30 min. h, Fold changes of surface ICOSL expression on human GC B cells stimulated with several concentrations of anti-CD40 for 4 and 8 hours, with medium control set as unit 1 (n=3). i, Bar plot showing survival of GC B in the presence of anti-CD40 (1 μg/ml) after 4 or 8 hours of stimulation (n=4). *p ≤ 0.05 and ***p ≤ 0.001; nonparametric Mann-Whitney test (U test).
Extended Data Fig. 8. Effect of ICOSL on CD40L presentation and reception in SLB model for TFH cell- GC B cell interaction.
a, Activated human T cells that express ICOS and CD40L were incubated with SLB containing ICAM-1 and UCHT1 (anti-CD3) as a basal condition with a ring of ICAM-1 surrounding a central cluster enriched in T cell receptor enriched extracellular vesicles by 15 minutes26. This condition resulted in low presentation of CD40L in punctate structures detected by anti-CD40L mAb that accumulated in the same central synapse with the TCR enriched extracellular vesicles. Addition of ICOSL the SLB resulted in strong central accumulation of fluorescent ICOSL with the TCR enriched extracellular vesicles, but no increase in CD40L presentation. Addition of CD40 the SLB resulted in a significant increase in CD40L accumulation, which we refer to as reception because its receptor dependent. When ICOSL and CD40 were added the reception of CD40L was further significantly enhanced over the level observed with CD40 alone. Thus, ICOSL ligation in the centre of the immunological synapse increases CD40L reception. All levels are shown in gray scale except CD40L panels, for which the pseudocolor scale is indicated. Scale bar 5 µm. b, Human TFH cells were incubated with SLB containing ICAM-1 and UCHT1 (anti-CD3). Addition of ICOSL resulted in increased accumulation of CgB at the synapse centre. Addition of CD40 did not further increased CgB accumulation.
Extended Data Fig. 9. Effect of ICOSL upregulation speed in the published and extended GC LEDA model and in the classical recycling model.
a, Characteristics of GC reactions in simulations with short (black) and long (colours) search phase for TFH help using the previously published LEDA model (see text). All tested variants (see legend box and text for details on the quantities) exhibit reduced and retarded output production while keeping affinity maturation unchanged. Mean (full lines) and standard deviation (shades) of 100 simulations. b, The LEDA model in Extended Data Fig. 9a was extended to allow for multiple short contacts between B and T cells and to explicitly represent ICOSL dynamics in B cells (see text for details). Characteristics of GC reactions in simulations with fast (black, grey) and slow (colours) ICOSL upregulation. All tested variants (see legend box and text for details on the quantities) exhibit reduced and retarded output production while keeping GC B cell affinity unchanged. Output affinity is enhanced in a subset of settings. Mean (full lines) and standard deviation (shades) of 100 simulations. c, The simulations in Extended Data Fig. 9b were repeated using the classical textbook recycling model with 80% of the selected B cells doing recycling and 20% of the selected B cells differentiating to output cells42. This replaced the LEDA model in Extended Data Fig. 9b. The simulations with short search periods for TFH help were repeated. Note that the overall output production is smaller in the classical recycling model43. The relative reduction of output in simulations with slow ICOSL upregulation is unchanged. Mean (full lines) and standard deviation (shades) of 100 simulations.
Extended Data Fig. 10. Dopamine derivative structure.
Diagram showing chemical structure of dopamine derivative after sample reconstitution with trifluoroacetic anhydride (TFAA) and trifluoroethanol (TFE).
Supplementary Material
Acknowledgments
We thank J. Meldolesi for electron microscopy analysis interpretation and to P. Podini for her technical assistance; M. Cook and E. Bartlett for critical reading of the manuscript; R. Cairella for his contribution to preparing histological samples; A. Wilson, A.-M. Hatch, A. Lopez, and E. Barry for assistance with obtaining tonsil samples; and D. Yu for valuable suggestions. We thank the Imaging and Cytometry Facility and the Biomolecular Research Facility at the John Curtin School of Medical Research for technical support. We acknowledge the contribution to this study made by the Oxford Centre for Histopathology Research and the Oxford Radcliffe Biobank, which are supported by the NIHR Oxford Biomedical Research Centre. C.G.V. is supported by fellowship, project and program grants from the Australian National Health and Medical Research Council. The Wellcome Trust supports M.L.D. and S.V. and European Research Council grant AdG670930 supports M.L.D. and D.S. Human Frontier Science Program (RGP0033/2015) supports M.M.H., M.L.D., C.G.V.
Footnotes
Data availability
The Source Data for the graphs in Figures and Extended Data Figures are provided in the online version of the paper and the source data for gels are provided in Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request.
Author contributions
C.D. and C.G.V. contributed equally to this work. I.P. performed most of the experiments and analysed the data. P.C., P.G. and H.V. helped with the experiments. M.P. contributed to data analysis. D.S. and S.V. performed SLB experiment and contributed to interpretation together with M.L.D. S.B. performed GC/MS/MS experiments. M.M-H. performed in silico modelling. H.M. performed 2-photon experiments and contributed to data analysis together with I.C.. M.G., M.L.D., M.M-H., M.P. and R.A.S. provided intellectual input, expertise and critical reading of the manuscript. I.P. and C.G.V. wrote the manuscript. C.G.V. supervised the project with D.C..
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Dustin ML, Colman DR. Neural and immunological synaptic relations. Science. 2002;298:785–789. doi: 10.1126/science.1076386. [DOI] [PubMed] [Google Scholar]
- 2.Kohm AP, Sanders VM. Norepinephrine: a messenger from the brain to the immune system. Immunology today. 2000;21:539–542. doi: 10.1016/s0167-5699(00)01747-3. [DOI] [PubMed] [Google Scholar]
- 3.McKenna F, et al. Dopamine receptor expression on human T- and B-lymphocytes, monocytes, neutrophils, eosinophils and NK cells: a flow cytometric study. Journal of neuroimmunology. 2002;132:34–40. doi: 10.1016/s0165-5728(02)00280-1. [DOI] [PubMed] [Google Scholar]
- 4.Musso NR, Brenci S, Setti M, Indiveri F, Lotti G. Catecholamine content and in vitro catecholamine synthesis in peripheral human lymphocytes. The Journal of clinical endocrinology and metabolism. 1996;81:3553–3557. doi: 10.1210/jcem.81.10.8855800. [DOI] [PubMed] [Google Scholar]
- 5.Bergquist J, Josefsson E, Tarkowski A, Ekman R, Ewing A. Measurements of catecholamine-mediated apoptosis of immunocompetent cells by capillary electrophoresis. Electrophoresis. 1997;18:1760–1766. doi: 10.1002/elps.1150181009. [DOI] [PubMed] [Google Scholar]
- 6.Bergquist J, Silberring J. Identification of catecholamines in the immune system by electrospray ionization mass spectrometry. Rapid communications in mass spectrometry : RCM. 1998;12:683–688. doi: 10.1002/(SICI)1097-0231(19980615)12:11<683::AID-RCM218>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Musso NR, Brenci S, Indiveri F, Lotti G. Acetylcholine-induced, calcium-dependent norepinephrine outflow from peripheral human lymphocytes. Journal of neuroimmunology. 1998;87:82–87. doi: 10.1016/s0165-5728(98)00057-5. [DOI] [PubMed] [Google Scholar]
- 8.Marino F, et al. Endogenous catecholamine synthesis, metabolism storage, and uptake in human peripheral blood mononuclear cells. Experimental hematology. 1999;27:489–495. doi: 10.1016/s0301-472x(98)00057-5. [DOI] [PubMed] [Google Scholar]
- 9.Cosentino M, et al. Human CD4+CD25+ regulatory T cells selectively express tyrosine hydroxylase and contain endogenous catecholamines subserving an autocrine/paracrine inhibitory functional loop. Blood. 2007;109:632–642. doi: 10.1182/blood-2006-01-028423. [DOI] [PubMed] [Google Scholar]
- 10.Nakano K, et al. Dopamine released by dendritic cells polarizes Th2 differentiation. International immunology. 2009;21:645–654. doi: 10.1093/intimm/dxp033. [DOI] [PubMed] [Google Scholar]
- 11.Shulman Z, et al. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular Helper T Cells. Annual review of immunology. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 14.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. Journal of immunology. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 15.Malosio ML, Giordano T, Laslop A, Meldolesi J. Dense-core granules: a specific hallmark of the neuronal/neurosecretory cell phenotype. Journal of cell science. 2004;117:743–749. doi: 10.1242/jcs.00934. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenbrunner U, et al. Sympathetic axons and nerve terminals: the protein composition of small and large dense-core and of a third type of vesicles. Neuroscience. 1990;37:819–827. doi: 10.1016/0306-4522(90)90111-g. [DOI] [PubMed] [Google Scholar]
- 17.Brauninger A, et al. Molecular analysis of single B cells from T-cell-rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood. 1999;93:2679–2687. [PubMed] [Google Scholar]
- 18.Braeuninger A, et al. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9337–9342. doi: 10.1073/pnas.94.17.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daubner SC, Le T, Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Archives of biochemistry and biophysics. 2011;508:1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meredith EJ, et al. Dopamine targets cycling B cells independent of receptors/transporter for oxidative attack: Implications for non-Hodgkin's lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13485–13490. doi: 10.1073/pnas.0605993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niemegeers CJ, Laduron PM. Pharmacology and biochemistry of haloperidol. Proceedings of the Royal Society of Medicine. 1976;69(suppl 1):3–8. doi: 10.1177/00359157760690S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casamayor-Palleja M, Khan M, MacLennan IC. A subset of CD4+ memory T cells contains preformed CD40 ligand that is rapidly but transiently expressed on their surface after activation through the T cell receptor complex. The Journal of experimental medicine. 1995;181:1293–1301. doi: 10.1084/jem.181.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, et al. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517:214–218. doi: 10.1038/nature13803. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe M, et al. Down-regulation of ICOS ligand by interaction with ICOS functions as a regulatory mechanism for immune responses. Journal of immunology. 2008;180:5222–5234. doi: 10.4049/jimmunol.180.8.5222. [DOI] [PubMed] [Google Scholar]
- 25.Liu YJ, et al. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342:929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- 26.Choudhuri K, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer-Hermann M. Overcoming the dichotomy of quantity and quality in antibody responses. Journal of immunology. 2014;193:5414–5419. doi: 10.4049/jimmunol.1401828. [DOI] [PubMed] [Google Scholar]
- 28.Meyer-Hermann M, et al. A theory of germinal center B cell selection, division, and exit. Cell reports. 2012;2:162–174. doi: 10.1016/j.celrep.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Tas JM, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotta R, Anderson RB, Kobayashi K, Newgreen DF, Young HM. Effects of tissue age, presence of neurones and endothelin-3 on the ability of enteric neurone precursors to colonize recipient gut: implications for cell-based therapies. Neurogastroenterol Motil. 2010;22:331–e386. doi: 10.1111/j.1365-2982.2009.01411.x. [DOI] [PubMed] [Google Scholar]
- 31.Luthje K, et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nature immunology. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- 32.Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2009;57:899–905. doi: 10.1369/jhc.2009.953612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa A, et al. The endoplasmic-sarcoplasmic reticulum of smooth muscle: immunocytochemistry of vas deferens fibers reveals specialized subcompartments differently equipped for the control of Ca2+ homeostasis. The Journal of cell biology. 1993;121:1041–1051. doi: 10.1083/jcb.121.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smythe GA, Edwards G, Graham P, Lazarus L. Biochemical diagnosis of pheochromocytoma by simultaneous measurement of urinary excretion of epinephrine and norepinephrine. Clinical chemistry. 1992;38:486–492. [PubMed] [Google Scholar]
- 35.Bergquist J, Tarkowski A, Ewing A, Ekman R. Catecholaminergic suppression of immunocompetent cells. Immunology today. 1998;19:562–567. doi: 10.1016/s0167-5699(98)01367-x. [DOI] [PubMed] [Google Scholar]
- 36.Slater EP, et al. Analysis by cDNA microarrays of gene expression patterns of human adrenocortical tumors. European journal of endocrinology / European Federation of Endocrine Societies. 2006;154:587–598. doi: 10.1530/eje.1.02116. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dustin ML, Starr T, Varma R, Thomas VK. Supported planar bilayers for study of the immunological synapse. Coligan John E, et al., editors. Current protocols in immunology. 2007;Chapter 18 doi: 10.1002/0471142735.im1813s76. Unit 18 13. [DOI] [PubMed] [Google Scholar]
- 40.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 41.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer-Hermann M, Deutsch A, Or-Guil M. Recycling probability and dynamical properties of germinal center reactions. J Theor Biol. 2001;210:265–285. doi: 10.1006/jtbi.2001.2297. [DOI] [PubMed] [Google Scholar]
- 43.Dustin ML, Meyer-Hermann M. Immunology. Antigen feast or famine. Science. 2012;335:408–409. doi: 10.1126/science.1218165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.