Abstract
The ability of cobalamin to coordinate different upper axial ligands gives rise to a diversity of reactivity. Traditionally, adenosylcobalamin is associated with radical-based rearrangements, and methylcobalamin with methyl cation transfers. Recently, however, a new role for adenosylcobalamin has been discovered as a light sensor, and a methylcobalamin-dependent enzyme has been identified that is suggested to transfer a methyl anion. Additionally, recent studies have provided a wealth of new information about a third class of cobalamin-dependent enzymes that do not appear to use an upper ligand. They function in reductive dehalogenations and epoxide reduction reactions. Finally, mechanistic details are beginning to emerge about the cobalamin-dependent S-adenosylmethionine radical enzyme superfamily for which the role of cobalamin has been largely enigmatic.
Graphical abstract
Introduction
Cobalamin (Cbl), often referred to as Nature’s most beautiful cofactor, is a complex organometallic compound that has a central cobalt ion (Figure 1). Organisms that require Cbl synthesize it using one of the largest characterized biosynthetic pathways in Nature (approximately 30 enzymatic steps) [1], uptake and salvage it from the environment using specialized machinery [2], or acquire Cbl from symbiotic relationships [3]. The expensive nature and reactivity of Cbl dictates the need for dedicated chaperones to assist in its delivery to target enzymes [4] and recent structural studies have provided molecular insight into how this process occurs [5]. Different flavors of Cbl arise from a diversity of ligands that can occupy the upper axial face with adenosyl- and methyl-substituents being the most common and best studied.
Figure 1.
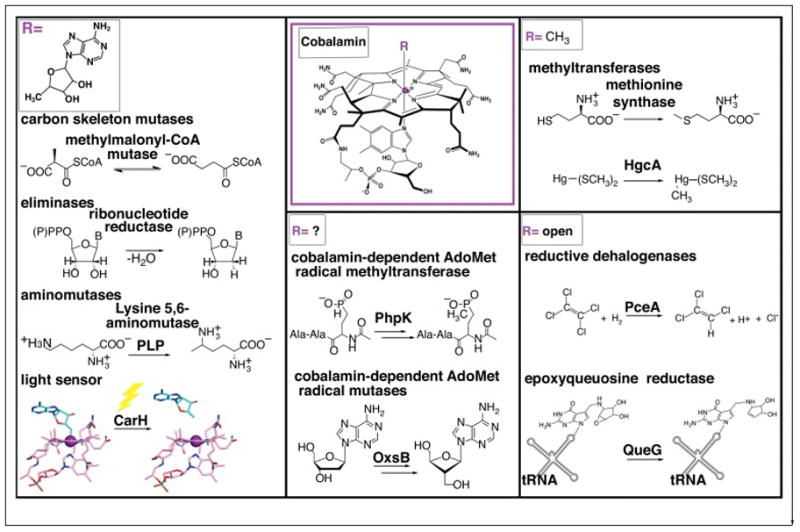
Classes of Cbl-dependent proteins as defined by the nature of the upper ligand. AdoCbl-dependent enzymes include carbon skeleton mutases, eliminases, and aminomutases. The latter of which also requires pyridoxyl phosphate (PLP). Recently, AdoCbl has been shown to be involved in light-dependent gene regulation of the carotenoid (Car) biosynthetic operon via the transcriptional regulator CarH, adding a new function for this flavor of Cbl cofactor. MeCbl-dependent methyl transferases include methionine synthase, which catalyzes the transfer of a methyl cation to homocysteine and the mercury methylase HgcA, which is thought to catalyze transfer of a methyl anion to a Hg2+-bis(thiolate) compound (represented as Hg-S(CH3)2), the most prevalent form of Hg2+ in methylmercury producers [17]. A third class of Cbl-dependent enzymes, which includes the reductive dehalogenase PceA and the queuosine biosynthetic enzyme QueG do not have an upper axial ligand and are denoted as “open”-Cbl-dependent enzymes. The final class is made up of enzymes that appear to require the machinery for both S-adenosylmethionine (AdoMet) radical chemistry and a Cbl cofactor. Examples include phosphinate methylase PhpK and oxetanocin-A biosynthetic enzyme OxsB. The role(s) of Cbl in this enzyme family is currently under investigation.
Adenosyl-Cbl (AdoCbl) is known for its use in radical-based enzymatic catalysis; reversible homolytic cleavage of the labile Co(III)-C bond results in the transient formation of a highly reactive 5′-deoxyadenosyl radical species (5′-dAdo•) that is capable of initiating a variety of rearrangement reactions (Figure 1). Methyl-Cbl (MeCbl)-dependent enzymes, on the other hand, typically cycle between the Co(III) and Co(I) states and transfer a methyl cation to nucleophilic substrates using heterolytic Co-C bond cleavage (Figure 1). Examples of methyltransferases include methionine synthase (MetH) and corrinoid iron-sulfur protein (CFeSP).
Proteins can bind Cbl both with the cofactor’s dimethylbenzimidazole (DMB) base serving as the lower ligand (“base-on”) and with the DMB displaced (“base-off”). In the latter case, a His residue contributed by an Asp-X-His-X-X-Gly motif typically serves as the lower axial ligand to Co (“base-off”,“His-on”) [6], although some enzymes bind Cbl without a lower ligand (“base-off”, “His-off”) [7,8]. However, the traditional notions of how proteins bind and utilize Cbl have been challenged by recent discoveries. Here we review the recent surprises that have placed Cbl in the spotlight again.
A newly discovered function for AdoCbl as a light sensor
An unexpected light-sensing function of AdoCbl was revealed through studies of the transcriptional regulator CarH, which exploits the light-sensitive Co-C bond of AdoCbl to sense environmental light conditions and up-regulate the transcription of systems that mitigate light-induced damage (i.e. reactive oxygen species and/or DNA damage). In the dark, AdoCbl-bound CarH forms a tetramer, which binds DNA and represses transcription of carotenoid biosynthetic genes in bacteria such as Myxococcus xanthus (Figure 2) [9,10]. In response to sunlight, the Co-C bond of AdoCbl cleaves, causing the CarH tetramer to fall apart and dissociate from DNA, initiating transcriptional activation of the genes responsible for production of light-protective carotenoids (Figure 2) [9,10]. In this manner, bacteria only make carotenoids when light conditions dictate.
Figure 2.
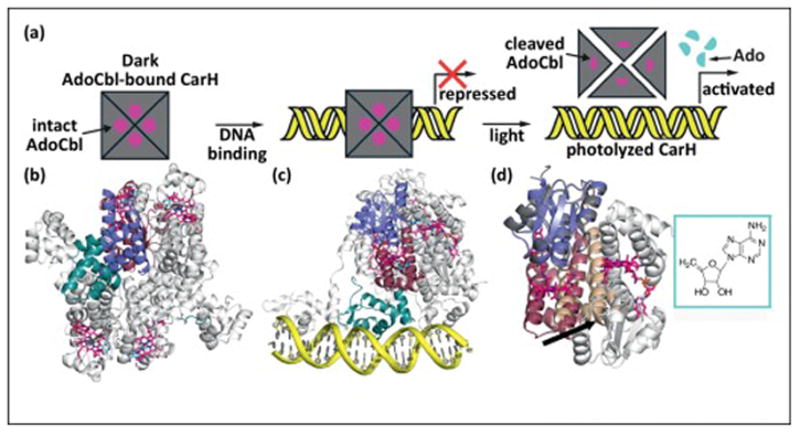
AdoCbl functions as a light sensor in the light-dependent regulation of carotenoid biosynthesis. (a) Cartoon showing steps involved in the light-dependent regulation of the carotenoid operon by CarH. (b) Structure of the AdoCbl-bound CarH tetramer that is formed in the dark [9]. Each CarH monomer is composed of a DNA-binding domain (teal), a helical bundle (raspberry) and a Cbl-binding domain (purple) [9]. Monomers form head-to-tail dimers, and two dimers form the tetramer (other protomers in gray). AdoCbl is displayed as sticks with cyan and pink carbons for the Ado group and Cbl, respectively. (c) Structure of the CarH tetramer (colored as in panel a) bound to the carotenoid biosynthetic operon (yellow) in the dark, blocking transcription. (d) Comparison of light and dark state CarH structures. Upon exposure to sunlight, the light-sensitive Co-C bond of AdoCbl cleaves, causing a shift in the helix-bundle domain (raspberry to wheat domain movement), breaking apart the CarH dimer interface (promoter at interface in gray) and dissembling the tetramer and activating transcription [9]. Light dependent cleavage of AdoCbl results in formation of 4′,5′-anhydroadenosine [11] (cyan, boxed).
AdoCbl photochemistry appears to have adapted to safeguard the use of this cofactor as a light sensor [11,12]. Instead of producing 5′-dAdo• upon light exposure, the nonreactive molecule 4′,5′-anhydroadenosine is formed (Figure 2d) [11]. 4′,5′-anhydroadenosine differs from 5′-dAdo• by one electron and one proton; its mechanism of formation awaits discovery [13].
New Cbl-binding motifs discovered in mercury-methylating enzymes
The bacterial two-gene cluster responsible for production of methylmercury from inorganic mercury has been identified [14]. These genes (hgcAB) encode a putative corrinoid protein, HgcA, and a 2-[4Fe-4S] cluster ferredoxin, HgcB [14]. The N-terminal domain of HgcA has sequence homology to CFeSP, which binds a “base-off, His-off” Cbl and is also involved in the methylation of a metal. For CFeSP, the metal is nickel and the nickel is part of the A-cluster of acetyl-CoA synthase; methylation of nickel is transient, en route to acetyl-CoA production [15].
Homology modeling of HgcA using the CFeSP structure suggests that HgcA will also bind a “base-off” Cbl, but may utilize a unique axial cysteine ligand from a conserved Asn-(Val/Ile)-Trp-Cys93-Ala-(Ala/Gly)-Gly-Lys motif [14]. Consistent with this proposal, mutation of Cys93 shifts the Aqua-Cbl-bound HgcA UV-Vis spectra [14] and leads to inactive enzyme in an in vivo assay [16].
Cys ligation is unprecedented in Cbl enzymes and would be expected to change cofactor reactivity. Based on density functional theory calculations, Co-Cys ligation is proposed to facilitate transfer of a methyl group from MeCbl to Hg2+ through either homolytic (transfer of a methyl radical species) or heterolytic (transfer of a methyl anion) Co-C bond cleavage, with the latter being favored [17]. This anionic methyl transfer to an electrophilic substrate is unparalleled as Cbl-dependent methyl-carriers typically transfer a methyl group as a methyl cation to a nucleophilic substrate. However, this proposal is reminiscent of a theory put forward in the 1960s that alkyl-Cbls might function as Grignard reagents for biosynthetic reactions [18]. Following an anionic methyl transfer, Co, which would remain in the Co(III) state needs to be re-methylated for the next catalytic cycle. It is currently hypothesized that HgcB serves as a reductant in generating the Co(I) state, which could be remethylated using some unidentified methyl donor [14].
Exciting Cbl chemistry without an upper ligand
Recent advances [19–23] confirm the existence of a third class of Cbl-dependent enzymes that use a form of Cbl that lacks an upper ligand. In fact, the open-faced nature of Cbl may be key to the chemistry performed (see below). Reductive dehalogenases were previously proposed to form this third class [24,25], but they are now joined by epoxyqueuosine (oQ) reductases (QueGs) [20,22,26,27], and CblCs, the enzymes responsible for decyanation/dealkylation of Cbl [21]. In addition to lacking an upper ligand, Cbl bound to these enzymes also lacks a lower ligand. To prevent water coordination at the lower axial position, access to Co is blocked by either the Cbl nucleotide tail or by a non-coordinating side-chain (Figure 3a, d) [19–23]. In addition to Cbl, reductive dehalogenases and QueG enzymes bind two [4Fe-4S] clusters (Figure 3c).
Figure 3.
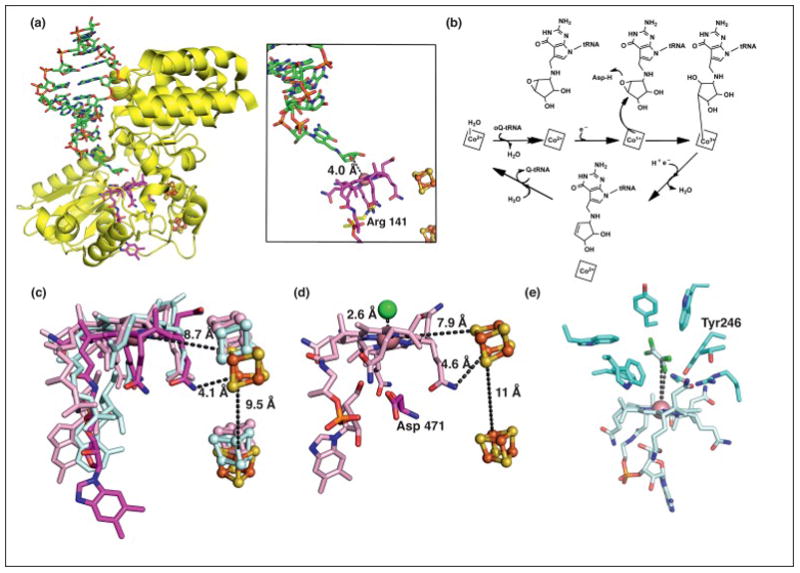
Structure and mechanism of a third class of Cbl-dependent enzymes that does not use Ado- or Me-Cbl. (a) Structure of QueG (yellow) with an oQ-modified tRNATyr anticodon stem loop bound (displayed as sticks with green carbons, left panel) [20]. The modified RNA base is inserted into a cavity in the protein, binding directly above Co of Cbl (displayed as sticks with dark pink carbons, right panel) [20]. Arg 141 (yellow carbons) of QueG blocks access to the lower axial face of Cbl. (b) The proposed mechanism of QueG. Nucleophilic attack of Co(I)-Cbl species on the oQ substrate forms an organocobalt adduct that can be converted to product and cob(II)alamin through reduction and protonation [20]. (c) An overlay of Cbls and two [4Fe-4S] clusters (orange and yellow spheres) from QueG (carbons in dark pink) and the reductive dehalogenase enzymes PceA (cyan) and NpRdhA (light pink). PceA harbors a norpseudo-B12 cofactor, which is characterized by a lower axial adenine base and a hydrogen atom at position 176 rather than a DMB base and methyl group, respectively, as found in Cbl. (d) The crystal structure of NpRdhA reveals a chloride binding site directly above Cbl [23]. Asp 471 blocks the lower face of Cbl. (e) The crystal structure of the TCE-bound PceA reveals that the distance between the substrate C and Cl atoms to Co measure 5.8- and 4.7-Å, respectively [19]. These distances do not support a direct Co-substrate interaction without invoking a conformational change. TCE is tightly packed in the active site by several bulky residues, which likely gate formation of a covalent adduct and contribute to substrate selectivity [19]. TCE is shown in two overlaid orientations as captured in the structure.
All enzymes in the class perform fascinating chemistry. QueG catalyzes reduction of epoxyqueuosine to form the RNA nucleoside queuosine (Q) that is present in the wobble position of certain tRNAs [26]. A recent structure of QueG from Bacillus subtilis co-crystallized with an oQ-modified tRNATyr anticodon stem loop reveals that Q binds directly above Cbl (Figure 3a) [20]. These structural data, along with electrochemical data that show the [4Fe-4S] clusters have redox potentials sufficient to generate the Co(I)-Cbl state, are consistent with a mechanism in which QueG catalyzes the conversion of oQ to Q through nucleophilic attack of Co(I)-Cbl on the substrate epoxide (Figure 3b–c) [20].
Reductive dehalogenases allow anaerobic microorganisms to use organohalide compounds as terminal electron acceptors during respiration [28]. The reductive dehalogenase from Nitratireductor pacificus (NpRdhA) catalyzes the reduction of ortho-halogenated phenolic compounds [23], whereas a reductive dehalogenase from Sulfurospirillum multivorans (PceA) catalyzes the reduction of tetrachloroethene (PCE) and trichloroethene (TCE) [19], both carcinogenic industrial solvents.
An oxidative addition mechanism has been proposed for NpRdhA involving formation of a Co-halide bond; electron density has been observed consistent with a chloride ion bound 2.6-Å above the Co (Figure 3d) and electron paramagnetic resonance spectroscopy has identified a direct halogen-Co interaction [23]. A close arrangement of substrate and Cbl would be expected with oxidative addition, and in the absence of a substrate-bound structure, modeling and molecular dynamic simulations have come to different conclusions about substrate placement [23,29].
A structure with substrate TCE is available for PceA, and the halide-Co distance appears too long at 4.7-Å for an oxidative addition mechanism (Figure 3d,e) [19]. Instead, it has been suggested that PceA requires Cbl for a new type of radical-based mechanism; dissociative one-electron transfer from the Co(I)-Cbl to substrate, forming a Co(II)-Cbl species [19,25,30–32]. Subsequent elimination of a chloride anion would generate a substrate-based radical species that could recombine with Co(II)-Cbl or be reduced and protonated [19,25,30–32]. Although steric constraints of the active site appear to disfavor the former, the latter could be achieved through electron donation by the either Cbl or the proximal [4Fe-4S] cluster and protonation via the conserved Tyr246 [19]. A possible binding pocket for the released halide ion was observed ~2.9-Å above the Co from a soaking experiment with ammonium iodide [19].
Whether more data will yield a unified mechanism for reductive dehalogenases is not clear. It may be that the nature of the substrate, halogenated aromatic (as in NpRdhA) versus halogenated alkenyl compounds (as in PCE) affords different mechanistic possibilities. Regardless, all current mechanistic proposals invoke unprecedented chemistry for Cbl in an enzymatic system.
An emerging superfamily of Cbl-dependent S-adenosylmethionine radical enzymes
Previous sequence analyses of S-adenosylmethionine (AdoMet) radical enzymes revealed that in addition to having the AdoMet radical [4Fe-4S] cluster-binding motif, some family members additionally had an N-terminal domain with sequence similarity to Cbl-binding proteins [33]. Specifically, these enzymes were shown to contain the “base-off” consensus motif, but lack the characteristic “His-on” motif [33]. Notably, similar to AdoCbl, a [4Fe-4S] cluster-ligated molecule of AdoMet can be used to catalyze radical chemistry through formation of 5′-dAdo• [34,35]. Likewise, similar to MeCbl, AdoMet can also be used as a biological methyl donor. Today this class of enzymes, which pair what appear to be redundant cofactors, has approximately 7000 annotated members implicated in the biosynthetic pathways of compounds including bialaphos, bacteriochlorophyll, fosfomycin, oxetanocin-A, and many others. Indeed, as predicted based on sequence, several of these enzymes have been shown to require Cbl for chemistry [36–43] and one member, TsrM, has been shown spectroscopically to bind Cbl in a “base-off”, “His-off” conformation [44]. Despite the large family size, only a small number of these enzymes have been isolated and characterized; in addition to the arduous oxygen and light sensitive cofactors, several have been reported to be insoluble [36,39–41].
Many of the Cbl-dependent AdoMet radical enzymes catalyze methylation reactions at challenging (i.e. non-nucleophilic) substrate centers [36–39,45–49], whereas other members appear to catalyze radical rearrangements [43,50]. Thus far, five of the functionally characterized Cbl-dependent AdoMet radical enzymes catalyze methylation reactions at unactivated carbon and phosphorus centers. Of particular interest is the reaction catalyzed by PhpK, which is responsible for the formation of a C-P bond linkage in a bialaphos precursor. This challenging methylation as well as other methylation reactions catalyzed by members of this enzyme class have been hypothesized to require both Cbl and AdoMet as shown in Figure 4a [36,37,39–41,48,49,51,52]. Unlike the traditional Cbl-dependent methyltransferase reactions, these enzymes are hypothesized to catalyze methylation of a substrate following homolytic Co-C bond cleavage of MeCbl; precedent for transfer of a methyl radical species comes from Cbl model chemistry [53].
Figure 4.
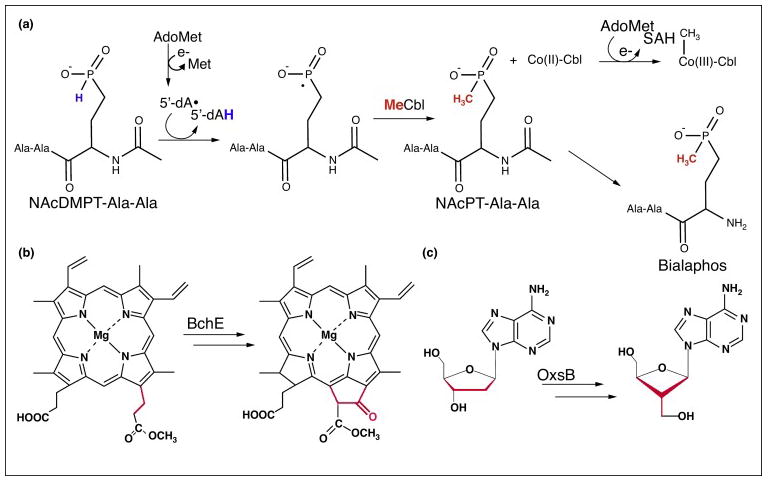
A sample of the reactions catalyzed by Cbl-dependent AdoMet radical enzymes. (a) The proposed mechanism for the Cbl-dependent AdoMet radical enzyme PhpK, which functions in the biosynthetic pathway of the commercially available herbicide bialaphos [51]. In the first step, AdoMet is reductively cleaved to generate 5′-dAdo•, which abstracts a hydrogen atom from the N-acetyldemethylphosphinothricin (NAcDMPT)-Ala-Ala substrate. The resulting radical intermediate reacts with MeCbl to form the methylated product N-acetylphosphinothricin (NAcPT)-Ala-Ala. Notably, this reaction formally involves the transfer of a methyl radical from MeCbl and regeneration of the active enzyme is thought to involve reductive methylation of Co(I)-Cbl using a second molecule of AdoMet. (b) BchE is a radical cyclase that catalyzes the conversion of magnesium protoporphyrin IX monomethyl ester to divinyl-protochlorophyllide, forming the fifth ring of bacteriochlorophyll. (c) OxsB is hypothesized to catalyze a radical mediated ring contraction of adenosine or an adenosine derivative (shown here as 2′-deoxyadenosine) in the biosynthesis of oxetanocin-A [50]. Similar to the reaction catalyzed by BchE, this reaction does not appear to require a methylation step.
The requirements of both Cbl and AdoMet by BchE and OxsB, which don’t appear to catalyze methylation reactions, remain enigmatic (Figure 4b–c). BchE and OxsB are part of the biosynthetic pathways for bacteriochlorophyll and antiviral agent oxetanocin-A [54], respectively. For these enzymes, even fewer mechanistic details are available; in vivo BchE assays showed accumulation of the BchE substrate, magnesium protoporphyrin IX monomethyl ester, under Cbl-depleted conditions establishing the Cbl-requirement of the enzyme [43,55]. There are currently two proposed mechanisms for how BchE could employ AdoMet radical chemistry and some form of Cbl to generate the fifth ring of bacteriochlorophyll; one mechanism requires the [4Fe-4S] cluster, AdoMet, and presumably three molecules of AdoCbl to catalyze cyclization [43]. However, the expensive AdoCbl requirement and the inefficient use of the AdoMet radical machinery has led to a very different proposal that instead requires the [4Fe-4S] cluster, three molecules of AdoMet, and hydroxo-cobalamin for cyclization [56]. In the case of OxsB, the substrate is unknown, but it has been suggested that OxsB could catalyze a radical-mediated ring contraction of adenosine or an adenosine derivative to produce oxetanocin-A [50].
Whereas the enzymes of this class are clearly evolutionarily related, it is still unclear whether all of the members will employ radical chemistry and whether each member will use Cbl and AdoMet similarly. As the “His-on” motif is absent for Cbl-binding, it is not known how these enzymes will bind Cbl (and whether all members will use the same form). Furthermore, it remains to be shown if the enzymes that don’t appear to catalyze methylation reactions will require both cofactors for activity. As there are currently no structural data for this class of enzymes, it remains unknown whether AdoMet, the [4Fe-4S] cluster, and Cbl will be located within a single active-site, a requirement of the mechanistic proposal in Figure 4a. Although more research is needed, the reactions catalyzed by these enzymes suggest that novel Cbl chemistry is likely to be involved.
Final Thoughts
Although Cbl is a well-studied cofactor, recent studies show that we still have much to learn about the reactivity and functions of Cbl-containing proteins. All the proteins highlighted in this review are likely to employ unprecedented reaction mechanisms for Cbl-dependent enzymes, and only time will tell what type of chemistry the Cbl-dependent AdoMet radical enzymes will utilize. It is likely that Cbl will remain in the spotlight for many years to come.
Acknowledgments
We thank Tsehai Grell for helpful discussions and Marco Jost for assistance in preparation of Figure 2. This work was supported by National Institute of Health Grants R01-GM69857 (C.L.D.) and F32-GM108189 (J.B.-R.). C.L.D. is a Howard Hughes Medical Institute Investigator.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Scott AI, Roessner CA. Biosynthesis of cobalamin (vitamin B(12)) Biochem Soc Trans. 2002;30:613–620. doi: 10.1042/bst0300613. [DOI] [PubMed] [Google Scholar]
- 2.Bassford PJ, Jr, Kadner RJ. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J Bacteriol. 1977;132:796–805. doi: 10.1128/jb.132.3.796-805.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 4.Padovani D, Labunska T, Banerjee R. Energetics of interaction between the G-protein chaperone, MeaB, and B12-dependent methylmalonyl-CoA mutase. J Biol Chem. 2006;281:17838–17844. doi: 10.1074/jbc.M600047200. [DOI] [PubMed] [Google Scholar]
- •5.Jost M, Cracan V, Hubbard PA, Banerjee R, Drennan CL. Visualization of a radical B12 enzyme with its G-protein chaperone. Proc Natl Acad Sci U S A. 2015;112:2419–2424. doi: 10.1073/pnas.1419582112. This work presents structural characterization of a fusion between an AdoCbl-dependent isobutyryl-CoA mutase and its G-protein chaperone. The structures presented here provide the first insight into the necessary conformational changes for loading a precious molecule of AdoCbl in an enzyme active site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 7.Svetlitchnaia T, Svetlitchnyi V, Meyer O, Dobbek H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc Natl Acad Sci U S A. 2006;103:14331–14336. doi: 10.1073/pnas.0601420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kung Y, Ando N, Doukov TI, Blasiak LC, Bender G, Seravalli J, Ragsdale SW, Drennan CL. Visualizing molecular juggling within a B12-dependent methyltransferase complex. Nature. 2012;484:265–269. doi: 10.1038/nature10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••9.Jost M, Fernandez-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY, Kang G, Padmanabhan S, Elias-Arnanz M, Drennan CL. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature. 2015;526:536–541. doi: 10.1038/nature14950. This work provides the molecular basis for repurposing AdoCbl as a light sensor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •10.Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. Initial in vitro and in vivo characterization of CarH; this paper laid the foundation for all subsequent CarH papers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jost M, Simpson JH, Drennan CL. The Transcription Factor CarH Safeguards Use of Adenosylcobalamin as a Light Sensor by Altering the Photolysis Products. Biochemistry. 2015;54:3231–3234. doi: 10.1021/acs.biochem.5b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutta RJ, Hardman SJ, Johannissen LO, Bellina B, Messiha HL, Ortiz-Guerrero JM, Elias-Arnanz M, Padmanabhan S, Barran P, Scrutton NS, et al. The photochemical mechanism of a B12-dependent photoreceptor protein. Nat Commun. 2015;6:7907. doi: 10.1038/ncomms8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chemaly SM. New light on vitamin B-12: The adenosylcobalamin-dependent photoreceptor protein CarH. S Afr J Sci. 2016;112:39–47. [Google Scholar]
- ••14.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, et al. The genetic basis for bacterial mercury methylation. Science. 2013;339:1332–1335. doi: 10.1126/science.1230667. The authors of this paper identify the two-gene cluster responsible for bacterial mercury methylation by D. desulfuricans ND132 and Geobacter sulfurreducens, resolving decades of investigation on how microbes produce the potent neurotoxin methylmercury from inorganic mercury. [DOI] [PubMed] [Google Scholar]
- 15.Barondeau DP, Lindahl PA. Methylation of carbon monoxide dehydrogenase from Clostridium thermoaceticum and mechanism of acetyl coenzyme A synthesis. J Am Chem Soc. 1997;119:3959–3970. [Google Scholar]
- 16.Smith SD, Bridou R, Johs A, Parks JM, Elias DA, Hurt RA, Jr, Brown SD, Podar M, Wall JD. Site-directed mutagenesis of HgcA and HgcB reveals amino acid residues important for mercury methylation. Appl Environ Microbiol. 2015;81:3205–3217. doi: 10.1128/AEM.00217-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Riccardi D, Beste A, Smith JC, Parks JM. Mercury methylation by HgcA: theory supports carbanion transfer to Hg(II) Inorg Chem. 2014;53:772–777. doi: 10.1021/ic401992y. [DOI] [PubMed] [Google Scholar]
- 18.Ingraham LL. B12 Coenzymes: Biological Grignard Reagents. Ann N Y Acad Sci. 1964;112:713–720. doi: 10.1111/j.1749-6632.1964.tb45049.x. [DOI] [PubMed] [Google Scholar]
- ••19.Bommer M, Kunze C, Fesseler J, Schubert T, Diekert G, Dobbek H. Structural basis for organohalide respiration. Science. 2014;346:455–458. doi: 10.1126/science.1258118. A series of crystal structures provides insight into how the enzyme PceA catalyzes the reductive dechlorination of toxic organohalide compounds. The reductive dehalogenase described here is hypothesized to use an open faced-Cbl in a novel radical-based mechanism. [DOI] [PubMed] [Google Scholar]
- ••20.Dowling DP, Miles ZD, Kohrer C, Maiocco SJ, Elliott SJ, Bandarian V, Drennan CL. Molecular basis of cobalamin-dependent RNA modification. Nucleic Acids Res. 2016;44:9965–9976. doi: 10.1093/nar/gkw806. This paper reveals molecular insight into a new Cbl function; the structure of QueG cocrystallized with an oQ-modified tRNATyr anticodon stem loop bound and corresponding electrochemical data are consistent with a reaction mechanism that involves the formation of a covalent Cbl-tRNA intermediate species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutmos M, Gherasim C, Smith JL, Banerjee R. Structural basis of multifunctionality in a vitamin B12-processing enzyme. J Biol Chem. 2011;286:29780–29787. doi: 10.1074/jbc.M111.261370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne KA, Fisher K, Sjuts H, Dunstan MS, Bellina B, Johannissen L, Barran P, Hay S, Rigby SE, Leys D. Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification. J Biol Chem. 2015;290:27572–27581. doi: 10.1074/jbc.M115.685693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••23.Payne KA, Quezada CP, Fisher K, Dunstan MS, Collins FA, Sjuts H, Levy C, Hay S, Rigby SE, Leys D. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature. 2015;517:513–516. doi: 10.1038/nature13901. A combination of X-ray crystallography and EPR spectroscopy in this paper reveals a Co-halide interaction in the reductive dehalogenase NpRdhA. Notably, the reaction described here is formally an oxidative addition of a halide atom to Co and would represent a new prototype of Cbl chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 25.Shey J, van der Donk WA. Mechanistic studies on the vitamin B-12-catalyzed dechlorination of chlorinated alkenes. J Am Chem Soc. 2000;122:12403–12404. [Google Scholar]
- •26.Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A. 2011;108:7368–7372. doi: 10.1073/pnas.1018636108. This paper describes the discovery of sequence conservation between QueG and the reductive dehalogenase family. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miles ZD, Myers WK, Kincannon WM, Britt RD, Bandarian V. Biochemical and Spectroscopic Studies of Epoxyqueuosine Reductase: A Novel Iron-Sulfur Cluster- and Cobalamin-Containing Protein Involved in the Biosynthesis of Queuosine. Biochemistry. 2015;54:4927–4935. doi: 10.1021/acs.biochem.5b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelton DR, Tiedje JM. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic Acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietra F. Uptake of Organohalide Pollutants, and Release of Partially Dehalogenated Products, by NpRdhA, a ‘Base-Off’ Cob(II)alamin-Dependent Reductive Dehalogenase from a Deep Sea Bacterium. A Molecular Dynamics Investigation. Chem Biodivers. 2015;12:1945–1953. doi: 10.1002/cbdv.201500195. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz RP, Wolf J, Habel A, Neumann A, Ploss K, Svatos A, Boland W, Diekert G. Evidence for a radical mechanism of the dechlorination of chlorinated propenes mediated by the tetrachloroethene reductive dehalogenase of Sulfurospirillum muftivorans. Environ Sci Technol. 2007;41:7370–7375. doi: 10.1021/es071026u. [DOI] [PubMed] [Google Scholar]
- 31.Glod G, Angst W, Holliger C, Schwarzenbach RP. Corrinoid-mediated reduction of tetrachloroethene, trichloroethene, and trichlorofluoroethene in homogeneous aqueous solution: Reaction kinetics and reaction mechanisms. Environ Sci Technol. 1997;31:253–260. [Google Scholar]
- 32.Schumacher W, Holliger C, Zehnder AJ, Hagen WR. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 1997;409:421–425. doi: 10.1016/s0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- 33.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frey PA. In: Comprehensive Natural Products II Chemistry and Biology. Mander LL, H-W, editors. Vol. 7. Elsevier; 2010. pp. 501–546. [Google Scholar]
- 35.Magnusson OT, Reed GH, Frey PA. Characterization of an allylic analogue of the 5′-deoxyadenosyl radical: an intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry. 2001;40:7773–7782. doi: 10.1021/bi0104569. [DOI] [PubMed] [Google Scholar]
- •36.Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, LeVieux J, Liu HW. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. J Am Chem Soc. 2013;135:8093–8096. doi: 10.1021/ja312641f. This paper elucidates the in vitro activity of the Cbl-dependent AdoMet radical enzyme GenK, which catalyzes methylation of an unactivated carbon center in the biosynthetic pathyway of the gentamicin antibiotic. This paper establishes that two equivalents of AdoMet are required per reaction cycle and that the methyl group of AdoMet is incorporated into product. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •37.Marous DR, Lloyd EP, Buller AR, Moshos KA, Grove TL, Blaszczyk AJ, Booker SJ, Townsend CA. Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis. Proc Natl Acad Sci U S A. 2015;112:10354–10358. doi: 10.1073/pnas.1508615112. The authors present the characterization of a Cbl-dependent AdoMet radical enzyme ThnK, which catalyzes two sequential methylation reactions in the biosynthetic pathway of the carbapenem antibiotic, thienamycin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamigiri K, Hidaka T, Imai S, Murakami T, Seto H. Studies on the biosynthesis of bialaphos (SF-1293) 12. C-P bond formation mechanism of bialaphos: discovery of a P-methylation enzyme. J Antibiot (Tokyo) 1992;45:781–787. doi: 10.7164/antibiotics.45.781. [DOI] [PubMed] [Google Scholar]
- •39.Werner WJ, Allen KD, Hu K, Helms GL, Chen BS, Wang SC. In vitro phosphinate methylation by PhpK from Kitasatospora phosalacinea. Biochemistry. 2011;50:8986–8988. doi: 10.1021/bi201220r. This work details the first purification of a Cbl-dependent AdoMet radical enzyme and additionally includes the first demonstration that any Cbl-dependent AdoMet radical enzyme can catalyze the transfer of a methyl group from MeCbl to substrate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen KD, Wang SC. Initial characterization of Fom3 from Streptomyces wedmorensis: The methyltransferase in fosfomycin biosynthesis. Arch Biochem Biophys. 2014;543:67–73. doi: 10.1016/j.abb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen KD, Wang SC. Spectroscopic characterization and mechanistic investigation of P-methyl transfer by a radical SAM enzyme from the marine bacterium Shewanella denitrificans OS217. Biochim Biophys Acta. 2014;1844:2135–2144. doi: 10.1016/j.bbapap.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierre S, Guillot A, Benjdia A, Sandstrom C, Langella P, Berteau O. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat Chem Biol. 2012;8:957–959. doi: 10.1038/nchembio.1091. [DOI] [PubMed] [Google Scholar]
- 43.Gough SP, Petersen BO, Duus JO. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc Natl Acad Sci U S A. 2000;97:6908–6913. doi: 10.1073/pnas.97.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••44.Blaszczyk AJ, Silakov A, Zhang B, Maiocco SJ, Lanz ND, Kelly WL, Elliott SJ, Krebs C, Booker SJ. Spectroscopic and Electrochemical Characterization of the Iron-Sulfur and Cobalamin Cofactors of TsrM, an Unusual Radical S-Adenosylmethionine Methylase. J Am Chem Soc. 2016 doi: 10.1021/jacs.5b12592. This paper includes a rigorous spectroscopic analysis of how the Cbl-dependent AdoMet radical enzyme TsrM binds both Cbl and AdoMet cofactors. [DOI] [PubMed] [Google Scholar]
- 45.Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 47.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westrich L, Heide L, Li SM. CloN6, a novel methyltransferase catalysing the methylation of the pyrrole-2-carboxyl moiety of clorobiocin. Chembiochem. 2003;4:768–773. doi: 10.1002/cbic.200300609. [DOI] [PubMed] [Google Scholar]
- 49.Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem Commun (Camb) 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 50.Ruszczycky MW, Ogasawara Y, Liu HW. Radical SAM enzymes in the biosynthesis of sugar-containing natural products. Biochim Biophys Acta. 2012;1824:1231–1244. doi: 10.1016/j.bbapap.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: a tale of two SAMS. Acc Chem Res. 2012;45:555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Donk WA. Rings, radicals, and regeneration: The early years of a bioorganic laboratory. J Org Chem. 2006;71:9561–9571. doi: 10.1021/jo0614240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosimann H, Kräutler B. Methylcorrinoids Methylate Radicals-Their Second Biological Mode of Action? Angew Chem Int Ed Engl. 2000;39:393–395. doi: 10.1002/(sici)1521-3773(20000117)39:2<393::aid-anie393>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Morita M, Tomita K, Ishizawa M, Takagi K, Kawamura F, Takahashi H, Morino T. Cloning of oxetanocin A biosynthetic and resistance genes that reside on a plasmid of Bacillus megaterium strain NK84-0128. Biosci Biotechnol Biochem. 1999;63:563–566. doi: 10.1271/bbb.63.563. [DOI] [PubMed] [Google Scholar]
- 55.Gray MJ, Escalante-Semerena JC. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci U S A. 2007;104:2921–2926. doi: 10.1073/pnas.0609270104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booker SJ. Anaerobic functionalization of unactivated C-H bonds. Curr Opin Chem Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]


