Abstract
We performed cytosine methylation sequencing on genetically diverse AML patients and found leukemic DNA methylation patterning is primarily driven by non-promoter regulatory elements and CpG shores. Enhancers displayed stronger differential methylation than promoters, consisting predominantly of hypomethylation. AMLs with dominant hypermethylation featured greater epigenetic disruption of promoters, while those with dominant hypomethylation displayed greater disruption of distal and intronic regions. Mutations in IDH and DNMT3A had opposing and mutually exclusive effects on the epigenome. Notably, co-occurrence of both mutations resulted in epigenetic antagonism, with most CpGs affected by either mutation alone no longer affected in double mutant AMLs. Importantly, this epigenetic antagonism precedes malignant transformation and can be observed in pre-leukemic LSK cells from Idh2R140Q or Dnmt3aR882H single and, Idh2R140Q/Dnmt3aR882H double mutant mice. Notably, IDH/DNMT3A double mutant AMLs manifested upregulation of RAS signaling signature and displayed unique sensitivity to MEK inhibition ex vivo as compared to AMLs with either single mutation.
Keywords: Acute Myeloid Leukemia, AML, Epigenetics, DNA Methylation, Enhancers
INTRODUCTION
Acute myeloid leukemia (AML) is a genetically and biologically heterogeneous disease that features a relative paucity of somatic mutations compared to other tumor types (1). In contrast, AML patients exhibit widespread disruption of cytosine methylation patterning (1, 2). These specific DNA methylation profiles have been exploited to classify AML into 16 different epigenetically defined subtypes that were partially linked to genetic lesions (3, 4) and enabled discovery of novel leukemia driving mechanisms (2–4). All of these studies focused primarily on cytosine methylation patterning at promoter areas and CpG islands. Yet, more recent studies in cancer epigenetics have highlighted potential roles for other types of genomic elements, such as enhancers and CpG shores, in contributing to perturbed epigenetic patterning in cancer (5–7). Enhancers are distal regulatory elements that often become active or inactive in a tissue specific manner to mediate particular developmental and functional processes (8). Enhancer activity status depends on the presence of specific histone modifications and other epigenetic marks including cytosine modifications. Enhancers are increasingly shown to play critical roles in myelopoiesis and leukemogenesis (9–11). Recent studies implicate epigenetic enhancer dysfunction through aberrant cytosine methylation as a contributing factor for development of AML in mouse models, for example in the case of Evi1 leukemias (9, 10) or mutant TET2 in combination with FLT3 internal tandem duplications (ITD) (12). Hence it is of great interest to explore whether these elements are indeed a prominent and perhaps even defining feature of epigenetic patterning in human primary AML.
One potential driving force for epigenetic reprogramming in AML is the occurrence of somatic mutations affecting proteins that regulate cytosine methylation. DNMT3A, which encodes a de novo DNA methyltransferase is affected by somatic mutations in approximately 25% of AML cases (1). Evidence to date would appear to indicate that certain DNMT3A mutants could function in a dominant negative manner to disrupt active DNMT3A tetramers (13–15). DNMT3A mutations occur during the preleukemic phase of AML pathogenesis (16, 17), attesting to a potential critical role for aberrant epigenetic reprogramming in malignant transformation of hematopoietic cells. The IDH1 and IDH2 loci encode metabolic enzymes that are also mutated in 15–20 % of AML cases (1, 18). Mutant IDH1 and IDH2 proteins generate the oncometabolite 2-hydroxyglutarate. This inhibits dioxygenases such as the TET family enzymes, which are involved in DNA demethylation (19), resulting in a DNA hypermethylation phenotype in AMLs carrying these mutations (4). Murine models of DNMT3A and IDH mutations have revealed that aberrant methylation driven by these lesions is not restricted to promoter regions but instead targets a significant proportion of intergenic and intronic regions (20, 21).
To better understand the nature of aberrant epigenetic programming in AML we utilized enhanced reduced representation bisulfite sequencing (ERRBS) on a panel of primary AML cases with detailed cytogenetic, molecular and epigenetic information (2). The expanded coverage and base-pair resolution of ERRBS allowed us to evaluate cytosine methylation changes at higher resolution and with greater breadth than previously possible. We evaluate the contribution of individual gene mutations to these epigenetic patterns, and specifically focus on the effect of co-occurring epigenetic mutations.
RESULTS
Comprehensive DNA methylation profiling extending beyond promoter regions robustly captures AML biology
AML can be classified into biologically distinct subtypes using gene promoter methylation patterns (2, 3). However, increasing evidence points toward a key regulatory role for DNA methylation beyond promoter regions (6, 7). We hypothesized that higher resolution methylation profiling using next-generation bisulfite sequencing would better define the basis for aberrant epigenetic programming in AML. We therefore performed ERRBS (22) on 119 adult AML patients selected from a cohort that had been previously studied using the promoter-centric HELP microarray assay and classified into 16 epigenetically defined clusters (2). Samples were randomly selected from within each of the original 16 epigenetically defined clusters and curated to include examples of the key somatic mutations currently understood to be associated with AML pathogenesis (summarized in Table 1).
Table 1.
Patient Characteristics
| Gender | N | (%) | |
|---|---|---|---|
|
|
|||
| Male | 66 | (55.46%) | |
| Female | 53 | (44.54%) | |
|
|
|||
| Age | |||
|
|
|||
| < 20 | 8 | (6.72%) | |
|
|
|||
| 20–40 | 38 | (31.93%) | |
|
|
|||
| 40–60 | 59 | (49.58%) | |
|
|
|||
| >= 60 | 14 | (11.76%) | |
|
|
|||
| Median | 44 | ||
|
|
|||
| FAB | |||
|
|
|||
| M0 | 7 | (5.88%) | |
|
|
|||
| M1 | 29 | (24.37%) | |
|
|
|||
| M2 | 29 | (24.37%) | |
|
|
|||
| M3 | 4 | (3.36%) | |
|
|
|||
| M4 | 21 | (17.65%) | * 1 M4E |
|
|
|||
| M5 | 23 | (19.33%) | |
|
|
|||
| NA | 5 | (4.2%) | * includes 4 ‘AML non M3’, 1 RAEB MDS |
|
|
|||
| Cytogenetics | |||
|
|
|||
| Normal | 60 | (50.42%) | |
|
|
|||
| inv(16) | 10 | (8.4%) | |
|
|
|||
| t(8;21) | 11 | (9.24%) | |
|
|
|||
| t(15;17) | 3 | (2.52%) | |
|
|
|||
| t(v;11q23) | 2 | (1.68%) | |
|
|
|||
| del 5/7q | 6 | (5.04%) | |
|
|
|||
| del9q | 2 | (1.68%) | |
|
|
|||
| Complex | 4 | (3.36%) | |
|
|
|||
| tri 8 | 2 | (1.68%) | |
|
|
|||
| Other | 12 | (10.08%) | |
|
|
|||
| NA | 5 | (4.2%) | |
|
|
|||
| Cytogenetic risk | |||
|
|
|||
| Favorable | 23 | (19.33%) | |
|
|
|||
| Intermediate | 76 | (63.87%) | |
|
|
|||
| Poor | 13 | (10.92%) | *includes 1 favorable + unfavorable |
|
|
|||
| NA | 6 | (5.04%) | *unavailable karyotype |
|
|
|||
| CEBPa abnormalities | |||
|
|
|||
| Silenced | 6 | (5.04%) | |
|
|
|||
| Double mutation | 12 | (10.08%) | |
|
|
|||
| Single mutation | 1 | (0.84%) | |
|
|
|||
| IDH mutation | |||
|
|
|||
| IDH1 | 10 | (8.4%) | |
|
|
|||
| IDH2 | 10 | (8.4%) | |
|
|
|||
| Other alterations | |||
|
|
|||
| NPM1 | 43 | (36.13%) | |
|
|
|||
| FLT3 | 48 | (40.34%) | |
|
|
|||
| DNMT3A | 27 | (22.69%) | |
|
|
|||
| EVI1 overexpression | 8 | (6.72%) | |
|
|
|||
| TET2 | 18 | (15.13%) | |
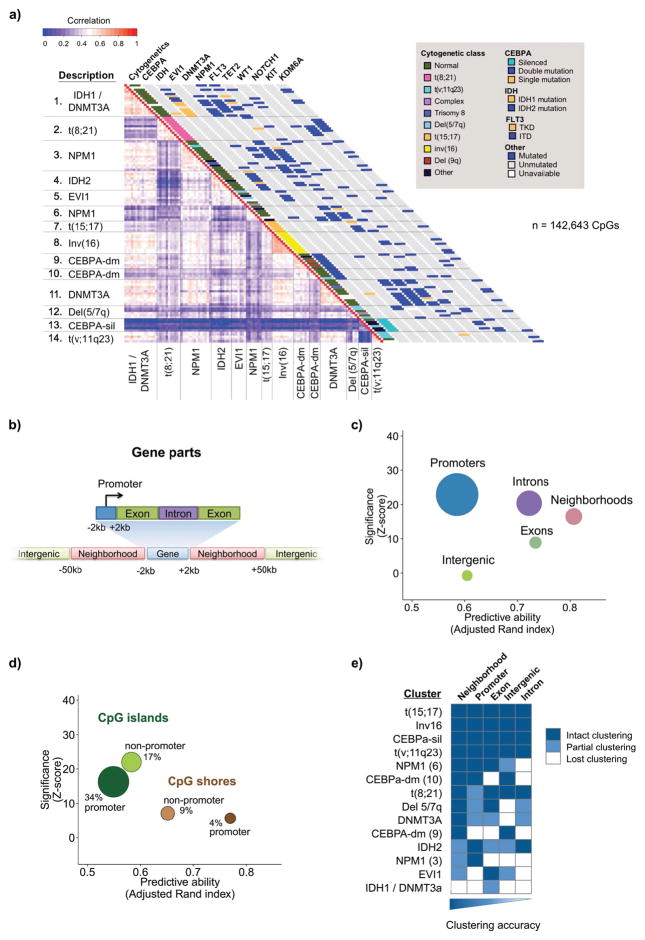
Unsupervised analysis by hierarchical clustering using Euclidean distance and based on the most variable CpGs across all patients (n = 142,643) yielded 14 clusters at a distance threshold that minimizes the number of clusters without merging samples with distinct cytogenetic or mutation profiles (Figure 1a). Clusters characterized by inv16, t(8;21), t(15;17), t(v;11q23), and CEBPA silencing remained identical to those specified by HELP. In contrast, cases that originally fell into a single cluster harboring patients with CEBPA double mutation (CEBPA-dm) were instead split into two distinct groups by ERRBS, one which was almost entirely cytogenetically normal (cluster 10), and another including cases with both normal karyotype and cytogenetic abnormalities (cluster 9). Similarly, IDH1/2 mutant cases, which were previously mainly grouped into two clusters including both IDH1 and IDH2 mutant cases (4), were now grouped by ERRBS profiling into a predominantly IDH1-mutant cluster (cluster 1) in which all mutant cases also harbored DNMT3A mutations, and a second cluster exclusively carrying IDH mutations without co-occurring DNMT3A mutations, almost all of which were IDH2 mutations (Table S1). The remaining clusters were enriched for a combination of DNMT3A, NPM1, and FLT3 abnormalities. These data indicate that comprehensive methylome sequencing more precisely links genetic lesions to perturbations in cytosine methylation patterning than previous, promoter-centric approaches (1–3).
Figure 1. Epigenetically defined classification of AML.
A: Representation of hierarchical clustering results based on ERRBS data using a correlation matrix heatmap. ERRBS defines AML subtypes with distinct molecular and cytogenetic characteristics. Groups were defined using a hierarchical clustering approach (see supplemental methods) and labeled according to their dominant distinguishing molecular and cytogenetic features. The lower triangular heatmap represents the correlation between the most divergent CpGs represented in all samples (n=142,643). Cytogenetic and specific molecular features are represented in the diagonal bars on the right. B: Schematic representation of the genomic compartments used in our analyses. C and D: relative predictive ability (x-axis) of each compartment using the adjusted Rand index to compare the clustering produced using the most divergent CpGs within each compartment to those using all highly variable CpGs. The significance of the finding for each compartment is shown on the y axis as a z score comparing the adjusted Rand index of the compartment to 500 iterations of the Rand index for the same number of CpGs selected randomly. E: Heatmap representation of the robustness of clustering for CpGs within each genomic compartment.
To further and more comprehensively assess the link between cytosine methylation signatures and somatic mutations we performed additional targeted resequencing of 49 genes implicated in myeloid malignancies (Table S2). Using a series of multinomial linear regression models (see details in methods section), we then quantified the ability of individual mutations, genetic rearrangements or known epigenetic abnormalities to specify epigenetic clustering. In addition to inv16, t(8;21), t(15;17) and t(v;11q23), only CEBPA-dm, NPM1, DNMT3A, and IDH2 mutations, or CEBPA gene silencing provided additional significant predictive ability (Figure S1a). No additional mutation or pair of mutations (including those affecting FLT3) contributed additional predictive power (Figure S1b). Notably, the presence of an IDH1 mutation, which was cluster defining only in combination with DNMT3A mutation, did not emerge as an independent predictor of clustering. Therefore, even when performing more comprehensive methylome analysis, only a subset of somatic mutations can be linked to specific cytosine methylation patterning effects. TET2 mutation status also did not emerge as a cluster-defining lesion, instead being distributed among several clusters.
Differential methylation at non-promoter gene regulatory regions is the primary driver of AML epigenetic subtypes
The genomic coverage of ERRBS extends beyond promoters to cover inter- and intragenic CpGs, as well as beyond CpG islands to cover also CpG shores and distal intergenic regions (22). In order to determine which genomic elements have the greatest influence on determining cluster-defining epigenetic patterning we first annotated CpGs covered by ERRBS into the following categories: (i) ‘promoter’ – CpGs +/−2 kb from gene transcription start sites (TSS); (ii) ‘gene neighborhood’ – CpGs between 2 kb and 50 kb away from the TSS or transcription end site; (iii) ‘downstream’ – CpGs +/− 2 kb from transcription end sites; (iv) ‘intergenic’ – CpGs greater than 50 kb from the nearest gene; (v) ‘exons’ – CpGs within any annotated exon; (vi) ‘introns’ – CpGs between exons (Figure 1b). 950,953 common CpGs with coverage ranging between 10–400X were uniformly captured in all 119 samples. Among these common CpGs, 44.7% were located within promoters, 16.1% within gene neighborhoods, 7.9% within intergenic regions, 1.5% within the downstream window, 9.2% within non-first exons and, 20.6% within non-first introns (Figure S2a). From a CpG island-centric point of view, 42.6% of CpGs were at CpG islands, while 9.7% were at CpG shores and 47.6% were located elsewhere (Figure S2b). Taken together these CpGs captured 82.6% of all RefSeq promoters, 91.6% of gene neighborhoods, 88.9% of intergenic regions, 16.7% of non-first exons, 33.9% of non-first introns, as well as 93.2% of all CpG islands, and 72.6% of CpG island shores (Figure S2c). Overall, these CpGs represent an extensive sampling of the epigenome.
To determine which of these genomic regions contributes the most to epigenetic patterning in AML, we used the adjusted Rand index to compare clustering using only CpGs located at the various genomic regions to that of the overall clustering. We found that CpGs located in gene neighborhoods were best able to accurately recapitulate epigenetic clusters despite representing only 15% of the CpGs used for the unsupervised clustering analysis. When compared to the same number of CpGs selected from the genome at random, we found this prediction to be significantly better than chance (p< 0.01, z-test). Other genomic subsets also predicted clustering better than chance, but with poorer cluster specification as indicated by their lower adjusted Rand indices (Figure 1c). While promoters classified patients poorly, the subset of CpGs in promoters within CpG shores performed far better than CpGs within CpG islands, underlining that CpG shores and not islands may be the principal sites for contributing to epigenetic identity in AML (Figure 1d). Next we sought to determine whether this general observation was equally true across all clusters or whether certain AML subtypes were best captured by specific compartments. A cluster-by-cluster performance analysis for each genomic compartment revealed that gene neighborhoods were indeed most accurate in predicting cluster assignments in most cases (10/14). However they were only partially able to capture the IDH1/2 clusters, one of the NPM1 clusters, and the EVI1 cluster. This may reflect the fact that differential methylation in IDH mutant cases tends to occur relatively nearer to transcriptional start sites (22). Overall, the remaining genomic compartments showed weaker clustering accuracy compared to gene neighborhoods (Figure 1e). The only AML subtypes captured uniformly by all genomic regions were those with t(15;17), inv(16), t(v;11q23), and those with CEBPA silencing. These findings suggest that oncogenic drivers primarily disrupt cytosine methylation at gene neighborhood regions, which are enriched in regulatory elements such as enhancers and transcription factor binding sites.
Active enhancers exhibit focal enrichment of differential methylation in AML
To determine the contribution of enhancer CpGs to epigenetic cluster identity, we determined the location of hematopoietic stem and progenitor cell (HSPC) enhancers using ChIP-seq datasets generated in human mobilized CD34+ cells (23) based on H3K4me1 and H3K27ac peaks in the absence of a H3K4me3 peak or an annotated promoter. Active enhancers were distinguished from poised enhancers by the presence of an H3K27ac peak. Enhancer-associated CpGs were predominantly within introns and gene neighborhoods, with 19.4% of active enhancers captured located at gene neighborhoods and 48.0% within introns (Figure S2d). Gene neighborhoods were notably enriched for both active and poised enhancer CpGs (p < 0.01 for each, Fisher exact test).
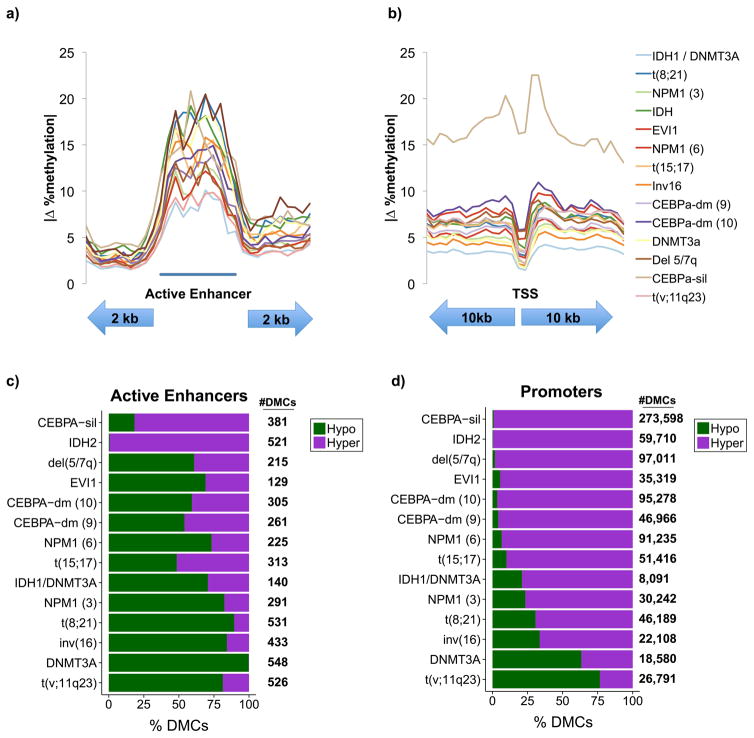
To determine if enhancers are generally disrupted by leukemic cytosine methylation patterning we specifically interrogated methylation change within active HSPC enhancers. We found that these loci exhibited focal methylation changes in AMLs when compared to normal CD34+ cells, and that the average enhancer methylation change at all covered CpGs was greater than surrounding sequences (Figure 2a; S3a, b). This pattern was in sharp contrast to promoters, which exhibited little methylation change relative to surrounding sequences (Figure 2b; S3c, d). These changes were not equal in nature in all clusters, and included both some with increases in average methylation and some with decreases in average methylation (Figure S4a). In absolute terms, the changes at promoters were both less marked and more uniform in nature across the different clusters. In addition, the change consisted almost exclusively of gains in methylation in areas surrounding the TSS with little to no change at the TSS itself (Figure S4b).
Figure 2. Differential methylation at regulatory regions.
A: average absolute change in CpG methylation within active enhancer regions and flanking areas for each cluster. The region shown was analyzed in 30 bins, 10 for each flank and 10 for the active enhancer. All CpGs within covered annotated enhancers are included. B: average absolute change in CpG methylation within promoter regions with the upstream and downstream flanks represented by 15 bins each. All covered CpGs within promoters are included. C–D: stacking bar plots illustrating the proportions of hyper and hypomethylated CpGs in each cluster relative to CD34+ controls.
At differentially methylated CpGs, the directionality of methylation change within these two compartments also differed. Among CpGs exhibiting significant differential methylation in AML (≥ 25% methylation difference, Beta-binomial q ≤ 0.01) relative to normal HSPC controls, the majority of clusters showed a predominance of hypomethylation at active enhancers with the exception of CEBPA-silenced and IDH2 mutant clusters. In contrast, promoters were generally hypermethylated, with the exception of DNMT3A and MLL fusion leukemias (Figure 2c, d). Therefore, aberrant cytosine methylation of HSPC enhancers is a defining feature of AML epigenetic patterns, with directionality of methylation dependent on driver oncogenes, while gene promoters are involved to a lesser extent.
Epigenetically defined AML patient clusters exhibit distinct regional distribution patterns and directionality
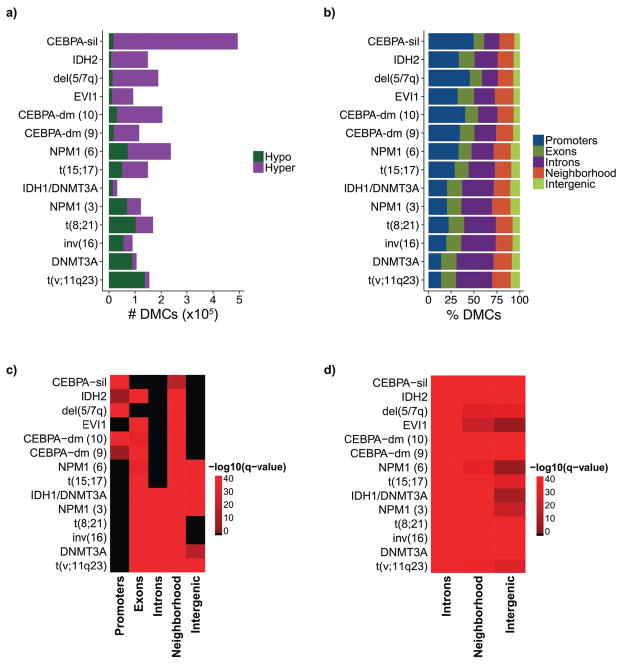
To characterize the extent to which cytosine methylation is perturbed in each epigenetically defined AML subtype we performed supervised analyses comparing the DNA methylation profiles of the AML cases in each cluster to ERRBS performed in healthy human bone marrow CD34+ HSPC controls (n=22). In all but one cluster, differentially methylated CpGs (DMCs) (≥ 25% methylation change, Beta-binomial q-value ≤ 0.01) represented less than 10% of CpGs covered by ERRBS (Figure 3a and S4c). The CEBPA-silenced cluster was the only outlier, with nearly 19% of covered CpGs hypermethylated relative to CD34+ controls. Notably, the cluster defined by coexistent DNMT3A and IDH1 mutations contained the fewest total DMCs (Figure 3a and S4d).
Figure 3. Genomic distribution of differential methylation relative to CD34+ controls.
A: Bar plots illustrating the overall number of hyper and hypomethylated CpGs for each cluster sorted from highest to lowest percentage of hypermethylated CpGs relative to CD34+ controls. B: Stacking bar plots illustrating the proportional distribution of differentially methylated CpGs in each genomic compartment. C: Heatmap illustrating the enrichment of differentially methylated CpGs in each genomic compartment. D: Heatmap illustrating the enrichment of differentially methylated CpGs within HSPC enhancers in different genomic compartments.
Next we evaluated the distribution of differential methylation in each cluster of AML patients (Table S3). Gene neighborhoods were the only genomic region enriched for DMCs in all clusters (p < 0.01 for all, Fisher exact test). However, DMCs within clusters characterized by a predominantly hypermethylated signature, such as the CEBPA-silenced cluster, the CEBPA-dm clusters, and del (5/7q), were preferentially enriched at promoter regions (p < 0.01 for each, Fisher exact test). In contrast, DMCs within clusters characterized by a predominantly hypomethylated signature, such as the DNMT3A mutant and t(v;11q23) clusters, were preferentially enriched at distal intergenic and intronic regions (p < 0.01 for each, Fisher exact test) (Figure 3b and 3c). Interestingly, less than half of the clusters showed enrichment of differential methylation at promoter regions (Figure 3c, S3d), but all showed significant enrichment at HSPC enhancers, regardless of genomic compartment (Figure 3d). Altogether these data underline the markedly diverse nature of epigenetic programs occurring in AML and again point to enhancers and other non-promoter regulatory regions as the critical disease defining sites for epigenetic dysregulation.
AMLs with co-occurring DNMT3A and IDH mutations exhibit epigenetic antagonism
Clusters 4 and 11 capture the hyper and hypomethylated profiles for IDH2 and DNMT3A mutations respectively, consistent with the known molecular mechanisms of these two mutant proteins (4, 13, 15, 21, 24). However, AML patients with dual IDH and DNMT3A mutations exhibited a markedly distinctive cytosine methylation pattern from patients in either cluster 4 or 11. Among these patients, 8/11 (73%) exhibited dual IDH1/DNMT3A mutations so this cohort will be referred to as ‘IDH1/DNMT3A’. Indeed, IDH1/DNMT3A patients manifested the smallest degree of differential methylation of any cluster, with only 31,882 DMCs relative to CD34+ controls. Of these, 47.3% of DMCs (15,072 CpGs) were hypomethylated and 52.7% (16,810 CpGs) were hypermethylated (Figure 3a). In striking contrast, DNMT3A mutations alone (cluster 11) resulted in 105,939 DMCs, 84.8% of which were hypomethylated, while isolated IDH2 mutations (cluster 4) led to a predominantly hypermethylated profile with 148,961 DMCs, 93.9% of which were hypermethylated.
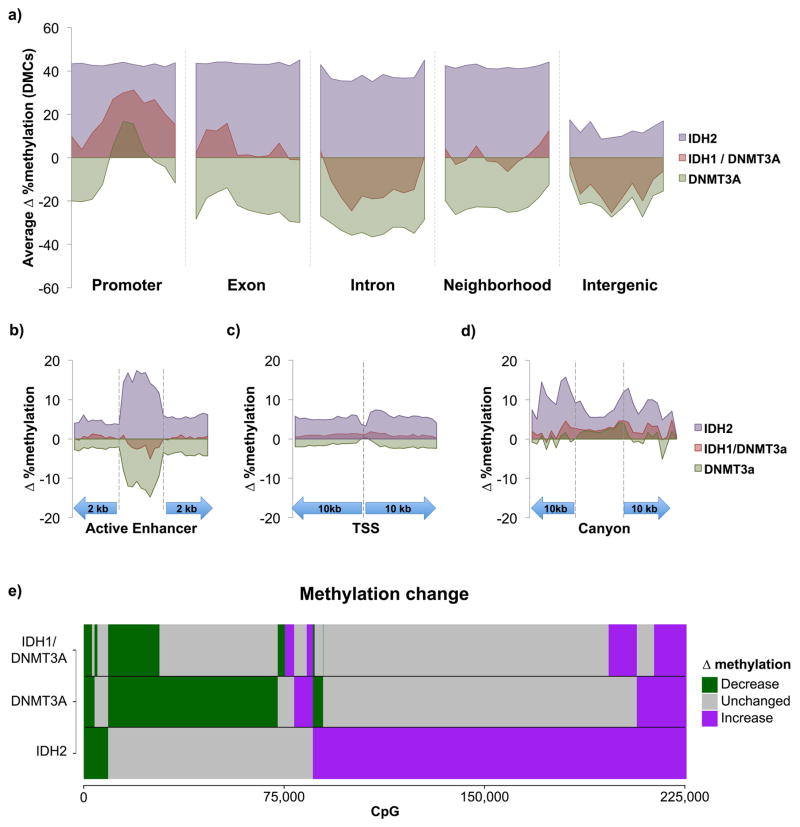
To better understand the nature of these differences we compared and contrasted the regional distribution of differential methylation in DNMT3A, IDH2 or IDH1/DNMT3A patients (Table S4). Whereas IDH2 mutants exhibited an average DMC methylation increase >40% above normal HSPC controls at neighborhoods, promoters and gene bodies, DNMT3A mutant cases manifested ~20% loss of methylation at gene neighborhoods and bodies, and little effect on promoters. In marked contrast, IDH1/DNMT3A double mutants completely lost the characteristic differential methylation of either IDH2 or DNTM3A at gene neighborhoods. IDH1/DNMT3A also manifested attenuated IDH effects at promoters and exons and, attenuation of the DNMT3A effect at introns and intergenic regions (Figure 4a).
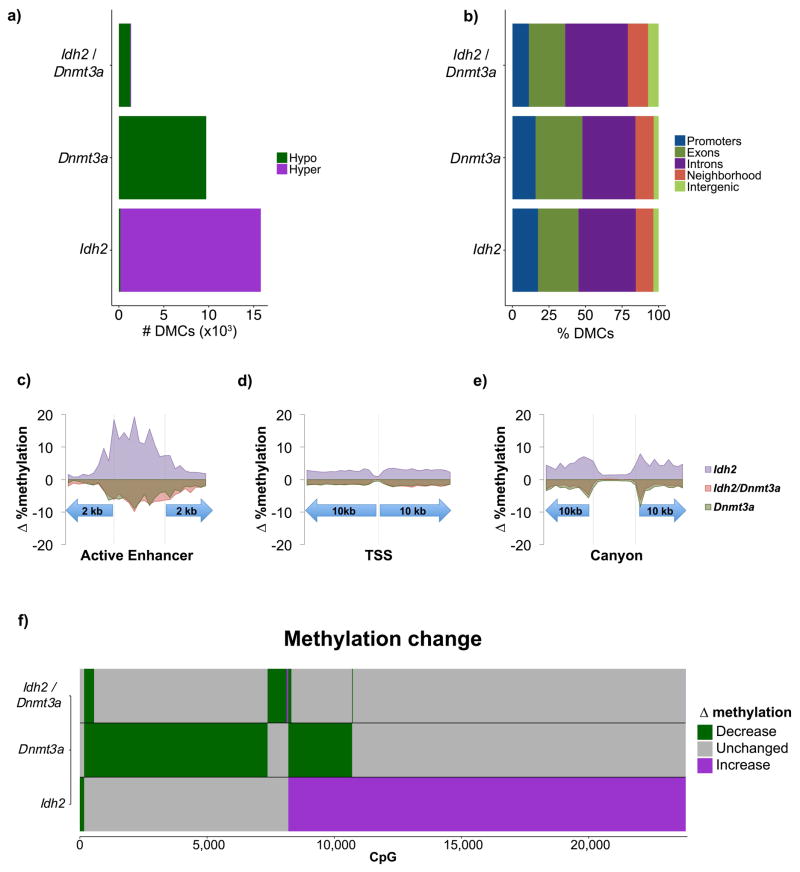
Figure 4. Antagonism of IDH and DNMT3A mutations in AML.
A; Average methylation change at differentially methylated CpGs within each mutation category across promoters, exons, introns, gene neighborhoods, and intergenic regions. B–D: Average overall methylation changes in AML relative to CD34+ controls at all covered CpGs within active enhancers, promoters, and canyons. E: Bar plots illustrating the type of methylation change relative to CD34+ controls at individual CpGs in IDH2, DNMT3A, and IDH1/DNMT3A mutant AMLs.
We next examined active enhancers in more detail since, as shown above, these regions represent a dominant site of epigenetic variability in AML. This analysis revealed that IDH2 mutant cases manifested overall enhancer hypermethylation when all covered CpGs were considered whereas DNMT3A mutant cases exhibited predominant enhancer hypomethylation. However, there was very little deregulation of enhancer cytosine methylation in IDH1/DNMT3A cases (Figure 4b). At promoter regions, differential methylation at all covered CpGs exhibited focal restriction centered at the transcription start site in all 3 subtypes. Regions flanking the TSS once again manifested overall hypermethylation at all covered CpGs in the IDH2 AMLs, whereas there was a mild degree of hypomethylation in the DNMT3A mutants. In contrast, the IDH1/DNMT3A had very little effect on promoters with almost complete lack of the IDH2 promoter hypermethylation effect (Figure 4c).
This analysis demonstrates that the overall gain in regional cytosine methylation seen in IDH2 AMLs and the general loss of cytosine methylation observed in DNMT3A AMLs was profoundly attenuated in the IDH1/DNMT3A double mutants. Specifically, these two mutations have opposing effects on gene neighborhoods and enhancers, yet both of these effects are mostly lost in IDH1/DNMT3A double mutant AML. Enhancers serve as platforms for the action of transcription factors. Using a bioinformatic approach we observed that differentially methylated HSPC enhancers in IDH2 AMLs were enriched for SMAD3, RXRA, and USF2, all of which contain strong ‘CG’ motifs (Figure S5A), whereas DNMT3A mutant cases were enriched for SMAD3 and REL NF-kappa-B motifs (Figure S5B). Even though IDH1/DNMT3A double mutant cases manifest much less enhancer perturbation, there was still some enrichment for NR2C2 motifs, which were not present in IDH or DNMT3A mutant cases (Figure S5C). All of these transcription factors are expressed in these leukemias suggesting that they could be in some way involved or affected by aberrant cytosine methylation (Figure S5D). As a complementary approach we examined available ChIP-seq data from leukemia and hematopoietic cells (Figure S5E). All three sets of patients featured enrichment for TAL1, GATA1, RUNX1 and others at differentially methylated enhancers. However, GATA2, CEBPA, CEBPB, PRAME, NFYA and TCF7L2 although uniquely enriched in both single mutant AMLs, were missing from the double mutant leukemias, which in turn did not manifest enrichment for any unique set of ChIP-seq transcription factor binding sites.
Mouse hematopoietic stem cells have been observed to contain large domains of unmethylated cytosines containing binding sites of transcription factors involved in hematopoiesis (termed ‘canyons’) (25). To determine whether these regulators of hematopoiesis are perturbed in AML, we identified 207/1104 of these regions with analogous sites in the human genome covered by our ERRBS data. IDH2 mutant AMLs exhibited relative hypomethylation at these canyons when compared to neighboring regions, although these canyons were still hypermethylated as compared to CD34+ controls. Notably, both DNMT3A only and IDH1/DNMT3A cases also showed mild hypermethylation at these canyons relative to normal HSPC controls, though this was much more attenuated than that seen in IDH2 cases. (Figure 4d). Hence while IDH and DNMT3A mutations also exert distinct effects on cytosine methylation at canyons, these seem to trend towards hypermethylation in AMLs relative to normal controls.
Given the opposing effects of these two mutations on methylation profiles, we sought to determine whether these mutations targeted the same CpG sites with opposing results, or whether each targets a distinct set of CpGs. We found that when compared to control HSPCs, AMLs with mutant IDH2 and DNMT3A for the most part target distinct CpGs sites (Figure 4e), and the CpGs that become hypermethylated in IDH2 AMLs are different from those that become hypomethylated in DNMT3A mutant AMLs. Strikingly, even though these CpG sites do not overlap, a majority of the uniquely hypermethylated cytosines in IDH2 and uniquely hypomethylated cytosines in DNMT3A did not manifest differential methylation compared to control HSPCs in IDH1/DNMT3A double mutant AMLs (Figure 4e). These findings indicate that even though IDH2 and DNMT3A individually target a different set of CpG sites, the pattern of methylation change in IDH1/DNMT3A mutants largely reflects antagonistic actions of IDH1 and DNMT3A mutations at most CpGs, while at the same time maintaining a subset of the respective hyper and hypomethylation effects of each allele. The majority of DNMT3A mutations in both DNMT3A alone and the IDH1/DNMT3A cases were at the R882 position (Table S1), thus the expected impact of mutant DNMT3A on the epigenome in these two clusters is similar in both groups.
IDH2 and DNMT3A directly induce epigenetic antagonism in pre-leukemic hematopoietic stem cells
We next sought to determine whether DNMT3A and IDH2 are directly responsible for the epigenetic antagonism observed in human patients, or whether these changes are simply a late consequence of malignant transformation. For this purpose we collected LSK cells from conditional Mx1-Cre;Dnmt3aR878H knock-in mice, which corresponds to the human DNMT3A R882 mutation (26), six months after inducing recombination. Notably, DNA methylation analysis by ERRBS revealed a similar hypomethylated distribution as seen in human DNMT3A mutant blasts, even through at this time point Dnmt3aR878H knock-in mice do not manifest any hematologic phenotype. Similarly, DNA methylation profiling of LSK cells from Idh2R140Q knock-in mice (27) collected at six months after recombination presented a similar aberrantly hypermethylated profile as seen in human AMLs carrying IDH1/2 mutations. Again these mice do not manifest hematologic phenotypes at this timepoint. Finally, in order to determine whether the co-occurrence of both mutations results in epigenetic antagonism, we generated a double knock-in with Dnmt3aR878H/Idh2R140Q and performed ERRBS profiling on LSK cells at six months after recombination. Importantly, at this timepoint, these, Dnmt3aR878H/Idh2R140Q mice also did not manifest a hematologic phenotype. The observed cytosine methylation profile in this genetically engineered model accurately reflected the epigenetic antagonism in the human double mutant AMLs, with Dnmt3AR878H/Idh2R140Q LSKs exhibiting far fewer differentially methylated CpGs than either Dnmt3AR878H or Idh2R140Q (Figure 5a, Table S5). The changes observed both at specific genomic regions as well as at individual CpGs for each allele paralleled the behavior seen in the human AML counterparts (Figure 5b as compared to Figure 3b). Active enhancers, exhibited focal enrichment of methylation variability with Idh2R140Q LSKs exhibiting gain of methylation and Dnmt3aR878H LSKs exhibiting focal loss of methylation (Figure 5c). At promoters, Idh2R140Q and Dnmt3aR878H LSKs again exhibited hyper- and hypomethylation respectively with a restriction in methylation variability at the TSS (Figure 5d). Within canyons, there was a relative restriction in methylation variability, with immediate canyon flanking sequences exhibiting hyper- and hypomethylation in Idh2R140Q and Dnmt3aR878H respectively (Figure 5e). While the absolute number of perturbed CpGs are minimal in number in the Idh2R140Q/Dnmt3aR878H double knock-in mice, there was a tendency for these few cytosines to be less methylated (Figure 5c–e). Similar to the pattern observed in their AML counterparts, differential methylation in the Idh2R140Q and Dnmt3aR878H mice occurred in largely different genomic compartments, and the majority of CpGs in the double mutant mice remained unchanged relative to normal controls (Figure 5f as compared to Figure 4e). These findings confirm that the epigenetic antagonism observed in human IDH/DNMT3A double mutant AMLs is directly linked to these alleles. The data also underscore that these epigenetic changes occur early on in preleukemic cells as a consequence of the expression of the mutant alleles and preceding the phenotypic changes associated with complete transformation of the cells.
Figure 5. Mouse model of IDH2, DNMT3A, and IDH2/DNMT3A AMLs.
A: Number of differentially methylated CpGs within Idh2R140Q, Dnmt3AR878H, and Idh2R140Q/Dnmt3AR878H mutant cohorts compared to normal LSK controls. B–D: Average overall methylation change in each cohort relative to normal controls within active enhancers, promoters, and canyons. E: Bar plots illustrating the type of methylation change relative to normal LSK controls in Idh2R140Q, Dnmt3AR878H, and Idh2R140Q/Dnmt3AR878H cohorts. F: Bar plots illustrating the type of methylation change relative to normal LSK controls at individual CpGs in Idh2R140Q, Dnmt3AR878H, and Idh2R140Q/Dnmt3AR878H mutant mice.
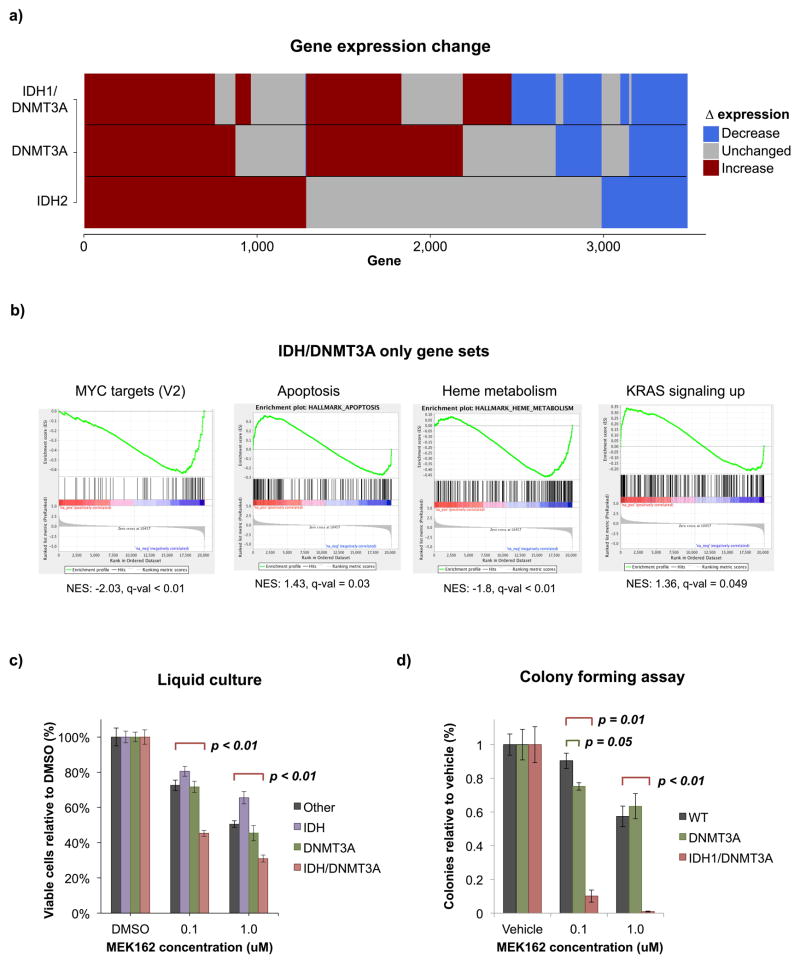
Transcriptome analysis reveals unique expression profile of IDH/DNMT3A double mutant AMLs, suggesting increased sensitivity to MEK inhibition
Although antagonistic in their effects on cytosine methylation, DNMT3A and IDH mutations must still cooperate to drive the leukemic phenotype. To determine how this might occur, we next evaluated the gene expression profiles of these AML patients. When compared to normal HSPC controls, IDH1/DNMT3A mutant AMLs contained 2,524 differentially expressed genes (≥ 1.5 fold change, adjusted p ≤ 0.05). Of these, only 535 genes were uniquely differentially expressed in the IDH1/DNMT3A cluster, with the remaining genes being shared with the DNMT3A and IDH cluster patients (Figure 6a). Gene set enrichment analysis (GSEA) using the Hallmark MSigDB gene sets was performed to identify sets of genes unique to IDH1/DNMT3A cases. The gene sets uniquely enriched are suggestive of competing pro- and anti-proliferative stimuli. KRAS signaling (NES=1.36, FDR q-value=0.048), which favors cell growth, appears to be opposed by the downregulation of certain MYC targets (V2: NES=−2.03, FDR q-value < 0.01) as well as upregulation of apoptosis-associated genes including multiple caspases (NES=1.43, FDR q-value-0.034). Normal hematopoietic processes such as heme metabolism are ultimately downregulated (NES=−1.8, FDR q-value < 0.01) (Figure 6b). Given the unique upregulation of the RAS signaling gene set in the double mutant AMLs, we hypothesized that these leukemias might be uniquely sensitive to inhibition of this signaling cascade. We tested this using a liquid culture assay in which primary AMLs carrying either single IDH, single DNMT3A, double IDH/DNMT3A mutations or AMLs with neither of these two mutations, were cultured in the presence of cytokines for 7 days and the MEK inhibitor MEK162 was added to the culture every 48 hours. Cells with co-occurrence of IDH/DNMT3A mutations showed significantly greater sensitivity to MEK inhibition than any of the other genotypes (p < 0.01) (Figure 6c). This observation was further validated in an orthogonal assay on methylcelullose in which number of total colonies formed by either primary AMLs or normal mononuclear cells from cord blood were scored after cells were cultured for 14 days in the presence of the inhibitor (Figure 6d). While IDH single mutants did not form colonies in this assay, once again IDH1/DNMT3A cases were more sensitive to RAS signaling inhibition via MEK inhibition than either DNMT3A, IDH2, or other types of AML (Figure 6c,d). Taken together, these data suggest that IDH and DNMT3A mutations each result in mixed pro- and anti-proliferative gene expression signatures, which is further perturbed in IDH1/DNMT3A cases in a manner that is at least partly mediated by RAS signaling and not dependent on aberrant cytosine methylation.
Figure 6. IDH1/DNMT3A expression cooperativity.
A: Bar plots illustrating the type of expression change at individual genes in IDH2, DNMT3A, and IDH1/DNMT3A mutant AMLs. B: GSEA of hallmark gene sets uniquely enriched in IDH1/DNMT3A patients. C: Liquid culture assessing the sensitivity of IDH1/DNMT3A AMLs compared to those with IDH mutations or DNMT3A mutations alone. A set of AMLs containing neither mutation was also assessed (Other). Results are displayed as percentages relative to DMSO treated cells with error bars showing the standard error of the mean for 3 biological replicates each performed in 3 technical replicates. Significant changes from ‘other’ samples assessed by one-tailed t-test are shown. D: Colony forming assay assessing the sensitivity of IDH1/DNMT3A AMLs to those with DNMT3A single mutations and cord blood samples (WT) in biological duplicate with 3 technical replicates per sample. Results are displayed in % colonies relative to WT with error bars showing the standard error of the mean. Significant changes from WT assessed by one-tailed t-test are shown.
DISCUSSION
Epigenetic dysregulation has emerged as one of the hallmarks of AML. Broad epigenetic changes occur during hematopoietic differentiation, and disruption of this process accompanied by proliferation of immature myeloid forms underlies the development of AML. The diversity of epigenetic signatures occurring in AML is indicative of multiple underlying mechanisms leading to the development and maintenance of the leukemic state. Herein, through the use of base pair resolution DNA methylation sequencing that represents the genome far more thoroughly than previous microarray studies we reveal the basic nature of the specific and distinct epigenetic programs that occur in AML. Although microarray profiling focused on promoters was sufficient to identify biological subtypes with distinct responses to therapy, significant heterogeneity remained, and several of the resulting categories did not have characteristic mutation profiles (2). We show here that when methylation profiling is extended into gene neighborhoods and intergenic regions, we are better able to classify AMLs and clarify their linkage to mutation profiles.
A major finding in this study is that methylation changes in the gene neighborhood region is better able to predict epigenetic subtype than changes within genes, promoters, or even CpG islands or CpG island shores. In addition, we found that differential methylation appears to particularly affect enhancers that are active in CD34+ HSPCs. In contrast, promoters show a general restriction in differential methylation. Thus, in spite of a relative paucity of somatic mutations, AMLs manifest profound epigenetic disruptions of enhancers that likely reflect differences in the mechanisms that drive malignant transformation. Given that many of the genetic lesions in AML directly or indirectly perturb hematopoietic master regulatory transcription factors, and that these factors largely function through enhancer binding, it is appealing to speculate that the observed distinctive enhancer profiles that distinguish AMLs from each other reflect perturbation of different combinations of transcription factors. Hence the epigenetic pattern of these AMLs may serve as an imprint of events that occur as hematopoietic stem/progenitor cells undergo stepwise transformation. It is also worth noting that epigenetic patterning is more tightly linked to differential methylation of CpG shores and not CpG islands, suggesting that alterations of CpG islands are more stochastic in AML and not as reflective of how specific somatic mutations perturb the epigenome. Finally, it is not clear whether the observed enhancer changes are simply the reflection of the transformation process, or whether their differential methylation contributes in any way to maintaining the malignant phenotype. Our data point to the need to carefully map epigenetic marks in experimental models of malignant transformation to explore such questions.
Another striking result of this study was the unexpected epigenetic antagonism between co-occurring IDH1 and DNMT3A mutation. In addition to our cohort, these mutations exhibit co-occurrence in other AML cohorts (TCGA: log odds 1.64, p < 0.01, Papaemmanuil et al: FWER < 0.05) (1, 28). Our study allows the direct comparison of the methylomes of IDH mutant and DNMT3A mutant AMLs, and we show that these cases contain opposing cytosine methylation profiles with differential methylation occurring at largely distinct CpGs in different genomic compartments. IDH2 mutations result in hypermethylated DMCs that uniformly extend throughout the gene and gene neighborhood compartments. In contrast, DNMT3A mutations result in hypomethylation within the gene body, gene neighborhood, and intergenic compartments at a largely distinct set of CpGs. Co-occurrence of both IDH1 and DNMT3A mutations results in a broad ‘loss’ of differential methylation from both compartments. Given that AML develops in a stepwise manner, these data do not exclude that whichever mutation occurred first did not impact and reprogram the epigenome as per the effect of each mutation. Indeed, in mouse models of IDH or DNMT3A mutation we observe similar effects to what we see in humans in which IDH and DNMT3A are independent of each other (4, 20, 21). It is also important to note that even if cytosine methylation does not appear perturbed, other chromatin marks such as 5′ hydroxymethylation or histone modifications might be affected.
Regardless, our data still point towards functional cooperation between mutant IDH1 and DNMT3A as indicated by gene expression signatures implicating upregulation of KRAS signaling, downregulation of certain MYC targets, and increased in-vitro sensitivity to MEK inhibition. This pattern is not the result of simple KRAS mutation in IDH1/DNMT3A cases, which do not co-occur in this cohort. In addition, recent murine models have shown that DNMT3A R882 mutations specifically cooperate with NRAS to promote leukemogenesis via concomitant specific DNA hypomethylation and gain of active histone modifications (29). In this context, we speculate that this finding might indicate a vulnerability of these leukemias to therapeutic targeting of signaling pathways linked to RAS. In summary this first large scale study of genome wide cytosine methylation patterning underlines the power of the epigenome to reveal insights into the mechanism of action of oncogenes and point to putative unexpected actionable targets in specific sets of AML patients.
Interestingly, the presence of a TET2 mutation did not emerge as a cluster-defining lesion as did IDH2 and IDH1 despite the fact that TET2 and IDH mutations both exert their effect on the epigenome through deregulation of the DNA demethylation pathway (4, 21, 24). This is consistent with prior studies (4, 30) and may be due to compensation by other TET enzymes. While IDH1 and IDH2 mutations inhibit all TET enzymes, the functional effect of TET2 deficiency alone might produce a more modest phenotype.
Methods
Patient Samples
The AML patient samples analyzed in this study were collected at Erasmus University Medical Center (Rotterdam, The Netherlands) from 1990–2008 and processed as previously described (31, 32). All samples consisted of >90% blasts. The samples used for analysis were chosen from our prior study using HELP analysis of DNA methylation (2), with 5–10 patient samples selected per epigenetic cluster based on sample availability. Twenty-two normal CD34+ bone marrow specimens were obtained from AllCells (Alameda, CA). In addition, 6 AML samples were obtained from the Memorial Sloan Kettering Hematologic Oncology Tissue Bank for colony forming assays. Two cord blood controls were obtained from the New York Blood Center. This research was approved by the institutional review boards at Weill Cornell Medical College, University of Michigan Medical School, Erasmus University Medical Center, and Memorial Sloan Kettering and written donor informed consent was obtained in accordance with the Declaration of Helsinki.
High-throughput methylation profiling
ERRBS was performed as previously described (22, 33). Briefly, DNA was isolated using standard phenol-chloroform extraction and digested with MspI, followed by end repair, A-tailing, and ligation of methylated Illumina adapters. Fragment lengths between 250–400bp were gel-isolated, treated with sodium bisulfite, and PCR amplified. Libraries were then sequenced on an Illumina HiSeq2000 per manufacturer’s recommended protocol for 50 bp single-end read runs. Image capture, analysis and base calling was performed using Illumina’s CASAVA 1.7. Alignment was performed using Bowtie version 0.12.7 and Bismark version 0.5.4 (34) after adapter trimming using FAR version 2.15 as part of the Weill Cornell Epigenomics Epigenomics bisseq pipeline (https://pbtech-vc.med.cornell.edu/git/thk2008/bisseq). Raw aligned reads and methylated base calls for CpGs with 10 – 400× coverage were deposited in GEO (GSE86952).
Gene expression analysis
Gene expression analysis for this cohort using Affymetrix Human Genome 133 Plus2.0 GeneChips has been published previously (32) (GEO accession number: GSE6891). For this study, raw data were re-processed using the HGU133plus2.0 BrainArray annotation version 17.0.0 (35), and differential expression was evaluated using limma version 3.22.7. Loci with ≥ 1.5 fold change relative to CD34+ controls and an adjusted P value of ≤ 0.05 were considered significant. Multiple probes mapping to the same locus were summarized using the median probe expression.
Identification and analysis of genetic lesions
Targeted amplicon gene enrichment was performed on all samples and 12 normal controls with a ThunderStorm instrument (RainDance Technologies) using a custom myeloid malignancy gene panel sequenced by the manufacturer. 260-bp paired end read sequencing was performed using Illumina chemistry (MiSeq v3) in 3 batches with 48 samples multiplexed per lane. Demultiplexed reads were mapped to the human reference genome (hs37d5) using BWA MEM. Post-alignment primer trimming was performed using amptools followed by mutation calling using FreeBayes. Variants were annotated using SnpEff. Amplicons displaying a non-zero median allele frequency in controls were dropped, and mutations were called for lesions displaying > 5% variant allele frequencies with > 10 reads representing the variant allele and having > 100× coverage at the locus.
Identification of mutations in NPM1, FLT3, IDH1, IDH2, DNMT3A, CEBPA, and FLT3, overexpression of EVI1, and silencing of CEBPA were performed as described in prior studies (2, 4, 36–41).
To quantify the predictive ability of each mutation relative to translocation data and all other individual mutations, we built multinomial logistic regression models containing (1) each pair of features, (2) the set of translocations alone, and (3) each pair of features adjusted for translocation status. To measure the goodness of fit of each model, we used the Akaike Information Criterion (42). To compare models in (3) to model (2), we calculated the probability of minimizing information loss using exp((AICtranslocation − AICmutation+translocation)/2). This measurement allows the calculation of the probability that the model including mutation data minimizes information loss relative to one containing translocation data alone
Analysis of differential methylation
The base-resolution methylation state of AML and CD34+ control samples relative to the human genome (hg19) was quantified using Bismark (version 0.5.4) after which CpGs with 10–400× coverage were selected. Differential methylation between epigenetic AML clusters and CD34+ controls was identified using a corrected beta-binomial test performed by methylSig (version 0.1.12) (43) with a minimum of 3 CpGs per group required to call differential methylation. Differentially methylated CpGs (DMCs) were called for any CpG with a q-value ≤ 0.01 and absolute methylation change ≥ 25%. Individual CpGs within genes were associated with the overlapping gene(s) or nearest gene, with promoter > exon > intron > neighborhood > intergenic priority using ChIPseeqer 2.1 (44) and a set of custom PERL and Python scripts (see supplementary methods). CpGs falling within the TSS +/− 2kb window were associated with promoters. CpGs falling within the TES +/− 2kb window were labeled as ‘downstream’. CpGs overlapping any annotated exon were associated with exons. CpGs within introns and not associated with either exons or promoters were associated with introns. Exons and introns were also separated into first exon/first intron and other exon/other intron categories. CpGs falling within the region extending from 2kb – 50kb from either the TSS or TES were termed ‘neighborhoods’. Regions ≥ 50kb from the nearest TSS or TES were termed ‘intergenic’. A 1:1 annotation scheme was used in which every CpG was assigned a single genomic compartment.
Unsupervised analysis and generation of clusters
Hierarchical clustering of the samples was performed using a Euclidean distance measurement applied to the subset of CpGs covered in all AML samples with standard deviation values in the top 15% of those surveyed (n=142,643). Dendrograms were constructed using Ward’s method, and clusters were produced by choosing the highest possible cutoff that did not merge well-characterized translocation-based AML samples with other samples. We then labeled the clusters according to their dominant distinct molecular characteristics, requiring that > 50% of samples in a cluster exhibit the cluster-defining feature.
Analysis of CpG predictive ability
Predictive ability of CpGs within a genomic subset was assessed by comparing the clusters produced by CpGs in a given subset to those produced when all CpGs were used. The same 15% most variable CpGs genome-wide were used for each subset and overall. This comparison was quantified using the adjusted rand index (45), and the significance of each annotation’s prediction was compared to 500 random shufflings of the CpGs reported by ERRBS in this study using a Z test.
Feature-centered DMC analysis
Feature-centered DMC enrichment analysis was performed using a custom set of PERL scripts (see supplementary methods). For enhancer and other feature-centered enrichment analyses, a set of virtual bins was created with the central third containing CpGs within the feature and the outer two thirds containing flanking CpGs. For all analyses in this study, 30 bins were used with flanking regions ranging from 50bp to 10kb. Significance testing was performed by comparing the number of DMCs falling within active enhancers to those in the 10kb flanking regions using a Fisher exact test. For analysis of fixed genomic loci such as transcription start sites, an analogous approach was used with the 30 bin window centered on the feature position.
IDH1/2 and DNMT3A cohort analysis
In order to analyze the interaction of IDH and DNMT3A mutations, patients with IDH1, IDH2, or DNMT3A mutations were identified irrespective of their cluster assignment. These patients were divided into groups with an IDH mutation alone, DNMT3A mutation alone, or an IDH mutation co-occurring with a DNMT3A mutation. Gene expression was analyzed as above by comparing each group to CD34+ normal controls using limma. Identification of differentially methylated CpGs was also performed as above by comparing each group to CD34+ controls using methylSig. Gene set enrichment analysis was performed using GSEA version 2.2.2.3 via comparison to the hallmark gene sets deposited in the Molecular Signatures Database version 5.2 (46).
To identify de-novo motifs contained within differentially methylated active enhancers, HOMER version 4.8.3 was used to analyze active enhancers with differentially methylated CpGs. Enrichment was assessed relative to active enhancers with non-differentially methylated CpGs and filtered for motifs containing strong CG signatures. To identify transcription factor binding site enrichment using experimentally generated ChIP-seq data, we used a subset of the ReMap database (47) generated from myeloid-derived cell lines (K562, CD34 derived, SKNO1, monocyte derived, macrophage derived, U937, APL derived, erythroid derived, hematopoietic stem cell derived, Kasumi1, embryonic stem cell derived, AML derived, hematopoietic progenitor derived, AML derived, NB4, THP1). We pooled data by transcription factor, and then assessed for enrichment in differentially methylated active enhancers compared to those with non-differentially methylated CpGs. Enrichment was assessed using Fisher’s exact test.
Murine model of IDH2/DNMT3A antagonism
All mice were housed at the Memorial Sloan Kettering Cancer Center animal facility with all procedures being approved by the Institutional Animal Care and Use Committee. Rosa26:CreERT2 mice were obtained from Jackson labs and were crossed to mice harboring a Cre-activatable knock-in, Idh2R140Q mutation (27) as well as mice harboring a Cre-activatable knock-in Dnmt3aR878H mutation (26). These crosses generated Idh2 single mutants, Dnmt3a single mutants, and Idh2-Dnmt3a double mutants. Bone marrow was isolated from 8–12 week old mice and transplanted into lethally irradiated CD45.1 hosts as previously described (26). Briefly, mice were euthanized and bone marrow was flushed from femurs and tibia followed by red blood cell lysis with ACK (ammonium-chloride-potassium) buffer. 1×106 mononuclear cells were injected via tail vein into lethally irradiated (950 cGγ) CD45.1 hosts. Engraftment was monitored by peripheral blood chimerism of CD45.2 every 4 weeks. After engraftment, mice were administered Tamoxifen via oral gavage (8 mg).
Mice were euthanized at indicated time points for bone marrow isolation from femur and tibia. Cells were ACK-lysed and then lineage depleted (Stem Cell 19756). Cells were incubated for 1 hour at 4°C with the following antibodies (CD34 RAM34 FITC, CD45.2 104 PE, cKIT 2B8 APC, CD16/32 93 AlexFluor700, Sca-1 D7 PE-Cy7, and Lineage cocktail APC-Cy7). Cells were washed with PBS+2%BSA and death was monitored with DAPI. Flow cytometric analysis was performed on a BD LSR Fortessa and sorting was performed on a BD FACS Aria III at the MSKCC Flow Cytometry Core Facility. Single live, Cd45.2+, Lineage−, Sca-1+, cKIT+ (LSK) cells were sorted into lysis buffer from Gentra Pure Blood Kit (Qiagen), and DNA was isolated for enhanced reduced representation bisulfite sequencing (ERRBS). DNA methylation differences were assessed using methylSig as above in which the IDH2, DNMT3A, and IDH2/DNMT3A were compared to normal LSK controls.
In vitro colony forming assays of primary IDH1/DNMT3A AML samples
Human cord blood mononuclear cells (CB-MNCs) and bone marrow mononuclear cells (BM-MNCs) from non-RAS mutant IDH2, DNMT3A, and IDH1/DNMT3A patients in biological duplicate and technical triplicate (8 total) were thawed and were seeded at 2.0×105 cells per well in triplicate into methylcellulose medium (Methocult, H4230, STEMCELL Technologies) with GM-CSF 10ng/mL and DMSO only, 0.1uM MEK162, and 1uM MEK162, obtained from Chemietek (Indianapolis, IN).,. Plates were placed into an incubator at 37°C and 5% CO2 for 14 days after which colony counts were obtained.
Liquid culture of primary IDH/DNMT3A AML samples
Primary human acute myeloid leukemia cells were derived from bone marrow or peripheral blood and blasts and mononuclear cells from three patients with IDH2 mutations without concurrent DNMT3A mutation, three with DNMT3A mutations without concurrent IDH1/2 mutation, two with IDH1/DNMT3A dual mutations, and one with IDH2/DNMT3A dual mutation. Two of the double mutant samples had IDH1 mutations and one had an IDH2 mutation. Cells were purified by Ficoll–Hypaque (Nygaard) centrifugation and cryopreserved. After thawing, cells were left to recover overnight in Iscove’s Modified Dulbecco’s Medium (IMDM)(Life Technologies) supplemented with 20% BIT 9500 serum substitute (Stem Cell Technologies), 16.7 μg/ml human low density lipoproteins (EMD Millipore), 55 μM beta-mercaptoethanol (Gibco) and a cocktail of recombinant human cytokines (G-CSF 20 ng/ml, GM-CSF 20 ng/ml, IL3 20 ng/ml, IL6 20 ng/ml, FLT3 ligand 50 ng/ml and SCF 50 ng/m; (Stem Cell Technologies). The next day, 6×104 cells were plated in 200ul of fresh medium in 96-well plates in the presence of either MEK162 (1 uM), MEK162 (0.1 uM) or DMSO vehicle control, all in technical triplicates. On day 2 and day 4, fresh MEK162 or vehicle was added. On day 6, viability and cell count were assessed by flow cytometry using staining with 7AAD (BD Biosciences). Data were acquired on a FACS Canto flow cytometer (BD Biosciences) and analyzed using FlowJo software v10.0.8 (FlowJo).
Supplementary Material
SIGNIFICANCE.
AML is biologically heterogeneous with subtypes characterized by specific genetic and epigenetic abnormalities. Comprehensive DNA methylation profiling revealed that differential methylation of non-promoter regulatory elements is a driver of epigenetic identity, that gene mutations can be context-dependent, and that co-occurrence of mutations in epigenetic modifiers can result in epigenetic antagonism.
Acknowledgments
We would like to acknowledge the Weill Cornell Medicine Epigenomics Core and the Weill Cornell Medicine Applied Bioinformatics Core, which performed sample processing, ERRBS, and initial bioinformatic sample processing. We would like to thank Caroline Sheridan, Tak Lee, and Yaseswini Neelamraju for technical support.
JG would like to acknowledge Matt Teater, for his advice and feedback during the preparation of this manuscript.
Funding Sources: JLG is funded by T32CA009207, OAG by K99CA178191, AMM is funded by LLS SCOR 7006-13, and R01 CA198089. MEF is funded by 1R01HL126947-01 and LLS QFC-3154-14. RA received funding from the Dutch Cancer Society “koningin Wilhelmina Fonds” and from the Lady Tata Foundation. FEG-B is funded by NCI K08CA169055 and American Society of Hematology ASHAMFDP-20121 award under the ASH-AMFDP partnership with the Robert Wood Johnson Foundation.
Footnotes
Disclosure of potential conflict of interest: No potential conflicts of interest.
Author contributions: JG analyzed the data and wrote the manuscript, DH, RA, TQ analyzed data, FGB contributed to data analysis and contributed to data interpretation, OAG provided data and contributed to data interpretation, BW, HK, RB, SR, MF, and AA performed experiments, JA and LD contributed data, PJMV contributed key specimens and data, MG contributed key specimens, RL contributed to data interpretation, OE oversaw data analysis and contributed to data interpretation, RD, AM and MEF conceived the study, oversaw data analysis, contributed to data interpretation and wrote the manuscript.
References
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullinger L, Ehrich M, Dohner K, Schlenk RF, Dohner H, Nelson MR, et al. Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115:636–42. doi: 10.1182/blood-2009-03-211003. [DOI] [PubMed] [Google Scholar]
- 4.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu Y, Lennartsson A, Gaidzik VI, Deneberg S, Karimi M, Bengtzen S, et al. Differential methylation in CN-AML preferentially targets non-CGI regions and is dictated by DNMT3A mutational status and associated with predominant hypomethylation of HOX genes. Epigenetics. 2014;9:1108–19. doi: 10.4161/epi.29315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer cell. 2016;29:922–34. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature genetics. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer cell. 2014;25:415–27. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avellino R, Havermans M, Erpelinck C, Sanders MA, Hoogenboezem R, van de Werken HJ, et al. An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation. Blood. 2016;127:2991–3003. doi: 10.1182/blood-2016-01-695759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer cell. 2015;27:502–15. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer cell. 2014;25:442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–9. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem. 2012;287:30941–51. doi: 10.1074/jbc.M112.366625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20:1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. The New England journal of medicine. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guryanova OA, Lieu YK, Garrett-Bakelman FE, Spitzer B, Glass JL, Shank K, et al. Dnmt3a regulates myeloproliferation and liver-specific expansion of hematopoietic stem and progenitor cells. Leukemia. 2016;30:1133–42. doi: 10.1038/leu.2015.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS genetics. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nature genetics. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nature medicine. 2016;22:1488–95. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih AH, Meydan C, Shank K, Garrett-Bakelman FE, Ward PS, Intlekofer A, et al. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discovery. 2017 doi: 10.1158/2159-8290.CD-16-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England journal of medicine. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, et al. Epigenetic Perturbations by Arg882-Mutated DNMT3A Potentiate Aberrant Stem Cell Gene-Expression Program and Acute Leukemia Development. Cancer cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–55. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. The New England journal of medicine. 2004;350:1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 32.Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–4. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, et al. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J Vis Exp. 2015:e52246. doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–54. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 37.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, van Oosterhoud S, van Putten WL, Valk PJ, et al. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 38.Abu-Duhier FM, Goodeve AC, Wilson GA, Gari MA, Peake IR, Rees DC, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol. 2000;111:190–5. doi: 10.1046/j.1365-2141.2000.02317.x. [DOI] [PubMed] [Google Scholar]
- 39.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol. 2001;113:983–8. doi: 10.1046/j.1365-2141.2001.02850.x. [DOI] [PubMed] [Google Scholar]
- 40.Lugthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck CA, Valk PJ, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–37. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 41.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–31. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 42.Burnham KP, Anderson DR, Burnham KP. Model selection and multimodel inference: a practical information-theoretic approach. 2. New York: Springer; 2002. [Google Scholar]
- 43.Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30:2414–22. doi: 10.1093/bioinformatics/btu339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubert L, Arabie P. Comparing partitions. Journal of Classification. 1985;2:193–218. [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2015;43:e27. doi: 10.1093/nar/gku1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.