Abstract
This study assesses a novel bile solubility test and MALDI-TOF for the differentiation of Streptococcus pneumoniae from other mitis group streptococci, including differentiation of S. pneumoniae from Streptococcus pseudopneumoniae. Eighty-four species verified mitis group isolates were subjected to our bile solubility test (which measures and calculates the differences of absorbance in the test tube containing 10% sodium deoxycholate versus a blank control tube, after incubation for 10 minutes at 36 °C using a spectrophotometer) and MALDI-TOF MS (both the standard result output and by visual spectra evaluation). Applying a calculated optimal cut-off absorbance-value of 2.1, differentiated S. pneumoniae from all but one other mitis group streptococci (one S. mitis isolate generated an OD-value above 2.1). MALDI-TOF score value identification identified correctly 46 S. pneumoniae and 4 S. pseudopneumoniae but misidentified 16 other mitis group strains. Visual spectra evaluation correctly identified all S. pneumoniae and S. pseudopneumoniae strains but misidentified 13 other mitis group strains. The bile solubility test based on spectrophotometric reading described in this study can differentiate S. pneumoniae from other Streptococcus species. Combining the bile solubility test and the MALDI-TOF spectra results provide a correct identification of all S. pneumoniae and S. pseudopneumoniae isolates.
Introduction
Worldwide, Streptococcus pneumoniae (pneumococci) infections cause high morbidity and mortality among children and elderly1. Invasive pneumococcal disease (IPD) is one of the most frequent types of bacteraemia and meningitis in infants globally as well as in Denmark2, 3. It is therefore essential to be able to identify S. pneumoniae easily and rapidly4, 5.
The detection and differentiation of the mitis group and in particular between the two species S. pneumoniae and Streptococcus pseudopneumoniae can be a very difficult task and often requires several different methods4, 5. Before reaching the point of pneumococcal serotyping identification, it is important to correctly identify S. pneumoniae from other mitis group streptococci. In general, methods such as the bile solubility test, optochin susceptibility, and colony morphology have been the methods of choice for species differentiation5, 6. In recent years, other methods, such as MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) identification based on spectra and molecular methods have been described and used for streptococcal species identification7, 8. However these methods have also been shown to have problems with false identification results9–11.
The bile solubility test is generally considered to be an accurate test for differentiating S. pneumoniae from other mitis group streptococci, including S. pseudopneumoniae 4, 5. The test results from the method is however difficult to interpret, particularly because it is based on a subjective human evaluation5. While the MALDI-TOF procedure is considered a more simple method of identification and is used in many clinical laboratories, correct identification of mitis group streptococci (S. pneumoniae and S. pseudopneumoniae in particular) is still a difficult task11.
In this study we describe a bile solubility test, which is not based on subjective human evaluation. The described bile solubility test uses a densitometer and establishes a standardized bacterial concentration and provides an optical density value (OD-value) for the bile solubility test. Furthermore, this study presents data establishing a cut-off OD-value which can differentiate S. pneumoniae from other mitis group streptococci, including S. pseudopneumoniae. We also evaluate species identification based on both MALDI-TOF score values and MALDI-TOF visual spectra evaluation. Both methods are tested against species verified isolates belonging to the mitis group. Finally we present a recommendation of how to identify/differentiate species within the mitis group in a clinical setting.
Materials and Methods
Clinical isolates
A total of 84 mitis group strains were tested, of which 47 were S. pneumoniae strains (22 serotype positive and 25 confirmed nontypeable pneumococci), nine strains were S. pseudopneumoniae, 20 strains were Streptococcus mitis, two strains were Streptococcus oralis, four strains were Streptococcus sanguinis, and two strains were Streptococcus australis.
Twenty-four strains including the species S. pneumoniae, S. mitis and S. pseudopneumoniae were obtained from the strain collection described by Kilian et al.12.
Fifteen non-capsular pneumococcal strains were obtained from The Centers for Disease Control and Prevention, Atlanta GA, USA. The isolates were identified using the PCR procedure described by Park et al.13.
Thirty strains including the species S. pneumoniae, S. pseudopneumoniae, S. mitis, S. oralis, S. sanguinis, and S. australis were obtained from the NSR laboratory, Statens Serum Institut (SSI). The isolates were identified using multilocus sequence analysis (MLSA)14 and the 16S rRNA-based molecular procedures12, 15.
Thirteen strains were capsular strains (Sanger strains) described by Bentley et al.16, representing the serotypes included in the PCV 13.
Two reference strains, S. pneumoniae ATCC49619 and S. pseudopneumoniae CCUG49455, were also included.
Three S. mitis strains were confirmed to have the lytA gene by molecular sequence procedures described in previous studies12, 15, two strains were from the strain collection described by Kilian et al.12 and one strain was obtained from the NSR laboratory, Statens Serum Institut.
The bile solubility testing
An inoculum was prepared from colonies obtained from overnight incubated blood agar plates (36 °C) in a tube with 2 ml of saline adjusting the cell density to a density of McFarland 4 using a spectrophotometer (Biomerieux, Densimat, Italy). The suspension was then divided equally into two tubes each containing each 1 ml. 200 µl of 10% sodium deoxycholate was added to the test tube and 200 µl of saline was added to the control tube. Both tubes were incubated for 10 minutes at 36 °C (stationary), before the absorbance of both were measured spectrophotometrically. The difference in absorbance of the test tube versus the control tube was calculated as an OD-value. A negative difference between the test tube and the blank control was set to 0.0 OD-value.
Twenty-five isolates consisting of S. pneumoniae (11 strains), S. pseudopneumoniae (2 strains) and S. mitis (12 strains) were tested four times on average, by three different people, over a six-month period.
MALDI-TOF MS identification
MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) (Bruker Daltonics; Compass 1.4, Version 3.4, Build 3.4.76.0) was performed on strains transferred directly from bacterial colonies. Species identification by MALDI-TOF MS was based either on the standard MALDI-TOF score value (Biotyper database version MBT 6903 MSP Library (#1829023)) or on a visual inspection for the presence/absence of peak pairs of m/z 2625, 2911, 2937.5, 5253, 5824, 5877 and 6955 as described by Werno et al.7.
Data analysis
Data were analysed using GraphPad Prism version 5 (GraphPad Software) for descriptive statistical analysis. All calculations of median and confidence interval (CI) were performed using the R version 3.2.4 for Windows (http://www.r-project.org/). All negative calculated OD-values were set to 0.0 OD. The Wilcoxon Rank-Sum (Mann-Whitney U) test in R was used to calculate P-values with C.I. P < 0.05 was considered significant. The Receiver Operating Characteristic (ROC) in R (pROC)17 was used to calculate the ROC values and C.I. values. For the optimal cut-off values were employed “closest.topleft” defined as the optimal threshold point closest to the top-left of the graph showing perfect sensitivity (X-axis) or specificity (Y-axis)17.
Results
Supplementary Tables 1 and 2 present specific and detailed bile and MALDI-TOF data for each tested strain
Depicted in Table 1 are the OD-values obtained from the bile solubility test when applied to the strains included in the study.
Table 1.
Mean OD-values for the species tested in the study. The species annotation of strains was based on MALDI-TOF output and spectra. For detailed results per species, see supplementary Table 1.
| Strain ID | Number of strains | OD Bile Solubility test | Median OD-value with CI |
|---|---|---|---|
| Range | |||
| S. pneumoniae, typeable | 22 | 2.6–3.4 | 3.0 (95% CI: 2.9–3.1) |
| S. pneumoniae, nontypeable | 25 | 2.3–3.3 | 2.9 (95% CI: 2.8–3.1) |
| S. pneumoniae, all isolates | 47 | 2.3–3.4 | 3.0 (95% CI: 2.9–3.1) |
| S. pseudopneumoniae | 9 | 0.3–2.0 | 1.8 (95% CI: 1.0 – 1.8) |
| 1 S. mitis | 20 | 0–2.55 | 0.1 (95% CI: 0.1–0.3) |
| 2Other Streptococcus species | 8 | 0 – 0.3 | 0.1 (95% CI: 0.00–0.3) |
1Three strains were autolysin (lytA) positive.
2Four S. sanguinis, two S. australis, two S. oralis.
The S. pneumoniae bile test showed a median OD-value of 3.0 (95% CI: 2.9–3.1) within the range of 2.6–3.4 with no significant difference (P = 0.28 95% CI: −0.1–0.25) between typeable S. pneumoniae (median OD value = 3.0 (95% CI: 2.9–3.1)) and non-serotypeable S. pneumoniae (median OD value = 2.9 (95% CI: 2.8–3.1)) (Table 1).
S. pseudopneumoniae strains showed a median OD-value 1.8 (95% CI: 1.0–1.8) within the range of 0.3–2.0 (Table 1).
S. mitis showed a median OD-value of 0.1 (95% CI: 0.1–0.3) within the range of 0–2.55. Of the 20 tested S. mitis strains, three isolates showed a high OD-value (above 1.0 OD). These three isolates were all found to contain the autolysin gene. The remaining Streptococcus species (S. oralis, S. sanguinis, S. australis) showed a median OD-value of 0.1 (95% CI: 0.0–0.3) within the range of 0.0–0.3 (Table 1).
Species identification based on MALDI-TOF score values was performed on all strains (Table 2). All pneumococcal isolates with a serotypeable capsule were correctly identified, while one non-capsular isolate was incorrectly identified as S. pseudopneumoniae (Table 2). Based on the score value, four of the nine S. pseudopneumoniae strains were correctly identified as S. pseudopneumoniae, while the remaining five were incorrectly identified as either S. oralis or S. pneumoniae (Table 2). Since we have not evaluated the standard MALDI-TOF database output regarding the remaining species tested in the study, correct species identification cannot be expected, but instead provides suggested annotations which need further confirmation. Of the 20 S. mitis strains, the score value correctly identified 10 of the 20 tested isolates, while the rest were scored as six S. oralis, one S. pseudopneumoniae and three S. pneumoniae. All S. oralis and S. sanguinis strains were correctly identified by the MALDI-TOF score value.
Table 2.
Comparison of species identification by official (Score value) and visual interpretations of MALDI-TOF seven profile peaks, with species annotation based on the m/z values described by Werno et al.7.
| Confirmed ID | m/z value | Test results based on score value | Test results based on visual evaluation value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Profile | 2625 | 2911 | 2937.5 | 5253 | 5824 | 5877 | 6955 | Score tested/correct | Visual tested/correct |
| S. pneumoniae (typeable) | − | − | + | − | − | + | − | 22/22 | 22/22 |
| S. pneumoniae (nontypeable) | − | − | + | − | − | + | − | 25/241 | 25/25 |
| S. pseudopneumoniae | + | − | + | + | − | +/− | − | 9/42 | 9/9 |
| S. mitis | + /− | +/− | +/− | +/− | +/− | +/− | +/− | 20/103 | 20/144 |
| S. oralis | − | + | − | − | + | − | +/− | 2/2 | 2/15 |
| S. australis | − | − | − | − | − | − | − | 2/06 | 2/07 |
| S. sanguinis | − | − | − | +/− | − | − | +/− | 4/4 | 4/08 |
+: only strains with peaks were found.+/−: strains both with and without peaks were detected. −: no strains with peaks were detected.
1One specimen identified as S. pseudopneumoniae.
2Four isolates identified as S. pneumoniae, one ID as S. oralis.
3Six isolates identified as S. oralis, three isolate ID as S. pneumoniae, one isolate ID as S. pseudopneumoniae.
4Three isolates identified as S. pneumoniae, three S. mitis/S. oralis.
5One isolate identified as S. mitis/S. oralis.
6Both isolates identified as S. parasanguinis.
7Both isolates could not be identified.
8All isolates could not be identified.
The species identification based on the MALDI-TOF score value only showed correct species identification if the top ten listed species with a score value above 1 was the same species. If more species were recommended with a score value above 1, misidentification was observed (Table 2, Supplementary Table 1).
Evaluating strain identification based on visual inspection of the seven peaks from the MALDI-TOF spectra7 showed a more precise identification of some of the tested isolates (Table 2). All pneumococcal isolates, both capsular and non-capsular isolates, showed peaks at m/z 2937.5 and m/z 5877 respectively, which are the characteristic peaks for pneumococcal isolates. Of the remaining streptococcal isolates three S. mitis isolates showed only peaks at m/z 2937.5 and m/z 5877 and could therefore not be differentiated from the S. pneumoniae isolates. All nine S. pseudopneumoniae were correctly identified based on the spectra, where eight of the nine isolates showed a peak at m/z 2625, 2937.5, 5253 and 5877, while one isolate did not show a peak at m/z 5877. This peak combination is described as characteristic for S. pseudopneumoniae and was not observed for any other tested isolates (Table 2, Supplementary Table 1).
By combining the bile OD-values and the MALDI-TOF results based on the evaluation of the seven peaks (Table 1, Table 2, Supplementary Table 1), we were able to identify correctly all S. pneumoniae isolates, if the bile OD-value was 2.1 or above and the isolate showed a peak at m/z 2937.5 and m/z 5877. All S. pseudopneumoniae were also correctly identified if the bile OD-value was between 0.9 and 2.1 and the isolate showed peaks at m/z 2625, 2937.5 and 5253.
The remaining tested strains belonging to the mitis group could not be differentiated into species based on evaluating the MALDI-TOF peaks and the bile OD-values without false negative identifications.
Table 3 and Fig. 1 present the optimal cut-off value and the Area Under the Curve (AUC) for differentiating all the tested strains, based on their bile solubility OD-values. Choosing a cut-off OD-value of 2.1 showed an excellent AUC of 100% (95% CI 100–100), which showed no false negative OD-values for nontypeable S. pneumoniae, while one S. mitis strain (OD-value of 2.7) showed OD-values above a cut-off value of 2.1 (Fig. 1). The median bile OD-value for nontypeable S. pneumoniae (median OD value = 2.9) differed significantly from the median bile OD-value for S. pseudopneumoniae (OD value = 1.8) (P = 0.0005, 95% CI 0.8–1.7).
Table 3.
Area under the curve (AUC) and the optimal cut-off values calculated by using “closest topleft”.
| Group 1 (n) | Group 2 (n) | % AUC (95% CI) | Differences of OD-values from the two groups | Optimal cut-off value |
|---|---|---|---|---|
| P value (95% CI) | “closest.topleft” | |||
| S. pneumoniae nontypeable (25) | S. pseudopneumoniae (Highest values) (9) | 100 (100 – 100) | P = 0.00001 (1.1–1.8) | 2.13 (100.0% specificity. 100% sensitivity). |
| S. pseudopneumoniae (9) | S. mitis (20) | 90.83 (79.38 – 100) | P = 0.0005 (0.8–1.7) | 0.9 (66.67% specificity. 65.0% sensitivity). |
| S. pseudopneumoniae (9) | S. mitis (20). S. sanguinis (4). S. australis (2). S. oralis (2) | 92.66 (83.6 – 100) | P = 0.0001 (0.9–1.7) | 0.9 (66.67% specificity. 78.57% sensitivity). |
Figure 1.
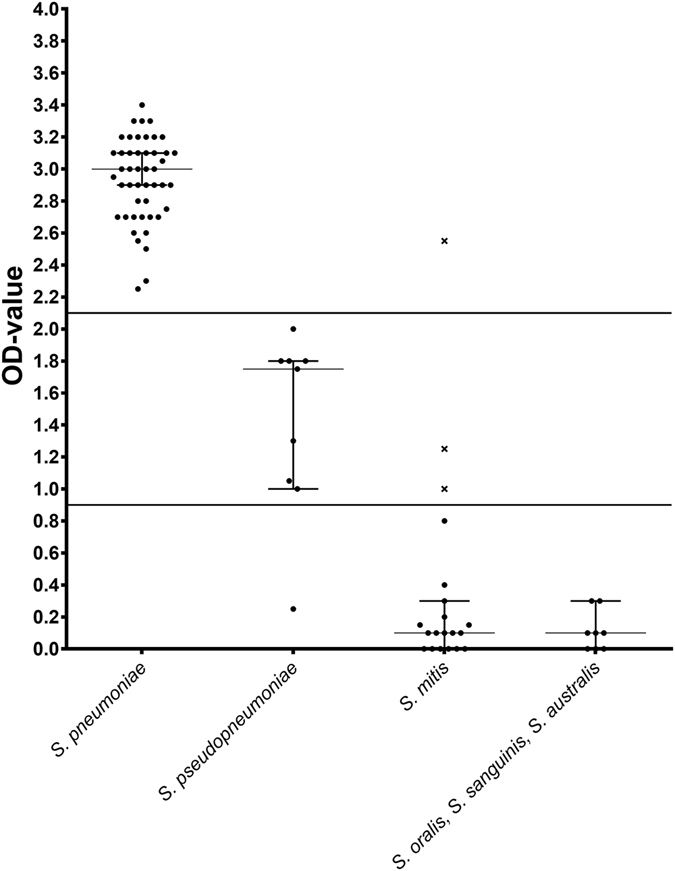
The median OD-value (95% CI) for six Streptococcus species. Lines indicate the calculated cut-off OD-values of 2.1 and 0.9. The three S. mitis strains containing the autolysin (lytA) gene are indicated by an ‘x’.
Choosing a cut-off OD-value of 0.9 for differentiating S. pseudopneumoniae from S. mitis, showed a good AUC of 90.83% (95% CI 79.38–100). The median OD-value for S. pseudopneumoniae (median OD value = 1.8) differed significantly from the median bile OD-value for S. mitis (OD value = 0.1) (P = 0.0005, 95% CI 0.8–1.7).
Choosing a cut-off OD-value of 0.9 for differentiating S. pseudopneumoniae from the other streptococcal species tested (S. oralis, S. sanguinis, S. australis) showed a good AUC of 92.66% (95% CI 83.6–100). The median bile OD-value for S. pseudopneumoniae (OD value = 1.8) differed significantly from the median bile OD-value for other strains (S. oralis, S. sanguinis, S. australis) (OD value = 0.1) (P = 0.0001, 95% CI 0.9–1.7).
Comparing the median bile OD-value for S. mitis (OD value = 0.1) with the combined median bile OD-value for other strains (S. oralis, S. sanguinis, S. australis) (OD value = 0.1) showed no significant difference, P = 0.4 and a CI of (95%, CI −0.1–0.2). The AUC was not calculated.
Table 4 and Supplementary Table 2 summarise the reproducibility of the data from repeated OD values of 25 strains of S. pneumoniae, S. pseudopneumoniae, and S. mitis. The number of tests is limited and therefore a statistical test has not been performed on the results. The isolates were tested for their bile OD-value repeatedly over approximately 6 months by different persons. It was found that the variation of the OD values in general for pneumococci was within the range of 0.6, although one strain showed a range of 1.1. The percentage of coefficient of variation (CV) for the tested S. pneumoniae isolates showed low variation, with the highest % CV found to be 17.74. Two S. pseudopneumoniae strains showed a range of 0.4. The highest % CV for the two strains was 75.9.
Table 4.
Repeated testing of 25 strains over time and performed by different persons. OD-values below 0 are considered as 0.0. For detailed data see supplementary Table 2.
| Number of tests | Total range of test (OD-values) | Median OD-value | % CV | ||
|---|---|---|---|---|---|
| S. pneumoniae | ATCC49619 | 7 | 2.9–3.5 (0.6) | 3.3 | 6.86 |
| S. pneumoniae Non-capsular | Kilian/13725 | 4 | 2.2–3.3 (1.1) | 2.85 | 17.74 |
| S. pneumoniae Non-capsular | Kilian/14860 | 4 | 2.9–3.3 (0.4) | 3.1 | 5.89 |
| S. pneumoniae Non-capsular | Kilian/A39557 | 4 | 2.1–2.5 (0.4) | 2.3 | 7.938 |
| S. pneumoniae Non-capsular | Kilian/A8061 | 4 | 2.4–2.8 (0.4) | 2.6 | 6.28 |
| S. pneumoniae Non-capsular | Kilian/A12931 | 4 | 3–3.6 (0.6) | 3.15 | 8.16 |
| S. pneumoniae Non-capsular | Kilian/A9003 | 4 | 2.9–3.2 (0.3) | 3.0 | 4.16 |
| S. pneumoniae Non-capsular | Kilian/A7890 | 4 | 2.8–3.1 (0.3) | 2.9 | 4.30 |
| S. pneumoniae Non-capsular | Kilian/A2009 | 4 | 2–2.5 (0.5) | 2.2 | 9.97 |
| S. pneumoniae Non-capsular | Kilian/A4708 | 4 | 2.8–3.4 (0.6) | 3.05 | 8.96 |
| S. pneumoniae Non-capsular | Kilian/A39363 | 4 | 2.4–2.9 (0.5) | 2.75 | 8.00 |
| S. pseudopneumoniae | Kilian/SK1516 | 4 | 1.2–1.4 (0.2) | 1.4 | 7.41 |
| S. pseudopneumoniae | Kilian/SK674 | 4 | 0 – 0.4 (0.4) | 0.25 | 75.90 |
| S. mitis | Kilian/SK642 | 4 | 0–0 (0) | 0 | 0 |
| S. mitis | Kilian/SK637 | 4 | 0–1.2 (1.2) | 0.1 | 164.13 |
| S. mitis | Kilian/SK271 | 4 | 0 – 0.2 (0.2) | 0.05 | 127.66 |
| S. mitis | Kilian/SK142 | 4 | 0–0.2 (0.2) | 0 | 200.0 |
| S. mitis (Autolysin gene) | Kilian/SK564 | 4 | 0.4-1.3 (0.9) | 1 | 48.65 |
| S. mitis | Kilian/SK137 | 4 | 0–0.2 (0.2) | 0 | 200.0 |
| S. mitis | Kilian/SK1126 | 4 | 0–0.2 (0.2) | 0 | 200.0 |
| S. mitis (Autolysin gene) | Kilian/SK597 | 4 | 0.2–1.4 (1.2) | 0.7 | 78.88 |
| S. mitis | Kilian/SK608 | 4 | 0–0.3 (0.3) | 0.05 | 141.42 |
| S. mitis | Kilian/SK321 | 4 | 0–0 (0) | 0 | 0 |
| S. mitis | Kilian/SK113 | 4 | 0–0.3 (0.3) | 0.05 | 141.4 |
| S. mitis | Kilian/SK578 | 2 | 0–0 (0) | 0 | 0 |
The remaining strains tested showed large differentiations in the tested OD-values.
Discussion
Since S. pneumoniae can cause severe human infections18, rapid and reliable species identification is essential5, 7, 11. However, differentiating S. pneumoniae from other mitis group streptococci (in particular the relatively newly described S. pseudopneumoniae 19) can be a very difficult task and often requires several different methods4, 5.
Many studies show that identification of S. pneumoniae based on only a single test method will give a bias of either false positive or false negative identifications and thereby a possible overestimation of a species5, 9, 20. Therefore, it is generally recommended to use several different methods for species identification9, 20, with the WHO recommended culture based pneumococcal identification in combination with both the optochin susceptibility and bile solubility test6. Conversely, Yahiaoui et al. recently recommended a new identification protocol, excluding the optochin susceptibility test, and instead recommending only the use of the bile solubility test for species identification21.
In this study we have presented a new objective and optimized the procedure for the bile solubility test, because it is generally considered to be one of the main standard tests for the identification of S. pneumoniae 5, 6. The advantages of using the bile solubility test is that it is a simple and reliable test4, which can be used in low tech laboratory facilities. The disadvantage of the bile solubility test generally used is that the test results are based on a visual interpretation, and the results are therefore imperilled to human subjective evaluation5. By using a densimat to measure both the amount of bacteria used for the bile solubility test and the final OD-value of the performed test, we removed the need for human evaluation and provide results solely based on instrumental measurement. Because the presented bile solubility procedure is based on instrumental measurement and not human evaluation, it was important to evaluate the test against strains verified for their identification. We did not test our bile procedure with the bile test based on visual evaluation, as the test results from a visually based bile test is very difficult to reproduce from one laboratory to another. The bile solubility test data presented in this study show that it is possible to obtain information (species identification as S. pneumoniae/S. pseudopneumoniae/other mitis group streptococci) from the OD-values of streptococcal species. Finding an OD-value of 2.1 or above will indicate that the species is correctly identified as S. pneumoniae, while finding an isolate with an OD-value in the range of 0.9 to 2.1 (Table 3) will suggest that it is an S. pseudopneumoniae. An OD-value below 0.9 will suggest that it is an isolate belonging to either S. mitis, S. oralis, S. sanguinis, or S. australis. A small study (Table 4) furthermore showed that the test was quite robust with only minor time and person variations on the OD-value.
The lytA gene (encoding autolysin) is said to be characteristic of S. pneumoniae and an indication of bile solubility; however, the presence of lytA has also been reported in other mitis group streptococci including S. pseudopneumoniae and S. mitis 9, 19. In this study we found that the three S. mitis isolates showing an OD-value within the range of 1.3 to 2.7 all contained the gene for autolysin (Fig. 1, Supplementary Table 1). Although the data are currently very limited, an unusually high OD-value observed for an S. mitis strain could possibly provide a marker for the presence of the lytA gene.
We also tested all the characterized isolates included in this study using MALDI-TOF. We found that species identification based on MALDI-TOF score value could only be recommended if the top ten species with a score value above 1 were the same species. If more species were recommended with a score value above 1, misidentification was observed. Further improvement of the MALDI-TOF database will improve the species identification and reduce misidentification including those observed in this study11, 22, 23. Instead of using the MALDI-TOF score value and including a visual evaluation of the MALDI-TOF spectra, we found that it was possible to identify both S. pneumoniae and S. pseudopneumoniae correctly by evaluating the seven peaks described by Werno et al.7. We found that three isolates of S. mitis were identified as S. pneumoniae. These three S. mitis strains showed identical peaks, characteristic for S. pneumoniae (Supplementary Table 1). We did not see any false positive identification of S. pseudopneumoniae. Using spectres can therefore be recommended for the identification of particularly S. pseudopneumoniae and S. pneumoniae strains; however, species identification based on the seven peaks is limited as it cannot be used to identify species such as S. sanguinis, where the MALDI-TOF score values correctly identified all four isolates.
Based on the results of this study we recommend a study workflow, utilising both the MALDI-TOF and bile solubility tests as presented in Fig. 2 as a means to test clinical mitis group isolates for correct annotation of S. pneumoniae and S. pseudopneumoniae strains. However, it has to be emphasised that the Fig. 2 flowchart is only based on the isolates tested in this study.
Figure 2.

A description of species identification based on MALDI-TOF and Bile solubility test for clinical isolates from the mitis group.
Species identification of isolates within the mitis group using the flowchart (Fig. 2) will be within the capacity of many clinical laboratories. For many clinical laboratories is it not a possibility to use molecular methods with a high enough specificity to correctly identify mitis group species due to time and expenses11. However it has to be emphasized that if the requirement described in the flowchart cannot be achieved for an isolate, other tests are needed for species identification6. As additional information, it has recently been recommended that the optochin test used by many diagnostic laboratories today as a part of routine identification of S. pneumoniae, is not used for the identification of S. pneumoniae 21. The presented methods can therefore be seen as alternatives to the optochin test.
Finally, it has to be stated by the authors that to obtain a fully correct identification of a streptococcal species within the mitis group, it is recommended to perform a molecular identification as described by Scholz et al.15. This identification procedure using NGS is, however, not within the capacity of many clinical settings.
It is advised that before using the described bile test procedure, it is recommended to evaluate the bile test against your own verified mitis group strains and the bile test procedure performed in the laboratory.
In conclusion this study presents data for a new objective bile solubility test based on instrumental measurement and not human interpretation. We present cut-off OD-values that can be used for differentiating/discriminating S. pneumoniae and S. pseudopneumoniae from other mitis group streptococci. Moreover, we confirmed the valuable use of visual MALDI-TOF spectra evaluation (as presented by Werno et al.7) for the identification of S. pneumoniae and S. pseudopneumoniae. We found that a combination of the identification data from these two tests provided a high rate of accurate identification of S. pneumoniae and S. pseudopneumoniae. We have presented a protocol (Fig. 2) using the combination of the MALDI-TOF test and the bile solubility test, which is very well suited for routine identification in clinical microbiology laboratory settings due to its quick and simple procedure.
Electronic supplementary material
Supplementary table 1, Supplementary table 2
Acknowledgements
Mogens Kilian is acknowledged for letting us use his strains of Streptococcus species and for helping in confirming the species identification of isolates from Statens Serum Institut. Monja Hammer, Karina Jans, and Kirsten Burmeister are acknowledged for their skilled laboratory work and input to this study. We acknowledge the Danish Departments of Clinical Microbiology for submitting invasive pneumococcal isolates for national surveillance throughout the study period.
Author Contributions
H.C.S. designed the study, analysed the data and drafted the manuscript. R.F. and K.F. analysed and reviewed the data, contributed to the manuscript and critically revised the manuscript. All authors have approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07772-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Murray J, et al. Global invasive bacterial vaccine-preventable diseases surveillance–2008-2014. MMWR Morb. Mortal. Wkly. Rep. 2014;63:1159–62. [PMC free article] [PubMed] [Google Scholar]
- 2.Howitz M, Hartvig Christiansen A, Harboe ZB, Mølbak K. Surveillance of bacterial meningitis in children under 2 y of age in Denmark, 1997–2006. Scand. J. Infect. Dis. 2008;40:881–887. doi: 10.1080/00365540802325914. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien KL. PCV13 impact evaluations: the obvious and the unpredicted. Pediatr. Infect. Dis. J. 2013;32:264–265. doi: 10.1097/INF.0b013e3182787f89. [DOI] [PubMed] [Google Scholar]
- 4.Dubois D, Segonds C, Prere MF, Marty N, Oswald E. Identification of clinical Streptococcus pneumoniae isolates among other alpha and nonhemolytic streptococci by use of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J. Clin. Microbiol. 2013;51:1861–1867. doi: 10.1128/JCM.03069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wessels E, Schelfaut JJ, Bernards AT, Claas EC. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J. Clin. Microbiol. 2012;50:1171–1177. doi: 10.1128/JCM.06609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satzke C, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2014;32:165–179. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 7.Werno AM, Christner M, Anderson TP, Murdoch DR. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012;50:2863–2867. doi: 10.1128/JCM.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satzke, C. et al. A Multi-Centre Comparative Study to Identify the Best Serotyping Methods for Examining Pneumococcal Carriage in Vaccine Evaluation Studies. PLoS. Med. 12, 10.1371/journal.pmed.1001903 (2015). [DOI] [PMC free article] [PubMed]
- 9.Simões Alexandra S, et al. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 2016;85:141–148. doi: 10.1016/j.diagmicrobio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Jauneikaite, E. et al. Current methods for capsular typing of Streptococcus pneumoniae. J. Microbiol. Methods. 113, 41–49. Review (2015). [DOI] [PubMed]
- 11.Harju I, et al. Improved Differentiation of Streptococcus pneumoniae and Other S. mitis group Streptococci by MALDI Biotyper Using an Improved MALDI Biotyper Database Content and a Novel Result Interpretation Algorithm. J. Clin. Microbiol. 2017;55:914–922. doi: 10.1128/JCM.01990-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilian M, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS. ONE. 2008;3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park IH, Geno KA, Sherwood LK, Nahm MH, Beall B. Population-based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes. PLoS. One. 2014;15:e97825. doi: 10.1371/journal.pone.0097825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop, C.J. et al. Assigning strains to bacterial species via the internet. BMC. Biol. 26, 10.1186/1741-7007-7-3 (2009). [DOI] [PMC free article] [PubMed]
- 15.Scholz CF, Poulsen K, Kilian M. Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence-based microbiome studies. J. Clin. Microbiol. 2012;50:1968–73. doi: 10.1128/JCM.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley, S. D. et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS. Genet. 2, 10.1371/journal.pgen.0020031 (2006). [DOI] [PMC free article] [PubMed]
- 17.Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC. Bioinformatics. 12, 10.1186/1471-2105-12-77 (2011). [DOI] [PMC free article] [PubMed]
- 18.Slotved HC, Dalby T, Hoffmann S. The effect of pneumococcal conjugate vaccines on the incidence of invasive pneumococcal disease caused by ten non-vaccine serotypes in Denmark. Vaccine. 2016;34:769–774. doi: 10.1016/j.vaccine.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 19.Arbique JC, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 2004;42:4686–4696. doi: 10.1128/JCM.42.10.4686-4696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter SS, et al. Accuracy of phenotypic methods for identification of Streptococcus pneumoniae isolates included in surveillance programs. J. Clin. Microbiol. 2008;46:2184–2188. doi: 10.1128/JCM.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yahiaoui RY, den Heijer CD, Wolfs P, Bruggeman CA, Stobberingh EE. Evaluation of phenotypic and molecular methods for identification of Streptococcus pneumoniae. Future Microbiol. 2016;11:43–50. doi: 10.2217/fmb.15.124. [DOI] [PubMed] [Google Scholar]
- 22.van Prehn J, van Veen SQ, Schelfaut JJ, Wessels E. MALDI-TOF mass spectrometry for differentiation between Streptococcus pneumoniae and Streptococcus pseudopneumoniae. Diagn. Microbiol. Infect. Dis. 2016;85:9–11. doi: 10.1016/j.diagmicrobio.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Angeletti S, et al. Viridans Group Streptococci clinical isolates: MALDI-TOF mass spectrometry versus gene sequence-based identification. PLoS. One. 2015;10:e0120502. doi: 10.1371/journal.pone.0120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1, Supplementary table 2


