ABSTRACT
Antibiotic resistance has emerged globally as one of the biggest threats to human and animal health. Although the excessive use of antibiotics is recognized as accelerating the selection for resistance, there is a growing body of evidence suggesting that natural environments are “hot spots” for the development of both ancient and contemporary resistance mechanisms. Given that pharmaceuticals can be entrained onto agricultural land through anthropogenic activities, this could be a potential driver for the emergence and dissemination of resistance in soil bacteria. Using functional metagenomics, we interrogated the “resistome” of bacterial communities found in a collection of Canadian agricultural soil, some of which had been receiving antibiotics widely used in human medicine (macrolides) or food animal production (sulfamethazine, chlortetracycline, and tylosin) for up to 16 years. Of the 34 new antibiotic resistance genes (ARGs) recovered, the majority were predicted to encode (multi)drug efflux systems, while a few share little to no homology with established resistance determinants. We characterized several novel gene products, including putative enzymes that can confer high-level resistance against aminoglycosides, sulfonamides, and broad range of beta-lactams, with respect to their resistance mechanisms and clinical significance. By coupling high-resolution proteomics analysis with functional metagenomics, we discovered an unusual peptide, PPPAZI 4, encoded within an alternative open reading frame not predicted by bioinformatics tools. Expression of the proline-rich PPPAZI 4 can promote resistance against different macrolides but not other ribosome-targeting antibiotics, implicating a new macrolide-specific resistance mechanism that could be fundamentally linked to the evolutionary design of this peptide.
IMPORTANCE Antibiotic resistance is a clinical phenomenon with an evolutionary link to the microbial pangenome. Genes and protogenes encoding specialized and potential resistance mechanisms are abundant in natural environments, but understanding of their identity and genomic context remains limited. Our discovery of several previously unknown antibiotic resistance genes from uncultured soil microorganisms indicates that soil is a significant reservoir of resistance determinants, which, once acquired and “repurposed” by pathogenic bacteria, can have serious impacts on therapeutic outcomes. This study provides valuable insights into the diversity and identity of resistance within the soil microbiome. The finding of a novel peptide-mediated resistance mechanism involving an unpredicted gene product also highlights the usefulness of integrating proteomics analysis into metagenomics-driven gene discovery.
KEYWORDS: agriculture, antibiotic resistance, biosolids, functional metagenomics, manure, soil microbiology
INTRODUCTION
Antimicrobial resistance is a serious threat to human and animal health on a global scale, with bacteria now resistant to the “last-resort” antibiotics, including carbapenems and polymyxins (1–4). It is estimated that without any international-level efforts to revert our current situation, antibiotic resistance would kill an additional 10 million people and cost the global economy 100 trillion U.S. dollars by 2050 (5). To safeguard the future of medicines required for treating otherwise fatal infections, there is an urgent need to advance our understanding of the many mechanisms that are exploited by bacteria to promote resistance.
The ancestral origin of clinical resistance is largely unknown and remains an subject that is elusive to both the medical and research communities. In the perspective of evolutionary genetics, bacteria are capable of adapting to stressful conditions through chromosomal mutations and/or acquisition of genetic materials that can provide them with survival advantages. As such, exposure to antibiotics is a key evolutionary driver for the development of various resistance mechanisms, including, but not limited to, enzymatic drug modification, alteration of drug targets, reduced cell membrane permeability, and transporters-mediated drug efflux (6). These resistance mechanisms are typically encoded by the so-called antibiotic/antimicrobial resistance genes (ARGs) and can be disseminated among microorganisms through horizontal gene transfer. The use of antibiotics will accelerate the selection for ARGs-carrying bacterial pathogens in clinical settings (7). Likewise, anthropogenic inputs of antibiotics into the environment through wastewater effluent, agricultural use of manure and biosolids, and aquaculture will increase the diversity and size of the environmental ARG reservoir, contributing to the spread of antibiotic resistance (8–10).
Antibiotic resistance is ancient and prevalent in the natural environment, and soil is a significant reservoir of ARGs (11, 12). Soil is a very complex and dynamic environmental matrix, typically containing billions of microorganisms and thousands of bacterial species in each kilogram (13). With such microbial diversity and richness, it is not surprising that soil harbors a large diversity of ARGs (14). In fact, recent studies analyzing soil samples collected from sites geographically or temporally distant from human activities recovered not only genetic material orthologous to known ARGs found in contemporary pathogenic bacteria but also novel resistance mechanisms that have potential clinical implications (15–17).
In the present study, we sought to explore the ARG content of agricultural soil from an experimental farm located in London, Canada, by using functional metagenomics. Soil samples collected from antibiotic-amended and control field plots (18, 19) were mined for ARGs that can promote resistance against clinically important antimicrobial agents. We report here the identification of 34 new ARGs and the functional characterization of some of these resistance determinants. We uncovered several previously unknown resistance genes, notably, one that encodes a novel peptide-associated macrolide resistance mechanism.
RESULTS
To explore the content of antibiotic resistance genes (ARGs) borne by soil microorganisms, three metagenomic fosmid libraries were constructed by cloning soil microbial DNA into Escherichia coli (Table 1). These libraries encompassed over 36 Gb of metagenomic DNA, equivalent to the combined size of about 7,200 E. coli genomes, assuming an average genome size of 5 Mb. Phenotypic selection for resistance against 12 antimicrobial agents belonging to six different antibiotic classes recovered ca. 250 fosmid clones from these metagenomic libraries. After restriction pattern analyses were performed to reduce clonality among candidates selected by the same antibiotic, retransformation confirmed at least 44 individual fosmids that are capable of conferring antibiotic resistance. High-throughput fosmid sequencing also revealed selection redundancy of using different and (un)related antibiotics, implying that some of the recovered fosmids can promote multidrug resistance. Of the 31 nonredundant fosmids that can confer resistance, 28 had their metagenomic DNA insert sequence resolved to completion (ranging from 24.2 to 43.9 kb in size), whereas the remaining three were partially assembled into separated contigs (8.8 to 26.2 kb) (see Data Set S1 in the supplemental material).
TABLE 1.
Metagenomic fosmid libraries constructed using DNA extracted from antibiotic-exposed and control field plot soils
| Library name | No. of annual applications (expt period) | Antibiotic composition (application concn,a mg kg−1 soil) | No. of clones | Avg insert size (kb) | Cloned bases (Mb) |
|---|---|---|---|---|---|
| UTC | 0 (2010–2014) | None | 111,000 | 28 | 3,100 |
| MACRO | 5 (2010–2014) | Erythromycin (10), azithromycin (10), clarithromycin (10) | 593,000 | 30 | 17,900 |
| SCT | 16 (1999–2014) | Sulfamethazine (1, 10), chlortetracycline (1, 10), tylosin (1, 10) | 488,000 | 31 | 15,100 |
| Total | 1,192,000 | 36,100 |
For the SCT treatment, the annual application concentration for each drug was increased from 1 to 10 mg kg−1 soil since 2005.
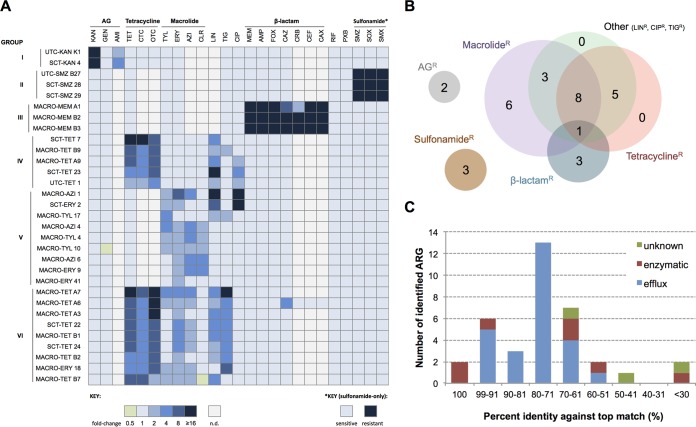
Based on the retransformant's antibiotic susceptibility profiles (Fig. 1A; see Table S1 in the supplemental material), the fosmid-conferred resistance against aminoglycosides, sulfonamides, and most of the observed β-lactams was not associated with co- or cross-resistance against other classes of antibiotics (Fig. 1B). On the contrary, all tetracycline-resistant fosmid clones displayed reduced susceptibility toward at least one additional class of antibiotic, including macrolides (Fig. 1B). Among the nine clones that exhibited a classic multidrug resistance phenotype (group VI), resistance against a third-generation cephalosporin, ceftazidime (CAZ), was detected only in the MACRO-TET A6 clone (Fig. 1A). Macrolide-specific resistance was observed in six fosmid clones, with MACRO-TYL 10 also demonstrating mildly enhanced sensitivity toward an aminoglycoside (Fig. 1A).
FIG 1.
Antibiotic susceptibility profiles and resistance phenotypes of selected soil metagenomic fosmid clones and distributions of their associated antibiotic resistance genes (ARGs). (A) The MICs of antibiotics (except sulfonamides) for 31 nonredundant metagenomic fosmid clones were determined using the broth microdilution method and are presented as fold change relative to those for a control strain. Light gray indicates that the MIC not determined (n.d.). For detecting sulfonamide resistance, serial dilutions of culture were spotted onto agar plates supplemented with the appropriate sulfonamide antibiotic. (B) Resistance phenotypes of the soil metagenomic clones. Each number represents the number of unique clones that displayed resistance against antibiotics belonging to the labeled drug classes. (C) Distribution of putative ARGs based on their predicted resistance mechanism and sequence homology against best matches found by blastX.
Using random transposon mutagenesis, 36 putative ARGs were identified from the resistance-associated fosmids (Table 2). Functional annotations suggested that the majority of the identified ARGs encode efflux-mediated mechanisms, while two known and five presumed enzymatically based resistance determinants were also uncovered (Fig. 1C). Additional open reading frame (ORFs) that showed no apparent linkage with any prominent resistance mechanisms were affiliated with resistance against macrolides (Table 2). To ascertain their ARG identity and to gain insight into the underlying resistance principles encoded by some of these soil-derived resistance determinants, a subcloning strategy was employed with the corresponding characterization findings presented in the following sections.
TABLE 2.
Putative ARGs identified from resistant metagenomic fosmid clones
| Group and fosmid | ORF no. | Description of best blastX match (bacterial species or strain) | Predicted mechanism | Sequence identity (%) | Sequence coverage (%) | Accession no. |
|---|---|---|---|---|---|---|
| I | ||||||
| UTC-KAN K1 | B5 | APH(3′) family aminoglycoside O-phosphotransferase (Bacteria, multispecies) | Enzymatic | 100 | 99 | WP_000338412.1 |
| SCT-KAN 4 | 7 | Aminoglycoside N(6′)-acetyltransferase (Chloroflexi bacterium OLB14) | Enzymatic | 57 | 99 | KXK13532.1 |
| II | ||||||
| UTC-SMZ B27 | 20 | Dihydropteroate synthase (“Candidatus Peribacteria” bacterium GW2011_GWB1_54_5) | Enzymatic | 62 | 95 | KKW39307.1 |
| SCT-SMZ 28 | 17 | Sulfonamide-resistant dihydropteroate synthase Sul2 (Bacteria, multispecies) | Enzymatic | 100 | 99 | WP_001043260.1 |
| SCT-SMZ 29 | A6 | Sulfonamide-resistant dihydropteroate synthase Sul2 (Bacteria, multispecies) | Enzymatic | 100 | 99 | WP_001043260.1 |
| III | ||||||
| MACRO-MEM A1 | 27 | Subclass B3 metallo-beta-lactamase (Duganella zoogloeoides) | Enzymatic | 69 | 89 | WP_019923984.1 |
| MACRO-MEM B2 | 21 | Beta-lactamase class B (bacterium enrichment culture clone cep-06) | Enzymatic | 98 | 99 | AIQ86556.1 |
| MACRO-MEM B3 | 5 | Beta-lactamase class B (bacterium enrichment culture clone cep-06) | Enzymatic | 98 | 99 | AIQ86556.1 |
| IV | ||||||
| SCT-TET 7 | 6 | Bcr/CflA family drug resistance efflux transporter (Rhizobium sp. strain YS-1r) | Efflux | 95 | 97 | WP_037146735.1 |
| MACRO-TET B9 | 32 | Transmembrane secretion effector (uncultured bacterium) | Efflux | 70 | 97 | AIA12822.1 |
| MACRO-TET A9 | 2 | Transmembrane secretion effector (uncultured bacterium) | Efflux | 79 | 99 | AIA15249.1 |
| SCT-TET 23 | 25 | Drug resistance transporter, EmrB/QacA subfamily (“Candidatus Saccharibacteria” bacterium GW2011_GWC2_48_9) | Efflux | 52 | 97 | KKW00845.1 |
| UTC-TET 1 | 29 | Drug resistance transporter EmrB/QacA subfamily (Ktedonobacter racemifer) | Efflux | 64 | 96 | WP_007922578.1 |
| V | ||||||
| MACRO-AZI 1 | B14 | MFS antibiotic efflux pump (uncultured bacterium) | Efflux | 71 | 94 | AIA18803.1 |
| SCT-ERY 2 | 14 | MFS antibiotic efflux pump (uncultured bacterium) | Efflux | 76 | 99 | AIA18803.1 |
| MACRO-TYL 17 | 12 | MATE family efflux transporter (Thermobaculum terrenum) | Efflux | 61 | 97 | WP_012876167.1 |
| MACRO-AZI 4 | 28 | None found | —c | — | — | — |
| ppp AZI 4a | None found | Unknown | — | — | — | |
| MACRO-TYL 4 | 18 | 30S ribosomal protein S21 (“Candidatus Uhrbacteria” bacterium GW2011_GWA2_53_10) | Unknown | 49 | 89 | KKW32987.1 |
| MACRO-TYL 10 | 16 | GTPase HflX (Simkania negevensis) | Unknown | 64 | 96 | WP_013943053.1 |
| MACRO-AZI 6 | 2 | RND transporter (Acidovorax sp. strain Root275) | Efflux | 91 | 99 | WP_057227541.1 |
| 3 | RND transporter (Acidovorax sp. strain Root275) | Efflux | 98 | 98 | WP_057227539.1 | |
| MACRO-ERY 41 | 17 | Major facilitator superfamily (uncultured bacterium) | Efflux | 66 | 93 | AIA17006.1 |
| VI | ||||||
| MACRO-TET A7 | 45 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 83 | 99 | AIA14488.1 |
| 46 | ABC-2-type transporter (uncultured bacterium) | Efflux | 86 | 98 | AIA14415.1 | |
| MACRO-TET A6 | 11 | Penicillin-binding protein (Streptomyces avermitilis) | Enzymatic | 29 | 79 | WP_037649119.1 |
| 12 | ABC-2-type transporter (uncultured bacterium) | Efflux | 96 | 99 | AIA10928.1 | |
| 13b | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 96 | 95 | AIA10927.1 | |
| MACRO-TET A3 | 7 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 82 | 96 | AIA14162.1 |
| 8b | ABC-2-type transporter (uncultured bacterium) | Efflux | 77 | 98 | AIA14382.1 | |
| SCT-TET 22 | 4 | ABC-2-type transporter (uncultured bacterium) | Efflux | 76 | 99 | AIA14382.1 |
| 5 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 78 | 95 | AIA15370.1 | |
| MACRO-TET B1 | 12b | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 80 | 99 | AIA14577.1 |
| 13 | ABC-2-type transporter (uncultured bacterium) | Efflux | 80 | 99 | AIA15441.1 | |
| SCT-TET 24 | 23 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 80 | 99 | AIA14577.1 |
| 24 | ABC-2-type transporter (uncultured bacterium) | Efflux | 80 | 99 | AIA15441.1 | |
| MACRO-TET B2 | 32 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 76 | 99 | AIA14488.1 |
| 33 | ABC-2-type transporter (uncultured bacterium) | Efflux | 73 | 98 | AIA14382.1 | |
| MACRO-ERY 18 | 17 | ABC-2-type transporter (uncultured bacterium) | Efflux | 77 | 99 | AIA13096.1 |
| 16 | Daunorubicin resistance ABC transporter, ATP-binding protein (uncultured bacterium) | Efflux | 79 | 96 | AIA13097.1 | |
| MACRO-TET B7 | 12 | MATE efflux family protein (uncultured bacterium) | Efflux | 79 | 99 | AIA12080.1 |
An alternative, oppositely oriented ORF present within ORF28 of MACRO-AZI 4.
ORF not directly specified by transposon mutagenesis but found within the same transposon-disrupted operon.
—, not available.
Efflux-mediated resistance.
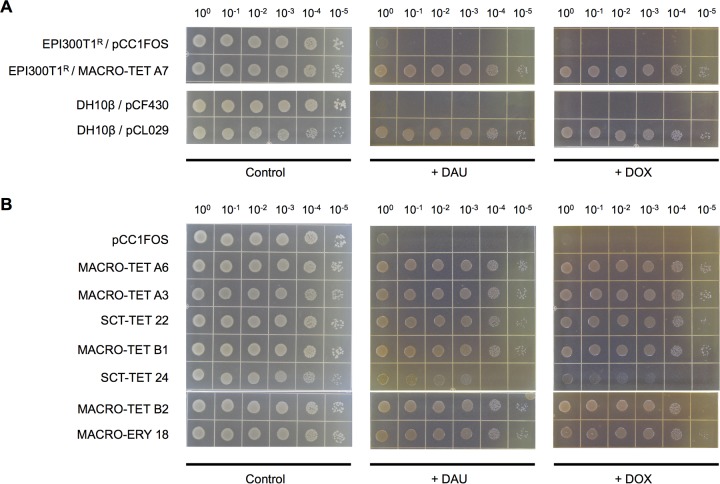
Individual versus combinational subcloning of two putative ATP-binding cassette (ABC) transporter-encoding ORFs identified from fosmid MACRO-TET A7 (i.e., orf-45TET A7 and orf-46TET A7) revealed a corequisite of these two ARGs for conferring multidrug resistance (Table 3). The restored antibiotic sensitivity of transposon mutants that had either one of the two transporter-encoding ARGs inactivated supports the notion that these two ARGs constitute a single functional efflux system. Susceptibility testing against the anticancer drugs daunorubicin (DAU) and doxorubicin (DOX) further demonstrated the capability of this ABC multidrug efflux pump to protect the bacterial host against the anthracycline class of antibiotics (Fig. 2A). It was discovered that homologues of this two-component efflux system of MACRO-TET A7 were present in seven other multidrug-resistant fosmid clones (Table 2), all of which displayed similar reductions in susceptibility toward both DAU and DOX (Fig. 2B).
TABLE 3.
Corequirement of two ABC transporter-encoding genes identified in fosmid MACRO-TET A7 for multidrug resistance
| E. coli strain | Vector | Genotypea |
MIC (μg/ml)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| orf-45TET A7 | orf-46TET A7 | TET | CTC | OTC | TYL | ERY | AZI | LIN | TIG | ||
| DH10β | pCF430 | − | − | ND | ND | ND | 2,048 | 512 | 32 | 128 | 4 |
| pCL027 | + | − | ND | ND | ND | 2,048 | 512 | 32 | 128 | 4 | |
| pCL028 | − | + | ND | ND | ND | 2,048 | 512 | 32 | 128 | 4 | |
| pCL029 | + | + | ND | ND | ND | 8,192 | 2,048 | 128 | 256 | 16 | |
| EPI300T1R | pCC1FOS | − | − | 4 | 64 | 4 | 2,048 | 256 | 16 | 256 | 1 |
| MACRO-TET A7 | + | + | 64 | 512 | 64 | 8,192 | 1,024 | 64 | 1,024 | 16 | |
| MACRO-TET A7/Δorf-45::Tn | − | + | 4 | 64 | 4 | 2,048 | 256 | 16 | 256 | 1 | |
| MACRO-TET A7/Δorf-46::Tn | + | − | 4 | 64 | 4 | 2,048 | 256 | 16 | 256 | 1 | |
+, present; −, absent or transposon disrupted.
ND, not determined. MIC values higher than that for the corresponding empty-vector control are in bold. AZI, azithromycin; CTC, chlortetracycline; ERY, erythromycin; LIN, lincomycin; OTC, oxytetracycline; TET, tetracycline; TIG, tigecycline; TYL, tylosin.
FIG 2.
Resistance against the anticancer drugs daunorubicin (DAU) and doxorubicin (DOX) mediated by soil metagenome-derived multidrug ABC transporter genes in E. coli. Five microliters of serially diluted overnight E. coli cultures with the starting OD600 normalized to 0.5 was spotted onto LB agar plates supplemented with 100 μg/ml of DAU (+ DAU) or DOX (+ DOX). LB agar plates with no anticancer drug (Control) were also included. (A) Strains carrying fosmid MACRO-TET A7, pCF430 derivatives containing the ABC transporter-encoding orf-45TET A7 and orf-46TET A7 (pCL029), or the appropriate empty-vector controls. (B) EPI300T1R strains with fosmid carrying close homologues of the multidrug ABC transporter genes identified in MACRO-TET A7.
Enzymatic resistance mechanisms: drug or target modifications.
ARGs identical to the well-characterized resistance determinants aph-3′, which encodes an aminoglycoside O-phosphotransferase (20), and sul2, which encodes a type II sulfonamide-resistant variant of dihydropteroate synthase (DHPS) (also known as FolP) (21), were uncovered (Table 2). Both ARGs were identified within genomic neighborhoods where elements implicated in lateral gene transfer (e.g., insertion sequence IS903, transposase, integrase, and conjugative transfer protein) and homologous recombination (e.g., DNA helicases and restriction endonuclease) were in close proximity (see Data Set S1 and Table S2 in the supplemental material).
Expression of orf-7KAN 4 derived from the fosmid clone SCT-KAN 4 was confirmed to promote resistance against three clinically significant aminoglycosides not only in E. coli but also in the opportunistic pathogen Pseudomonas aeruginosa (Table 4). Given its sequence similarity (∼60% shared identity) to an aminoglycoside 6′-N-acetyltransferase (Table 2), these results suggest that orf-7KAN 4 encodes a potentially new member of this aminoglycoside-modifying enzyme family, which confers antibiotic resistance through drug inactivation.
TABLE 4.
New aminoglycoside resistance determinant, orf-7KAN 4, found in fosmid SCT-KAN 4 can confer antibiotic resistance to both E. coli and P. aeruginosa
| Species and strain | Vector | MIC (μg/ml)a |
||
|---|---|---|---|---|
| GEN | AMI | KAN | ||
| E. coli DH10β | pCF430 | 2 | 4 | 4 |
| pCL030 (pCF430::orf-7KAN 4) | 4 | 16 | 64 | |
| P. aeruginosa PAO1 | pUCP19 | 2 | 4 | 256 |
| pCL031 (pUCP19::orf-7KAN 4) | 16 | 256 | 2,048 | |
MIC values higher than that for the corresponding empty-vector control are in bold. AMI, amikacin; GEN, gentamicin; KAN, kanamycin.
The broad-spectrum β-lactam resistance determinants recovered from the meropenem-selected fosmid clones (Fig. 1A, group III) were both inhibited by EDTA but not by the classic serine-type β-lactamase inhibitor clavulanic acid (Table 5), in agreement with their predicted functional entities as metallo-β-lactamases (Table 2). The extensive β-lactam resistance phenotype was also recapitulated in E. coli and P. aeruginosa strains carrying the subcloned orf-27MEM A1, which was found proximal to additional β-lactamase-related ARG-like genetic elements in MACRO-MEM A1 (Data Set S1 and Table S2).
TABLE 5.
Soil metagenome-derived metallo-β-lactamases conferring broad-spectrum β-lactam resistance
| Species and strain | Vector | MIC (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CRB | FOX | CAZ | CEF | CAX | MEM | MEM + CLVb | MEM + EDTAc | ||
| E. coli | ||||||||||
| EPI300T1R | pCC1FOS | 8 | 16 | 16 | 2 | 0.5 | 0.125 | 0.03 | 0.03 | 0.03 |
| MACRO-MEM A1 | 256 | 32 | 256 | 16 | 128 | 64 | 32 | 32 | 0.03 | |
| MACRO-MEM B2 | >1,024 | >2,048 | 256 | 64 | 16 | 8 | 16 | 16 | 0.03 | |
| DH10β | pCF430 | 2 | 8 | 16 | 2 | 0.5 | 0.125 | 0.015 | ND | ND |
| pCL032 (pCF430::orf-27MEM A1) | 64 | 16 | 256 | 16 | 32 | 16 | 8 | ND | ND | |
| P. aeruginosa PAO1 | pRK415 | 256 | 32 | 1,024 | 4 | 64 | 8 | 0.25 | ND | ND |
| pCL033 (pRK415::orf-27MEM A1) | 2,048 | 32 | 2,048 | 32 | 512 | 256 | 256 | ND | ND | |
MIC values higher than that for the corresponding empty-vector control are in bold. AMP, ampicillin; CAX, cefotaxime; CAZ, ceftazidime; CEF, ceftiofur; CLV, clavulanic acid; CRB, carbenicillin; FOX, cefoxitin; MEM, meropenem. ND, not determined.
CLV was included at 4 μg/ml.
EDTA was included at 5 mM.
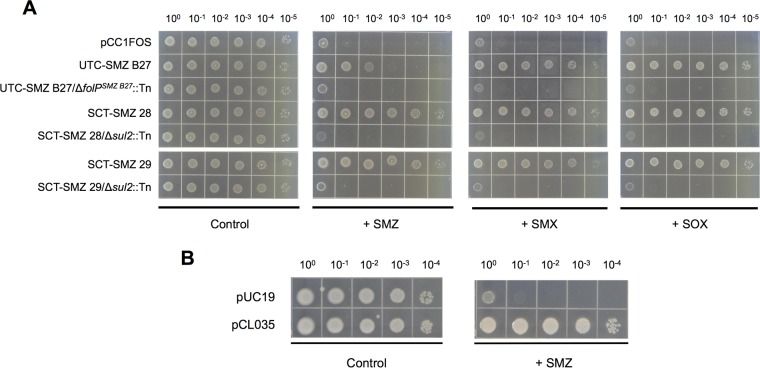
A novel sulfonamide resistance gene, here designated folPSMZ B27, was discovered from the fosmid clone UTC-SMZ B27. FolPSMZ B27 shares 35% sequence identity with the E. coli DHPS (22), but, reminiscent of the case for the two sul2-carrying fosmid clones, folPSMZ B27-expressing E. coli cells were recalcitrant to the bacteriostatic effect of sulfonamide antibiotics (Fig. 3), which is mediated through the inhibition of dihydropteroate synthase (DHPS)-dependent folate biosynthesis. Based on its ability to confer growth competence to a thymidine-auxotrophic folP-knockout E. coli strain (23) under a thymidine-deficient condition (Fig. 4B and C), it was verified that folPSMZ B27 encodes a functional DHPS. The ability to withstand the antagonistic effect of sulfonamides specific to DHPSs (Fig. 4D and E) also implies that FolPSMZ B27 represents an insensitive variant of these enzymes.
FIG 3.
Resistance against sulfonamide antibiotics mediated by soil metagenome-derived ARGs folPSMZ B27 and sul2. Five microliters of serially diluted overnight E. coli EPI300T1R (A) or DH10β (B) cultures with starting OD600 normalized to 0.5 were spotted onto Iso-Sensitest agar plates supplemented with 2,000 μg/ml of sulfamethazine (+ SMZ), 1,000 μg/ml of sulfamethoxazole (+ SMX), or 1,000 μg/ml of sulfisoxazole (+ SOX). Iso-Sensitest agar plates with no sulfonamide added (Control) were also included. (A) pCC1FOS-derived metagenomic fosmid clones and their respective transposon insertion mutants. (B) E. coli strains carrying pUC19 or its folPSMZ B27-expressing derivative pCL035.
FIG 4.
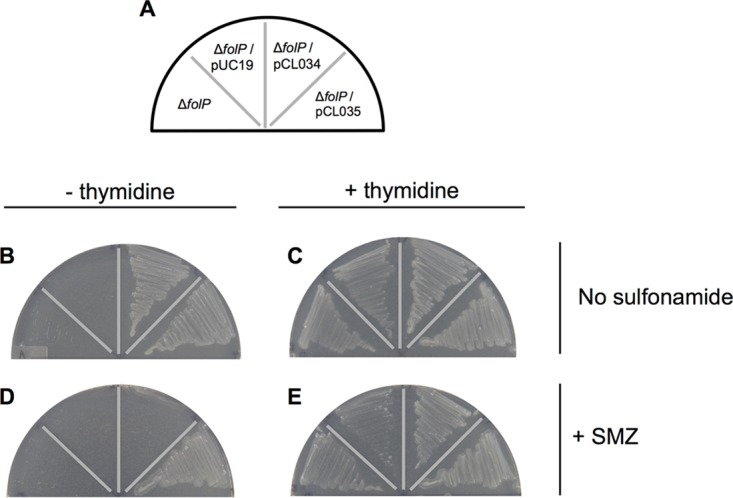
Growth complementation and sulfonamide tolerance conferred by folPSMZ B27 to a thymidine-auxotrophic folP-knockout E. coli strain. Single colonies of the C600 ΔfolP::Kmr strain (ΔfolP) and its derivatives transformed with pUC19, the prototypic folPE. coli-expressing pCL034, and the folPSMZ B27-expressing pCL035 were streaked according to the plate map shown (A) onto Iso-Sensitest agar without (B) or with (C) the supplement of 200 μg/ml of thymidine. For assessing sulfonamide susceptibility, 2,000 μg/ml of sulfamethazine (+ SMZ) was included in separated thymidine-deficient (D) and thymidine-amended (E) plates.
By selecting for MACRO-TET A6-derived transposon mutants with restored CAZ sensitivity, a new cephalosporin resistance determinant, ORF11 (here designated pbpTET A6), was identified adjacent to an ABC multidrug transporter-encoding operon (i.e., ORF12 and -13) (Table 2). PBPTET A6 consists of putative N-terminal transglycosylase and C-terminal transpeptidase domains, resembling the bifunctional class A penicillin-binding proteins (PBPs) (24). Subcloning of pbpTET A6 confirmed the ability of this ARG-encoding product to render E. coli resistant to selected third- and fourth-generation cephalosporins (Table 6). Introduction of pbpTET A6 into P. aeruginosa further demonstrated its versatility and potential in promoting resistance against the same spectrum of antibiotics in a clinically significant pathogen (Table 6).
TABLE 6.
Cephalosporin resistance in E. coli and P. aeruginosa conferred by a probable penicillin-binding protein gene homologue, pbpTET A6, recovered from a multidrug-resistant fosmid clone
| Species and strain | Vector | Genotypea |
MIC (μg/ml)b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pbpTET A6 | orf-12TET A6 | CAZ | CAX | FEP | TET | CTC | OTC | TYL | ERY | AZI | LIN | TIG | ||
| E. coli | ||||||||||||||
| EPI300T1R | pCC1FOS | − | − | 2 | 0.125 | 1 | 4 | 64 | 4 | 2,048 | 256 | 16 | 256 | 1 |
| MACRO-TET A6 | + | + | 8 | 0.125 | 1 | 32 | 256 | 64 | 4,096 | 512 | 32 | 512 | 4 | |
| MACRO-TET A6/Δorf-12::Tn | + | − | 8 | ND | ND | 4 | 64 | 4 | 2,048 | 256 | 16 | 256 | 1 | |
| MACRO-TET A6/ΔpbpTET A6::Tn | − | + | 2 | ND | ND | 32 | 256 | 64 | 4,096 | 512 | 32 | 512 | 4 | |
| DH10β | pUC19 | − | − | 2 | 0.125 | 1 | ND | ND | ND | ND | ND | ND | ND | ND |
| pCL050 | + | − | 32 | 0.5 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | |
| P. aeruginosa PAO1 | pUCP19 | − | − | 4 | 16 | 1 | ND | ND | ND | ND | ND | ND | ND | ND |
| pCL051 | + | − | 128 | 32 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | |
+, present; −, absent or transposon disrupted.
ND, not determined. MIC values higher than that for the corresponding empty-vector control are in bold. AZI, azithromycin; CAZ, ceftazidime; CAX, cefotaxime; CTC, chlortetracycline; ERY, erythromycin; FEP, cefepime; LIN, lincomycin; OTC, oxytetracycline; TET, tetracycline; TIG, tigecycline; TYL, tylosin.
Unknown macrolide resistance mechanisms.
The MACRO-TYL 10-derived ORF-16TYL 10 has 64% sequence identity to the GTPase HflX of a Chlamydia-related bacterium, Simkania negevensis, and promoted mild resistance against macrolides when subcloned into different E. coli hosts (Table 7). Intriguingly, expression of ORF-16TYL 10 in strain AG100A, a macrolide-sensitive derivative of AG100 that is devoid of the significant multidrug resistance-nodulation-division (RND) efflux system component AcrAB (25), did not result in any appreciable level of macrolide resistance. Consistent with this, the MACRO-TYL 10-associated reduction of macrolide susceptibilities was also compromised by the RND pump inhibitor phenyl-arginine-β-naphthylamide (PAβN). Yet, orf-16TYL 10 expression did not provide any resistance against other ribosome-targeting antibiotics, including tetracycline, lincomycin, and chloramphenicol (Fig. 1A and Table 7), despite some of them being excellent substrates of the AcrAB efflux pump (26). Collectively, these observations rule out a general upregulation of AcrAB's efflux activity in the orf-16TYL 10-expressing cell and indicate that ORF-16TYL 10 constitutes a new macrolide-specific resistance mechanism which is somehow dependent on the macrolide extrusion mediated by AcrAB.
TABLE 7.
Reduced macrolide susceptibility conferred by orf-16TYL 10 is dependent on the efflux activity of RND multidrug pump AcrAB in E. coli
| E. coli strain | Vectora | Genotypeb |
MIC (μg/ml)c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| orf-16TYL 10 | acrAB | TYL | ERY | AZI | CLN | GEN | LIN | CAM | ||
| EPI300T1R | pCC1FOS | − | + | 2,048 | 256 | 16 | 256 | 8 | 256 | ND |
| MACRO-TYL 10 | + | + | 4,096 | 512 | 32 | 512 | 4 | 256 | ND | |
| pCC1FOS (+ PAβN) | − | + | 16 | 4 | 1 | 2 | 8 | ND | ND | |
| MACRO-TYL 10 (+ PAβN) | + | + | 16 | 4 | 1 | 2 | 4 | ND | ND | |
| DH10β | pUC19 | − | + | 2,048 | 256 | 16 | 256 | 2 | 256 | 4 |
| pCL036 | + | + | 4,096 | 1,024 | 32 | 512 | 2 | 256 | 4 | |
| pCL037 | + | + | 4,096 | 1,024 | 32 | 512 | 2 | 256 | 4 | |
| AG100 | pUC19 | − | + | 2,048 | 128 | 16 | 128 | 4 | ND | ND |
| pCL036 | + | + | 4,096 | 256 | 32 | 256 | 4 | ND | ND | |
| AG100A | pUC19 | − | − | 64 | 8 | 4 | 8 | 4 | ND | ND |
| pCL036 | + | − | 64 | 8 | 4 | 8 | 4 | ND | ND | |
+ PAβN, the RND pump inhibitor phenyl-arginine-β-naphthylamide was included at 25 μg/ml. pCL036 and pCL037 are identical except that the orf-16TYL 10-carrying insert fragment was cloned in opposite orientations.
−, absent; +, present.
MIC values higher and lower than that of the corresponding empty-vector control are in bold and in italic, respectively. AZI, azithromycin; CAM, chloramphenicol; CLN, clarithromycin; ERY, erythromycin; GEN, gentamicin; LIN, lincomycin; TYL, tylosin. ND, not determined.
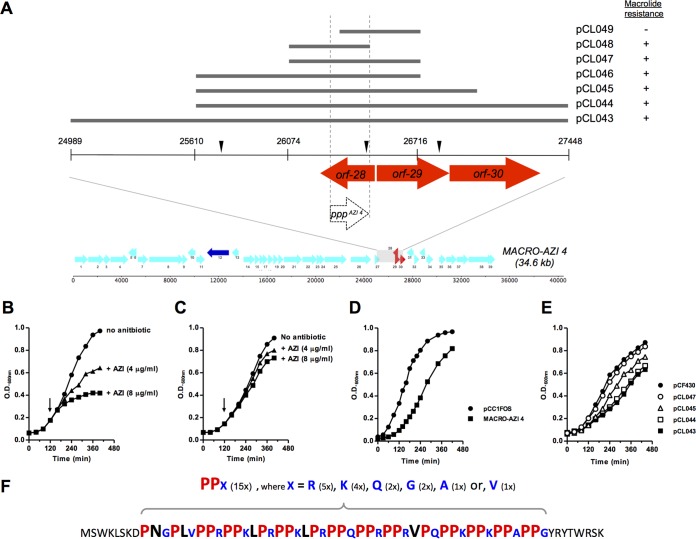
From the macrolide-resistant fosmid MACRO-AZI 4, transposon mutagenesis identified the in silico-predicted orf-28AZI 4, orf-29AZI 4, and orf-30AZI 4 as potential resistance determinants (Fig. 5A). A blastX homology search against the NCBI nonredundant (nr) protein database using these three theoretical ORFs as queries resulted in no significant similarity found for orf-28AZI 4 and two hypothetical proteins for orf-29AZI 4 and orf-30AZI 4. Through constructing a series of pCF430-based subclones (pCL043 to pCL047) (Fig. 5A), it was deduced that the macrolide resistance, which is independent of the RND pump-mediated efflux, was attributed solely to the region encompassing orf-28AZI 4 (Table 8). Compared to the control strain (Fig. 5B), the pCL047 transformant was clearly less susceptible to the growth-inhibitory effect of azithromycin (Fig. 5C). Furthermore, the growth deficiency associated with MACRO-AZI 4 (Fig. 5D) and only certain subclones (Fig. 5E) failed to correlate with the generally observed macrolide-resistant phenotype (Fig. 5A and Table 8).
FIG 5.
Characterization of soil metagenome-derived fosmid MACRO-AZI 4 and identification of a novel macrolide resistance determinant, the PPPAZI 4 gene. (A) Schematic presentation of the subcloned fragments derived from fosmid MACRO-AZI 4. The approximate locations of individual transposon insertions found on fosmid MACRO-AZI 4 that restored macrolide sensitivity are indicated by inverted black triangles. Colored arrows depict the in silico-predicted ORFs, while the white arrow represents an alternative ORF, the PPPAZI 4 gene. Macrolide susceptibilities of subclones are stated as sensitive (−) or resistant (+) based on result in Table 8. (B and C) Azithromycin susceptibilities of the pCF430 (B)- and pCL047 (C)-carrying DH10β strains. Cell growth in LB medium at 37°C with mild agitation was monitored by measuring the OD600 every 20 to 30 min using a spectrophotometer. Arrows indicated when azithromycin was introduced. (D and E) Growth impairment associated with subcloned MACRO-AZI 4 insert fragments. EPI300T1R (D) and DH10β (E) strains carrying the indicated vector were grown in LB medium, without any antibiotic supplements, exactly as described above. (F) Amino acid sequence of the polyproline peptide encoded by the PPPAZI 4 gene. PPX motifs are in bold and colored in red (proline residues), blue (variable residues [X]), and black (nonconserved residues). Numbers in parentheses specify the number of occurrences of the indicated residue or motif.
TABLE 8.
Macrolide susceptibility of the metagenomic fosmid clone MACRO-AZI 4 and its derivatives carrying different subcloned fragments
| Strain | Vectora | Genotypeb |
MIC (μg/ml)c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| orf-28AZI 4 | orf-29AZI 4 | orf-30AZI 4 | TYL | ERY | AZI | CLN | TLT | JOS | ||
| EPI300T1R | pCC1FOS | − | − | − | 2048 | 256 | 16 | 256 | ND | ND |
| MACRO-AZI 4 | + | + | + | 8,192 | 512 | 64 | 512 | ND | ND | |
| pCC1FOS (+ PAβN) | − | − | − | 16 | 4 | 1 | 2 | ND | ND | |
| MACRO-AZI 4 (+ PAβN) | + | + | + | 64 | 8 | 2 | 4 | ND | ND | |
| DH10β | pCF430 | − | − | − | 2,048 | 512 | 32 | 512 | 128 | 512 |
| pCL043 | + | + | + | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL044 | + | + | + | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL045 | + | + | − | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL046 | + | − | − | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL047 | + | − | − | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL048 | −* | − | − | 8,192 | 2,048 | 128 | 1,024 | 512 | 1,024 | |
| pCL049 | − | − | − | 2,048 | 512 | 32 | 512 | 128 | 512 | |
+ PAβN, the RND pump inhibitor phenyl-arginine-β-naphthylamide was included at 25 μg/ml.
−, absent; +, present; *, a partial sequence of orf-28AZI 4 corresponding to the PPPAZI 4 gene is present.
ND, not determined. MIC values higher than that for the corresponding empty-vector control are in bold. AZI, azithromycin; CLN, clarithromycin; ERY, erythromycin; JOS, josamycin; TLT, telithromycin; TYL, tylosin.
To verify the expression of any hypothetical protein that could be related to resistance, total proteomic analyses were performed to search for fosmid-specific proteins in the MACRO-AZI 4 fosmid clone and its resistant derivatives (pCL043 and pCL047). Unexpectedly, no peptide fragment corresponding to the theoretical translational product of orf-28AZI 4 was detected (see Data Set S2 in the supplemental material). Instead, a 61-residue-long polyproline peptide, here designated PPPAZI 4, encoded by an alternative reading frame present within orf-28AZI 4 (Fig. 5A) was consistently expressed in all three resistant clones. Closer examination of this unusual proline-rich peptide revealed that the bulk of PPPAZI 4 is composed of 15 direct-repeats of an (im)perfect “PPX” motif (Fig. 5F) and that it shares no homology with proteins in the NCBI nr database. While the macrolide-resistant phenotype and PPPAZI 4 expression were fully recapitulated in cells carrying pCL048, deletion of the first 38 bp of the PPPAZI 4 gene and its upstream sequence from pCL047 (yielding pCL049) resulted in no detectable PPPAZI 4 expression and, most importantly, a complete loss of resistance (Fig. 5A and Table 8). To examine the selectivity of PPPAZI 4-associated resistance, susceptibility against two additional and structurally distinct macrolides, telithromycin (TLT) (a 14-member ring ketolide) and josamycin (JOS) (a 16-member ring), was also examined. Again, reduced susceptibility against TLT and JOS was observed with only the PPPAZI 4-expressing cells and not their nonexpressing counterparts, consistent with the case for other macrolide antibiotics. Taking the results together, although the mechanism underlying this peptide-associated macrolide resistance is currently unclear (see Discussion for proposed mechanisms), PPPAZI 4 expression is evidently linked to general resistance against macrolides, implicating the PPPAZI 4 gene as a novel ARG.
DISCUSSION
The present study set out to uncover ARGs from soil exposed to antibiotics heavily used in human medicine or food animal production. The idea of mining this antibiotic-amended agricultural soil for novel ARGs is justified not only by the clinical significance of these antimicrobial agents (27) and their high potential for soil exposure through manure or biosolid fertilization but also by the fact that the soil microbial communities displayed adaptability and responsiveness toward these specific drug exposures in recent studies (18, 19, 28). Using functional metagenomics, a number of ARGs were discovered, most of which encode (multi)drug transporters of all five superfamilies. This finding is consistent with those of others (29, 30), supporting drug efflux as one of the most prominent strategies employed by environmental bacteria to resist antibiotics that are either produced by other soil microorganism or introduced anthropogenically. The identification of new antibiotic-inactivating enzymes that can promote resistance against aminoglycosides and β-lactams in both E. coli and P. aeruginosa reinforces the notion that soil is a significant reservoir of as-yet-unknown ARGs that could seriously exacerbate clinical resistance if recruited by human pathogens. Furthermore, the proteomics-driven discovery of a potentially new peptide-mediated resistance mechanism against macrolides underscores the value of deploying high-resolution analytical proteomic technology in functional metagenomics studies.
We demonstrated substrate polyspecificity in a homologous group of ABC efflux systems that can confer resistance against a spectrum of structurally distinct antibiotics and anticancer drugs. These probable two-component transporters all display moderate sequence homology (30 to 50% shared identity) against the DrrAB multidrug efflux system of the DAU-/DOX-producing Streptomyces peucetius (31) and are predicted to originate from bacteria of diverse phyla, including Actinobacteria, Firmicutes, and Proteobacteria (see Table S3 in the supplemental material). While the identification of novel DrrAB homologues in different bacterial taxa is not unexpected, the ligand promiscuity of these functionally conserved, but sequence-wise diverse, ABC transporters (DrrAB included) raises insightful questions of what drove the development of polyspecific substrate recognition and whether these transporters have shared evolutionary history with similar multidrug efflux system found in now-pathogenic organisms.
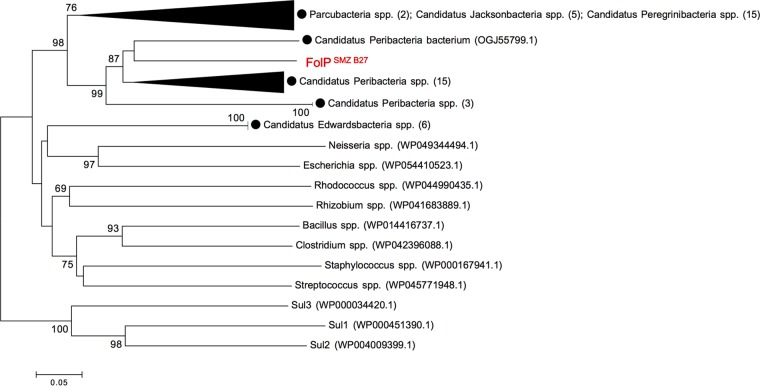
The discovery of folPSMZ B27 from the metagenome of soil with no history of exposure to the synthetic sulfonamide antibiotics is intriguing. FolPSMZ B27 is distantly related (∼30% shared sequence identity) to three highly prevalent sulfonamide-resistant DHPS alternatives, Sul1, Sul2, and Sul3 (32), and shows the greatest homology with deduced DHPSs of several uncultured bacteria belonging to the candidate phyla radiation (CPR) group (Fig. 6). CPR organisms are estimated to constitute >15% of the bacterial domain and have relatively small genomes that lack numerous biosynthetic pathways (33). It is therefore tempting to speculate that some bacterial members of this largely uncharacterized phylum might possess alternative folate-biosynthetic mechanisms (e.g., novel DHPS enzyme variants) that are naturally insensitive to the antagonistic effect of sulfonamide. If so, such kinds of underappreciated metabolic pathways might offer biological clues to the ancestry of specific resistance determinants (e.g., FolPSMZ B27 and PBPTET A6), which can act incidentally against artificial antimicrobial agents.
FIG 6.
Neighbor-joining tree of aligned amino acid sequences of FolPSMZ B27 and bacterial dihydropteroate synthases (DHPSs). The alignment was performed with prototypic DHPSs and sulfonamide-resistant DHPS variants (Sul1, Sul2, and Sul3) using the ClustalW program within MEGA 7. Genera and species are followed by their GenBank accession numbers in parentheses. DHPSs from the candidate phyla radiation (CPR) group bacteria are specified with black circles. Some of the branches were grouped to improve the visibility of the tree (the number of sequences grouped is indicated in parentheses). Bootstrap values were calculated as a percentage of 1,000 replicates, and those above 60% are shown at the branching points. The sequence of FolPSMZ B27 obtained in this study is depicted in red. Scale bar = 0.05 change/site.
In a genomic context, some of the ARGs are syntenic to elements that indicate lateral gene transfer and acquisition (Table S2). Apart from the most striking case seen with sul2, three unrelated ARGs (pbpTET A6, a metallo-β-lactamases gene, and a transporter gene) were found in close proximity to genes encoding homologues of two bacterial nonhomologous end-joining (NHEJ) complex components, the Ku and ligase D (LigD) proteins (Table S2). NHEJ carries out DNA double-strand break repair and was discovered first in eukaryotes and subsequently in bacteria (34). The association of ARGs with a putative NHEJ apparatus hints at possible horizontal gene transfer events. Although Ku and LigD are not universal among the bacterial domain, ARG acquisition by similar recombination-independent mechanism has also been reported in E. coli (35). Collectively, these observations suggest that the end-joining DNA repair mechanism might play a role in the spread of environmental ARGs.
While the molecular principles underlying the new macrolide resistance determinants reported have yet to be fully elucidated, the evidence here is consistent with the ribosome being central to these resistance mechanisms. ORF-16TYL 10 has substantial homology to the ribosome-associated GTPase HflX (36). HflX can interact with the peptidyl transferase center (PTC) and nascent peptide exit tunnel (NPET) of the ribosome to induce dissociation of 30S and 50S subunits in a nucleotide-dependent manner, thereby rescuing stalled ribosomes during cellular stress (37). Interestingly, both the NPET-binding macrolides (e.g., erythromycin) and the PTC-binding antibiotics (e.g., chloramphenicol) can inhibit the ribosome-induced GTPase activity of HflX (38). Hence, we hypothesize that ORF-16TYL 10 functions as an alternative ribosome-splitting factor to alleviate the macrolide-mediated translational arrest. We postulate that ORF-16TYL 10 might be able to better “sense” the drug-induced translational stalling event at the PET (and presumably not at the PTC) and/or to withstand the inhibitory effect of macrolides on its ribosome-induced GTPase activity. The dissociation of macrolide-bound ribosome promoted by ORF-16TYL 10 is believed to reset the stalled ribosome to allow productive translation. Concerning the apparent AcrAB dependency of the ORF-16TYL 10-conferred resistance, observations highly similar to ours, in which the macrolide resistance caused by ribosomal mutation was “masked” by drug efflux pump deficiency, have been reported for several bacteria, including E. coli (39). This so-called “drug target mutation masking” effect in response to efflux pump inhibition is fundamentally linked to growth bistability and is favored by factors including high-affinity drug binding to its target and high target abundance within bacterial cells (39), both of which are applicable in the case of macrolide antibiotics and their ribosomal target. As the exact mechanism remains to be unraveled, our data lean toward a conceivable theory of ORF-16TYL 10 acting on the ribosome target, instead of selectively modulating the efflux pump activity, to confer macrolide-specific resistance in an efflux-dependent manner.
An exciting discovery in this study is the macrolide resistance associated with a novel polyproline peptide, PPPAZI 4, which is encoded by an alternative ORF not recalled by the gene prediction algorithm. Together with its exceptionally high proline content, the repeated appearance of diproline motifs in PPPAZI 4 suggests that this small peptide functions in the context of macrolide-induced translational arrest. Sequences containing consecutive proline residues (especially with three or more) can trigger significant stalling of the bacterial ribosome, which is effectively rescued by the translation elongation factor EF-P (40–43). The likelihood of a nascent peptide becoming arrested at its diproline motif is determined by the identity of residues flanking the proline pair (41, 44–46). In contrast to strong stalling sequences (e.g., PPP), PPR and RPP motifs (where R = Arg) confer weak stalling activity, as their translation has little reliance on the activity of EF-P (41, 45). Furthermore, in vivo PPK (where K = Lys) is translationally arrested only when preceded by an acidic residue (44). These findings illustrate the context-specific nature of EF-P-dependent stalling of diproline motifs. Strikingly, PPPAZI 4 consists of 15 (im)perfect PPX motifs in direct tandem arrangement, with Arg (R) and Lys (K) being most frequently found between diproline sequences and no PPP motif present (Fig. 5F). Our preliminary data have shown that PPPAZI 4 can be detected in both an efp deletion mutant (47) and its parental strain carrying plasmid pCL047 and that the macrolide resistance associated with PPPAZI 4 expression is EF-P independent (see Data Set S2 and Table S4 in the supplemental material). Taken together, these results suggest that the sequence of PPPAZI 4 is evolutionarily optimized to ensure its proper synthesis, probably by minimizing the appearance of any strong EF-P-alleviated ribosomal stalling signals, for a reason(s) that could be related to macrolide resistance.
How can the effective translation of PPPAZI 4 be linked to macrolide resistance? We believe our finding is reminiscent of the previously reported peptide-mediated resistance, in which the synthesis of a pentapeptide-encoding ORF renders both Gram-positive (48) and Gram-negative (49) bacteria resistant against erythromycin. Lovmar et al. (50) proposed a cis-acting model implicating a specific interaction between the nascent pentapeptide and the tunnel-bound macrolide, which can lead to erythromycin removal from its ribosomal binding site upon peptidyl-tRNA release at the end of translation cycle. However, PPPAZI 4 is significantly longer than the described pentapeptides. Also, unlike the macrolide selectivity of different resistant pentapeptides, PPPAZI 4 can promote resistance against broad range of macrolides, suggesting an alternative mechanism of action. Notably, PPPAZI 4 shares the first four residues with one of the ketolide-resistant pentapeptides (MSWKI) (51). We speculate that this N terminus sequence facilitates the initial threading of the PPPAZI 4 nascent peptide through the macrolide-obstructed NPET of the ribosome, analogous to how appending the N-terminal sequence of the H-NS protein to the OsmC protein, whose syntheses are erythromycin resistant and erythromycin sensitive, respectively, allowed the latter to evade the macrolide-induced translational attenuation (52). Although the mere presence of a consensus pentapeptide sequence at the N termini of longer peptides did not confer erythromycin resistance (48), PPPAZI 4 comprises a unique multi-PPX segment that has Arg (R) and Lys (K) as the dominant spacer residues and Leu (L) as the major proline substitute in the imperfect PPX motifs. These amino acids are thought to give peptides a high propensity to adopt a distinctive secondary structure called the type II polyproline (PPII) helix (53). In structural terms, the PPII helix is a left-handed helix that is relatively open reading frame and flexible and has no internal hydrogen bonding (54). It is plausible that formation of the PPII helical structure, after the initial threading through of the PPPAZI 4 nascent peptide, can eject the antibiotic, resulting in ribosomal “cleaning” to restore ribosomal translation. This hypothetical model is somewhat analogous to the previously proposed “bottle brush” theory (51), with the exception that PPPAZI 4 can confer cis-acting resistance against general, instead of specific, macrolide members through the complete synthesis of its polyproline segment. The lack of antibiotic resistance in subclone that expressed a truncated version of the PPPAZI 4 gene encoding only the first 25 amino acids (data not shown) is in support of the theory that the multi-PPX sequence is key to the peptide's ability to confer macrolide resistance. Certainly, given that PPII helices are implicated in diverse molecular functions, including cell signaling, transcription, cell motility, and bacterial pathogenesis (54), we cannot exclude the possibility that PPPAZI 4 may possess other biological functions that can render bacteria less susceptible to macrolide antibiotics.
MATERIALS AND METHODS
Additional method details are provided in Text S1 in the supplemental material.
Soil treatments and sampling.
Soil samples were collected in fall 2014 from two series of antibiotic-amended (and control) field plots established at the Agriculture and Agri-Food Canada (AAFC) experimental farm in London, Canada (18, 19). The drug application history of the three groups of field plots used in the present study, namely, UTC (untreated control), MACRO (macrolides), and SCT (agricultural antibiotics), is presented in Table 1.
Soil DNA extraction and metagenomic clone library construction.
High-molecular-weight metagenomic DNA was extracted from 100 g of sieved soil using an indirect extraction method as described by to Liles et al. (55). Pulse-field gel electrophoresis (PFGE) was used to size fractionate the DNA onto a 1% agarose gel. Nucleic acid with the chosen size of 25 to 70 kb was electroeluted from the excised gel block and concentrated by ethanol precipitation. Three individual metagenomic libraries were constructed using a pCC1FOS fosmid library production kit (Epicentre) with the DNA derived from the aforementioned field plots (UTC, MACRO, and SCT) and the E. coli host strain EPI300-T1R. Fosmid clones from each library were aseptically scraped from the plates, pooled into subpools of ca. 6,000 clones ml−1, and stored at −80°C in LB broth supplemented with 12.5 μg ml−1 chloramphenicol and 15% (vol/vol) glycerol. To estimate the average insert size of each library, fosmid DNA was extracted from 30 randomly selected clones, followed by NotI digestion and PFGE analysis (56).
Selection for antibiotic-resistant metagenomic fosmid clones.
Each library was replicated by inoculating scrapes of frozen subpool stocks into 50 ml of LB broth supplemented with 12.5 μg ml−1 chloramphenicol for 2 h at 37°C. Ten- and 100-fold-diluted library cultures were plated separately onto LB agar or Iso-Sensitest agar plates supplemented with one of 12 antibiotics. Colonies formed after the initial 16 to 18 h of incubation at 37°C, together with those, if any, that appeared after further incubation at room temperature for 24 h were subjected to clonality assessment. To preclude any spontaneous mutation of the E. coli host strain that might be (co)selected for during the selection process, fosmid DNA with a unique NotI restriction profile was reelectrotransformed into EPI300-T1R cells to confirm the fosmid-associated antibiotic resistance phenotypes.
Fosmid sequencing and identification of putative ARGs.
To resolve the sequence of cloned metagenomic DNA, resistance-associated fosmid targets were sequenced using a MiSeq (multiplex) sequencing system (Illumina). To identify putative antibiotic resistance genes (ARGs) harbored on each resistance selected fosmid, transposon mutagenesis was performed using either the EZ-Tn5 <KAN-2> or <TET-1> insertion kit (Epicentre). Transposon-bearing fosmid DNA isolated from mutants with restored susceptibility toward the selection antibiotic was sequenced to locate the transposon insertion site using a transposon-specific primer set.
Subcloning of putative ARGs.
The bacterial strains and plasmids used in this study are summarized in Table 9. For E. coli, selected ORFs (including the predicted promoters) were cloned into pUC19 or pCF430 using PCR-amplified and enzyme-restricted fragments derived from fosmid DNA and introduced into the DH10β strain, unless otherwise specified. The PCR conditions and primer pairs used are provided in Table 10. To construct pCL049, pCL047 was digested with KpnI to remove a 263-bp fragment encompassing the 5′ end of the PPPAZI 4 gene, followed by purification and religation of the remaining DNA fragment. For Pseudomonas aeruginosa, DNA insert fragments were extracted from the pUC19- or pCF430-based constructs by restriction digestion and recloned into the P. aeruginosa-compatible plasmid pUCP19 or pRK415, followed by electrotransforming into the PAO1 strain.
TABLE 9.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| EPI300T1R | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL (Strr) nupG trfA tonA | Epicentre |
| DH10β | Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 relA1 endA1 araD139 Δ(ara-leu)7697 fhuA galK16 galE15 e14− rpsL (Strr) nupG rph spoT1 | New England BioLabs |
| C600 ΔfolP::Kmr | C600 derivative with insertion of a 1,250-bp kanamycin resistance gene and deletion of 330 bp within the folP coding region; Kanr | 23 |
| AG100 | K-12 argE3 thi-1 rpsL xyl mtl Δ(gal-uvrB) supE44 | 25 |
| AG100A | ΔacrAB::Tn903 derivative of AG100; Kanr | 25 |
| BW25113 | Wild-type parental strain used for the Keio collection | 47 |
| MW1014 | Δefp derivative of Keio collection strain with kanamycin resistance cassette removed | 47 |
| P. aeruginosa strain | ||
| K767 | PAO1 prototroph (wild type) | 58 |
| Plasmids | ||
| pCC1FOS | Host fosmid vector for metagenomic library; Camr | Epicentre |
| pUC19 | E. coli cloning vector; Ampr | 59 |
| pCF430 | Broad-host-range cloning vector with araC-PBAD cassette; Tetr | 60 |
| pUCP19 | Broad-host-range P. aeruginosa cloning vector; Ampr | 61 |
| pRK415 | P. aeruginosa-E. coli shuttle cloning vector; Tetr | 62 |
| pCL027 | pCF430 derivative carrying orf-45TET A7 of fosmid MACRO-TET A7 | This study |
| pCL028 | pCF430 derivative carrying orf-46TET A7 of fosmid MACRO-TET A7 | This study |
| pCL029 | pCF430 derivative carrying orf-45TET A7 and orf-46TET A7 of fosmid MACRO-TET A7 | This study |
| pCL030 | pCF430 derivative carrying orf-7KAN 4 of fosmid SCT-KAN 4 | This study |
| pCL031 | pUCP19 derivative carrying orf-7KAN 4 of fosmid SCT-KAN 4 | This study |
| pCL032 | pCF430 derivative carrying orf-27MEM A1 of fosmid MACRO-MEM A1 | This study |
| pCL033 | pRK415 derivative carrying orf-27MEM A1 of fosmid MACRO-MEM A1 | This study |
| pCL034 | pUC19 derivative carrying wild-type folP of E. coli | G. Swedberg, unpublished |
| pCL035 | pUC19 derivative carrying folPSMZ B27 of fosmid UTC-SMZ B27 | This study |
| pCL036 | pUC19 derivative carrying orf-16TYL 10 of fosmid MACRO-TYL 10 | This study |
| pCL037 | pUC19 derivative carrying orf-16TYL 10 of fosmid MACRO-TYL 10 in opposite-orientation | This study |
| pCL043 | pCF430 derivative carrying 2,460-bp fragment encompassing orf-28AZI 4, orf-29AZI 4, and orf-30AZI 4 of fosmid MACRO-AZI 4 | This study |
| pCL044 | pCL043 derivative carrying 1,839-bp fragment encompassing orf-28AZI 4, orf-29AZI 4, and orf-30AZI 4 of fosmid MACRO-AZI 4 | This study |
| pCL045 | pCL044 derivative carrying 1,385-bp fragment encompassing orf-28AZI 4 and orf-29AZI 4 of fosmid MACRO-AZI 4 | This study |
| pCL046 | pCL045 derivative carrying 1,107-bp fragment encompassing orf-28AZI 4 of fosmid MACRO-AZI 4 | This study |
| pCL047 | pCL046 derivative carrying 643-bp fragment encompassing orf-28AZI 4 of fosmid MACRO-AZI 4 | This study |
| pCL048 | pCL047 derivative carrying 391-bp fragment encompassing PPPAZI 4 gene of fosmid MACRO-AZI 4 | This study |
| pCL049 | pCL047 derivative missing a 263-bp, KpnI-restricted fragment | This study |
| pCL050 | pUC19 derivative carrying pbpTET A6 of fosmid MACRO-TET A6 | This study |
| pCL051 | pUCP19 derivative carrying pbpTET A6 of fosmid MACRO-TET A6 | This study |
Camr, chloramphenicol resistant; Ampr, ampicillin resistant; Kanr, kanamycin resistant; Tetr, tetracycline resistant.
TABLE 10.
PCR primers and extension times used in this study
| Plasmid | Primer | Oligonucleotide sequence (5′→3′)a | Extension time (s) |
|---|---|---|---|
| pCL027 | TET A7-abc1-XbaI-F | GAGCGTCTAGATTTGACCGATCTTCCGTGG | 40 |
| TET A7-abc1-KpnI-R | GGATCAGGTACCGAAGTCGAGATAGCTGATGC | ||
| pCL028 | TET A7-abc2-XbaI-F | GAGACTCTAGACGATACGGCTTACCAGTCTGC | 40 |
| TET A7-abc2-KpnI-R | GATCAGGTACCGTTACCAAGCAGTAGCAGCAG | ||
| pCL029 | TET A7-abc1-XbaI-F | 75 | |
| TET A7-abc2-KpnI-R | |||
| pCL030 | KAN 4-aac6′-XbaI-F | GATGTCTAGAGTTGCACAAACGTCTCGCAC | 30 |
| KAN 4-aac6′-KpnI-R | GCATGGTACCGAGCAGGTTAAGCAGTGG | ||
| pCL032 | MEM A1-mbl-XbaI-F | AGCGATCTAGAGTGGGTGCGACAG | 30 |
| MEM A1-mbl-KpnI-R | GCAAGGTACCGAAACTGGGACATGGATG | ||
| pCL035 | SMZ B27-folP-XbaI-F | CAGTTCTAGATACCAGTATCCCAACCGCGTC | 30 |
| SMZ B27-folP-EcoRI-R | GGACCTGAATTCGCACTTGCAGCTCTTG | ||
| pCL036 | TYL 10-hflX-XbaI-F | GCACCTCTAGACGTTGCTTTTGCTGCCAAG | 45 |
| TYL-10-hflX-SalI-R | GACTGTCGACGAACTTCATCAGCCGCTC | ||
| pCL037 | TYL 10-hflX-SalI-F | CGACTGTCGACGTTGCTTTTGCTGCCAAG | 45 |
| TYL 10-hflX-XbaI-R | CGACATCTAGAGAACTTCATCAGCCGCTC | ||
| pCL043 | AZI 4-2460-XbaI-F | GACGTCTAGAGCAAATCGCCTATCCAACGC | 75 |
| AZI 4-2460-PstI-R | GCATCTGCAGTCTCGCACACCACTCACTATC | ||
| pCL044 | AZI 4-1839-XbaI-F | CGAACTCTAGACTAAGGTCCAGGAGGAAAC | 50 |
| AZI 4-2460-PstI-R | |||
| pCL045 | AZI 4-1839-XbaI-F | 40 | |
| AZI 4-1385-PstI-R | GCATCTGCAGGCGCTTTATAGATTCCCAATCG | ||
| pCL046 | AZI 4-1839-XbaI-F | 30 | |
| AZI 4-1107-PstI-R | GCATCTGCAGCGGTTTTCTGGCTAACACG | ||
| pCL047 | AZI 4-643-XbaI-F | CGAGGTCTAGAGCATTATTGGGTACTTGAC | 30 |
| AZI 4-1107-PstI-R | |||
| pCL048 | AZI 4-643-XbaI-F | CGAGGTCTAGAGCATTATTGGGTACTTGAC | 15 |
| AZI 4-391-PstI-R | GCATCTGCAGTCATTTAGATCTCCAAGTA | ||
| pCL050 | TET A6-pbp-XbaI-F | GGACGTCTAGAATCGCATCCTTAACAGGGTG | 75 |
| TET A6-pbp-EcoRI-R | GGACGGAATTCACCCTGTCCTGAAGGCATTTG |
Restriction endonuclease cleavage sites are underlined. Repeated primer sequences are not shown.
Antibiotic susceptibility assays.
The susceptibility of E. coli and P. aeruginosa to antibiotics was assessed using the 2-fold serial microtiter broth dilution method (57) and the MICs recorded after 18 h of incubation at 37°C. To evaluate sulfonamide and anthracycline susceptibility, serial dilutions of overnight cultures were spotted onto Iso-Sensitest agar (for sulfonamides) or LB agar (for anthracyclines) plates supplemented with the appropriate antibiotic, followed by 24 h of incubation at 37°C.
Proteomics analysis.
Briefly, E. coli cultures with final optical densities at 600 nm (OD600) of ca. 2 were pelleted, resuspended in 50 mM ammonium bicarbonate, and sonicated on ice using a Q125 sonicator (Qsonica). After centrifugation, 100 μg of protein was reduced, alkylated, and digested with trypsin in the presence of RapiGest surfactant (Waters Corp) The tryptic peptides were resolved using an Easy-nLC 1000 nano system with the Acclaim C18 PepMap column (75 μm by 15 cm) coupled to a Q-Exactive Orbitrap (Thermo-Fisher Scientific). The tandem mass spectrometry (MS/MS) scans were searched against the target/reverse proteome of E. coli MG1655 (Proteome ID UP0000625, accessed April 2016) with an additional 39 annotated gene products derived from fosmid MACRO-AZI 4. A second search was completed using additional amino acid sequences deduced from all six possible reading frames (including PPPAZI 4) in the subcloned fragment of pCL043.
Accession number(s).
All sequences reported in this paper have been deposited in GenBank (accession no. KY705323 to KY705356) and the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by competitive funding awarded through the Growing Forward 2 program of AAFC. C.H.-F.L. was supported through the NSERC Fellowship in Government Laboratories program.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank A. Heather and L. Moe for providing pCF430, G. Swedberg for providing pCL034 and the ΔFolP strain, and W. Navarre and M. Ibba for providing strains BW25113 and MW1014. We also thank K. Poole for his kind gift of various E. coli and P. aeruginosa strains and plasmids used in this study. We acknowledge M. Sumarah and R. Austin for their assistance in performing proteomic analysis and high-throughput fosmid sequencing, respectively. We are especially grateful to all members of E.T.'s team and the LRDC farm staff for their technical assistance.
C.H.-F.L and E.T. conceived and designed the study. C.H-F.L, K.V.E., and J.R. performed experiments. C.H.-F.L, K.V.E., S.G., J.R., and E.T. analyzed data. C.H.-F.L, J.R., and E.T. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00989-17.
REFERENCES
- 1.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization. http://www.who.int/drugresistance/documents/surveillancereport/en/ Accessed 3 March 2017.
- 2.Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN, Flamm RK. 2016. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY antimicrobial surveillance program during 2014-2015. Antimicrob Agents Chemother 60:5623–5624. doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Neill J. May 2016. Tackling drug-resistant infections globally: final report and recommendations. Rev Antimicrob Resist https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf.
- 6.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 7.Cantón R, Morosini M-I. 2011. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 8.Durso LM, Cook KL. 2014. Impacts of antibiotic use in agriculture: what are the benefits and risks? Curr Opin Microbiol 19:37–44. doi: 10.1016/j.mib.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Huijbers PMC, Blaak H, de Jong MCM, Graat EAM, Vandenbroucke-Grauls CMJE, de Roda Husman AM. 2015. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 10.Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM, Williams AP. 2013. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis 13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 11.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 12.D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 13.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesme J, Cécillon S, Delmont Tom O, Monier J-M, Vogel Timothy M, Simonet P. 2014. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol 24:1096–1100. doi: 10.1016/j.cub.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 15.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2008. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 16.Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD. 2012. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One 7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 18.Topp E, Chapman R, Devers-Lamrani M, Hartmann A, Marti R, Martin-Laurent F, Sabourin L, Scott A, Sumarah M. 2013. Accelerated biodegradation of veterinary antibiotics in agricultural soil following long-term exposure, and isolation of a sulfamethazine-degrading Microbacterium sp. J Environ Qual 42:173–178. doi: 10.2134/jeq2012.0162. [DOI] [PubMed] [Google Scholar]
- 19.Topp E, Renaud J, Sumarah M, Sabourin L. 2016. Reduced persistence of the macrolide antibiotics erythromycin, clarithromycin and azithromycin in agricultural soil following several years of exposure in the field. Science Total Environ 562:136–144. doi: 10.1016/j.scitotenv.2016.03.210. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updates 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rådström P, Swedberg G. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob Agents Chemother 32:1684–1692. doi: 10.1128/AAC.32.11.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sköld O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist Updates 3:155–160. doi: 10.1054/drup.2000.0146. [DOI] [PubMed] [Google Scholar]
- 23.Fermér C, Swedberg G. 1997. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J Bacteriol 179:831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 25.Okusu H, Ma D, Nikaido H. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elkins CA, Nikaido H. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J Bacteriol 184:6490–6498. doi: 10.1128/JB.184.23.6490-6499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO. 2012. Critically important antimicrobials for human medicine, 3rd revision World Health Organization, Geneva, Switzerland. [Google Scholar]
- 28.Cleary DW, Bishop AH, Zhang L, Topp E, Wellington EMH, Gaze WH. 2016. Long-term antibiotic exposure in soil is associated with changes in microbial community structure and prevalence of class 1 integrons. FEMS Microbiol Ecol 92:fiw159. doi: 10.1093/femsec/fiw159. [DOI] [PubMed] [Google Scholar]
- 29.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G. 2014. Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson MK, Forsberg KJ, Dantas G. 2015. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 9:207–216. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Sharma M, Kaur P. 2014. The DrrAB efflux system of Streptomyces peucetius is a multidrug transporter of broad substrate specificity. J Biol Chem 289:12633–12646. doi: 10.1074/jbc.M113.536136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyrsch ER, Roy Chowdhury P, Chapman TA, Charles IG, Hammond JM, Djordjevic SP. 2016. Genomic microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front Microbiol 7:843. doi: 10.3389/fmicb.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 34.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, di Fagagna FD, Devine AKM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 35.Chayot R, Montagne B, Mazel D, Ricchetti M. 2010. An end-joining repair mechanism in Escherichia coli. Proc Natl Acad Sci U S A 107:2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain N, Dhimole N, Khan AR, De D, Tomar SK, Sajish M, Dutta D, Parrack P, Prakash B. 2009. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem Biophys Res Commun 379:201–205. doi: 10.1016/j.bbrc.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Mandava CS, Cao W, Li X, Zhang D, Li N, Zhang Y, Zhang X, Qin Y, Mi K, Lei J, Sanyal S, Gao N. 2015. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol 22:906–913. doi: 10.1038/nsmb.2956. [DOI] [PubMed] [Google Scholar]
- 38.Coatham ML, Brandon HE, Fischer JJ, Schümmer T, Wieden H-J. 2016. The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res 44:1952–1961. doi: 10.1093/nar/gkv1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fange D, Nilsson K, Tenson T, Ehrenberg M. 2009. Drug efflux pump deficiency and drug target resistance masking in growing bacteria. Proc Natl Acad Sci U S A 106:8215–8220. doi: 10.1073/pnas.0811514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 41.Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. 2013. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci U S A 110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 43.Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci U S A 110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elgamal S, Katz A, Hersch SJ, Newsom D, White P, Navarre WW, Ibba M. 2014. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet 10:e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lassak J, Wilson DN, Jung K. 2016. Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol Microbiol 99:219–235. doi: 10.1111/mmi.13233. [DOI] [PubMed] [Google Scholar]
- 46.Starosta AL, Lassak J, Peil L, Atkinson GC, Virumäe K, Tenson T, Remme J, Jung K, Wilson DN. 2014. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res 42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hersch SJ, Wang M, Zou SB, Moon K-M, Foster LJ, Ibba M, Navarre WW. 2013. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. mBio 4:e00180-13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenson T, DeBlasio A, Mankin A. 1996. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci U S A 93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novikova SI, Bushueva AM, Trachuk LA, Konstantinova GE, Serkina AV, Hoischen C, Gumpert J, Chestukhina GG, Mankin A, Shevelev AB. 2000. Introduction of a mini-gene encoding a five-amino acid peptide confers erythromycin resistance on Bacillus subtilis and provides temporary erythromycin protection in Proteus mirabilis. FEMS Microbiol Lett 182:213–218. doi: 10.1111/j.1574-6968.2000.tb08897.x. [DOI] [PubMed] [Google Scholar]
- 50.Lovmar M, Nilsson K, Vimberg V, Tenson T, Nervall M, Ehrenberg M. 2006. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem 281:6742–6750. doi: 10.1074/jbc.M511918200. [DOI] [PubMed] [Google Scholar]
- 51.Tripathi S, Kloss PS, Mankin AS. 1998. Ketolide resistance conferred by short peptides. J Biol Chem 273:20073–20077. doi: 10.1074/jbc.273.32.20073. [DOI] [PubMed] [Google Scholar]
- 52.Kannan K, Vázquez-Laslop N, Mankin Alexander S. 2012. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 53.Brown AM, Zondlo NJ. 2012. A propensity scale for type II polyproline helices (PPII): aromatic amino acids in proline-rich sequences strongly disfavor PPII due to proline-aromatic interactions. Biochemistry 51:5041–5051. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- 54.Adzhubei AA, Sternberg MJE, Makarov AA. 2013. Polyproline-II helix in proteins: structure and function. J Mol Biol 425:2100–2132. doi: 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, Handelsman J. 2008. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl Environ Microbiol 74:3302–3305. doi: 10.1128/AEM.02630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wichmann F, Udikovic-Kolic N, Andrew S, Handelsman J. 2014. Diverse antibiotic resistance genes in dairy cow manure. mBio 5:e01017. doi: 10.1128/mBio.01017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amsterdam D. 1996. Susceptibility testing of antimicrobials in liquid media, p 52–111. In Lorian V. (ed), Antibiotics in laboratory medicine, 4th ed Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 58.Masuda N, Ohya S. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother 36:1847–1851. doi: 10.1128/AAC.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 60.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 61.Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 62.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.