ABSTRACT
Hydrologic exchange plays a critical role in biogeochemical cycling within the hyporheic zone (the interface between river water and groundwater) of riverine ecosystems. Such exchange may set limits on the rates of microbial metabolism and impose deterministic selection on microbial communities that adapt to dynamically changing dissolved organic carbon (DOC) sources. This study examined the response of attached microbial communities (in situ colonized sand packs) from groundwater, hyporheic, and riverbed habitats within the Columbia River hyporheic corridor to “cross-feeding” with either groundwater, river water, or DOC-free artificial fluids. Our working hypothesis was that deterministic selection during in situ colonization would dictate the response to cross-feeding, with communities displaying maximal biomass and respiration when supplied with their native fluid source. In contrast to expectations, the major observation was that the riverbed colonized sand had much higher biomass and respiratory activity, as well as a distinct community structure, compared with those of the hyporheic and groundwater colonized sands. 16S rRNA gene amplicon sequencing revealed a much higher proportion of certain heterotrophic taxa as well as significant numbers of eukaryotic algal chloroplasts in the riverbed colonized sand. Significant quantities of DOC were released from riverbed sediment and colonized sand, and separate experiments showed that the released DOC stimulated respiration in the groundwater and piezometer colonized sand. These results suggest that the accumulation and degradation of labile particulate organic carbon (POC) within the riverbed are likely to release DOC, which may enter the hyporheic corridor during hydrologic exchange, thereby stimulating microbial activity and imposing deterministic selective pressure on the microbial community composition.
IMPORTANCE The influence of river water-groundwater mixing on hyporheic zone microbial community structure and function is an important but poorly understood component of riverine biogeochemistry. This study employed an experimental approach to gain insight into how such mixing might be expected to influence the biomass, respiration, and composition of hyporheic zone microbial communities. Colonized sands from three different habitats (groundwater, river water, and hyporheic) were “cross-fed” with either groundwater, river water, or DOC-free artificial fluids. We expected that the colonization history would dictate the response to cross-feeding, with communities displaying maximal biomass and respiration when supplied with their native fluid source. By contrast, the major observation was that the riverbed communities had much higher biomass and respiration, as well as a distinct community structure compared with those of the hyporheic and groundwater colonized sands. These results highlight the importance of riverbed microbial metabolism in organic carbon processing in hyporheic corridors.
KEYWORDS: 16S rRNA gene, biogeochemistry, biomass, composition, deterministic selection, dissolved organic carbon, hyporheic corridor, microbial communities, respiration, riverine
INTRODUCTION
The interface between river water and groundwater (i.e., the hyporheic zone) represents a zone of elevated microbial metabolism and biogeochemical cycling in riverine ecosystems. During periods of high river flow, fluid exchange between groundwater and river water can occur outside the local riverbed hyporheic zone, extending into and beyond the riparian zone. The term hyporheic corridor (HC) (1) is used to encompass this broader ecotone, which represents a temporally variable domain where groundwater and river waters mix in response to hydrologic dynamics. The HC is an example of a hot moment (2, 3), where the mixing of river water and groundwater constituents occurs in high flow zones in which river water-groundwater exchange is most intense. The HC is a zone of confluence of organic materials and nutrients that has the potential to remove nitrogen inputs and to process large amounts of organic carbon (4) as well as environmental pollutants and even microbial pathogens (5). A scientific understanding of the linkage between microbial communities and biogeochemical cycling within the HC is fundamental to forecasting how these critical systems will respond to dynamic changes in watershed and local-scale hydrology.
General theories for mechanisms that control the composition of microbial communities were recently expanded by Vellend (80) and Hanson et al. (7), detailing the key components as speciation, organism dispersal, ecological selection, and ecological drift. These factors have been further classified into two classes, deterministic and stochastic. Deterministic selection is imposed by the environmental conditions that a microbial community is exposed to (8–10), whereas stochastic selection occurs due to perturbations, such as probabilistic dispersal, random birth-death events, and unpredictable disturbances (11). Determining how deterministic versus stochastic selection processes influence microbial community assembly is still poorly understood, and few studies have investigated these influences simultaneously. Stegen et al. (12–14) conducted such an analysis on subsurface microbial communities at the U.S. Department of Energy Hanford 300 Area site in eastern Washington, USA (see Materials and Methods). Broadly speaking, the results suggested that microbial community assembly was controlled by deterministic selection linked to environmental conditions, dispersal related to hydrological transport, and random ecological drift as a result of stochastic population shifts. In another recent study at the Hanford 300 Area site, Stegen et al. (15) found that during river water intrusion into the Columbia River HC, there was a loss of dissolved organic carbon (DOC) linked to microbial metabolism in hyporheic sediments. This phenomenon triggered a shift from stochastic to deterministic selection and was associated with elevated abundances of microbial taxa that can degrade a broad suite of organic compounds. Parallel work by Graham et al. (16, 17) provided further evidence for the importance of hydrologic mixing-induced deterministic selection in controlling Hanford 300 Area HC microbial community composition and linked such processes to changes in rates of microbial metabolism. However, these studies were performed in situ; hence, the impact of dynamic mixing versus homogeneous environmental selection (where the selective environment remains spatially homogeneous within each successional stage [18]) is unknown.
This study employed an experimental approach to gain insight into how river water-groundwater mixing might be expected to impact Hanford 300 Area HC microbial communities. As discussed in the report by Graham et al. (16), the Hanford Reach of the Columbia River represents a model system in which to integrate community ecology and microbial metabolism in a hydrobiogeochemical context. The biomass, composition, and metabolic activity of attached communities from three different habitats were investigated, including ones obtained (via in situ colonization of sterile sand [see Materials and Methods]) from groundwater, riverbed, and hyporheic zone environments. To simulate river water-groundwater mixing events, each microbial community was “cross-fed” with either sterile groundwater, sterile river water, or sterile DOC-free artificial versions of these fluids. Key to the experimental design was the exclusion of dispersal effects and the limitation of stochastic processes to disturbance during the isolation and sampling of the colonized sand. Microbial biomass (ATP content), respiratory activity (resazurin reduction), and community composition (16S rRNA gene amplicon sequencing) were followed over time to assess the temporal responses to cross-feeding.
Several prior studies have demonstrated habitat-driven selection of hyporheic microbial communities, including ones in systems with different hydrology, substratum properties, organic matter inputs, or gradients in heavy-metal contamination (19–26). Based on this work and recent microbial community assembly studies in the Hanford 300 Area HC (15–17), deterministic selection imposed by the various DOC sources/amounts in the three different habitats was expected to lead to distinct microbial assemblages in the colonized sand materials. Stegen et al. (15) showed (using high-resolution Fourier transform-ion cyclotron resonance mass spectrometry) significant differences in the properties of DOC among groundwater, river water, and hyporheic zone fluids, which were correlated with both microbial respiration and community composition. We hypothesized that the microbial community response to experimental manipulation during the laboratory incubations would positively reflect such deterministic selection, such that groundwater-derived microbial communities would show increased biomass and metabolism when exposed to filtered natural groundwater compared with that when exposed to filtered river water (or artificial carbon-free groundwater), whereas riverbed microbial communities would have higher biomass and metabolism when given filtered river water than when given filtered groundwater (or artificial carbon-free river water). The response of hyporheic zone microbial communities was an unknown, since that habitat experiences frequent mixing of groundwater and river water during flow reversals (see below). A second hypothesis was that microbial communities in the microcosms would undergo a temporal shift in response to cross-feeding, i.e., a community exposed to a nonnative fluid source would become similar to communities present in the corresponding nonnative community. Experimental evaluation of these hypotheses provides new insight into the connection between habitat-driven deterministic selection and biogeochemical function in a large river HC.
RESULTS
Microbial biomass and respiration.
The initial cross-feeding experiment (CF1) showed that the biomass (ATP content) and aerobic respiratory activity (resazurin reduction) of the riverbed sand (RBS) community were consistently higher than those of communities associated with the other two sand types irrespective of the fluid source (Fig. 1). This was true even for the artificial river water (ARW) and artificial groundwater (AGW) reactors that received negligible DOC (<0.1 ppm) input. Similar results were observed in the second experiment (CF2) (see Fig. S6 in the supplemental material). No consistent increases or decreases in biomass values related to feed solution were observed throughout the experiments. The groundwater sand (GWS) and piezometer sand (PZS) reactors that received river water (RW) with higher DOC content (ca. 1.2 ppm) showed very little separation in biomass and respiration from those receiving groundwater (GW; ca. 0.3 ppm DOC), AGW, or ARW. Aerobic respiration rates remained relatively stable throughout both experiments, except for the RBS reactors, where a decreasing trend was observed over time (Fig. 1E; see also Fig. S6E). The same relative order of biomass and respiration for the different sand types was observed in the desorption/cross-feeding (DCF) experiment (Fig. 2), with RBS showing much higher values than PZS and GWS, and PZS showing higher values than GWS. Biomass values in reactors exposed to sediment desorbed (Sed) DOC or RBS DOC were not systematically higher than in reactors receiving RW or ARW (Fig. 2A to C). However, respiration rates were elevated in GWS and (to a lesser extent) PZS exposed to Sed DOC and RBS DOC compared with those exposed to RW or ARW (Fig. 2E and F).
FIG 1.
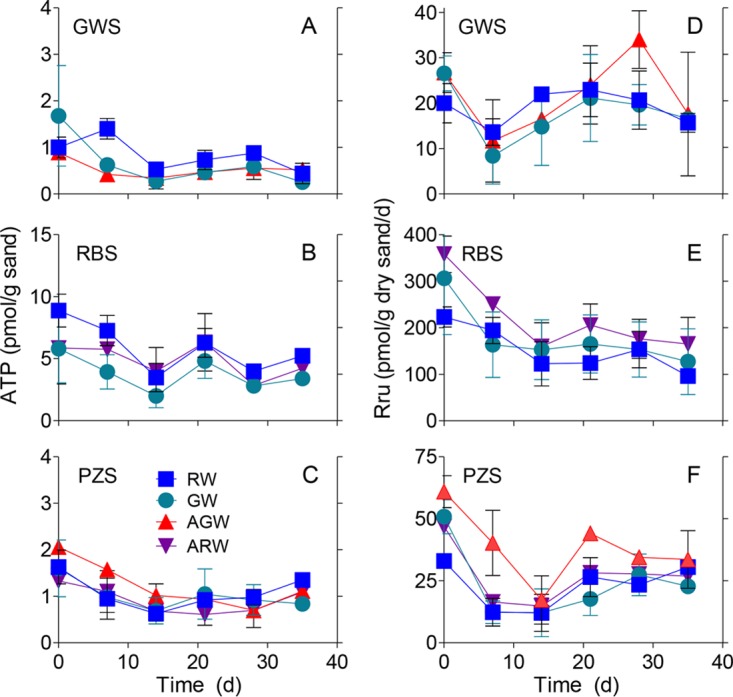
ATP contents and rates of resazurin transformation to resorufin (Rru) for the groundwater (A, D), riverbed (B, E), and piezometer (C, F) sand colonized reactors for cross-feed experiment 1. Each data point shows the mean ± SD or range from triplicate or duplicate reactors.
FIG 2.
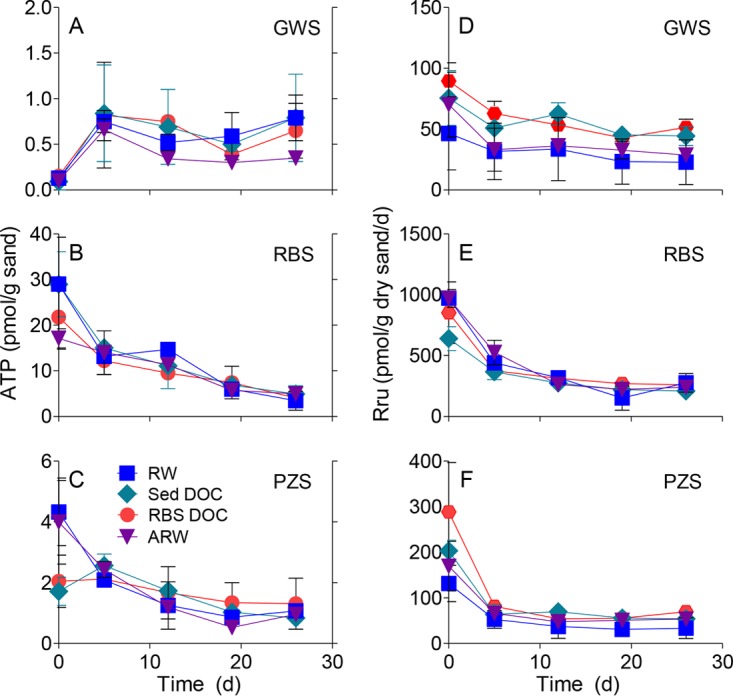
ATP contents (A to C) and rates of resazurin transformation to resorufin (Rru) (D to F) for the desorption/cross-feed experiment.
A statistical linear mixed modeling (LMM) comparison of the two replicate cross-feeding experiments using the lme4 package in R (Table 1) confirmed that sand type was the greatest source of variation for both biomass and respiration. F tests with P values lower than 0.05 were considered significant. Time was a significant factor only for CF2 microbial respiration rates (which decreased with time); this was not the case for CF2 microbial biomass or for CF1 respiration. The significance of year for the biomass data was due to CF2 (2015) having higher overall values than CF1 (2014). Although fluid type was a statistically significant factor for the combined CF1/CF2 biomass data and the CF2 respiration data, these differences neither supported nor refuted the overall hypothesis of this study (see below). LMM results for the DCF experiment revealed that sand type and time were again the most significant sources of variation for biomass content. For respiration, fluid type was a marginally significant source of variation, with sand type and time still the leading sources of variation.
TABLE 1.
Linear mixed model analysis of variance for the cross-feeding experiments
| Dependent variable | Factor | df | F value | P value |
|---|---|---|---|---|
| Biomass | Year | 1 | 131.63 | 2.20E−16 |
| CF1 and CF2 | Sand | 2 | 287.67 | 2.20E−16 |
| Water | 3 | 3.77 | 0.01 | |
| Time | 1 | 2.56 | 0.11 | |
| Respiration CF1a | Sand | 2 | 44.89 | 2.95E−14 |
| Water | 3 | 0.73 | 0.54 | |
| Time | 1 | 3.74 | 0.06 | |
| Respiration CF2a | Sand | 2 | 156.56 | 5.43E−13 |
| Water | 3 | 6.62 | 2.70E−03 | |
| Time | 1 | 81.22 | 1.67E−08 | |
| Biomass DCF | Sand | 2 | 86.58 | 7.10E−09 |
| Water | 3 | 1.39 | 0.29 | |
| Time | 1 | 82.26 | 9.94E−13 | |
| Respiration DCF | Sand | 2 | 62.09 | 5.16E−08 |
| Water | 3 | 4.07 | 0.027 | |
| Time | 1 | 30.11 | 1.07E−06 |
Respiration data for CF1 contained several measurements that were just above the detection limit. Such data were not present in the CF2 data; thus, the two data sets are modeled separately.
Pairwise comparisons revealed a few cases where fluid type had a statistically significant impact on microbial biomass or respiration. For the two cross-feeding experiments (see Table S1), biomass values showed small (typically less than 2-fold) but significant differences among water types that were, in general, unexpected relative to the working hypothesis of the study that a given sand type would perform best when provided fluid from its native environment. For example, for CF2, the biomass for RBS receiving GW was higher than for RBS receiving RW, and GWS receiving RW was higher than GWS receiving GW. Likewise, for respiration in CF2, rates were higher for GWS with RW than with GW. Other significant differences were related to differential responses to AGW or ARW compared with those to nonartificial fluids. For the DCF experiment (see Table S2), GWS reactors had significantly higher biomass when exposed to Sed DOC and RBS DOC than when exposed to ARW. Also, both GWS and PZS respiration rates were significantly higher in the presence of Sed DOC and RBS DOC than in the presence of RW.
DOC dynamics.
DOC data for CF1 and CF2 showed an unexpected trend for the RBS reactors (Fig. 3), where DOC concentrations exceeded the values expected for the given feed stocks. These results indicate that DOC was released into solution from the RBS between sampling points. PZS reactors containing GW and AGW displayed smaller but detectable amounts of DOC release. By contrast, GWS and PZS reactors containing RW showed a slight loss of DOC. A substantial release of DOC from RBS was again observed in the DCF experiment (Fig. 4). A loss of DOC occurred in GWS exposed to RW, Sed DOC, and RBS DOC; likewise, PZS exposed to Sed DOC and RBS DOC showed a loss of DOC.
FIG 3.
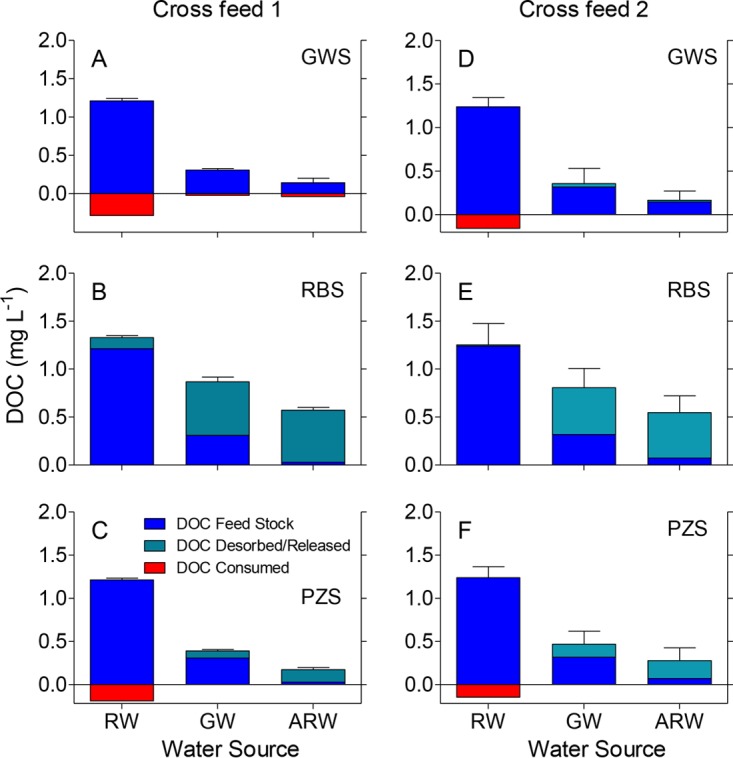
DOC concentrations in the fluid phase of groundwater (A, D), riverbed (B, E), and piezometer (C, F) sand reactors from the two replicate cross-feed experiments. The green and red sections show the release or consumption of DOC based on the difference between the reactor DOC and the expected DOC content of the feed stock solution (blue sections). Values are the means ± SDs from 3 time points from triplicate reactors for experiment 1, and the means ± SDs from 6 time points from triplicate reactors for experiment 2.
FIG 4.
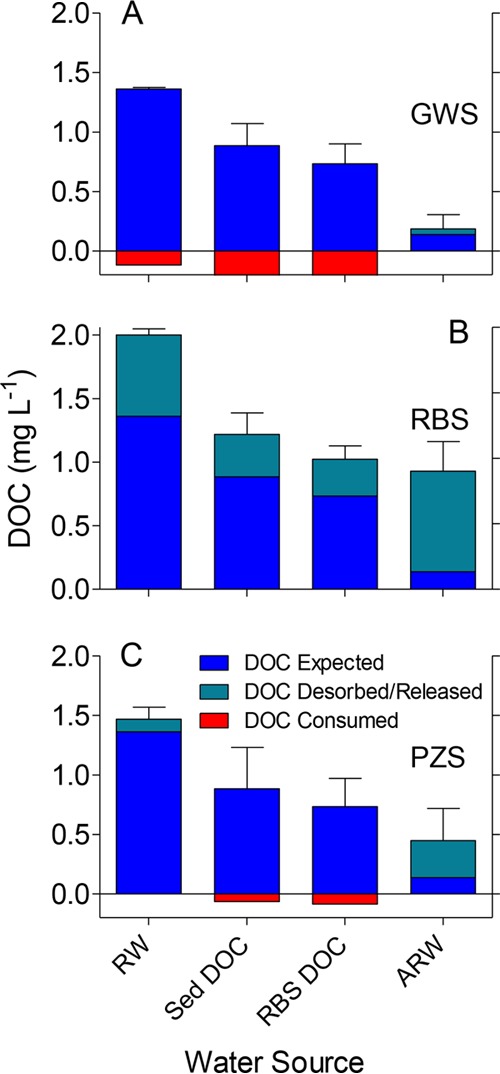
DOC concentrations in the fluid phase of riverbed (A), groundwater (B), and piezometer (C) sand reactors for the desorption/cross-feed experiment. The green and red sections show the release or consumption of DOC based on the difference between the reactor DOC and the DOC content of the feed stock solution (indicated on x axis). Values show means ± SDs from 3 time points from triplicate reactors.
Microbial community composition.
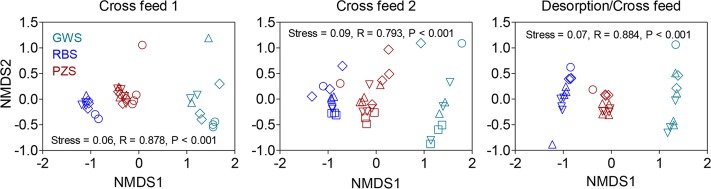
Nonmetric dimensional scaling (NMDS) analysis was conducted on the 16S rRNA gene amplicon libraries to assess differences in microbial community structure among the different sand reactor types for the three cross-feeding experiments. Libraries from different fluid treatments were grouped together for each time point during each experiment, because analysis of similarity (ANOSIM) showed that (with the exception of GWS in the DCF experiment [see below]) fluid type did not significantly influence the community composition (Table 2). The results revealed large differences in community structure among the different reactor types for each of the three experiments (Fig. 5). Although the RBS and PZS communities grouped closer to each other than either did to the GWS, ANOSIM nevertheless showed that the RBS and PZS communities were distinct (Table 2). This finding is not surprising in light of the difference in alpha diversity among the sand reactor communities (see Table S3).
TABLE 2.
ANOSIM of microbial community compositions across different sand and water types for the three cross-feeding experiments
| Experiment | Factor | R value | P value |
|---|---|---|---|
| CF1 | Sand typea | 0.878 | 0.001 |
| CF2 | Sand type | 0.793 | 0.001 |
| DCF | Sand type | 0.884 | 0.001 |
| CF1 | Water type (GWS)b | 0.115 | 0.216 |
| CF1 | Water type (RBS) | 0.120 | 0.140 |
| CF1 | Water type (PZS) | 0.090 | 0.317 |
| CF2 | Water type (GWS) | −0.096 | 0.233 |
| CF2 | Water type (RBS) | −0.062 | 0.706 |
| CF2 | Water type (PZS) | −0.187 | 0.968 |
| DCF | Water type (GWS) | 0.579 | 0.024 |
| DCF | Water type (RBS) | −0.235 | 0.987 |
| DCF | Water type (PZS) | 0.196 | 0.148 |
Pooled across all time points and water types.
Pooled across all time points for each water type.
FIG 5.
NMDS analysis of microbial community composition for the cross-feeding and desorption/cross-feed experiments. R and P values in each panel are results of ANOSIM of 16S rRNA gene amplicon dissimilarity. Results for reactors with different fluid sources are shown by the same symbol color because ANOSIM showed that fluid type did not significantly influence community composition (see Table 2). Different time points (days) are shown with the following symbols. For cross-feed 1: ○, 0; ♢, 14; △, 28; ▽, 35. For cross-feed 2: ○, 0; ♢, 7; △, 21; ▽, 35; □, 41. For desorption/cross-feed: ○, 0; ♢, 5; △, 19; ▽, 26.
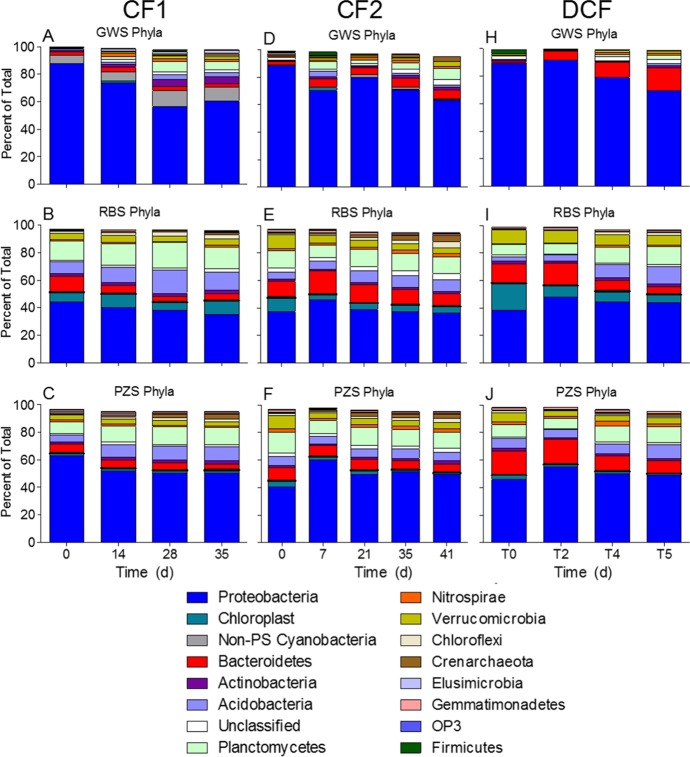
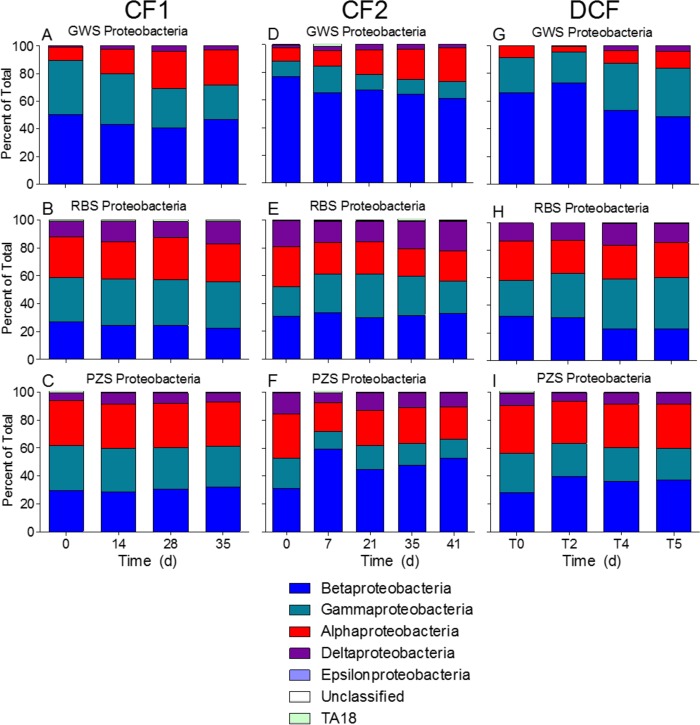
A phylum-level analysis revealed major differences in sand reactor community composition for each of the three experiments (Fig. 6 and Table 3). We found it insightful to approach this analysis by examining Proteobacteria and non-Proteobacteria phyla in two separate groupings. Proteobacteria were more abundant in the GWS (70 to 75% of total reads) than in the RBS (50 to 55%) and PZS (40 to 45%) reactors, particularly in CF2 and DCF. However, non-Proteobacteria phyla increased over time in the GWS reactors, whereas the relative abundance of Proteobacteria versus non-Proteobacteria showed only minor changes over time in the RBS and PZS reactors. Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were the dominant proteobacterial classes in all reactors, with smaller but substantial amounts of Deltaproteobacteria in the RBS and PZS reactors (Fig. 7).
FIG 6.
Phylum-level microbial community compositions in the colonized sand reactors. Each bar shows the average result from 2 to 4 reactors that received different fluid sources during the incubation experiments. Results for reactors with different fluid sources were combined for each time point during each experiment because ANOSIM showed that fluid type did not significantly influence community composition (see Table 2). Chloroplast refers to the chloroplast from eukaryotic algae; non-PS Cyanobacteria refers to nonphotosynthetic cyanobacteria from the class ML635J-21 in the Greengenes and Silva 16S rRNA gene databases.
TABLE 3.
Relative abundance of selected phylaa summed across all water types for a given sand type for each of the three cross-feeding experiments
| Expt. | Phylum | Total reads (%) |
||
|---|---|---|---|---|
| GWS | RBS | PZS | ||
| CF1 | Proteobacteria | 72.3 | 39.8 | 53.3 |
| Acidobacteria | 1.9 | 11.7 | 8.8 | |
| Bacteroidetes | 3.0 | 6.7 | 5.5 | |
| Cyanobacteria | 8.3b | 9.4c | 2.1c | |
| Nitrospirae | 1.6 | 1.1 | 1.4 | |
| Planctomycetes | 3.4 | 15.5 | 11.5 | |
| Verrucomicrobia | 1.1 | 4.6 | 3.4 | |
| Crenarchaeota | 0.6 | 1.1 | 2.2 | |
| CF2 | Proteobacteria | 70.1 | 40.0 | 50.7 |
| Acidobacteria | 1.1 | 7.5 | 6.2 | |
| Bacteroidetes | 6.3 | 13.0 | 7.4 | |
| Cyanobacteria | 1.3b | 5.4c | 2.5c | |
| Nitrospirae | 1.8 | 1.7 | 2.2 | |
| Planctomycetes | 5.5 | 11.6 | 12.0 | |
| Verrucomicrobia | 2.1 | 5.5 | 4.9 | |
| Crenarchaeota | 2.3 | 2.1 | 2.1 | |
| DCF | Proteobacteria | 75.8 | 44.3 | 50.2 |
| Acidobacteria | 0.7 | 8.1 | 7.7 | |
| Bacteroidetes | 13.7 | 10.4 | 13.4 | |
| Cyanobacteria | 0.4b | 9.3c | 1.8c | |
| Nitrospirae | 1.1 | 0.9 | 2.5 | |
| Planctomycetes | 1.9 | 10.5 | 10.4 | |
| Verrucomicrobia | 1.3 | 8.1 | 4.4 | |
| Crenarchaeota | 0.1 | 0.2 | 0.9 | |
See Discussion for phyla.
Taxa were dominated (82 to 94%) by nonphotosynthetic taxa from class ML635J-21.
Taxa were dominated (70 to 94%) by chloroplasts of eukaryotic algae of the order Stramenopiles.
FIG 7.
Proteobacterial class composition in the colonized sand reactors. Each bar shows the average result from 2 to 4 reactors that received different fluid sources during the incubation experiments. Results for reactors with different fluid sources were combined for each time point during each experiment because ANOSIM showed that fluid type did not significantly influence community composition (see Table 2).
The families represented within the main proteobacterial classes and selected non-Proteobacteria phyla for each sand type were summarized for each experiment (see Tables S4 to S6). Based on the phylum-level breakdown in Fig. 6, the major non-Proteobacteria phyla selected for analysis included Acidobacteria, Bacteroidetes, Nitrospirae, Planctomycetes, Verrucomicrobia, and Thaumarchaeota. For the family-level analysis, the numbers of amplicon reads for each family were summed across all water types for a given sand type and divided by the total number of reads for the corresponding phylum. Only families that comprised ≥0.1% of total phylum reads were considered in the analysis.
The detailed treatment of the phylum/family-level analysis is provided in Text S2 in the supplemental material. A key result is that certain taxa were much more abundant in the RBS and (to a lesser extent) PZS reactors than in GWS, including non-Proteobacteria phyla with known heterotrophic physiologies (e.g., Acidobacteria, Planctomycetes, and Verrucomicrobia) and Deltaproteobacteria with unique metabolic properties (fermentative Syntrophobacteraceae, anaerobic respiratory Geobacteraceae, and various diverse heterotrophic taxa from the order Myxococcales). Another major distinction was the presence of different types of “Cyanobacteria” sequences in RBS and PZS versus GWS. A BLAST analysis revealed that the dominant Cyanobacteria operational taxonomic units (OTUs) in RBS (90 to 95% of total Cyanobacteria sequences) and PZS (72 to 84%) actually corresponded to chloroplasts from the order Stramenopiles, a large group of eukaryotic algae that includes diatoms. By contrast, the dominant (88 to 94%) Cyanobacteria OTU in GWS was related to class ML635J-21, which Soo et al. (27) showed is related to the Melainabacteria, a novel group of nonphotosynthetic (non-PS) Cyanobacteria recently identified in the human gut and in groundwater from Rifle, CO (28). The role of non-PS Cyanobacteria in GWS is unknown, whereas the chloroplast 16S rRNA gene sequences (as well as scanning electron microscope [SEM] images [see Fig. S7]) provide unambiguous evidence for the presence of eukaryotic algal biomass in the RBS and PZS.
Although ANOSIM indicated that fluid source, in general, did not have a significant influence on overall microbial community composition, we examined the major Proteobacteria and non-Proteobacteria taxa mentioned above to look for possible case-by-case differences in abundance as a function of fluid source. The search revealed no systematic variations in the relative abundance of different Proteobacteria classes in CF1 and CF2 (data not shown). For non-Proteobacteria in CF1 and CF2, GWS fed with RW showed modest increases in Planctomycetes and Verrucomicrobia over time (see Fig. S8). For DCF, a similar stimulation of Planctomycetes and Verrucomicrobia occurred in GWS reactors in response to RW, Sed DOC, and RBS DOC compared with that in response to ARW (see Fig. S9B and C). Even more striking was the severalfold increase in Bacteroidetes abundance in GWS in response to Sed DOC and RBS DOC (Fig. S9A). The latter results likely account for the significant ANOSIM R value for GWS in DCF (Table 2). No systematic variations in proteobacterial class abundance were observed for DCF (data not shown).
DISCUSSION
Habitat selection of attached HC microbial communities.
Although the sand packs did not reproduce in situ groundwater, riverbed, or hyporheic zone sediment properties, the sand is from the same geological (Hanford) formation and provided a clean substratum on which microbial community development as a function of in situ habitat could be assessed (16). NMDS analysis of the 16S rRNA gene amplicon libraries indicated that distinct attached microbial communities did in fact arise in the three different in situ colonization habitats (Fig. 5). The RBS and PZS were more diverse (see Table S3 in the supplemental material) and contained a much larger proportion of non-Proteobacteria taxa (Fig. 6). These results clearly demonstrate that the habitat imposed deterministic selection on attached microbial communities. This basic finding agrees with a prior high-throughput (pyrosequencing) 16S rRNA gene amplicon analysis of microbial communities in contrasting field and laboratory hyporheic zone sediments (29) and with a wealth of ongoing studies in various lotic ecosystems (see Battin et al. [30] for an excellent review). However, our results do not support the hypothesis that, because of deterministic selection during colonization, microbial communities would show maximal biomass and metabolic activity when exposed to their native fluid source as opposed to alternative DOC sources (Fig. 1 and 2; see also Fig. S6). Instead, it appears that colonized sand type, as a reflection of habitat-driven deterministic selection during colonization, exerted primary control on biomass and metabolic activity (see LMM results in Table 1). Although, in some cases, fluid source had a significant influence on biomass or respiration, the differences did not indicate that a given sand type had a higher biomass or respiration when provided with fluid from its native environment (see pairwise comparisons in Table S1). The overwhelming influence of in situ colonization habitat was also reflected by the fact that (with a few notable exceptions [see below]) microbial community composition generally showed no change in response to fluid source manipulation (see ANOSIM results in Table 2). Despite some internal reorganization of communities in the sand reactors (in particular GWS [see Fig. 6 and 7]), an ANOSIM showed that the three different sand type communities remained distinct (P values < 0.004) over time during each of the experiments (data not shown).
The patterns in the relative abundance of dominant taxa provide insight into potential linkages between community composition and the physiological function of those communities in the different HC habitats. A subset of these taxa was present in comparable numbers in all three habitats, whereas others showed major variations connected to differential organic matter input and metabolism. Metabolically diverse heterotrophic Proteobacteria (31) were abundant in all the colonized sands, which is consistent with other hyporheic microbial communities (22, 29, 30) as well as previous Hanford 300 Area studies (15–17), and indicates a central role of these organisms in organic carbon (OC) decomposition across the HC. The abundance of Pseudomonadaceae relative to all Gammaproteobacteria was particularly high in the GWS, which is consistent with a previous sand colonization study in Hanford 300 Area groundwater (32), where Pseudomonadaceae accounted for 60 to 80% of all Proteobacteria. In addition, incubation experiments with Hanford 300 Area sediments indicated a key role for Pseudomonadaceae and other heterotrophic taxa in OC metabolism (33). Non-Proteobacteria heterotrophs from phylum Bacteroidetes were also abundant in all sand types (3 to 14% of all reads across the three experiments), suggesting these organisms are well adapted to the diverse suite of OC sources present in the Hanford 300 Area HC (15). Likewise, organisms from the genus Nitrospira, a well-known aerobic nitrite-oxidizing taxon (34), accounted for a small percentage of total reads in each sand type and are likely to have been involved in nitrogen cycling across the HC. A similar conclusion applies also to the (presumed) aerobic ammonium-oxidizing Thaumarchaeota families Cenarchaeaceae and Nitrososphaeraceae (35), which is consistent with a recent study that revealed substantial numbers of Thaumarchaeota and inferred their role in nitrogen cycling within the Hanford 300 Area HC (16).
The distributions of other taxa showed clear differences across the three HC habitats. The dominance of aerobic predatory Bdellovibrionaceae (36) and bacteriolytic Kofleriaceae (37) within GWS Deltaproteobacteria suggest that these taxa likely thrive in Hanford 300 Area groundwater through consumption of other heterotrophic microbial biomass. The much higher abundance of other types of Deltaproteobacteria in RBS (and to a lesser extent PZS) than in GWS is an obvious distinction that is almost certainly related to in situ metabolic function. In particular, the presence of fermentative (e.g., Syntrophobacteraceae) and anaerobic respiratory (e.g., Geobacteraceae) organisms point to microbial populations in RBS and PZS that thrive under low O2 or anoxic conditions, e.g., in anaerobic microsites within an otherwise bulk aerobic environment. This idea is consistent with the conclusion that the high biomass and respiration of RBS and (to a lesser extent) PZS compared with those of GWS resulted from the input of fresh river-derived OC (see below) and with the results of an earlier study that provided geochemical evidence for anoxic conditions and cultivation-based evidence for the presence of considerable numbers of anaerobic respiratory taxa in Hanford 300 Area riverbed sediments (38). Among non-Proteobacteria phyla, organisms from the Acidobacteria, Planctomycetes, and Verrucomicrobia phyla were severalfold more abundant in RBS and PZS than in GWS (Table 3). The dominant taxa detected within these phyla (see Text S2 and Tables S4 to S6) were all relatives of heterotrophic organisms whose prominence points to enhanced (relative to GWS) OC decomposition in RBS and PZS reactors by these phyla.
Riverbed hot spot for organic carbon processing.
In addition to RBS displaying 5- to 10-fold higher microbial biomass and respiration than GWS and PZS (Fig. 1 and 2; see also Fig. S6), reactor fluid phase analyses indicated that DOC was released into solution from RBS during the incubation experiments (Fig. 3 and 4). We speculate that the released DOC originated from DOC and/or particulate organic carbon (POC) that accumulated in the RBS (e.g., via natural depositional and sorption processes at the riverbed surface) during in situ incubation. The input of fresh POC was evidenced by the presence of eukaryotic algal chloroplast sequences in the 16S rRNA gene amplicon libraries (Fig. 6), as well as from direct microscopic observation of diatom frustules in the RBS (Fig. S7). We were unable to detect (by high-temperature combustion in an elemental analyzer) differences in the POC content of the different colonized sands from CF1 (all values were near the detection limit of ca. 0.1% C [data not shown]); however, the presence of bioavailable OC in the RBS was clearly reflected by their much higher biomass and respiration rates. In addition, a visual inspection of the RBS reactors at the start of CF1 revealed the presence of a layer of fine-grained materials that settled onto the sand surface after mixing and the addition of filtered RW at the start of the experiment (see Fig. S10B). Such materials were absent in the GWS and present in much lower abundance in the PZS reactors (Fig. S10A and C). Although we cannot exclude the possibility that some of the fresh POC in RBS came from photosynthesis during colonization, the sand packs were contained within a perforated metal tube that remained covered with riverbed sand and gravel during in situ incubation. Based on this, we conclude that the algal biomass that entered the RBS likely came from the water column and/or from periphyton material (see Fig. S4C and D) that was mobilized by turbulence near the riverbed surface. Even if some of the algal biomass came from in situ photosynthesis within the buried tube-enclosed sand packs, it seems likely that such photosynthesis would also take place within the upper several centimeters of the native riverbed material. The key point is that fresh primary production entered into the upper several centimeters of the riverbed surface and had a major impact on microbial community composition and respiratory activity.
The release of DOC in the RBS cross-feeding reactors motivated us to test whether or not riverbed surface sediment and riverbed colonized sand would release DOC during short-term (24 h) “desorption” experiments and, if so, whether such released DOC would stimulate microbial biomass and metabolism in colonized sand packs. Previous studies have documented the influence of labile DOC addition on the composition and/or metabolism of stream hyporheic microbial communities (e.g., see references 24, 29, 39–41), although only one of these studies (29) benefited from the large-scale 16S rRNA gene amplicon sequencing approach employed here. Two potential mechanisms for DOC release during the desorption experiments include (i) actual desorption from surface binding sites on sediment/sand particles and (ii) release associated with POC decomposition. There is ample evidence for sorption of DOC to soil and sediment particle surfaces (3, 42–46) and for the release of sorbed DOC during hydrologic disturbance (47–49). Numerous studies have documented the release of DOC during POC decomposition in soils and sediments (e.g., see references 50–56). Although POC decomposition cannot be ruled out as a mechanism for DOC release, because the experiments were conducted at 4°C and for a relatively short time period, it seems likely that much of the DOC release was via desorption. Given the relatively long-term nature of the cross-feeding experiments compared with that of the desorption experiment, it is possible that at least some of the DOC released during the former originated from POC decay. Unfortunately, the temporal resolution of reactor fluid sampling during the cross-feeding experiments was insufficient to assess the time frame on which DOC was released from the RBS, i.e., whether it was released quickly via desorption after sampling and fluid replacement or more slowly as a result of POC decay. However, the key point is that the RBS contained sources of OC that were mobilized into solution during incubation.
The addition of desorbed DOC had no significant impact on microbial biomass or respiration in the RBS reactors (Fig. 2B and E). However, respiration rates in both the GWS and PZS reactors showed a significant positive response (2- to 3-fold increase) to freshly desorbed DOC sources compared with that to RW (Fig. 2E and F; see also Table S2). In addition, taxa such as Bacteroidetes, Planctomycetes, and Verrucomicrobia, which were abundant members of the non-Proteobacteria phyla in RBS and PZS, were increased in GWS in response to RW, Sed DOC, and RBS DOC (Fig. S7 and S8). The stimulation of such types of heterotrophic taxa by increased OC input agrees with results from hyporheic sediment column experiments where different levels of DOC input were used to assess the influence of OC loading on microbial assemblages (29), although the stimulated phyla differed, with Verrucomicrobia being the only major overlap. The responses by GWS and PZS to river-derived DOC input were also consistent with Stegen et al. (15) who showed that the rising river stage transported bioavailable DOC into the hyporheic zone, thereby stimulating respiration. Likewise, the stimulation of selected heterotrophs by DOC input agrees with the inference by Graham et al. (16) that river water intrusion into the Hanford 300 Area HC selected for an increased abundance of heterotrophic taxa (although not the same ones observed in this study). However, despite the microbial communities and biogeochemical responses of GWS and PZS to the freshly desorbed DOC, these respiration rates were far below those in the RBS reactors. This finding, together with the insignificant impact of desorbed DOC on RBS respiration rates, suggests that OC accumulation and metabolism played a key role in RBS biomass accumulation during in situ colonization and respiration during laboratory incubation.
The DCF results support the idea that the input of OC from the overlying riverbed was at least partially responsible for the relatively high biomass and respiration of the PZS compared with those of the GWS in the cross-feeding experiments. The PZS packs were incubated in the hyporheic zone ca. 1 m below the riverbed surface and therefore had less contact with materials in the river water. However, some fine-grained material was evident in the PZS reactors (Fig. S10C), which was less than in the RBS reactors but more than in the GWS reactors which showed no such layer (Fig. S10A). The PZS 16S rRNA gene amplicon libraries also contained detectable amounts of chloroplast sequences (Fig. 6). These results suggest that the near-surface hyporheic zone received at least some input of POC from the river, which could account for the presence of a microbial community more similar to RBS than GWS (Fig. 6 and 7). In the case of PZS, the algal biomass detected could not have come from in situ photosynthesis but rather must have entered the hyporheic zone via downward transport from the riverbed.
Biogeochemical implications.
The Columbia River represents a premier example of how river stage variations can lead to the large-scale mixing of river water and groundwater. Forecasting models predict an increase in large rain events (>50 mm/day) and total precipitation in North America (see reference 57 and references therein). Such increases in precipitation are expected to escalate the occurrence and intensity of river water-groundwater mixing events (58–60), leading to a higher potential for the development of hot spots and moments within the HC. Although progress has been made in linking microbial community dynamics with biogeochemistry in hyporheic zone environments (see references 16 and 15 and references therein), mixing-induced variations in microbial community structure and function remain a poorly understood component of riverine biogeochemical function (61).
The central goal of this study was to gain experimental insight (through fluid source manipulation, or “cross-feeding”) into how hydrologic exchange may be expected to influence the sizes, activities, and compositions of microbial communities within the nearshore hyporheic zone and the broader HC. The initial hypothesis underlying the work was related to the potential influence of deterministic selection imposed by DOC sources (during in situ colonization) on the community response to fluid source manipulation. However, our results did not concur and instead pointed to a different first-order impact of hydrodynamics on HC microbial communities, namely, the potential for depositional processes and hydrologic exchange to drive labile OC into the riverbed. This process led to much higher microbial biomass and respiration in colonized sand from the riverbed than in sand colonized in the hyporheic zone or in groundwater adjacent to the river. In retrospect, these results are perhaps not surprising, as it is well recognized that on a volume-normalized basis, by far the largest amount of riverine biogeochemical activity takes place either at or just below the riverbed surface (see reference 4 for a review). This is because the concentration and residence time of organic matter and associated microbial communities in riverbed sediments are typically several orders of magnitude greater than in the overlying water (62, 63). From a hydrological perspective, permeable riverbed sediments can be considered reactive sieves or filters which can capture entrained suspended particles and dissolved materials during river water infiltration (6, 30, 64, 65). Thus, riverbed sediments are a focal point for material carried by the river, concentrating both allochthonous and autochthonous organic matter (63, 66, 67). Such organic matter inputs provide electron donors to support an array of respiratory and fermentative reactions, as reflected by the distinct RBS microbial communities reported in this study and the results of an earlier cultivation-based study in Hanford 300 Area hyporheic sediments (38).
Microbial biomass and respiration in the PZS, though much lower than in the RBS, were higher than in GWS, and our experimental results (i.e., from the DCF experiment) suggest that DOC released from riverbed sediment and colonized sand could stimulate the respiration of PZS and GWS microbial communities. This finding agrees with those of seminal studies of hyporheic zone metabolism (59, 68, 69) and highlights a key biogeochemical implication of this study (illustrated in Fig. S11) that the input of POC and its degradation within the near-surface riverbed are likely to drive an influx of labile DOC into the hyporheic zone and potentially the broader HC, depending on the intensity of river water-groundwater exchange. Our results are consistent with the conclusion of Graham et al. (16) that microbial community structure and function in the Hanford 300 Area HC are the result of deterministic selective pressures that come into play during hydrologic exchange. It seems likely that the broad range of physiologies in the RBS and PZS communities means they have the potential to take advantage of frequent shifts in prevailing biogeochemical conditions, such as changes in OC supply and attendant shifts in redox conditions. By comparison, the GWS had a less diverse range of physiologies represented and was likely adapted to relatively stable in-shore biogeochemical conditions. In light of these observations, one might hypothesize that hyporheic zone (including riverbed) communities are more resilient to environmental perturbation than those in more stable groundwater systems. Further studies in HC environments such as the Hanford 300 Area are required to assess the relationship between environmental stability and microbial community/biogeochemical resilience in large river ecosystems. In particular, the potential influence of colonization time on the response of attached communities to environmental perturbation needs to be assessed over a range of time scales that encompasses those of the perturbations, e.g., the large seasonal shifts in hydrobiogeochemical conditions that are known to take place in the Hanford Reach of the Columbia River (15, 16) and other large river ecosystems.
MATERIALS AND METHODS
Hanford 300 Area HC.
The Hanford 300 Area site is located on the north side of the Hanford Reach of the Columbia River, a ca. 80-km free-flowing section of river below Priest Rapids Dam. At this location, the Columbia River flows through Pleistocene flood gravels of the Hanford Formation which are underlain by Miocene-Pliocene fluvial and lacustrine sediments of the Ringold Formation (70). The Hanford Formation sands and gravels have a high conductivity, and daily power generating operations at the Priest Rapids Dam can cause river stage fluctuations of >3 m within a 6-h period (71) and common vertical fluctuations of >1.5 m in a 24-h period (72). Larger seasonal changes in river stage can cause river water intrusion for extended periods of time, extending far into the groundwater zone traveling through underground areas of high conductivity known as paleochannels (73–75) (see Fig. S1 in the supplemental material). Because the river stage is almost always changing, the Hanford Reach HC is extensive and dynamic, with groundwater and river water mixing zones constantly changing spatially and temporally.
In situ sand pack incubation.
Sand pack microcosms were constructed using approximately 80 cm3 of Hanford Formation medium-grade sand (Central Pre-Mix, Pasco, WA) in a modified 2 in. by 4.5 in. 18 mesh (1 mm) stainless steel infusers (R.S.V.P. International, Inc.) plugged with Pyrex fiber glass (Corning, Inc.). The sand packs were combusted at 450°C for 8 h to remove any OC and to render them sterile. Sterile sand packs were incubated in situ for a 6-week period in three distinct locations (habitats) within the Hanford 300 Area corridor (see Fig. S2): (i) within inland groundwater zone well 2-32 (screened within the upper 15 m of the Hanford Formation) located ca. 200 m from the north edge of the river (denoted groundwater sand [GWS]), (ii) within hyporheic zone piezometer P3 in the riverbed (screened over the upper 2 m of sediment) ca. 5 m from the river edge (denoted piezometer sand [PZS]), and (iii) just below riverbed surface, a few meters from the river edge (denoted riverbed sand [RBS]). The photos in Fig. S4 illustrate the 300 Area near-shore environment, including the placement of piezometer T3 and the gravel-covered riverbed. The GWS and PZS packs were incubated in a vertical array attached to stainless steel cable. The RBS packs were contained within perforated (1 mm holes) stainless steel tubes, which were subsequently emplaced within a shallow (ca. 10-cm deep) trench dug into the riverbed and covered by a layer of riverbed gravel to prevent illumination. Periodic visual observation indicated that the tubes remained covered during the incubation period. The RBS packs were placed at an elevation below the seasonal low water level and remained under water throughout the colonization period.
Based on continuous (hourly) measurements of specific conductivity (SpC) in the groundwater well compared with that of river water (see Fig. S5), river water did not intrude into the groundwater well during the in situ sand pack colonization periods. Thus, the GWS microbial communities were not influenced by exposure to intruded river water during in situ colonization. This was important to the experimental design, because it ensured that any response of GWS microbial communities to experimental fluid source manipulation was not confounded by fluid source changes during in situ incubation. In contrast to the groundwater well, the piezometers experienced frequent changes in SpC because of river stage-driven flow reversals and associated river water-groundwater mixing (Fig. S4).
Water sampling.
Groundwater was collected from Hanford 300 Area well 2-32, and RW was collected adjacent to Hanford 300 Area piezometer P3. The fluids were collected on the same day that the sand packs were retrieved after their 6-week in situ incubation period. A peristaltic pump connected to 0.22-μm polyethersulfone Sterivex filters (Millipore Corp.) was used to collect water. Approximately 2 liters of fluid was passed through the filter prior to sample collection. The fluids were collected and stored in sterile acid-washed Nalgene 2-liter Teflon fluorinated ethylene propylene (FEP) bottles (Thermo Fisher). Testing showed that the containers did not leach significant amounts of DOC. All water feedstock solutions were stored at 4°C and measured for DOC at each time point to ensure no loss of DOC during storage. Although no loss of DOC was observed, we did not monitor the composition of the DOC or check for bacterial growth.
Colonized sand incubation experiments.
A conceptual overview of the colonized sand incubation experiments is provided in Fig. S3. Two replicate cross-feeding experiments were conducted with sand pack materials that were colonized in situ during September/October of 2014 or July/August of 2015. After the in situ colonization period, sand packs were retrieved and stored at 4°C. In the laboratory, replicate sand packs of each type were homogenized at room temperature. For cross-feed experiment 1 (CF1), 100-g portions of wet sand were placed in sterile (autoclaved) 150-ml glass jar reactors. The second cross-feed experiment (CF2) was conducted with 50-g portions of wet sand. Most of the reactors were prepared in triplicates, with a few in duplicates. Reactors were loosely capped but not sealed to allow for an aerobic environment and stored in the dark. Feedstock solutions consisted of filter-sterilized (0.2 μm) groundwater (GW), river water (RW), artificial river water (ARW), and artificial groundwater (AGW). The ARW and AGW compositions matched the inorganic contents of their natural counterparts but did not contain any DOC. A 30-ml volume of feed stock solution was added to each reactor at the start of the experiment. At evenly spaced time points over a 5- to 7-week period, all of the fluid phase was extracted with a carbon-free glass pipette and analyzed for DOC concentration determination. After fluid phase extraction, the sand was homogenized using a sterile metal spatula and weighed subsamples (0.5 to 1.5 g) were collected for ATP biomass, resazurin reduction, and 16S rRNA gene amplicon sequencing (see below). Reactors were then provided with fresh feed solution and stored in the dark at room temperature until the next sampling point. The volume of fluid added to the reactors decreased by 2 ml at each sampling point to maintain a constant sand-to-water ratio relative to the amount of sand left in the reactor.
A third experiment, denoted desorption/cross-feeding (DCF), was conducted in March/April 2015 to test if DOC released from riverbed sediment or RBS packs could influence microbial biomass, respiratory activity, and community structure. The concept underlying the DCF experiment was that deposition/accumulation of OC at the riverbed surface could lead to the release of DOC that could subsequently move into the HC and stimulate microbial metabolism during periods of groundwater intrusion, which happens on a daily basis in the Hanford 300 Area (Fig. S5). Sand types and incubation techniques were identical to those of the cross-feed experiments. The feed stock solutions used were river sediment desorbed DOC (denoted Sed DOC), RBS desorbed DOC (denoted RBS DOC), RW, and ARW. Desorbed DOC was obtained by adding 500 g of river sediment or RBS pack material to 500 ml of ARW. The sediment or sediment slurries were vortexed gently for 30 s and allowed to settle for 24 h at 4°C. After settling, the fluid phase was removed and passed through a GF/F filter (Whatman) to remove particulates. We assumed that by maintaining the slurries at 4°C (10 to 15°C lower than in situ temperatures), most of the released DOC came from desorption as opposed to microbial attack on particulate organic carbon (POC) in the sediment or sand. The released DOC solutions were stored at 4°C and tested at each time point to ensure no loss of DOC content throughout the duration of the experiment.
Analytical methods.
A detailed description of the analytical methods employed in this study is provided in Text S1. Briefly, DOC concentrations were determined with a high-temperature combustion carbon analyzer. ATP content and resazurin reduction were used as measures of microbial biomass and aerobic respiration, respectively. Recent studies have demonstrated the utility of resazurin reduction to assess aerobic respiration as a function of microbial biomass and community composition in stream and HC environments (16, 76). The LMM package in R (77) was used to analyze the biomass and respiration data. DNA was extracted from sand microcosm samples using standard commercial kits as previously described (32). 16S rRNA gene amplicon libraries (107 total) were generated using the 515F-806R universal prokaryotic primers (78). The amplicons were sequenced on an Illumina MiSeq instrument at Pacific Northwest National Laboratory. The sequences were processed through the QIIME pipeline (79). The software package Primer (version 7.0) was used for NMDS analysis of microbial community composition and for ANOSIM comparisons of community compositions among different reactor treatments. Dried colonized sand materials were examined using a Hitachi S-3400 variable pressure SEM.
Accession number(s).
Raw sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SAMN07191787 to SAMN07191821 (CF1), SAMN07191822 to SAMN07191859 (CF2), and SAMN07191860 to SAMN07191893 (DCF).
Supplementary Material
ACKNOWLEDGMENTS
We thank Emily Graham (PNNL) for reviewing the manuscript, and acknowledge Shaomei He and Jacqueline Meija (UW—Madison) for helpful discussions.
This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, Subsurface Biogeochemical Research Program through the SBR Scientific Focus Area at the Pacific Northwest National Laboratory (PNNL).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00260-17.
REFERENCES
- 1.Stanford JA, Ward JV. 1993. An ecosystem perspective of alluvial rivers: connectivity and the hyporheic corridor. J North Am Benthol Soc 12:48–60. doi: 10.2307/1467685. [DOI] [Google Scholar]
- 2.Gu CH, Anderson W, Maggi F. 2012. Riparian biogeochemical hot moments induced by stream fluctuations. Water Resour Res 48:W09546. doi: 10.1029/2011WR011720. [DOI] [Google Scholar]
- 3.McCracken KL, McDowell WH, Harter RD, Evans CV. 2002. Dissolved organic carbon retention in soils: comparison of solution and soil measurements. Soil Sci Soc Am J 66:563–568. doi: 10.2136/sssaj2002.5630. [DOI] [Google Scholar]
- 4.Trimmer M, Grey J, Heppell CM, Hildrew AG, Lansdown K, Stahl H, Yvon-Durocher G. 2012. River bed carbon and nitrogen cycling: state of play and some new directions. Sci Total Environ 434:143–158. doi: 10.1016/j.scitotenv.2011.10.074. [DOI] [PubMed] [Google Scholar]
- 5.Tufenkji N, Ryan JN, Elimelec M. 2002. The promise of bank filtration. Environ Sci Technol 36:422A–428A. [DOI] [PubMed] [Google Scholar]
- 6.Hatch CE, Fisher AT, Ruehl CR, Stemler G. 2010. Spatial and temporal variations in streambed hydraulic conductivity quantified with time-series thermal methods. J Hydrol (Amst) 389:276–288. doi: 10.1016/j.jhydrol.2010.05.046. [DOI] [Google Scholar]
- 7.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 8.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 9.Langenheder S, Székely AJ. 2011. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5:1086–1094. doi: 10.1038/ismej.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, Sloan WT. 2010. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci U S A 107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegen JC, Lin X, Fredrickson JK, Konopka AE. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Front Microbiol 6:370. doi: 10.3389/fmicb.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegen JC, Lin X, Konopka AE, Fredrickson JK. 2012. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6:1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stegen JC, Lin XJ, Fredrickson JK, Chen XY, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. 2013. Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stegen JC, Fredrickson JK, Wilkins MJ, Konopka AE, Nelson WC, Arntzen EV, Chrisler WB, Chu RK, Danczak RE, Fansler SJ, Kennedy DW, Resch CT, Tfaily M. 2016. Groundwater-surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nat Commun 7:11237. doi: 10.1038/ncomms11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham EB, Crump AR, Resch CT, Fansler S, Arntzen E, Kennedy DW, Fredrickson JK, Stegen JC. 2016. Coupling spatiotemporal community assembly processes to changes in microbial metabolism. Front Microbiol 7:1949. doi: 10.3389/fmicb.2016.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham EB, Crump AR, Resch CT, Fansler S, Arntzen E, Kennedy DW, Fredrickson JK, Stegen JC. 2017. Deterministic influences exceed dispersal effects on hydrologically-connected microbiomes. Environ Microbiol 19:1552–1567. doi: 10.1111/1462-2920.13720. [DOI] [PubMed] [Google Scholar]
- 18.Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. 2015. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci U S A 112:E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Febria CM, Beddoes P, Fulthorpe RR, Williams DD. 2012. Bacterial community dynamics in the hyporheic zone of an intermittent stream. ISME J 6:1078–1088. doi: 10.1038/ismej.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Febria CM, Hosen JD, Crump BC, Palmer MA, Williams DD. 2015. Microbial responses to changes in flow status in temporary headwater streams: a cross-system comparison. Front Microbiol 6:522. doi: 10.3389/fmicb.2015.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feris KP, Ramsey PW, Frazar C, Rillig M, Moore JN, Gannon JE, Holben WE. 2004. Seasonal dynamics of shallow-hyporheic-zone microbial community structure along a heavy-metal contamination gradient. Appl Environ Microbiol 70:2323–2331. doi: 10.1128/AEM.70.4.2323-2331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feris KP, Ramsey PW, Frazar C, Rillig MC, Gannon JE, Holben WE. 2003. Structure and seasonal dynamics of hyporheic zone microbial communities in free-stone rivers of the western United States. Microb Ecol 46:200–215. doi: 10.1007/BF03036883. [DOI] [PubMed] [Google Scholar]
- 23.Findlay RH, Yeates C, Hullar MAJ, Stahl DA, Kaplan LA. 2008. Biome-level biogeography of streambed microbiota. Appl Environ Microbiol 74:3014–3021. doi: 10.1128/AEM.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Findlay SEG, Sinsabaugh RL, Sobczak WV, Hoostal M. 2003. Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnol Oceanogr 48:1608–1617. doi: 10.4319/lo.2003.48.4.1608. [DOI] [Google Scholar]
- 25.Gao XQ, Olapade OA, Leff LG. 2005. Comparison of benthic bacterial community composition in nine streams. Aquat Microb Ecol 40:51–60. doi: 10.3354/ame040051. [DOI] [Google Scholar]
- 26.Mosher JJ, Findlay RH. 2011. Direct and indirect influence of parental bedrock on streambed microbial community structure in forested streams. Appl Environ Microbiol 77:7681–7688. doi: 10.1128/AEM.06029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Dennis PG, Steen JA, Parks DH, Tyson GW, Hugenholtz P. 2014. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol Evol 6:1031–1045. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, Ley RE. 2013. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Sharp JO, Saikaly PE, Ali S, Alidina M, Alarawi MS, Keller S, Hoppe-Jones C, Drewes JE. 2012. Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl Environ Microbiol 78:6819–6828. doi: 10.1128/AEM.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI. 2016. The ecology and biogeochemistry of stream biofilms. Nat Rev Microbiol 14:251–263. doi: 10.1038/nrmicro.2016.15. [DOI] [PubMed] [Google Scholar]
- 31.Kersters K, Devos P, Gillis M, Swings J, Vandamme P, Stackebrandt E. 2006. Introduction to the Proteobacteria, p 3–37. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed Springer, New York, NY. [Google Scholar]
- 32.Converse BJ, McKinley JP, Resch TC, Roden EE. 2015. Microbial mineral colonization across a subsurface redox transition zone. Front Microbiol 6:858. doi: 10.3389/fmicb.2015.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percak-Dennett EM, Roden EE. 2014. Geochemical and microbiological responses to oxidant introduction in reduced subsurface sediment from the Hanford 300 Area, Washington. Environ Sci Technol 48:9197–9204. doi: 10.1021/es5009856. [DOI] [PubMed] [Google Scholar]
- 34.Daims H. 2014. The family Nitrospiraceae, p 733–749. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: other major lineages of Bacteria and the Archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 35.Oshiki M, Satoh H, Okabe S. 2016. Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ Microbiol 18:2784–2796. doi: 10.1111/1462-2920.13134. [DOI] [PubMed] [Google Scholar]
- 36.Rotem O, Pasternak Z, Jurkevitch E. 2014. Bdellovibrio and like organisms, p 3–17. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 37.Garcia R, Müller R. 2014. The family Haliangiaceae, p 173–181. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 38.Moser DP, Fredrickson JK, Geist DR, Arntzen EV, Peacock AD, Li S-MW, Spadoni T, McKinley JP. 2003. Biogeochemical processes and microbial characteristics across groundwater-surface water boundaries of the Hanford Reach of the Columbia River. Environ Sci Technol 37:5127–5134. doi: 10.1021/es034457v. [DOI] [PubMed] [Google Scholar]
- 39.Foreman CM, Covert JS. 2003. Linkages between dissolved organic matter composition and bacterial community structure, p 343–362. In Findlay SEG, Sinsabaugh RL (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, New York, NY. [Google Scholar]
- 40.Sinsabaugh RL, Covert JS. 2003. Integrating dissolved organic matter metabolism and microbial diversity: an overview of conceptual models, p 426–454. In Findlay SEG, Sinsabaugh RL (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, New York, NY. [Google Scholar]
- 41.Wiegner TN, Kaplan LA, Ziegler SE, Findlay RH. 2015. Consumption of terrestrial dissolved organic carbon by stream microorganisms. Aquat Microb Ecol 75:225–237. doi: 10.3354/ame01761. [DOI] [Google Scholar]
- 42.Dunnivant FM, Jardine PM, Taylor DL, McCarthy JF. 1992. Transport of naturally-occurring dissolved organic carbon in laboratory columns containing aquifer material. Soil Sci Soc Am J 56:437–444. doi: 10.2136/sssaj1992.03615995005600020016x. [DOI] [Google Scholar]
- 43.Moore TR, Turunen J. 2004. Carbon accumulation and storage in mineral subsoil beneath peat. Soil Sci Soc Am J 68:690–696. doi: 10.2136/sssaj2004.6900. [DOI] [Google Scholar]
- 44.Perez MAP, Moreira-Turcq P, Gallard H, Allard T, Benedetti MF. 2011. Dissolved organic matter dynamic in the Amazon basin: sorption by mineral surfaces. Chem Geol 286:158–168. doi: 10.1016/j.chemgeo.2011.05.004. [DOI] [Google Scholar]
- 45.Remington SM, Strahm BD, Neu V, Richey JE, da Cunha HB. 2007. The role of sorption in control of riverine dissolved organic carbon concentrations by riparian zone soils in the Amazon basin. Soil Sci 172:279–291. doi: 10.1097/ss.0b013e318032ab46. [DOI] [Google Scholar]
- 46.Tao S, Lin B. 2000. Water soluble organic carbon and its measurement in soil and sediment. Water Res 34:1751–1755. doi: 10.1016/S0043-1354(99)00324-3. [DOI] [Google Scholar]
- 47.Koelmans AA, Prevo L. 2003. Production of dissolved organic carbon in aquatic sediment suspensions. Water Res 37:2217–2222. doi: 10.1016/S0043-1354(02)00581-X. [DOI] [PubMed] [Google Scholar]
- 48.Komada T, Reimers CE. 2001. Resuspension-induced partitioning of organic carbon between solid and solution phases from a river–ocean transition. Mar Chem 76:155–174. doi: 10.1016/S0304-4203(01)00055-X. [DOI] [Google Scholar]
- 49.Schelker J, Grabs T, Bishop K, Laudon H. 2013. Drivers of increased organic carbon concentrations in stream water following forest disturbance: separating effects of changes in flow pathways and soil warming. J Geophys Res Biogeosci 118:1814–1827. doi: 10.1002/2013JG002309. [DOI] [Google Scholar]
- 50.Bengtson P, Bengtsson G. 2007. Rapid turnover of DOC in temperate forests accounts for increased CO2 production at elevated temperatures. Ecol Lett 10:783–790. doi: 10.1111/j.1461-0248.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- 51.Blodau C, Roulet NT, Heitmann T, Stewart H, Beer J, Lafleur P, Moore TR. 2007. Belowground carbon turnover in a temperate ombrotrophic bog. Global Biogeochem Cycles 21:GB1021. doi: 10.1029/2005GB002659. [DOI] [Google Scholar]
- 52.Godde M, David MB, Christ MJ, Kaupenjohann M, Vance GF. 1996. Carbon mobilization from the forest floor under red spruce in the northeastern U.S.A. Soil Biol Biochem 28:1181–1189. doi: 10.1016/0038-0717(96)00130-7. [DOI] [Google Scholar]
- 53.Kalbitz K, Rupp H, Meissner R. 2002. N, P- and DOC-dynamics in soil and groundwater after restoration of intensively cultivated fens, p 99–116. In Broll G, Merbach W, Pfeiffer E-M (ed), Wetlands in central Europe. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 54.Lehman RM, Mills AL. 1994. Field evidence for copper mobilization by dissolved organic matter. Water Res 28:2487–2497. doi: 10.1016/0043-1354(94)90067-1. [DOI] [Google Scholar]
- 55.Planer-Friedrich B, Hartig C, Lissner H, Steinborn J, Suss E, Hassan MQ, Zahid A, Alam M, Merkel B. 2012. Organic carbon mobilization in a Bangladesh aquifer explained by seasonal monsoon-driven storativity changes. Appl Geochem 27:2324–2334. doi: 10.1016/j.apgeochem.2012.08.005. [DOI] [Google Scholar]
- 56.Zhao LYL, Schulin R, Weng LP, Nowack B. 2007. Coupled mobilization of dissolved organic matter and metals (Cu and Zn) in soil columns. Geochim Cosmochim Acta 71:3407–3418. doi: 10.1016/j.gca.2007.04.020. [DOI] [Google Scholar]
- 57.Trenberth KE. 2011. Changes in precipitation with climate change. Clim Res 47:123. doi: 10.3354/cr00953. [DOI] [Google Scholar]
- 58.Boulton AJ, Findlay S, Marmonier P, Stanley EH, Valett HM. 1998. The functional significance of the hyporheic zone in streams and rivers. Annu Rev Ecol Syst 29:59–81. doi: 10.1146/annurev.ecolsys.29.1.59. [DOI] [Google Scholar]
- 59.Jones JB Jr, Holmes RM. 1996. Surface-subsurface interactions in stream ecosystems. Trends Ecol Evol 11:239–242. doi: 10.1016/0169-5347(96)10013-6. [DOI] [PubMed] [Google Scholar]
- 60.Sophocleous M. 2002. Interactions between groundwater and surface water: the state of the science. Hydrogeol J 10:52–67. doi: 10.1007/s10040-001-0170-8. [DOI] [Google Scholar]
- 61.Marmonier P, Archambaud G, Belaidi N, Bougon N, Breil P, Chauvet E, Claret C, Cornut J, Datry T, Dole-Olivier MJ, Dumont B, Flipo N, Foulquier A, Gerino M, Guilpart A, Julien F, Maazouzi C, Martin D, Mermillod-Blondin F, Montuelle B, Namour P, Navel S, Ombredane D, Pelte T, Piscart C, Pusch M, Stroffek S, Robertson A, Sanchez-Perez JM, Sauvage S, Taleb A, Wantzen M, Vervier P. 2012. The role of organisms in hyporheic processes: gaps in current knowledge, needs for future research and applications. Ann Limnol 48:253–266. doi: 10.1051/limn/2012009. [DOI] [Google Scholar]
- 62.Findlay S. 2010. Stream microbial ecology. J North Am Benthol Soc 29:170–181. doi: 10.1899/09-023.1. [DOI] [Google Scholar]
- 63.Tank JL, Rosi-Marshall EJ, Griffiths NA, Entrekin SA, Stephen ML. 2010. A review of allochthonous organic matter dynamics and metabolism in streams. J North Am Benthol Soc 29:118–146. doi: 10.1899/08-170.1. [DOI] [Google Scholar]
- 64.Battin T, Sengschmitt D. 1999. Linking sediment biofilms, hydrodynamics, and river bed clogging: evidence from a large river. Microb Ecol 37:185–196. doi: 10.1007/s002489900142. [DOI] [PubMed] [Google Scholar]
- 65.Findlay S. 1995. Importance of surface-subsurface exchange in stream ecosystems: the hyporheic zone. Limnol Oceanogr 40:159–164. doi: 10.4319/lo.1995.40.1.0159. [DOI] [Google Scholar]
- 66.Cole JJ, Prairie YT, Caraco NF, McDowell WH, Tranvik LJ, Striegl RG, Duarte CM, Kortelainen P, Downing JA, Middelburg JJ, Melack J. 2007. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems 10:172–185. doi: 10.1007/s10021-006-9013-8. [DOI] [Google Scholar]
- 67.Webster JR, Benfield EF, Ehrman TP, Schaeffer MA, Tank JL, Hutchens JJ, D'Angelo DJ. 1999. What happens to allochthonous material that falls into streams? A synthesis of new and published information from Coweeta. Freshw Biol 41:687–705. doi: 10.1046/j.1365-2427.1999.00409.x. [DOI] [Google Scholar]
- 68.Jones JB., Jr 1995. Factors controlling hyporheic respiration in a desert stream. Freshw Biol 34:91–99. doi: 10.1111/j.1365-2427.1995.tb00426.x. [DOI] [Google Scholar]
- 69.Jones JB Jr, Fisher SG, Grimm NB. 1995. Vertical hydrologic exchange and ecosystem metabolism in a Sonoran Desert stream. Ecology 76:942–952. doi: 10.2307/1939358. [DOI] [Google Scholar]
- 70.Thorne PD, Bergeron MP, Williams MD, Freedman VL. 2006. Groundwater data package for Hanford assessments. PNNL-14573, Rev 1. Pacific Northwest National Laboratory, Richland, WA. doi: 10.2172/882976. [DOI] [Google Scholar]
- 71.Tiffan KF, Garland RD, Rondorf DW. 2002. Quantifying flow-dependent changes in subyearling fall Chinook salmon rearing habitat using two-dimensional spatially explicit modeling. N Am J Fish Manag 22:713–726. doi:. [DOI] [Google Scholar]
- 72.Waichler SR, Perkins WA, Richmond MC. 2005. Hydrodynamic simulation of the Columbia River, Hanford Reach, 1940–2004. PNNL-15226. Pacific Northwest National Laboratory, Richland, WA. [Google Scholar]
- 73.Arntzen EV, Geist DR, Dresel PE. 2006. Effects of fluctuating river flow on groundwater/surface water mixing in the hyporheic zone of a regulated, large cobble bed river. River Res Appl 22:937–946. doi: 10.1002/rra.947. [DOI] [Google Scholar]
- 74.Johnson TC, Slater LD, Ntarlagiannis D, Day-Lewis FD, Elwaseif M. 2012. Monitoring groundwater-surface water interaction using time-series and time-frequency analysis of transient three-dimensional electrical resistivity changes. Water Resour Res 48:W07506. doi: 10.1029/2012WR011893. [DOI] [Google Scholar]
- 75.Zachara JM, Chen XY, Murray C, Hammond G. 2016. River stage influences on uranium transport in a hydrologically dynamic groundwater-surface water transition zone. Water Resour Res 52:1568–1590. doi: 10.1002/2015WR018009. [DOI] [Google Scholar]
- 76.Stanaway D, Haggerty R, Benner S, Flores A, Feris K. 2012. Persistent metal contamination limits lotic ecosystem heterotrophic metabolism after more than 100 years of exposure: a novel application of the resazurin resorufin Smart Tracer. Environ Sci Technol 46:9862–9871. doi: 10.1021/es3015666. [DOI] [PubMed] [Google Scholar]
- 77.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 78.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.