Abstract
The RecFOR pathway has been shown to be essential for DNA repair through the process of homologous recombination in bacteria and, recently, to be important in the recovery of stalled replication forks following UV irradiation. RecO, along with RecR, RecF, RecQ and RecJ, is a principal actor in this fundamental DNA repair pathway. Here we present the three-dimensional structure of a member of the RecO family. The crystal structure of Deinococcus radiodurans RecO (drRecO) reveals possible binding sites for DNA and for the RecO-binding proteins within its three discrete structural regions: an N-terminal oligonucleotide/oligosaccharide-binding domain, a helical bundle and a zinc-finger motif. Furthermore, drRecO was found to form a stable complex with RecR and to bind both single- and double-stranded DNA. Mutational analysis confirmed the existence of multiple DNA-binding sites within the protein.
Keywords: Cys4 zinc-finger, DNA repair, OB fold, RecFOR pathway, RecO
Introduction
Maintaining genomic integrity is crucial in all organisms. DNA damage can be caused by many factors, for example, ultraviolet (UV) radiation, γ-radiation, chemical mutagens and even intrinsic errors during DNA replication. Deinococcus radiodurans tolerates ionising radiation at doses lethal to other organisms and is capable of surviving 5–30 kGy of ionising radiation (Minton, 1994), whereas most other organisms cannot survive doses greater than 50 Gy. Such massive radiation doses are estimated to induce several hundred double-strand breaks, thousands of single-strand gaps and about 1000 sites of DNA base damage per chromosome (Battista, 1997 and references therein). The two RecA-dependent DNA repair pathways (RecBCD and RecFOR), which normally operate independently of each other, are important in double-strand break repair and post-replication daughter-strand gap repair, respectively (Morimatsu and Kowalczykowski, 2003). The primary function of these two pathways in bacteria is the recombination-mediated repair of stalled or collapsed DNA replication forks (Cox, 2001). In D. radiodurans, RecD is the only protein in the RecBCD pathway that has so far been identified, whereas all members of the RecFOR pathway have been identified and annotated (Makarova et al, 2001). Consequently, it has been speculated that the RecFOR pathway might replace some of the RecBCD pathway function in D. radiodurans (Aono et al, 2003). It has also been shown that members of the RecFOR family of proteins can recover the viability of certain RecBCD mutants in Escherichia coli (Amundsen and Smith, 2003).
The RecFOR pathway comprises several proteins, for example, RecQ (3′-5′ helicase), RecJ (5′-3′ nuclease), and the RecF, RecO and RecR proteins, where the latter three, perhaps in a molecular complex, displace single-stranded DNA (ssDNA)-binding protein (SSB) and facilitate the production of a RecA-coated ssDNA filament (Amundsen and Smith, 2003). The RecFOR proteins have been found to be important in protecting the nascent lagging strand when replication forks are stalled on UV radiation damage sites (Chow and Courcelle, 2004). Mutants in any of the RecFOR proteins cause E. coli to become both hypersensitive to UV radiation and show extensive degradation in the nascent lagging strand. This degradation, in turn, is limited in RecF, RecO or RecR mutants, when in addition either RecJ or RecQ is inactivated (Chow and Courcelle, 2004). It has been shown that purified RecF, RecO and RecR proteins can form a complex in an apparent equimolar ratio (Umezu et al, 1993; Hegde et al, 1996), and that the E. coli RecO protein interacts physically with RecF, RecR and SSB (Umezu and Kolodner, 1994; Hegde et al, 1996). The E. coli RecO protein has also been found to have an ability to bind SSB-coated ssDNA and to anneal complementary ssDNA strands regardless of whether the DNA was preincubated with SSB or not (Kantake et al, 2002).
RecR from D. radiodurans (drRecR) has been found to form tetramers in a ring-like structure (Lee et al, 2004). A central hole in the tetramer has a diameter of 30 Å and is suited for binding double-stranded DNA (dsDNA). In the crystal structure of drRecR, two such tetramers are interlocked and it is therefore believed that RecR tetramers can open and close in order to bind DNA (Lee et al, 2004).
The RecFOR complex is specific for the dsDNA–ssDNA junction in the lagging strand of DNA at a stalled replication fork. Mapping this specificity onto the individual components of the RecFOR complex is crucial in understanding the ongoing processes at such sites. Obtaining biochemical and structural information about the involvement of RecO in DNA binding and its interactions with other DNA repair proteins is important. As a first step in understanding the structural aspects of its involvement in DNA repair, the crystal structure of D. radiodurans RecO (drRecO) has been determined. Mutational analysis indicates the regions of the protein that are involved in binding DNA, and reveals that there are multiple sites interacting with DNA, one of which appears to be a species-specific site. The crystal structure and mutational analysis of drRecO, in addition to the recently published structure of drRecR, serve as starting points for comprehension of DNA repair not only in D. radiodurans but also in a wider context.
Results
The structure of RecO from D. radiodurans
The crystal structure of drRecO was determined to 2.4 Å resolution by the single-wavelength anomalous dispersion (SAD) method. drRecO has dimensions of approximately 60 × 30 × 30 Å3 and is composed of an N-terminal oligonucleotide/oligosaccharide-binding (OB) fold region (Murzin, 1993), a three-helix bundle, a Cys4 zinc-finger motif and finally a group of four helices inserted between the three-helix bundle and the zinc-finger motif (Figure 1). There are two molecules per crystallographic asymmetric unit (a.s.u.) with a root-mean-square (r.m.s.) deviation between the two monomers of 0.68 Å for 220 aligned Cα atoms. The buried surface area for each monomer is on average 7.1% (825 Å2), suggesting that the two molecules per a.s.u. is a crystal packing effect. This is in agreement with a single peak at a size consistent with a monomer of drRecO in size exclusion chromatography and in accordance with chemical crosslinking experiments (data not shown). The N-terminal OB fold domain in drRecO consists of residues 1–79 in five highly curved β-strands (β1–β5; Figures 1 and 2), which form a β-barrel. There is normally a capping helix (or occasionally a coil) between β3 and β4 in the classic OB fold (Bochkarev and Bochkareva, 2004), which seals one end of the β-barrel. In drRecO, these residues are flexible and absent from electron density in both monomers (residues 41–45). The OB fold domain is followed by one turn of α-helix (α1) and three long α-helices (residues 86–144; α2–α4) stacked tightly into an antiparallel α-helical bundle. The helical bundle is followed by an unusual Cys4 zinc-finger motif (residues 153–176) with the sequence CX2CX16CX2C. A zinc atom, tetrahedrally coordinated by Cys153, Cys156, Cys173 and Cys176, is well defined in electron density in each monomer. Four additional helices (residues 184–239; α5–α8) are spatially inserted between the zinc-finger and the helical bundle.
Figure 1.
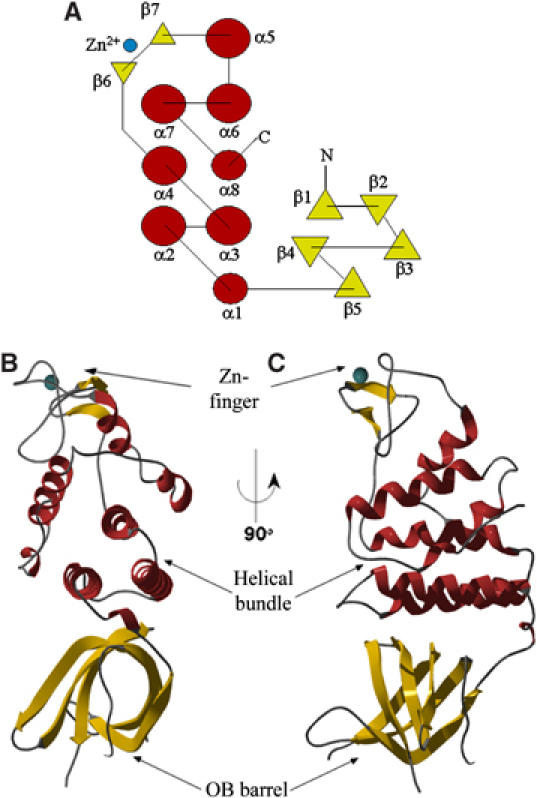
Topology sketch (A) and ribbon illustrations (B, C) of the secondary structure elements in drRecO. α-Helices are shown in red and β-strands in yellow. The zinc atom in the zinc-finger motif is illustrated as a cyan sphere. The illustration given in (C) is rotated 90° from the view in (B).
Figure 2.
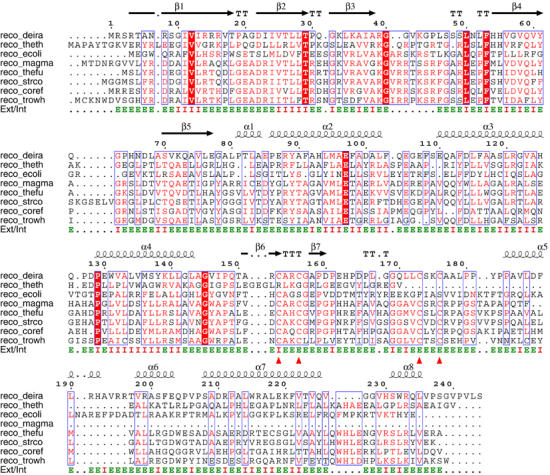
Sequence alignments of RecO from D. radiodurans (TrEMBL, Q9RW50), T. thermophilus (TrEMBL, Q72L07), E. coli (Swiss-Prot, P15027), Magnetospirillum magnetotacticum (NCBI, ZP_00048866), Thermobifida fusca (NCBI, ZP_00058304), Streptomyces coelicolor (Swiss-Prot, Q9L2H3), Corynebacterium efficiens (TrEMBL, Q8FNG1) and Tropheryma whipplei (TrEMBL, Q83MZ1). Secondary structure elements from the crystal structure of drRecO are indicated with spirals (α-helices) and arrows (β-strands) above each row of the sequence alignment. Amino-acid residues having water-accessible surfaces greater than 10 Å2 are indicated with ‘E' for external, otherwise indicated with ‘I' for internal. The four cysteine residues in the zinc-finger motif of drRecO are indicated with red triangles. Equivalent residues conserved in more than 60% (5) of the sequences are boxed and identical residues are shown on a red background.
Structural comparison to other proteins
drRecO has 25% sequence identity to Thermus thermophilus RecO and 17% to E. coli RecO. Sequence alignments of bacterial RecO proteins show that the conservation of the N-terminal part of the proteins is higher than for the C-terminus (Figure 2). Using only the OB fold domain as a search template, DALI (Holm and Sander, 1993) gave good scores for several nucleotide-binding proteins, with the best match being human replication protein A (RPA; Bochkarev et al, 1997), E. coli aspartyl-tRNA synthetase (Eiler et al, 1999) and SSB proteins from E. coli (Raghunathan et al, 2000) and Sulfolobus solfataricus (Kerr et al, 2003), followed by the breast cancer susceptibility gene 2 encoded protein (BRCA2), which also aligns well to the OB fold domain of drRecO.
RPA is the human equivalent of SSB and is a heterotrimer consisting of a total of six OB fold domains, of which four interact with ssDNA (Bochkareva et al, 2002 and references therein). A structural alignment where the DNA-bound structures of RPA and SSB are superimposed onto the OB fold of drRecO displays some similarities in the DNA-binding region. Within the OB fold, ssDNA binding is generally mediated by three distinct structural elements: the β1–β2 loop, the β4–β5 loop and the C-terminal part of β3, where the first provides positively charged residues to form ion-pair(s) with the ssDNA phosphodiester backbone and the latter two each provide one conserved aromatic moiety (Bochkarev and Bochkareva, 2004). These residues are illustrated in Figure 3, where it can be seen that, except for the β4–β5 loop, which is shorter in drRecO than in the other OB-loop-containing proteins, positively charged residues in drRecO occupy similar positions. In all the compared SSB proteins, there are at least two aromatic residues that have been shown to be important in ssDNA binding. In drRecO, no such aromatic residues can be found and there could be a tendency for substitution of aromatic residues in RPA/SSB into positively charged residues in drRecO. Among RecO proteins (Figure 2), R16 and K35 in drRecO are conserved in being positively charged, thus further strengthening this possibility.
Figure 3.
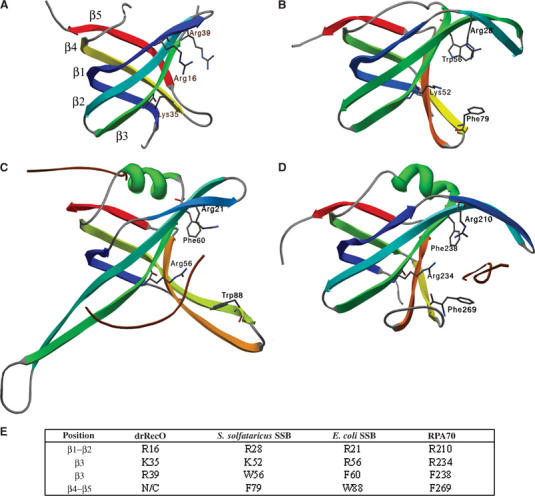
Structural comparison between some aligned OB fold domains. (A) drRecO, (B) S. solfataricus SSB (1O7I), (C) E. coli SSB (1EYG) and (D) human RPA70 (1JMC). All the displayed OB folds are coloured in rainbow colours ranging from blue at the N-terminus to red at the C-terminus. The nucleotide-interacting residues are generally found in the cleft on the right-hand side of the displayed domain. Residues in the β1–β2 loop and in the β4–β5 loop line this cleft. The OB fold domain in drRecO is shorter (81 residues including six missing residues in the β3–β4 loop) than any of the other structurally aligned proteins (85–108). Some of the semiconserved residues thought to be important in ssDNA binding are shown as sticks. In (C, D), the cocrystallised ssDNA is shown as brown coils. (E) Semiconserved residues between the four proteins, also illustrated in (A–D). N/C, not conserved.
Zinc-fingers have long been considered as important mediators of protein–DNA interactions (Branden and Tooze, 1991). Typically, they have the secondary structure succession β–β–α where the α-helix contacts DNA through major-groove interactions. In drRecO, the α-helix is not formed (Figure 4A). A few zinc-finger motifs with resemblance to drRecO were manually identified and aligned, among them being the zinc-finger from the crystal structures of drRecR (Lee et al, 2004) and the Sec23/24 heterodimer (Bi et al, 2002), where additional residues around the zinc coordination sphere (mainly the β–β segment) could be structurally aligned with the drRecO zinc-finger motif (conserved β-strands shown in dark blue in Figure 4A–C). The zinc-finger motif in the RecFOR family member RecQ from E. coli (Bernstein et al, 2003) lacks conservation of the β–β segment and is structurally dissimilar to RecO (Figure 4D). However, the importance of the zinc-finger motif in drRecO is questionable as the E. coli and T. thermophilus RecO proteins both lack the C-terminal zinc-finger motif found in many bacterial RecO proteins.
Figure 4.
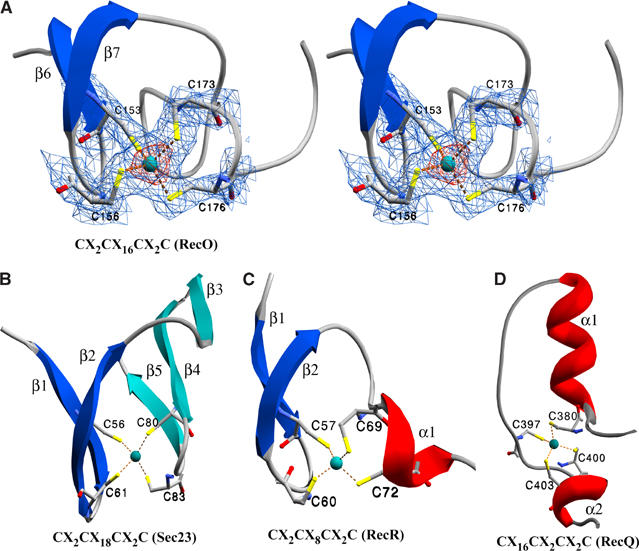
Comparison of Cys4 zinc-finger motifs. (A) Stereo presentation of the zinc-finger in drRecO. The four cysteine residues coordinating the zinc atom are illustrated in a ball-and-stick presentation and the zinc atom is shown as a cyan sphere. The electron density maps are 2mFo−DFC maps contoured at 2σ (blue) and 7σ (red), respectively. The cysteine residues and the zinc atom were all omitted from the refinement. The secondary structure elements are labelled as in Figure 2. (B–D) Zinc-finger motifs from Sec23 (1M2V), RecR (1VDD) and RecQ (1OYW). Secondary structure elements are numbered relative to the figures. β-Strands in dark blue are conserved.
Although the two most striking structural features of drRecO are the OB fold domain and the zinc-finger motif, the presence of the three-helix bundle and the C-terminal helical inserts between the OB fold domain and the zinc-finger should not be neglected. In particular, a patch in the α5–α6 region (190-RHAVRRTVR-200), which is not conserved among the aligned RecO proteins, is rich in positively charged residues, all of which are exposed to solvent and could be of importance in a species-specific function of drRecO (discussed later).
The RecOR complex
E. coli RecR is a dimer in solution and interacts with RecO (Umezu and Kolodner, 1994; Hegde et al, 1996). In E. coli, the two proteins are suggested to form a complex in a 1:1 molar ratio, presumably in the form of a 2:2 heterotetramer (Umezu and Kolodner, 1994). They have also been shown to be required in a 1:1 molar ratio for RecA to overcome inhibition by SSB (Umezu et al, 1993). drRecR has been shown to be a tetramer or an octamer in solution in a concentration-dependent manner (Lee et al, 2004). To investigate the nature of the RecOR complex in D. radiodurans, the proteins were expressed individually and copurified in a three-step purification process. As both proteins were tagged with an N-terminal histidine tag, multiple purification steps were used to ensure that a molecular complex was formed. On a final size exclusion chromatography step, the RecOR complex eluted as a single peak (elution volume: 12.1 ml) corresponding to a molecular weight of approximately 150–160 kDa (Figure 5). RecO and RecR (tetramer) alone eluted later from the size exclusion column at 17.1 and 13.5 ml, respectively, agreeing with their calculated molecular weights (26 and 95 kDa). Although monomeric drRecR (23.7 kDa) is smaller than drRecO (26.3 kDa), it migrates as being slightly larger than drRecO on an SDS–PAGE denaturing gel (Figure 5). The ratio of drRecR/drRecO in the RecOR complex was estimated by comparison with known amounts of individually purified drRecR and drRecO on SDS–PAGE, and was found to be in a 2:1 ratio, probably in the form of a 4:2 heterohexamer.
Figure 5.
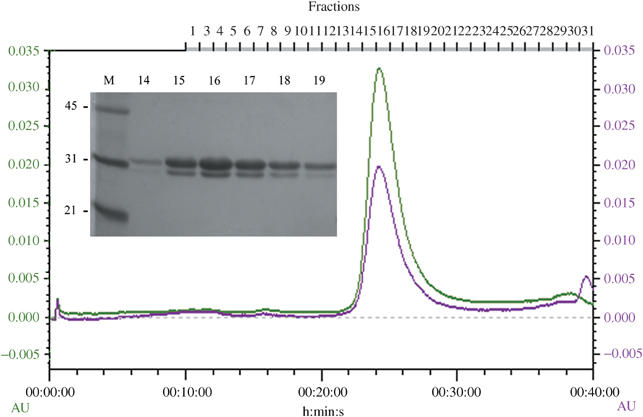
Chromatogram of the gel filtration step for the RecOR complex using a flow rate of 0.5 ml/min. The green and violet lines show the absorbance at 280 and 260 nm, respectively. The inset is the SDS–PAGE denaturing gel of the fractions as shown above the chromatogram. Molecular weight markers (M) are shown in kDa. drRecR (23.7 kDa) migrates as being slightly larger than drRecO (26.3 kDa) and the proteins are at an apparent 2:1 ratio in the RecOR complex.
DNA binding of recombinant drRecO
The DNA-binding properties of drRecO were investigated. Because of the high pI of drRecO, electrophoretic mobility shift assays (EMSA) were analysed by agarose gel electrophoresis in order to visualise migration towards both anode and cathode. The commonly used nondenaturing polyacrylamide bandshift assay did not allow the visualisation of both protein alone and protein–DNA complexes on the same gel within a pH range of 6.5–8.5. Two different approaches were thus used to study the DNA-binding properties of drRecO: (i) visualisation of a DNA bandshift using ethidium bromide staining and (ii) visualisation of a protein bandshift using Coomassie staining. For the first approach, drRecO was incubated with supercoiled plasmid DNA (pcDNA3.1) and the reaction mixtures were analysed by agarose gel electrophoresis (Figure 6A). The addition of drRecO to pcDNA3.1 leads to a dramatic shift of the DNA (Figure 6A, lanes 3 and 4). The addition of Mg2+ to the DNA in the absence of drRecO has no effect on migration (Figure 6A, lane 2). The presence of Mg2+ in lane 4 appears to enhance the binding of drRecO to plasmid DNA, since in its absence a smear of retarded DNA (Figure 6A, lane 3) was observed, as opposed to the well-defined band only just entering the gel when Mg2+ is added to the incubation buffer. This band clearly corresponds to a protein–DNA complex, since in a duplicated experiment when Coomassie stained for protein, a band was observed at the corresponding position (Figure 6B, lane 8). Moreover, the addition of increasing amounts of drRecO to pcDNA3.1 in the presence of Mg2+ resulted in a concentration-dependent shift of the DNA. The incubation of bovine serum albumin (BSA) with pcDNA3.1 did not shift the DNA (Figure 6A, lanes 9 and 10). The shifting of the supercoiled DNA into the well (Figure 6A, lanes 4 and 7) appears to be RecO-specific, while the appearance of a minor form of relaxed circular or linear plasmid DNA may be a result of a nonspecific interaction of protein with supercoiled DNA, since this band is also visible upon incubation of supercoiled DNA with BSA in the presence of Mg2+. To further investigate the DNA-binding ability of drRecO, a second approach was used in which EMSA was carried out at two different pH values (6.8 and 8.3) and analysed by Coomassie staining of the protein. Since drRecO has a high pI (9.2), at a pH of 8.3 with an overall slightly positive charge, the native protein migrates towards the cathode, whereas upon binding to DNA, the overall charge becomes negative and as a result the drRecO—DNA complexes migrate in the opposite direction, that is, towards the anode. At pH 6.8, however, the overall charge of drRecO is significantly more positive and the binding to DNA gives a gel retardation, rather than an inverted migration pattern, caused by a reduction in the overall positive charge of the complex relative to the protein alone. Clear protein bandshifts were observed at pH 8.3 and 6.8 upon incubation of drRecO with ssDNA and dsDNA oligonucleotides (Figure 6B, lanes 2, 3, 10 and 11). However, unlike in the case with supercoiled plasmid DNA, the addition of 40 mM Mg2+ to the reaction mixtures had no significant effect on DNA binding (Figure 6C, lanes 3 and 8). Similarly, the addition of Mg2+ to the protein alone did not alter its migration (Figure 6B, lane 5). In order to further characterise the nature of RecO–DNA interactions, the effect of adding either a metal chelator such as 50 mM Na-EDTA pH 8.0 (to disrupt the C-terminal zinc-finger) or 1 M NaCl (to interfere with ionic interactions) to the protein–DNA mixtures was studied. The analysis of drRecO by size exclusion chromatography in the presence of either 50 mM Na-EDTA pH 8.0 or 1 M NaCl revealed that the overall conformation of drRecO was maintained, as the elution volume of the protein was comparable to that in native conditions (data not shown). Moreover, the addition of either 50 mM Na-EDTA pH 8.0 or 1 M NaCl did not affect the migration of drRecO on the agarose gel (Figure 6C, lanes 11 and 12). The presence of EDTA in the incubation buffer markedly interferes with the binding of drRecO to ssDNA and dsDNA, resulting in a significantly reduced bandshift (Figure 6C, lanes 4 and 9). The resulting protein–DNA complexes are partially retained in the wells and display a more smeared pattern, suggesting that drRecO–DNA interactions are less stable when EDTA is added. A similar smearing is observed upon addition of a high level of salt (1 M NaCl) to the protein–DNA mixtures, although to a lesser extent (Figure 6C, lanes 5 and 10).
Figure 6.
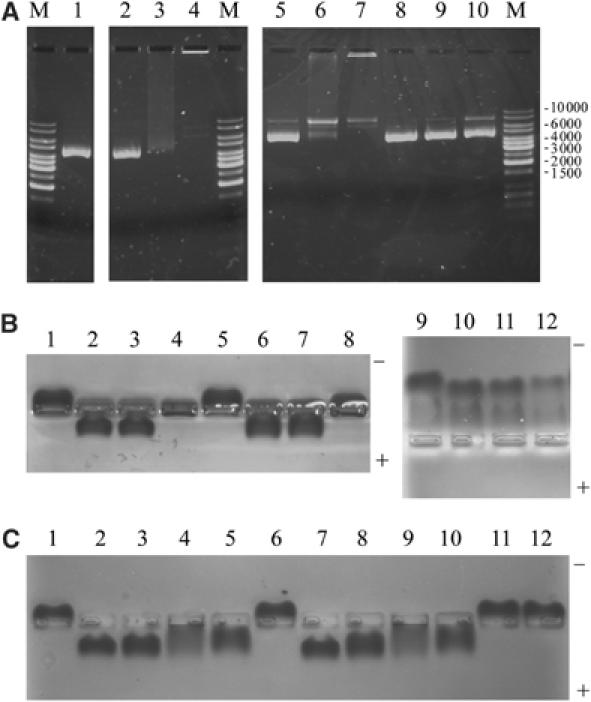
EMSA of drRecO. (A) Agarose gel electrophoresis staining with ethidium bromide for visualising DNA. Lane M, DNA size markers in base pairs; lane 1, 0.2 μg supercoiled plasmid DNA (pcDNA3.1); lane 2, pcDNA3.1+40 mM MgCl2; lane 3, pcDNA3.1+3 μg drRecO; lane 4, pcDNA3.1+drRecO+40 mM MgCl2; lanes 5–7, as for lane 4 but using 0.05, 0.5 and 2 μg drRecO, respectively; lane 8, as for lane 2; lane 9, pcDNA3.1+5 μg BSA; lane 10, pcDNA3.1+BSA+40 mM MgCl2. (B, C) Agarose gel electrophoresis, but Coomassie staining for protein. (B) Lanes 1–8 at pH 8.3. Lane 1, 5 μg drRecO; lane 2, drRecO+1 μg ssDNA; lane 3, drRecO+1 μg dsDNA; lane 4, drRecO+0.2 μg pcDNA3.1; lanes 5–8, as for lanes 1–4 but with 40 mM MgCl2 added; lanes 6–9, as for lanes 1–4, but at pH 6.8. (C) Lane 1, 5 μg drRecO; lane 2, drRecO+1 μg ssDNA; lane 3, drRecO+ssDNA+40 mM MgCl2; lane 4, drRecO+ssDNA+50 mM EDTA; lane 5, drRecO+ssDNA+1 M NaCl; lane 6, drRecO; lane 7, drRecO+1 μg dsDNA; lane 8, drRecO+dsDNA+40 mM MgCl2; lane 9, drRecO+dsDNA+50 mM EDTA; lane 10, drRecO+dsDNA+1 M NaCl; lane 11, drRecO+50 mM EDTA; lane 12, drRecO+1 M NaCl.
DNA binding of drRecO mutants
ssDNA and dsDNA were manually docked onto a molecular surface representation of drRecO (not shown). These models were used as a starting point for generating mutants of drRecO. Both single and multiple point mutants were designed to disrupt positively charged residues (mutations into Ala and/or Glu; see Figure 7A). Of the 16 drRecO mutants obtained, all were expressed, and 12 were soluble and successfully purified (Figure 7C). Based on limited proteolysis, all 12 purified mutants appeared to be folded correctly as they had similar degradation patterns to the wild-type protein (Figure 7D). The four insoluble mutants were K35E, R191A/H192A/R195A/R196A, R14A/R16A/R191A/H192A/R195A/R196A and R14A/R16A/K35A/R39A/R191A/H192A/ R195A/R196A. In order to investigate the DNA-binding abilities of the drRecO mutants, they were incubated with plasmid DNA as described previously for wild-type drRecO (Figure 6A), and the protein–DNA mixtures were analysed on agarose gels (Figure 7A and B). The mutants could be grouped into three major classes: (1) unaffected DNA binding, (2) partially disrupted DNA-binding capability and (3) loss of DNA binding (as summarised in Figure 8F). Upon binding to the plasmid DNA, wild-type drRecO and DNA are retained in the well (Figure 7A and B, lane 2). This is also the case for mutants K35A and the double-mutant R191A/H192A (lanes 5 and 11), suggesting that they have an unaffected binding to DNA. Similarly, mutant R14A/R16A is only very weakly affected by the mutation. On the other hand, for mutant R16E, and the multiple mutants K35E/R39E and K35E/R195E/R196E, DNA binding appears to be significantly impaired, since DNA and protein migrate in opposite directions (lanes 4, 6 and 14). The remaining six mutants display some residual DNA binding, as can be deduced from the smearing of both the DNA and the protein in Figure 7A and B (lanes 7–10, 12 and 13).
Figure 7.
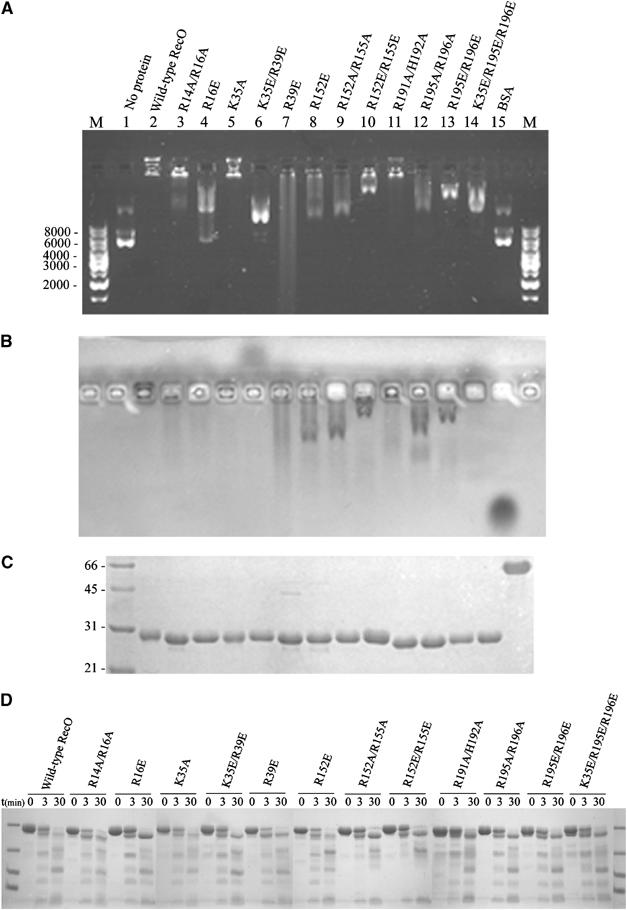
Characterisation of drRecO mutants. (A) Agarose gel electrophoresis staining with SYBR safe for visualising DNA. Lanes M, DNA size markers in base pairs; lane 1, 0.2 μg supercoiled plasmid DNA (pcDNA3.1); lane 2, pcDNA 3.1+40 mM MgCl2+5 μg drRecO; lanes 3–14, as for lane 2, but using 5 μg of the mutant indicated above the lanes; lane 15, as for lane 2, but using 5 μg BSA. (B) The gel from (A) but Coomassie staining for protein. (C) 15% SDS-PAGE gel of the wild-type and mutated drRecO used. Lane 1, broad-range molecular marker in kDa; lane 2, 3 μg wild-type drRecO; lanes 3–14, 3 μg of each of the mutants indicated above the lanes in (A); lane 15, 3 μg BSA. (D) Limited proteolysis of wild-type and mutated drRecO.
Figure 8.
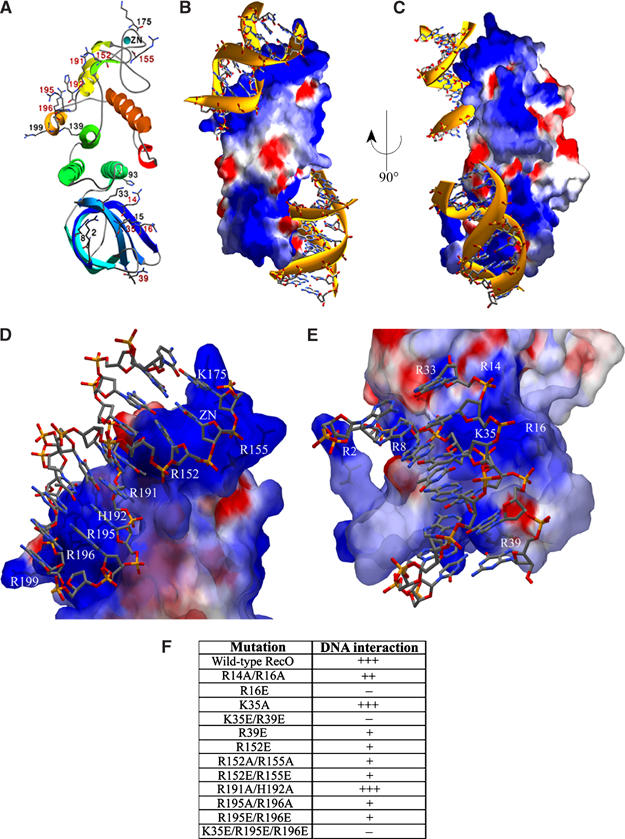
Models for dsDNA interacting with drRecO based on the DNA-binding studies and mutational analysis. In (A), the secondary structure succession is outlined in colours ranging from blue to red. Some positively charged residues are shown for comparison to positive regions seen in the estimated electrostatic surface potentials. Residues mutated in this study are labelled in red. The electrostatic surface potentials in (B–E) are contoured at ±3 kT/e, where red describes a negative and blue a positive potential. dsDNA interacting with drRecO is modelled as sticks in (B, C). Two alternative binding sites involving the OB barrel (bottom) and a positive patch (190-RHAVRRTVR-200) unique for drRecO ending at the zinc-finger (top) are shown. (D) Close-up of dsDNA modelled to interact with the positive patch unique to drRecO with positively charged residues labelled. (E) Close-up of the region in the OB barrel found to be important for dsDNA binding in drRecO. (F) Indication of how well the mutants of drRecO bind to DNA; +++, unaffected DNA-binding ability; +, reduced DNA-binding ability; −, loss of DNA-binding ability.
Discussion
drRecO is a multidomain protein. The OB fold and helical bundle are the most conserved regions among RecO proteins, suggesting that they may be responsible for carrying out vital and conserved functions shared between RecO proteins. While the three-helix bundle is well conserved, no clear function has been assigned to this domain. The tight packing of this helical bundle suggests that it may play a structural role, maintaining the OB fold and the zinc-finger motif in their respective orientations. This domain organisation has also been shown for the trimerisation core of human RPA, in which one helix is contributed from each subunit (Bochkareva et al, 2002 and references therein). The OB fold domain contributed by RPA14 and the zinc-finger motif in the C-terminal domain of RPA70 are at either ends of the protein complex with a three-helix bundle in the middle. The zinc-finger motif and the inserted helices are more weakly conserved within RecO proteins and may perform species-specific roles. Although RecO consists of well-characterised domains, assigning the exact function to each of these regions is not straightforward. Through purification of the RecOR complex, we have shown that drRecO is able to form a stable protein–protein complex. In addition, by EMSA and mutational analysis, we have both identified the ability of drRecO to interact with DNA and further indicated which regions in drRecO are important in protein–DNA interactions. Residues essential for drRecO to interact with dsDNA have been identified.
Protein–protein interactions
We have shown that drRecO and drRecR interact to form a tightly associated, stable complex. The size of the RecOR complex was estimated by size exclusion chromatography studies to be around 150 kDa. SDS–PAGE analysis of known amounts of the individually purified proteins and pure RecOR complex revealed a heterohexamer of four RecR molecules and two RecO molecules. The size of the RecOR complex would then be around 160 kDa (including the N-terminal sequence tags), suggesting that two RecO molecules interact with a tetramer of RecR.
It is well established that E. coli RecO interacts with both SSB and RecR (Umezu and Kolodner, 1994) as well as RecF (Hegde et al, 1996). In E. coli, Hegde et al (1996) have shown that a stable molecular complex can be formed between RecF, RecO and RecR in an apparent equimolar ratio. D. radiodurans RecF has a theoretical pI of 5.33 and is mostly acidic. This is also the case for D. radiodurans SSB, which has a pI of 5.34 and a duplication of the OB fold domain. An obvious common denominator between the proteins that RecO forms molecular complexes with is thus that RecR, SSB and RecF proteins are anionic, while RecO is a highly cationic protein. It is possible that the well-characterised protein–protein interactions (Umezu and Kolodner, 1994; Hegde et al, 1996) are largely dependent on differences in electrostatic potentials between the proteins involved. In addition, DNA itself is negatively charged and the DNA-binding properties of RecO would most likely be affected by interactions of RecO with its protein partners. The proposed ability of RecO to bind to multiple proteins simultaneously may also suggest that more than one region of RecO may be involved in these specific protein–protein interactions. Further biochemical and/or structural studies will be needed to fully map the different interaction sites on RecO.
Protein–DNA interactions
Non-sequence-specific protein–DNA interactions are predominantly either of hydrophobic nature, where aromatic or aliphatic residues contact DNA bases, or of an ionic nature, where polar or positively charged residues contact the negatively charged phosphate backbone. E. coli RecO has previously been shown to interact with both ssDNA and dsDNA (Luisi-DeLuca and Kolodner, 1994), and T. thermophilus RecO interacts with a single-stranded oligonucleotide (Aono et al, 2003). We show that drRecO also interacts with both ssDNA and dsDNA. The binding of drRecO to both ssDNA and dsDNA oligonucleotides was not affected by the addition of 40 mM Mg2+. However, in the case of RecO binding to supercoiled plasmid DNA, the presence of Mg2+ in the buffer did influence binding. Mg2+ is, however, known to strongly affect the conformation of supercoiled DNA (Adrian et al, 1990; Rybenkov et al, 1997a, 1997b), and this may explain the influence on the plasmid DNA-binding properties of RecO. Sequence alignments and secondary structure predictions of RecO proteins from other organisms indicate that the overall structure may be conserved even in the RecO proteins that lack the zinc-finger found in drRecO. This suggests that the zinc-finger motif in drRecO may serve a role mainly as a structural scaffold for maintaining the rigidity of the protein, rather than as a mediator of protein–protein or protein–DNA interactions. However, when EDTA is added in order to disrupt the zinc-finger (Figure 6C, lanes 4 and 9), the DNA binding is highly affected, although not totally abolished. The observed smeared pattern strongly suggests that although drRecO still interacts with DNA in the presence of EDTA, the interactions are altered, and presumably less stable than in its absence. Adding EDTA to the protein itself did not affect its migration on an agarose gel (Figure 6C, lane 11), and size exclusion chromatography of the protein in the presence of EDTA indicated that the monomeric state of the protein was maintained. The Cys4 zinc-finger motif in drRecO appears to be important in binding both ssDNA and dsDNA, although since some interaction is still observed in the presence of EDTA, it is not alone in being important for these interactions. When 1 M NaCl was added to drRecO at the same time as DNA (Figure 6C, lanes 5 and 10), reduced bandshifts were observed for both types of DNA, suggesting that ionic interactions are also important in protein–DNA binding.
Based on the results of the DNA-binding studies and structural comparison, preliminary models of ssDNA and dsDNA bound to drRecO were made (not shown). As the binding of both types of DNA was found to be affected by the addition of 50 mM EDTA, it seems likely that both ssDNA and dsDNA contact the part of the protein where the zinc-finger resides. Electrostatic surface potentials show drRecO to have two regions of pronounced positive charges: (i) a ridge in the α5–α6 region (190-RHAVRRTVR-200), which is close to the zinc-finger and is atypical among RecO proteins (there is a sequence insertion in drRecO; see Figure 2), is exposed to solvent and is rich in positively charged residues, and (ii) a region in the OB barrel, which appears to be conserved in RecO proteins and is rich in positively charged residues. In Figure 8B, the positive ridge runs diagonally on the upper left and the positive region in the OB barrel is in the lower half of the figure. As these regions both have pronounced positive charge and shape complementarity to DNA, they were thought to be equally well suited to interact with DNA. With the apparent conservative substitution from aromatic residues in SSB proteins to positively charged residues in RecO proteins (Figure 3), it seems likely that the OB barrel region is responsible for forming DNA interactions, which are conserved between RecO from different organisms, while the positive patch close to the zinc-finger may be involved in additional species-specific DNA interactions.
As both positive regions cover a large area, mutating positively charged residues into Ala may not be sufficient to abolish all DNA binding in these regions. Mutation into Glu was therefore chosen in some cases in order to generate repulsion between the mutant proteins and DNA, hopefully without compromising the stability of the protein. This assumption was supported by limited proteolysis on wild-type and mutant proteins, which showed the degradation patterns to be similar for all mutants compared to the wild-type protein (Figure 7D). For all proteins, there are similar fragments remaining after 30 min incubation with trypsin, indicating that all proteins are in a native state.
The residues mutated in this study are coloured in red in Figure 8A to illustrate their location in the structure. The result of the DNA-binding study (Figures 7 and 8F) shows that for many of the mutants, the DNA-binding affinity is altered compared to wild-type drRecO, while in some cases no residual DNA binding is observed. The mutations with the most pronounced effect on DNA binding were R16E, K35E/R39E and K35E/R195E/R196E, indicating that the OB fold region is particularly important for DNA binding in drRecO. The mutations with the least marked effect were all Ala mutations: K35A and the double mutants R191A/H192A and R14A/R16A. The remaining mutants, R39E, R152E, R152A/R155A, R152E/R155E, R195A/R196A and R195E/R196E, all appear to have residual binding capacity for DNA, although reduced compared to the wild-type drRecO. As single and double mutants in each of the two regions were found to reduce DNA binding of drRecO, it seems clear that there are multiple DNA-binding sites in drRecO and further that the binding capacity for DNA is somewhat stronger in the N-terminal OB fold region than in the positive ridge and zinc-finger region.
Based on these mutants, dsDNA was modelled to interact with both the positive ridge and zinc-finger region, and with the OB fold region (Figure 8D and E). There appears to be a pronounced shape and charge complementarity between the surfaces of drRecO and dsDNA. For the mutants along the positively charged ridge and zinc-finger region, all except for R191A/H192A display an impaired DNA-binding ability compared to wild-type drRecO. When studying the residues mutated in this region more closely, R152 and R195 appear to interact more closely with DNA, while the side chains of R191, H192 and R196 all point away from the modelled DNA and are probably not directly involved in protein–DNA contacts. Mutants R195A/R196A and R195E/R196E both display a significantly reduced DNA bandshift, suggesting that this part of the protein also binds DNA. Interestingly, the β6–β7 segment in the zinc-finger motif (Figure 4A) is also close to the modelled DNA, and as mutants R152E, R152A/R155A and R152E/R155E all seem to affect DNA binding to similar extents, these mutants may also be influencing the stability of the zinc-finger. The reduced DNA binding observed for R152 and R155 mutants is indeed very similar to that observed upon EDTA treatment of the wild-type RecO. Interestingly, for mutants R152A/R155A and R152E/R152E, the trypsin digestion pattern is slightly altered, which could indicate that the mutation of Arg155 into Ala or Glu either disrupts the trypsin-binding site or modifies the local structure in the zinc-finger region. This change can most likely be assigned to Arg155, since the R152E mutant resembles the degradation pattern of wild-type drRecO (Figure 7D).
For the mutants in the OB fold region, R16E and K35E/R39E seem to have the most drastic effects on DNA binding. In view of the dramatic effect caused by K35E in the K35E/R195E/R196E mutant compared to R195E/R196E (Figure 8A, lanes 13 and 14), R16 and K35 are suggested to be the major contributors to DNA binding in the OB fold. As the single R39E mutation is sufficient to reduce the stability of the protein–DNA complex (smearing of both the protein and DNA), R39 appears to be important as well, although perhaps to a lesser extent. These observations suggest that the OB fold of drRecO is critical for DNA binding. For mutations R14A/R16A and K35A, there appears to be a substantial residual charge complementarity in this region (a total of nine positively charged residues) in order to maintain a DNA-binding ability even in the mutant proteins.
Biological implication
The OB fold is found in a large number of DNA-binding proteins and biochemical and structural studies have revealed its role in nonspecific DNA binding (Bochkarev et al, 1997; Raghunathan et al, 2000; Kerr et al, 2003). Notably, the OB fold of RecO is structurally most similar to the OB folds of ssDNA-binding proteins (RPA, SSB, BRCA2), again supporting our finding that the OB fold is essential for drRecO–DNA interaction. The observation that conserved aromatic residues in the OB fold of RPA/SSB are not present in drRecO and to some extent have been replaced by positively charged residues (Figure 3) is intriguing in that the mode of DNA binding may be modified, as indeed has been suggested for human RAD52 (Singleton et al, 2002). RAD52 is a functional homologue of RecO, which facilitates the displacement of RPA by RAD51 (a RecA homologue) and it also anneals RPA-coated ssDNA with its complementary ssDNA (Kantake et al, 2002). The crystal structure of the N-terminal part of RAD52 reveals a ring-like structure with a deep groove along the surface of the protein where a set of well-conserved positively charged residues are positioned to interact with the ssDNA phosphodiester backbone in such a way that the DNA bases would be pointing outwards. It was suggested that in this way RAD52 could present the ssDNA to allow homologous pairing with a complementary strand and promote DNA strand annealing (Singleton et al, 2002). In our proposed DNA-binding model, the positive residues of drRecO exclusively interact with the negatively charged phosphate backbone of DNA and would therefore allow annealing to take place. In addition, the structural similarities found between BRCA2 and drRecO may reflect their functions in homologous recombination. While BRCA2 has been found to be involved in RAD51-mediated homologous recombination as a stimulator that helps to remove RPA and in the formation of the ssDNA–RAD51 nucleoprotein filament (Yang et al, 2002), there is clear evidence that RecO can displace SSB and facilitate the production of a RecA-coated ssDNA filament (Amundsen and Smith, 2003).
drRecO has been shown to interact with both ssDNA and dsDNA under physiological conditions, and also forms a stable complex with drRecR in what appears to be a heterohexamer composed of four drRecR molecules and two drRecO molecules. drRecR has also been reported to interact with ssDNA and dsDNA, but only when using high Mg2+ concentrations (Lee et al, 2004). As the RecFOR complex is specific for the ssDNA–dsDNA junction in a stalled replication fork, while the individual drRecR and drRecO proteins interact with both ssDNA and dsDNA, it appears that the formation of a molecular complex between the proteins is necessary in order to obtain the specificity for damaged DNA. Since all three protein partners have been shown to be equally important in this function (Chow and Courcelle, 2004), understanding the function of each of the proteins is a first step to a more generalised understanding of the nature of the RecFOR complex. The observation that drRecO can bind both ssDNA and dsDNA and that these interactions may occur simultaneously on different DNA-binding sites of RecO may reflect its central role at DNA replication forks. The ability of RecO to interact with both DNA and proteins is thus most likely essential for its function in DNA repair as a member of the RecFOR complex.
Conclusions
We have determined the three-dimensional structure of a member of the RecO family. The crystal structure reveals an unexpected domain organisation. The ability to interact with RecR and DNA is in agreement with the proposed role of RecO in the RecFOR DNA repair pathway. Mutational analysis of drRecO illustrated the parts of RecO specifically involved in DNA binding. This constitutes an essential step for a better understanding of the multiple functions outlined for this protein in the RecFOR DNA repair pathway. The unusual combination of structural features together with its surprising overall similarity to the heterotrimeric core of the eukaryotic ssDNA-binding protein RPA raises the possibility that drRecO may be an evolutionary ancestor of RPA. Higher order OB fold proteins display a wide variety of structural architectures, which may therefore result from successive gene duplication and domain shuffling events occurring during the course of evolution. The similarity between RecO and eukaryotic proteins such as RPA and also BRCA2 suggests that our understanding of DNA repair pathways in D. radiodurans may also be useful in clarifying the complex mechanisms involved in genome maintenance in higher eukaryotes.
Materials and methods
Cloning, expression and purification of RecO from D. radiodurans
The gene encoding drRecO (SwissProt: Q9RW50) was cloned into pDEST17 (Invitrogen) for expression with an N-terminal hexahistidine tag in collaboration with Protein'eXpert (Grenoble, France). Expression in BL21 Star(DE3)pLysS (Invitrogen) was induced for 4 h at 37°C and the soluble fraction of the lysed cells was loaded on a 5 ml HiTrap Ni-Chelating column (Amersham Biosciences) pre-equilibrated in 50 mM Tris–HCl pH 7.5, 150 mM NaCl and 5 mM β-mercaptoethanol (Buffer A). A salt-wash step using 50 mM Tris–HCl pH 7.5, 1 M NaCl and 5 mM β-mercaptoethanol (Buffer B) was applied prior to a linear gradient of 0–500 mM imidazole. Fractions containing drRecO were identified by SDS–PAGE. Pooled fractions were dialysed overnight into Buffer B, before size exclusion chromatography (Superdex 200 (10/30) column; Amersham Biosciences) in Buffer B. The pooled fractions of drRecO were concentrated to 7 mg/ml and stored at 4°C in Buffer B. Single and multiple point mutants were prepared using either the QuikChange-single- or QuikChange-multi-site-directed mutagenesis kits (Stratagene). The mutants were confirmed by DNA sequencing (EMBL GeneCore Facility), expressed in BL21 Star(DE3)pLysS cells and purified as described for wild-type RecO. For DNA-binding assays, wild-type and mutant RecO proteins were used directly after the Ni-chelating purification step. Limited proteolysis was performed on wild-type and mutant RecO proteins by incubating them with a 1:500 molar ratio of trypsin at room temperature for 0, 3 and 30 min.
Crystallisation and data collection
Crystals were prepared by mixing 1 μl of protein at 7 mg/ml with 1 μl of reservoir solution (either 0.2 M calcium acetate, 8% (w/v) PEG 20000, 8% (w/v) PEG 550 MME, buffered with 0.1 M Tris–HCl at pH 8.5, or 0.8 M sodium formate, 10% (w/v) PEG 8000, 10% (w/v) PEG 1000, buffered with 0.1 M Tris–HCl at pH 7.5). The crystals were thin rectangular plates and were cryoprotected in a solution of the reservoir solution with 20–30% (v/v) glycerol and flash-cooled at 100 K. All diffraction data were collected using beamlines ID14-1 and ID14-4 at the ESRF. A data set was collected on the fixed-wavelength beamline ID14-1 (λ=0.934 Å), to a resolution limit of 2.4 Å. Another crystal, diffracting to 2.7 Å resolution, was thereafter used for data collection at the peak of the zinc absorption edge (λ=1.2827 Å), at ID14-4.
Structure determination and refinement
The data set from the zinc absorption edge was indexed and integrated using MOSFLM (Leslie, 1992), followed by scaling, merging and conversion of the intensities to structure factors using the CCP4 programs SCALA and TRUNCATE (CCP4, 1994). The data set collected at ID14-1 was indexed, integrated and scaled using the XDS program package (Kabsch, 1993) and converted to structure factors using TRUNCATE. The data collection statistics are presented in Table I. The crystals were of space group C2. Using SOLVE (Terwilliger, 2002), in SAD mode, two zinc sites were identified. These two sites were used as input to SHARP (La Fortelle and Bricogne, 1997) and, after solvent-flattening, interpretable electron density maps to 2.7 Å resolution were produced. The NCS matrices were calculated and used as input to DM (CCP4, 1994). ARP/wARP (Perrakis et al, 1999) and RESOLVE (Terwilliger, 2002) built 423 and 292 out of a total of 530 residues in the two molecules in the a.s.u., respectively, of which 80 and 186 side chains were docked from the sequence. Cycles of refinement in REFMAC5 (CCP4, 1994) interspersed with manual rebuilding gave Rwork and Rfree of 22.5 and 26.7%, respectively. The statistics from phasing and refinement are shown in Table I. Atomic coordinates and structure factor data have been deposited in the Protein Data Bank with accession numbers 1W3S and 1W3Ssf, respectively.
Table 1.
Data collection, structure solution and refinement statistics
| Data set | Peak | Remote |
|---|---|---|
| Data collection statistics | ||
| Beamline | ID14-4 | ID14-1 |
| Wavelength (Å) | 1.2827 | 0.934 |
| Resolution range (Å) | 19.9–2.7 (2.85–2.70) | 48.5–2.4 (2.53–2.40) |
| Rsym (%) | 9.3 (48.1) | 6.2 (55.2) |
| Multiplicity | 8.3 (8.3) | 2.3 (2.3) |
| Mean I/σI | 17.8 (4.1) | 8.5 (1.7) |
| Completeness (%) | 99.7 (100.0) | 98.8 (99.7) |
| Anom. completeness (%) | 99.8 (100.0) | — |
| Space group | C2 | C2 |
| Unit cell parameters | ||
| a (Å) | 134.7 | 134.4 |
| b (Å) | 52.1 | 52.4 |
| c (Å) | 100.7 | 101.1 |
| β (deg) | 106.4 | 106.3 |
| Phasing statistics | ||
| No. of heavy atoms/a.s.u. | 2 Zn2+ | — |
| Rcullisa | 0.92 (0.73) | — |
| Phasing powera | 0.65 (1.37) | — |
| FOM SHARPa | 0.21 (0.35) | — |
| FOM DMb | 0.79 (0.86) | — |
| Refinement statistics | ||
| No. of atomsc | — | 3557/3532/23/2 |
| B-factorsc | — | 53/56/43/46 |
| Rfree (%) | — | 26.7 (36.0) |
| Rwork (%) | — | 22.5 (33.0) |
| Geometry | — | |
| Bonds (Å) | — | 0.010 |
| Angles (deg) | — | 1.324 |
| ESU (Å)d | — | 0.219 |
| aStatistics from SHARP phasing for all data to 2.7 Å; numbers in parentheses are for data with a high-resolution cutoff at 4 Å. | ||
| bFOM from DM with averaging using pre-solvent-flattened FOMs from SHARP as input. Otherwise as above. | ||
| cTotal/protein/water/zinc. | ||
| dEstimated overall coordinate error from REFMAC5 based on maximum likelihood. | ||
Production and purification of the RecOR complex
RecR from D. radiodurans (drRecR) was cloned and expressed in the same manner as outlined for drRecO. Cells expressing RecO and RecR were mixed and lysed together and the soluble fraction loaded on a 5 ml HiTrap Ni-Chelating column pre-equilibrated in Buffer A. A linear gradient of 25–500 mM imidazole was applied. Fractions containing both drRecO and drRecR were identified by SDS–PAGE, pooled and dialysed overnight into 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 5 mM β-mercaptoethanol (Buffer C), before being loaded on a MonoQ column (Amersham Biosciences) pre-equilibrated in Buffer C. A linear gradient of 0–50% Buffer B was run and fractions containing pure RecOR complex were thereafter pooled before a final size exclusion chromatography step using a Superdex 200 column equilibrated in Buffer A.
DNA-binding assay
Three types of DNA were used for the DNA-binding experiments: a single-stranded 21-mer oligonucleotide, a double-stranded 21-mer oligonucleotide (5′-TGCGAGGTCAAAGGTCACCTG-3′ and 5′-ACAGGTGACCTTTGACCTCGC-3′) and a supercoiled circular plasmid (pcDNA3.1; Invitrogen). The single-stranded oligonucleotide was obtained by boiling the double-stranded oligonucleotide for 10 min at 95°C followed by rapid cooling on ice. For the plasmid DNA–protein binding assay, 0.2 μg of pcDNA3.1 was incubated with 0, 0.05, 0.5, 2 or 3 μg of drRecO and with 5 μg of BSA, in the presence of either 0 or 40 mM MgCl2 for 2 h at 25°C. The RecO–plasmid DNA complexes were separated by 0.75% agarose gel electrophoresis in 2 × TAE buffer (80 mM Tris–acetate, 2 mM EDTA) and DNA was visualised by ethidium bromide staining. For the protein bandshift assay, two different agarose electrophoresis systems were used: one in 2 × TA (80 mM Tris–acetate) at pH 8.3 and another in 100 mM Tris–HCl at pH 6.8. In both cases, 5 μg of drRecO, which had been purified by size exclusion chromatography in Buffer A, was incubated with either no DNA, 1 μg of single- or double-stranded oligonucleotide DNA or 0.2 μg of plasmid DNA (pcDNA3.1) in 50 mM Tris–HCl pH 6.8. In order to study the nature of the protein–DNA interactions, 40 mM MgCl2 and 50 mM Na-EDTA pH 8.0 or 1 M NaCl were added to the DNA prior to adding protein. The RecO–DNA complexes were incubated for 2 h at 4°C, before being analysed on 1% agarose electrophoresis gels. To study the mutant RecO–DNA interactions, 0.2 μg of pcDNA3.1 was incubated with 5 μg of wild-type RecO, RecO mutants and BSA in the presence of 40 mM MgCl2 for 2 h at 25°C. The protein–DNA mixes were separated on a 0.7% agarose gel in 0.5 × TBE (50 mM Tris–borate, 0.5 mM EDTA) containing SYBR safe DNA stain (Invitrogen). The agarose gels were stained with Coomassie blue in order to visualise the protein bands.
Acknowledgments
This work is part of an ongoing in-house research project in the ESRF Macromolecular Crystallography Group. We acknowledge Dr Peter Barath, Dr Andrew McCarthy, Dr Elspeth Gordon, Dr Hanna-Kirsti S Leiros, Dr Raimond Ravelli and Dr Gordon A Leonard for valuable help and discussions.
References
- Adrian M, ten Heggeler-Bordier B, Wahli W, Stasiak AZ, Stasiak A, Dubochet J (1990) Direct visualization of supercoiled DNA molecules in solution. EMBO J 9: 4551–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Smith GR (2003) Interchangeable parts of the Escherichia coli recombination machinery. Cell 112: 741–744 [DOI] [PubMed] [Google Scholar]
- Aono S, Hartsch T, Schulze-Gahmen U (2003) Crystallization of a member of the recFOR DNA repair pathway, RecO, with and without bound oligonucleotide. Acta Crystallogr D 59: 576–579 [DOI] [PubMed] [Google Scholar]
- Battista JR (1997) Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol 51: 203–224 [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL (2003) High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J 22: 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: 271–277 [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E (2004) From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol 14: 36–42 [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385: 176–181 [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21: 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C, Tooze J (1991) Introduction to Protein Structure. New York: Garland Publishing Inc. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chow KH, Courcelle J (2004) RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J Biol Chem 279: 3492–3496 [DOI] [PubMed] [Google Scholar]
- Cox MM (2001) Historical overview: searching for replication help in all of the rec places. Proc Natl Acad Sci USA 98: 8173–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D (1999) Synthesis of aspartyl-tRNA(Asp) in Escherichia coli—a snapshot of the second step. EMBO J 18: 6532–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde SP, Qin MH, Li XH, Atkinson MA, Clark AJ, Rajagopalan M, Madiraju MV (1996) Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF–RecO–RecR complex in DNA repair and recombination. Proc Natl Acad Sci USA 93: 14468–14473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 24: 795–800 [Google Scholar]
- Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC (2002) Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc Natl Acad Sci USA 99: 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF (2003) Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J 22: 2561–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fortelle Ed, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW (2004) Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J 23: 2029–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, no. 26, Daresbury Laboratory, Warrington, UK
- Luisi-DeLuca C, Kolodner R (1994) Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J Mol Biol 236: 124–138 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ (2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 65: 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton KW (1994) DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol 13: 9–15 [DOI] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Murzin AG (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J 12: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Raghunathan S, Kozlov AG, Lohman TM, Waksman G (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol 7: 648–652 [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR (1997a) The effect of ionic conditions on the conformations of supercoiled DNA. I. Sedimentation analysis. J Mol Biol 267: 299–311 [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR (1997b) The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J Mol Biol 267: 312–323 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA 99: 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated structure solution, density modification and model building. Acta Crystallogr D 58: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD (1993) Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci USA 90: 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Kolodner RD (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem 269: 30005–30013 [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848 [DOI] [PubMed] [Google Scholar]


