Abstract
The recent discovery of ‘molecular subtypes’ in human primary colorectal cancer has revealed correlations between subtype, propensity to metastasize and response to therapy. It is currently not known whether the molecular tumor subtype is maintained after distant spread. If this is the case, molecular subtyping of the primary tumor could guide subtype-targeted therapy of metastatic disease. In this study, we classified paired samples of primary colorectal carcinomas and their corresponding liver metastases (n=129) as epithelial-like or mesenchymal-like, using a recently developed immunohistochemistry-based classification tool. We observed considerable discordance (45%) in the classification of primary tumors and their liver metastases. Discordant classification was significantly associated with the use of neoadjuvant chemotherapy. Furthermore, gene expression analysis of chemotherapy-exposed versus chemotherapy naive liver metastases revealed expression of a mesenchymal program in pre-treated tumors. To explore whether chemotherapy could cause gene expression changes influencing molecular subtyping, we exposed patient-derived colonospheres to six short cycles of 5-fluorouracil. Gene expression profiling and signature enrichment analysis subsequently revealed that the expression of signatures identifying mesenchymal-like tumors was strongly increased in chemotherapy-exposed tumor cultures. Unsupervised clustering of large cohorts of human colon tumors with the chemotherapy-induced gene expression program identified a poor prognosis mesenchymal-like subgroup. We conclude that neoadjuvant chemotherapy induces a mesenchymal phenotype in residual tumor cells and that this may influence the molecular classification of colorectal tumors.
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related mortality. The vast majority of CRC patients die from metastatic disease. At the time of presentation 20–25% of CRC patients already have metastatic disease and an additional ~35% will develop metastases during follow-up.1, 2, 3
The currently used clinical and pathological parameters have insufficient predictive power to identify the patients who are at risk of developing metastases. Recently, gene expression-based molecular classification studies have identified subgroups of human colon cancer.4, 5, 6, 7, 8, 9, 10, 11 Cross-comparison of these studies subsequently revealed the existence of four consensus molecular subtypes (CMS), which are characterized by differential activity of various signaling pathways.12 Interestingly, the molecular subtypes also differ in terms of prognosis. Notably, tumors of the ‘mesenchymal-like’ subtype (CMS4) have a shorter disease-free and overall survival due to higher metastatic potential and relative resistance to chemotherapy- and EGFR-targeted therapy.12, 13, 14 To enable implementation of the novel classification system in clinical practice, we recently developed an immunohistochemistry-based diagnostic test that distinguishes mesenchymal-like cancers from epithelial-like subtypes based on the expression of five markers in tumor cells (HTR2B, FRMD6, ZEB1, CDX2 and pancytokeratin).13 This test can be used for patient stratification and the development of subtype-directed therapies.
The molecular classification system and the novel diagnostic test are based on the analysis of primary colorectal tumors. It is currently unknown whether molecular subtypes are preserved in metastatic cancer, and whether classification of primary colorectal tumors can guide subtype-targeted therapy for metastatic disease. Dissemination and homing to a different organ environment requires adaptations by cancer cells to survive and proliferate, which could result in altered gene expression patterns. Moreover, neoadjuvant therapy is frequently used to downsize primary (rectal) tumors and liver metastases, and may change the constitution of the tumor bulk and affect gene expression in residual cancer cells.
Results and discussion
Neoadjuvant therapy of primary colorectal tumors is associated with a mesenchymal tumor subtype
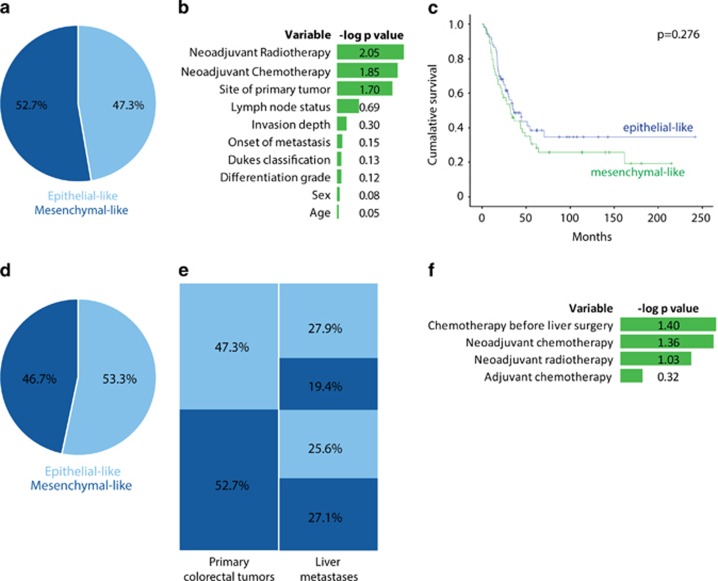
Paraffin-embedded tissue samples of the resection specimens of paired primary colorectal tumors and corresponding liver metastases were available of 129 patients, and were assembled into a tissue microarray. This cohort solely consists of patients with operable colorectal liver metastases. The tissue microarray is created of the resection specimens of both tumors of these patients. All patient characteristics are described in Table 1. In brief, the majority was male, had synchronous metastases and a moderately differentiated colon tumor. Tissue microarray sections were subsequently used for analysis of the expression of HTR2B, FRMD6, ZEB1, CDX2 and pancytokeratin by immunohistochemistry, as described previously.13 After digital analysis of the staining patterns of the primary tumors, 61 tumors were scored as epithelial-like (47.3%) and 68 were scored as mesenchymal-like (52.7%) (Figure 1a). In the current cohort—consisting exclusively of metastasized tumors—the percentage of mesenchymal-like tumors is approximately two-fold higher compared to studies on stage I–IV primary colorectal tumors.12 This is in line with previous analyses of the mesenchymal phenotype in two large cohorts of metastasized primary colorectal tumors.13, 15, 16 Mesenchymal-like tumors have a higher risk of recurrence, which explains their enrichment in these cohorts.
Table 1. Patient characteristics.
|
All tumors |
Epithelial-like |
Mesenchymal-like |
P-value | ||||
|---|---|---|---|---|---|---|---|
| n=129 | (%) | n=61 | (%) | n=68 | (%) | ||
| Age | 61.9 | (37–83) | 62.4 | (42–83) | 61.5 | (37–82) | |
| Sex | |||||||
| Female | 43 | 33.3 | 21 | 34.4 | 22 | 32.4 | 0.835 |
| Male | 86 | 66.7 | 40 | 65.6 | 46 | 67.6 | |
| Onset of metastasis | |||||||
| Synchronous | 76 | 58.9 | 37 | 60.7 | 39 | 57.4 | 0.703 |
| Metachronous | 53 | 41.1 | 24 | 39.3 | 29 | 42.6 | |
| Site of primary tumor | |||||||
| Right colon | 28 | 21.7 | 11 | 18 | 17 | 25 | 0.119 |
| Transverse colon | 4 | 3.1 | 0 | 0 | 4 | 5.9 | |
| Left colon | 13 | 10.1 | 7 | 11.5 | 6 | 8.8 | |
| Sigmoid | 39 | 30.2 | 25 | 41 | 14 | 20.6 | |
| Rectosigmoid | 4 | 3.1 | 1 | 1.6 | 2 | 2.9 | |
| Rectum | 41 | 31.8 | 16 | 26.2 | 25 | 36.8 | |
| Unknown | 1 | 0.8 | 1 | 1.6 | 0 | 0 | |
| Invasion depth | |||||||
| T2 | 9 | 7 | 6 | 9.8 | 3 | 4.4 | 0.506 |
| T3 | 96 | 74.4 | 45 | 73.8 | 51 | 75 | |
| T4 | 22 | 17.3 | 10 | 16.4 | 12 | 17.6 | |
| Unknown | 2 | 1.6 | 0 | 0 | 2 | 2.9 | |
| Lymph node status | |||||||
| N0 | 49 | 38 | 28 | 45.9 | 21 | 30.9 | 0.206 |
| N1 | 48 | 37.2 | 21 | 34.4 | 27 | 39.7 | |
| N2 | 29 | 22.5 | 11 | 18 | 18 | 26.5 | |
| Unknown | 3 | 2.3 | 1 | 1.6 | 2 | 2.9 | |
| Dukes classification | |||||||
| B1 | 5 | 3.9 | 3 | 4.9 | 2 | 2.9 | 0.733 |
| B2 | 20 | 15.5 | 8 | 13.1 | 12 | 17.6 | |
| C1 | 1 | 0.8 | 0 | 0 | 1 | 1.5 | |
| C2 | 27 | 20.9 | 14 | 23 | 13 | 19.1 | |
| D | 75 | 58.1 | 36 | 59 | 39 | 57.4 | |
| Unknown | 1 | 0.8 | 0 | 0 | 1 | 1.5 | |
| Differentiation | |||||||
| Well | 7 | 5.4 | 2 | 3.3 | 5 | 7.4 | 0.752 |
| Well–moderate | 2 | 1.6 | 1 | 1.6 | 1 | 1.5 | |
| Moderate | 78 | 60.5 | 37 | 60.7 | 41 | 60.3 | |
| Moderate–poor | 8 | 6.2 | 3 | 4.9 | 5 | 7.4 | |
| Poor | 11 | 8.5 | 7 | 11.5 | 4 | 5.9 | |
| Unknown | 23 | 17.9 | 11 | 18 | 11 | 16.2 | |
| Neoadjuvant radiotherapy | |||||||
| Yes | 32 | 24.8 | 9 | 14.8 | 23 | 33.8 | 0.009 |
| No | 95 | 73.6 | 52 | 85.2 | 43 | 63.2 | |
| Neoadjuvant chemotherapy | |||||||
| Yes | 13 | 10.1 | 2 | 3.3 | 11 | 16.2 | 0.013 |
| No | 114 | 88.4 | 59 | 96.7 | 55 | 80.9 | |
| Unknown | 2 | 1.6 | 0 | 0 | 2 | 2.9 | |
| Adjuvant chemotherapy | |||||||
| Yes | 45 | 34.9 | 20 | 32.8 | 25 | 36.8 | 0.506 |
| No | 81 | 62.8 | 41 | 67.2 | 40 | 58.8 | |
| Unknown | 3 | 2.3 | 0 | 0 | 3 | 4.4 | |
Patient characteristics of the classified patients. Age was compared via the Wilcoxon rank sum test, for all other variables the X2-test was used.
Figure 1.
Discordant classification of primary colorectal tumors and corresponding liver metastases. (a) The tissue microarray (TMA) was constructed from the resection specimens of primary colorectal tumors and the resection specimens of colorectal liver metastases of 129 patients. Tumor-rich areas were identified via haemotoxylin and eosin stainings and three cores of 0.6 mm were obtained per tumor type. Digital images of TMA immunohistochemically stained slides were obtained via an Aperio Scanscope XT system (Leica Biosystems, Wetzlar, Germany). These were automatically analyzed as described before.13 Cores with a random forest probability of 60% were scored as 'mesenchymal-like'. Patient subtypes were determined using majority consensus. Here a pie chart shows the distribution of epithelial-like and mesenchymal-like of the primary colorectal tumors in our paired tumor cohort. (b) Patient characteristics of epithelial-like tumors were compared to mesenchymal-like tumors. Age was compared via the Wilcoxon rank sum test, for all other variables the X2-test was used. Minus log10 P-values were calculated and are shown in the graph. Table 1 shows the detailed list of patient characteristics. (c) Kaplan–Meier survival curves of the overall survival after liver resection, calculated with a log-rank test (P=0.276). The blue line represents the patients with epithelial-like colorectal tumors and the green line represents patients with mesenchymal-like colorectal tumors. (d) Pie chart of the classification of liver metastases of our paired tumor cohort. (e) Relationship between the classification of primary colorectal tumors and the corresponding liver metastases. (f) The influence of chemotherapy on the classification of paired tumors. Neoadjuvant chemotherapy for the primary colorectal tumors or liver metastases and adjuvant therapy for the primary CRC were all univariate analyzed via the X2-test. Chemotherapy was considered neoadjuvant if it was given to downsize the tumor, primary or metastases, prior to the surgery. Adjuvant chemotherapy is chemotherapy given after the initial resection of the primary colorectal tumor. Concordant tumor pairs were depicted to discordant tumor pairs, which were separated in switching from epithelial-like to mesenchymal-like and vice versa. Minus log10 P-values were calculated and depicted in this graph.
Patient and tumor characteristics (including age, gender, onset of disease, invasion depth, lymph node status, Dukes classification and differentiation status) were equally distributed between the two groups of patients with epithelial-like and mesenchymal-like tumors. However, univariate analysis showed differences in tumor localization: mesenchymal-like tumors were more often located in the rectum, whereas epithelial-like tumors were predominantly located in the sigmoid (P=0.020). Furthermore, we found that patients with mesenchymal-like tumors had more frequently received neoadjuvant radiotherapy (P=0.009) and neoadjuvant chemotherapy (P=0.013) compared to patients with epithelial-like tumors (Figure 1b; Table 1). Multivariate analysis identified neoadjuvant chemotherapy as an independent predictor (P=0.012).
As neoadjuvant chemoradiation is mainly administered to patients with rectal cancer, tumor localization could be a confounding factor in the association between chemoradiation and the mesenchymal phenotype. However, rectal cancers treated with neoadjuvant chemo- and/or radiotherapy more often had a mesenchymal phenotype than untreated rectal cancers (71% versus 30%, respectively; P=0.027), indicating that neoadjuvant therapy is a predictor of the mesenchymal phenotype, independent of tumor localization. These findings are in line with previous results showing that post-treatment rectal tumors were mostly classified to the stroma-rich subtype,17 although this classification was based on stromal parameters.
Mesenchymal-like colorectal tumors (CMS4) have a poorer prognosis compared to the other three CMS.12 In this cohort, which consists solely of patients with operable metastatic colorectal tumors, a trend of survival disadvantage for mesenchymal-like tumors could be observed (P=0.276, Figure 1c). The hazard ratio after multivariate analysis is 1.441 (95% confidence interval (CI) 0.893–2.325, P=0.134).
Discordant molecular classification of primary colorectal tumors and their corresponding liver metastases is associated with neoadjuvant chemotherapy
The immunohistochemical diagnostic test was also applied to the liver metastases of the same patients: 69 metastases were scored as epithelial-like (53.3%) and 60 metastases were scored as mesenchymal-like (46.5%), which roughly corresponds to the subtype distribution of the primary tumors (Figure 1d). However, the classification of primary tumors and the corresponding liver metastases was concordant in only 71 pairs (55% (95% CI 46.3–63.7)), of which 36 were epithelial-like tumors (27.9% (95% CI 20.1–35.8)) and 35 were mesenchymal-like tumors (27.1% (95% CI 19.4–34.9)). Of the 58 discordant tumor pairs (45% (95% CI 36.3–53.7)), 25 primary colorectal tumors that were classified as epithelial-like (19.4% (95% CI 12.5–26.3)) gave rise to liver metastases that were classified as mesenchymal-like, and 33 mesenchymal-like colorectal tumors (25.6% (95% CI 18.0–33.2)) gave rise to epithelial-like liver metastases (Figure 1e). This frequently discordant molecular classification is in contrast with the high concordance that has been reported for mutations in driver genes in primary tumors and the corresponding metastases, for example, KRAS, BRAF and PIK3CA.18, 19 This finding is important as it indicates that molecular subtyping via immunohistochemistry of primary colorectal tumors cannot simply be extrapolated to classify metastatic disease.
We observed a significant correlation between the administration of chemotherapy prior to tumor resection (neoadjuvant chemotherapy) and discordant classification. More than half of the primary tumors that were exposed to neoadjuvant chemotherapy showed discordant classification of the tumor pairs. In the majority of the cases, this was a switch from a mesenchymal-like primary colorectal tumor, to an epithelial-like liver metastasis, rather than vice versa (P=0.044). Similarly, discordant classification was more common in patients who received chemotherapy prior to resection of liver metastasis compared to those who did not. In these cases a switch from an epithelial-like primary colorectal tumor (not exposed to chemotherapy) to a mesenchymal-like liver metastasis (exposed to chemotherapy) was predominant (P=0.040) (Figure 1f; Supplementary Table 1). These findings suggest that neoadjuvant chemotherapy influences cancer cell biology and drives colorectal tumors toward a more mesenchymal-like phenotype.
Molecular changes in response to chemotherapy have been described for other types of cancer. For example, neoadjuvant treatment in breast cancer is associated with gene expression changes and discordant molecular subtype classification in 38% of the cases.20, 21 Chemotherapy-induced changes in HER2 status are also frequently observed in breast cancer and have a potential impact on clinical management.22 Besides chemotherapy, the molecular subtype could also be influenced by the organ microenvironment and/or intra-tumor heterogeneity, and/or heterogeneity between distinct metastases. Indeed, we have recently found that there is extensive subtype heterogeneity among different regions of the same tumor in approximately half of the colorectal tumors analyzed.23 When selecting patients for subtype-targeted therapies, all factors influencing the tumor subtype should be taken into consideration.
Chemotherapy-surviving tumor cells express a mesenchymal gene signature that identifies aggressive CMS4-like tumors.
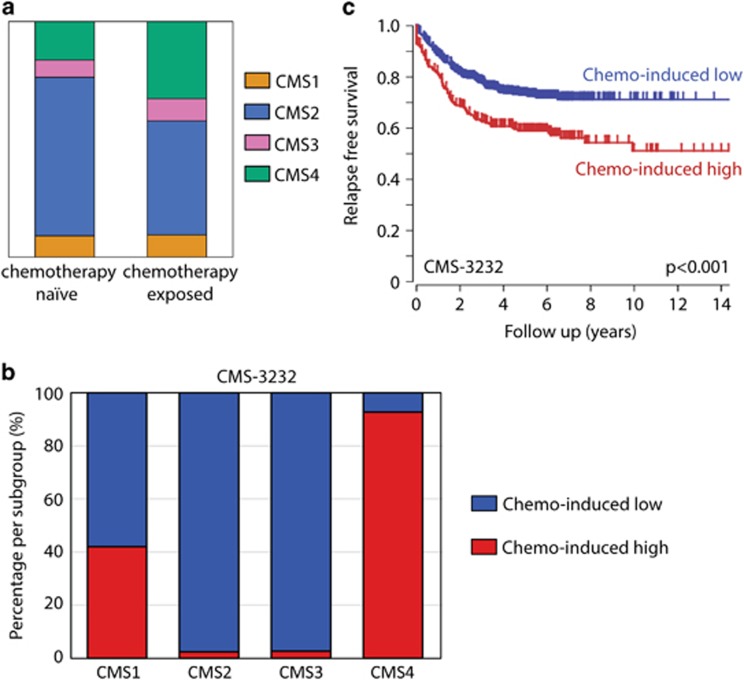
To further explore the relationship between neoadjuvant therapy and the mesenchymal phenotype, we classified liver metastases from a previously published data set24 with the CMS classifier.5, 12 In this same set of liver metastases, we compared chemotherapy-exposed liver metastases to (n=64) chemotherapy naive liver metastases (n=55). Chemotherapy-exposed metastases were more frequently classified as mesenchymal-like compared to the chemotherapy naive liver metastases (33% versus 16%, P=0.06, Figure 2a). In comparison, ~32% of liver metastases exposed to prior treatment in the Khambata-Ford et al.25 cohort were classified as the mesenchymal subtype.5 In an unbiased approach, we identified all genes that were higher expressed in chemotherapy-exposed liver metastases compared to chemotherapy naive liver metastases. These genes were used to cluster the primary tumors of two large cohorts into high and low subgroups. The tumor group expressing high levels of these genes was strongly enriched for mesenchymal-type tumors (CMS4) in the CMS-3232 cohort12 (Figure 2b). These patients had a significantly reduced relapse-free survival probability (Figure 2c).
Figure 2.
5-FU-based chemotherapy is associated with a mesenchymal tumor phenotype. (a) In the liver metastases data set two groups were made, chemotherapy naive and chemotherapy exposed, these were compared in distribution of the CMS classification. The bar graph shows chemotherapy before surgery is associated with an increased proportion of mesenchymal-type tumors (CMS4). The CMS subgroups are CMS1: orange, CMS2: blue, CMS3 pink, CMS4: green. (b) The chemotherapy-induced genes were used to cluster the tumors of the CMS cohorts into chemo-induced high and low subgroups (K-means option in R2, using a two group separation) based on single gene P-values. All tumors had previously been classified into molecular subtypes. The graphs show the distribution of the CMS subtypes within the chemo-induced high and low subgroups. The chemo-induced high subgroup is enriched for mesenchymal subtypes (CMS4). (c) Kaplan–Meier curves showing the differences in relapse-free survival between the chemo-induced high and low subgroups in the CMS-3232 cohort.
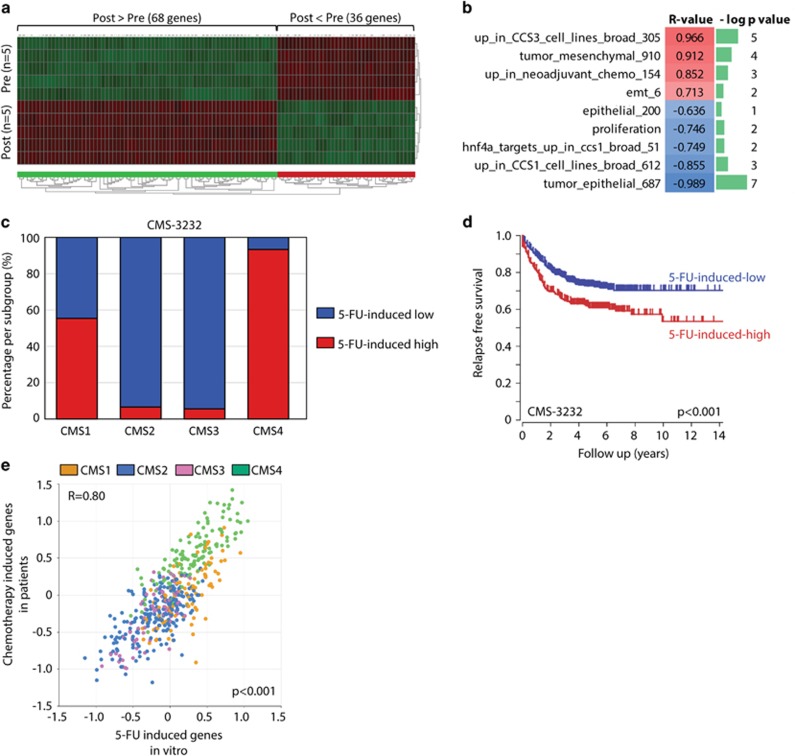
The vast majority of patients receiving neoadjuvant therapy prior to primary tumor resection are patients with rectal cancer. According to the Dutch guidelines, neoadjuvant chemotherapy in this patient category consists of oral capecitabine, a 5-fluorouracil (5-FU) prodrug, in combination with radiation therapy.26, 27 To study a potential causal relationship between chemotherapy and mesenchymal gene expression we exposed patient-derived colonospheres to six cycles of 5-FU. RNA was isolated from colonospheres prior to treatment (n=5) and from the surviving tumor cells after the last cycle (n=5). Gene expression profiling revealed that 5-FU treatment resulted in drastic changes in gene expression (Supplementary Table 2), with 68 significantly upregulated genes and 36 significantly downregulated genes (P<e−6; Figure 3a). Additionally, we found that expression of the 68 5-FU-induced genes was strongly correlated with the expression of previously published gene sets reflecting a mesenchymal tumor phenotype. These include (i) the core drivers of epithelia–mesenchymal transition, (ii) genes that are upregulated in liver metastases that have previously been exposed to chemotherapy, (iii) genes that are highly expressed in the tumor cell compartment of mesenchymal-like tumors28 and (iv) genes that are highly expressed in mesenchymal-like colorectal cancer cell lines in vitro.28 By contrast, the 68 5-FU-induced genes showed a strong negative correlation with (i) a gene set reflecting epithelial differentiation,28 (ii) genes that are highly expressed in the tumor cell compartment of epithelial-like tumors,28 (iii) genes that are highly expressed in epithelial-like colorectal cancer cell lines in vitro and (iv) the target genes of HNF4a, a master inducer of epithelial differentiation and suppressor of mesenchymal genes28 (Figure 3b).
Figure 3.
Chemotherapy induces mesenchymal gene expression in patient-derived colonospheres. (a) Liver metastasis-derived colonospheres were treated with 5-FU for six cycles. RNA was isolated from control (n=5) and 5-FU-treated cells (n=5), and were analyzed by gene expression profiling. The heat map shows all genes that were significantly (P<e−6) upregulated (68) or downregulated (36) in post-treatment tumor cells. See Supplementary Table 2 for a full list of genes. (b) Expression of the 5-FU-induced gene set (68 genes; Supplementary Table 2) was correlated with gene sets reflecting either an epithelial or a mesenchymal tumor phenotype in the data set of the same experiment. Correlations were assessed by using the ‘gene set versus gene sets’ option in the R2 genomics analysis and visualization platform. Gene sets reflecting a mesenchymal tumor cell phenotype positively correlate with the 5-FU-induced gene set (indicated in red), while gene sets reflecting an epithelial phenotype show a negative correlation (in blue). The P-values of the correlations are indicated as minus log10 P-values in green bars. (c) The 68 5-FU-induced genes were used to cluster the tumors of the CMS cohorts into 5-FU-induced high and low subgroups (K-means option in R2, using a two group separation) based on single gene P-values. All tumors had previously been classified into molecular subtypes. The graphs show the distribution of the CMS subtypes within the 5-FU-induced high and low subgroups. The 5-FU-induced high subgroup is enriched for mesenchymal subtypes (CMS4). (d) Kaplan–Meier curves showing the differences in relapse-free survival between the 5-FU-high and 5-FU-low subgroups in both cohorts. (e) Expression of the experimental-derived gene set of 5-FU-induced genes was compared to expression of genes upregulated in chemotherapy-exposed liver metastases by using the ‘relate 2 tracks’ option in the R2 genomics analysis and visualization platform. The XY-plot shows the correlation of the expression of both gene sets (R=0.80, P=2.0e−110) in the CIT subset of the CMS cohort. The CMS subgroups are CMS1: orange, CMS2: blue, CMS3 pink, CMS4: green.
To address this further, we used the 68 5-FU-induced genes to cluster the primary colorectal tumors of two large cohorts into two subgroups with high and low expression of the 5-FU-induced genes (K-means). The 5-FU-high groups were strongly enriched for mesenchymal-type tumors (CMS4) in the CMS-3232 cohort12 (Figure 3c) and had a significantly reduced relapse-free survival probability (Figure 3d). To further assess the relevance of the 5-FU-induced gene set derived from the in vitro experiments, we compared its expression to the genes that are upregulated in liver metastases of patients who were exposed to neoadjuvant chemotherapy (Supplementary Table 3). This revealed a strong correlation (R=0.80, P=2.0e−110) between the expression of both independently generated gene sets (Figure 3e). The tumors expressing the highest levels of 5-FU-induced genes (both the in vitro-derived and patient-derived signatures) were enriched in CMS4 (right upper quadrant), indicating that chemotherapy-induced genes are already highly expressed in treatment-naive mesenchymal-like primary colorectal tumors. There are three genes positively identifying mesenchymal tumor in the IHC test: HTR2B, FRMD6 and ZEB1. ZEB1 was not present on the arrays used and could thus not be evaluated. HTR2B and FRMD6 were both expressed to very low levels in this cohort (2log values lower than 4), for which we have no logical explanation. Indeed, in the CMS-3232 data set we observed a strong positive correlation between the chemotherapy-induced gene set and ZEB1, HTR2B and FRMD6, as expected, with ANOVA P-values of 3.2e−193, 6.8e−149 and 8.8e−315, respectively.
In this report we show that neoadjuvant therapy can influence molecular classification of primary colorectal tumors and liver metastases. Chemotherapy induces a shift toward a more mesenchymal-like phenotype. These results are in line with previous reports showing the treatment-resistant nature of mesenchymal-like tumors both in colon cancer14, 29, 30 and other types of cancer.31 Although chemotherapy has prolonged median overall survival of patients with metastatic CRC from ~6 months to over 2 years, tumor recurrence (almost) always ensues.15, 32 An in-depth analysis of the phenotype of residual tumor cells following chemotherapy will provide novel targets for therapy targeting residual disease. The results in the present study suggest that CMS4-targeted therapy may not only be effective in the treatment of CMS4-diagnosed tumors, but also in the adjuvant treatment of chemotherapy-surviving tumor residue of other CRC subtypes that gained a mesenchymal-like phenotype.
Acknowledgments
We would like to thank Dr J van Gorp from the Diakonessenhuis, Dr CA Seldenrijk from the Antonius Hospital in Nieuwegein and Dr J Meijer from the Rijnstate Hospital in Arnhem for their help providing patient material. Furthermore we thank D Castigliego of the Department of Pathology for the excellent technical assistance. KT was supported by grants from the PON foundation and ‘Vrienden UMCU’. IU, JMJ and KMG were supported by grants from the Dutch Cancer Foundation (IU (UU2014-6617), JMJ (UU2013-5865) and KMG (UU2010-4608)). The study was supported by KT, IU, JMJ, KMG from PON foundation, Dalijn foundation, Dirkzwager-Assink Fund, Gieskes-Strijbis Fund, Dutch Cancer Society UU2014-6617, UU2013-5865, UU2010-4608.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
The authors declare no conflict of interest.
Supplementary Material
References
- Chau I, Cunningham D. Treatment in advanced colorectal cancer: what, when and how? Br J Cancer 2009; 100: 1704–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol 2009; 27: 3109–3116. [DOI] [PubMed] [Google Scholar]
- Midgley R, Kerr D. Colorectal cancer. Lancet 1999; 353: 391–399. [DOI] [PubMed] [Google Scholar]
- Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 2013; 19: 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013; 19: 614–618. [DOI] [PubMed] [Google Scholar]
- Sadanandam A, Wang X, de Sousa EMF, Gray JW, Vermeulen L, Hanahan D et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle 2014; 13: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SC, Park YY, Park ES, Lim JY, Kim SM, Kim SB et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut 2012; 61: 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013; 10: e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer 2014; 134: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A, Beran G, Chresta CM, McWalter G, Pritchard A, Weston S et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics 2012; 5: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budinska E, Popovici V, Tejpar S, D'Ario G, Lapique N, Sikora KO et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol 2013; 231: 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015; 21: 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh A, Trumpi K, de Sousa EMF, Wang X, de Jong JH, Fessler E et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res 2016; 23: 387–398. [DOI] [PubMed] [Google Scholar]
- Song N, Pogue-Geile KL, Gavin PG, Yothers G, Kim SR, Johnson NL et al. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes: secondary analysis of NSABP C-07/NRG Oncology Randomized Clinical Trial. JAMA Oncol 2016; 2: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman M, Antonini NF, Douma J, Wals J, Honkoop AH, Erdkamp FL et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): a phase III randomised controlled trial. Lancet 2007; 370: 135–142. [DOI] [PubMed] [Google Scholar]
- Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 2009; 360: 563–572. [DOI] [PubMed] [Google Scholar]
- Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015; 47: 312–319. [DOI] [PubMed] [Google Scholar]
- Mao C, Wu XY, Yang ZY, Threapleton DE, Yuan JQ, Yu YY et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015; 5: 8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CB, Li F, Ma JT, Zou HW. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest 2012; 30: 741–747. [DOI] [PubMed] [Google Scholar]
- Magbanua MJ, Wolf DM, Yau C, Davis SE, Crothers J, Au A et al. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res 2015; 17: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintman M, Buus R, Cheang MC, Sheri A, Smith IE, Dowsett M. Changes in expression of genes representing key biologic processes after neoadjuvant chemotherapy in breast cancer, and prognostic implications in residual disease. Clin Cancer Res 2016; 22: 2405–2416. [DOI] [PubMed] [Google Scholar]
- Valent A, Penault-Llorca F, Cayre A, Kroemer G. Change in HER2 (ERBB2) gene status after taxane-based chemotherapy for breast cancer: polyploidization can lead to diagnostic pitfalls with potential impact for clinical management. Cancer Genet 2013; 206: 37–41. [DOI] [PubMed] [Google Scholar]
- Ubink I, Elias SG, Moelans CB, Laclé MM, van Grevenstein WMU, van Diest PJ et al. A novel diagnostic tool for selecting patients with mesenchymal-type colon cancer reveals intratumor subtype heterogeneity. J Natl Cancer Inst (e-pub ahead of print 1 August 2017; doi:10.1093/jnci/djw303). [DOI] [PubMed]
- Snoeren N, van Hooff SR, Adam R, van Hillegersberg R, Voest EE, Guettier C et al. Exploring gene expression signatures for predicting disease free survival after resection of colorectal cancer liver metastases. PLoS ONE 2012; 7: e49442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007; 25: 3230–3237. [DOI] [PubMed] [Google Scholar]
- Kacar S, Varilsuha C, Gurkan A, Karaca C. Pre-operative radiochemotherapy for rectal cancer. A prospective randomized trial comparing pre-operative vs postoperative radiochemotherapy in rectal cancer patients. Acta Chir Belg 2008; 108: 518–523. [DOI] [PubMed] [Google Scholar]
- Sauer R, Liersch T, Merkel S, Fietkau R, Hohenberger W, Hess C et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol 2012; 30: 1926–1933. [DOI] [PubMed] [Google Scholar]
- Vellinga TT, den Uil S, Rinkes IH, Marvin D, Ponsioen B, Alvarez-Varela A et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene 2016; 35: 5263–5271. [DOI] [PubMed] [Google Scholar]
- Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res 2006; 12(14 Pt 1): 4147–4153. [DOI] [PubMed] [Google Scholar]
- Tato-Costa J, Casimiro S, Pacheco T, Pires R, Fernandes A, Alho I et al. Therapy-induced cellular senescence induces epithelial-to-mesenchymal transition and increases invasiveness in rectal cancer. Clin Colorectal Cancer 2016; 15: 170–178.e3. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 2009; 106: 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JC, Bekaii-Saab TS, Cohn AL, Hurwitz HI, Kozloff M, Tezcan H et al. Treatment patterns and clinical outcomes in patients with metastatic colorectal cancer initially treated with FOLFOX-bevacizumab or FOLFIRI-bevacizumab: results from ARIES, a bevacizumab observational cohort study. Oncologist 2012; 17: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.