Laser trapping is used to manipulate single attached microtubules in vitro. Direct mechanical measurement shows that attachment of microtubule minus ends to yeast spindle pole bodies is extraordinarily strong.
Abstract
Centrosomes, or spindle pole bodies (SPBs) in yeast, are vital mechanical hubs that maintain load-bearing attachments to microtubules during mitotic spindle assembly, spindle positioning, and chromosome segregation. However, the strength of microtubule-centrosome attachments is unknown, and the possibility that mechanical force might regulate centrosome function has scarcely been explored. To uncover how centrosomes sustain and regulate force, we purified SPBs from budding yeast and used laser trapping to manipulate single attached microtubules in vitro. Our experiments reveal that SPB–microtubule attachments are extraordinarily strong, rupturing at forces approximately fourfold higher than kinetochore attachments under identical loading conditions. Furthermore, removal of the calmodulin-binding site from the SPB component Spc110 weakens SPB–microtubule attachment in vitro and sensitizes cells to increased SPB stress in vivo. These observations show that calmodulin binding contributes to SPB mechanical integrity and suggest that its removal may cause pole delamination and mitotic failure when spindle forces are elevated. We propose that the very high strength of SPB–microtubule attachments may be important for spindle integrity in mitotic cells so that tensile forces generated at kinetochores do not cause microtubule detachment and delamination at SPBs.
INTRODUCTION
The centrosome is the microtubule-organizing center of the cell, responsible for nucleation of microtubules and organization of the bipolar mitotic spindle. Centrosomes serve as mechanical hubs, subjected to force from interpolar microtubules (Dumont and Mitchison, 2009; Goshima and Scholey, 2010; van Heesbeen et al., 2014; Shimamoto et al., 2015), kinetochore microtubules (Liu and Lampson, 2009; Umbreit and Davis, 2012; Chacón et al., 2014), and astral microtubules (Morris, 2000; Laan et al., 2012; Nicholas et al., 2015). Mechanoregulation has been implicated in many cellular processes, including the tension-dependent stabilization of kinetochore–microtubule attachments during mitosis (Nicklas and Koch, 1969; Dewar et al., 2004; Cane et al., 2013; reviewed in Sarangapani and Asbury, 2014). However, the mechanical strength of centrosome–microtubule attachments is unknown, and the role of mechanical signals at centrosomes remains unclear.
The mammalian centrosome consists of a pair of centrioles surrounded by a matrix of proteins. This matrix, called the pericentriolar material, serves as a scaffold and anchors microtubule-nucleating proteins. The pericentriolar material exhibits fluid-like properties, with turnover and growth dynamics that have not yet been fully characterized (Woodruff et al., 2014, 2015). The complexity and dynamics of pericentriolar material make it difficult to understand how forces are transmitted through the mammalian centrosome structure. In contrast, the yeast centrosome, or spindle pole body (SPB), has been well characterized as a highly organized trilaminar structure embedded in the nuclear membrane (Moens and Rapport, 1971; Byers and Goetsch, 1975; O’Toole et al., 1999; reviewed in Jaspersen and Winey, 2004; Kilmartin, 2014).
While yeast SPBs are morphologically distinct from mammalian centrosomes, the microtubule-organizing and nucleation functions, as well as the essential protein components, are highly conserved across organisms (Stearns et al., 1991; Gillingham and Munro, 2000; Wang et al., 2010). The γ-tubulin small complex is essential for microtubule nucleation and caps microtubule minus ends in all organisms (Byers et al., 1978; Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000; Kollman et al., 2015). In yeast, the γ-tubulin small complex is tethered to the core of the SPB via interactions with Spc110 on the nuclear face and Spc72 on the cytoplasmic face (Figure 1A; Knop and Schiebel, 1997, 1998; Sundberg and Davis, 1997; Nguyen et al., 1998; Souès and Adams, 1998). Both yeast tethering proteins, Spc110 and Spc72, are essential, and both share homology with the mammalian proteins of similar function, pericentrin (kendrin) and CDK5RAP2 (centrosomin; Flory et al., 2000; Flory and Davis, 2003; Lin et al., 2014, 2015). Spc110 is also likely to enhance the microtubule-nucleation activity of the γ-tubulin small complex (Kollman et al., 2010; Lin et al., 2014; Lyon et al., 2016).
FIGURE 1:
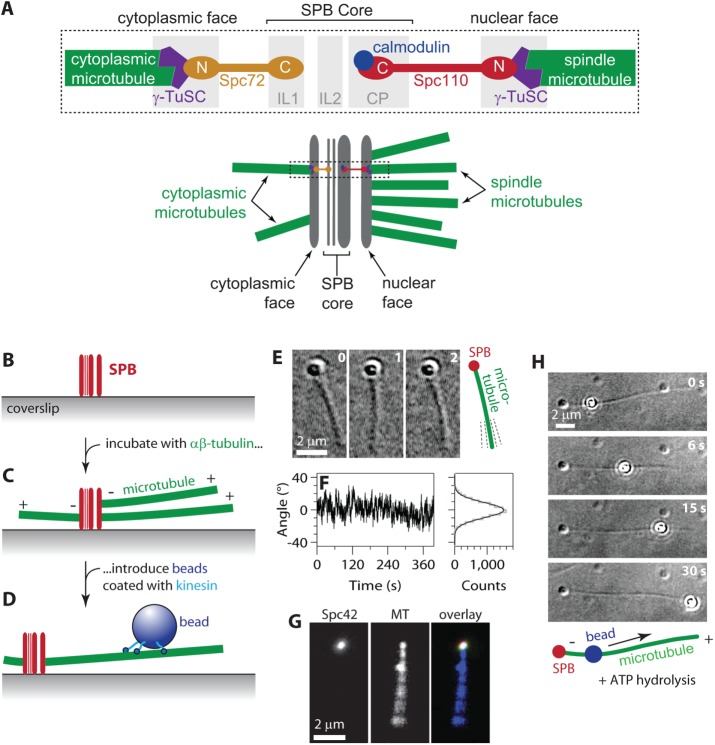
Reconstitution of SPB–microtubule attachments. (A) Arrangement of components within the SPB. Top, microtubule minus ends attach γ-tubulin small complexes (γ-TuSCs) on the nuclear and cytoplasmic faces of the SPB. Spc110 molecules tether γ-TuSCs on the nuclear side to the inner core of the SPB. Spc72 molecules tether γ-TuSCs on the cytoplasmic side. Bottom, in cells, ∼20 microtubules emanate from the nuclear side to form the intranuclear mitotic spindle. Only 2 or 3 emanate from the cytoplasmic side. Dashed box indicates the region depicted in the top diagram. (B–D) Assay for observing individual SPB–microtubule attachments. Purified SPBs adhere tightly to glass coverslips (B). After incubation with free αβ-tubulin to promote nucleation and growth, the SPBs are associated with microtubule minus ends (C). Beads decorated with kinesins form strong linkages to the microtubules (D). (E) Time-lapse VE-DIC images showing pivoting of a microtubule attached to a coverslip-anchored SPB. (F) Fluctuations in the orientation of an SPB-attached microtubule over time (left) and distribution of angles (right, gray histogram) fit with a normal distribution (right, black curve; SD, 9.4°). Additional examples of microtubule pivoting and statistical analyses are shown in Supplemental Figure S2. (G) An individual fluorescent SPB (Spc42-mCherry) bound to a microtubule (MT) and viewed by TIRF microscopy. Pivoting of the filament around its SPB-attached end is evident from blurring of its distal end. (H) In the presence of ATP, a kinesin-decorated bead walks toward the plus end of an SPB-nucleated microtubule. See also Supplemental Movie S1.
Because the molecular organization of the budding yeast SPB is understood more completely than that of other centrosomes, yeast SPBs are ideal for initial characterization of centrosome–microtubule attachment strengths. Their organization suggests clear hypotheses about which molecules bear load and transmit force through the organelle. We used our understanding of SPB structure and the genetic tractability of yeast to explore these hypotheses and report the first measurements of centrosome–microtubule attachment strengths.
RESULTS AND DISCUSSION
Reconstitution of SPB–microtubule attachments
The SPBs used in our experiments were isolated by affinity purification using a C-terminal tandem affinity purification (TAP) tag on Spc97 of the γ-tubulin small complex (Fong et al., 2016). These SPBs also contained Spc42 C-terminally tagged with mCherry for visualization in fluorescence experiments. Yeast strains carrying both tagged components in place of the untagged versions grew at normal rates, indicating that the tagged versions are functional. The presence of Spc110 and Spc97 in the isolated SPBs was verified by Western blot analysis (Supplemental Figure S1A). The presence of all SPB components was verified by mass spectrometry (Supplemental Table S1).
The SPBs were adhered sparsely to KOH-cleaned glass coverslips, and free bovine tubulin was introduced to nucleate microtubules from the SPBs. The nucleated microtubules were stabilized with Taxol, and then polystyrene microbeads, coated with conventional kinesin (a Kinesin-1 construct from Drosophila; see Materials and Methods), were allowed to bind the Taxol-stabilized microtubules (Figure 1, B–D). In the absence of ATP, the kinesin-coated beads formed strong, static attachments to the microtubules and served as handles through which to apply force to the microtubule–SPB interface (as described in the next section). We primarily used video-enhanced differential interference contrast (VE-DIC) imaging to identify the individual SPBs, which were distinctly visible as puncta on the coverslip surface, and to view the nucleated microtubules and the beads (Figure 1, E and H). To confirm that the puncta observed by DIC imaging were indeed SPBs, we identified putative SPBs that had nucleated microtubules by DIC and then examined the same fields of view by total internal reflection fluorescence (TIRF) microscopy. More than 99% of the microtubule-nucleating puncta identified by DIC also exhibited Spc42-mCherry fluorescence (149 of 150 puncta examined). Fluorescently labeled microtubules emanated end-on from the SPB puncta (Figure 1G).
In vivo, microtubules are anchored to centrosomes by their minus ends, while their plus ends extend outward, toward the chromosomes and the cell cortex. To determine whether this polarity was preserved in our in vitro assay, we added ATP to the buffer to activate the plus end–directed walking motility of the kinesin-coated beads on SPB-attached microtubules. After addition of ATP, we allowed the kinesin beads to walk freely along the microtubule, in the absence of externally applied tension (Figure 1H). In 97% of the cases, the beads moved away from the SPBs, toward the free (plus) ends (94 of 97 beads tested). Thus the microtubules were attached to the SPBs by their minus ends, as in the physiological situation.
In vivo, microtubules emanate from SPBs at a variety of angles, indicating that their minus ends are tethered through flexible links (Winey et al., 1995). This flexibility is biologically important, particularly during early mitosis, when pivoting of SPB-associated microtubules allows them to be efficiently captured by kinetochores (Kalinina et al., 2013). Flexibility of the SPB-–microtubule interface is probably also important for establishing a bipolar spindle from a monopolar initial condition because pivoting of microtubules emanating from side-by-side poles is probably required to form antiparallel cross-links (O’Toole et al., 1999; Blackwell et al., 2017). In our in vitro assay, the SPB-nucleated microtubules pivoted about their point of attachment to the SPB, fluctuating around a stable mean orientation (Figure 1, E and F, and Supplemental Figure S2). This behavior shows that their orientation was somewhat constrained yet flexible enough for thermal energy to drive deflections, consistent with the flexible tethering seen in vivo (O’Toole et al., 1999; Kalinina et al., 2013).
Development of a laser trap assay to measure SPB–microtubule attachment strength
Once we verified that our in vitro experimental setup reconstituted biologically relevant SPB–microtubule attachments, we sought to measure their strength using a laser trap. We first selected an SPB that had nucleated a single microtubule to which a kinesin-coated bead had attached. Then the laser trap was controlled by a computer to apply precise tensile forces to the bead, directed away from the SPB (Figure 2, A and B). We operated the trap in a force-ramp mode, in which the force is initially held constant (preload regime; Figure 2C) and then automatically increased at a constant rate (5 pN/s) in an attempt to rupture the SPB–microtubule interface (ramp regime; Figure 2C). In prior work, we used similar force-ramp protocols to measure the rupture strengths of attachments between recombinant kinetochore complexes and microtubule tips (Franck et al., 2010; Tien et al., 2010) and also between native yeast kinetochore particles and dynamic microtubule tips (Akiyoshi et al., 2010; Sarangapani et al., 2014). However, we found that the strength of microtubule attachments to SPBs was much stronger than to kinetochores. Their very high strength posed additional technical challenges for measuring their rupture strength.
FIGURE 2:
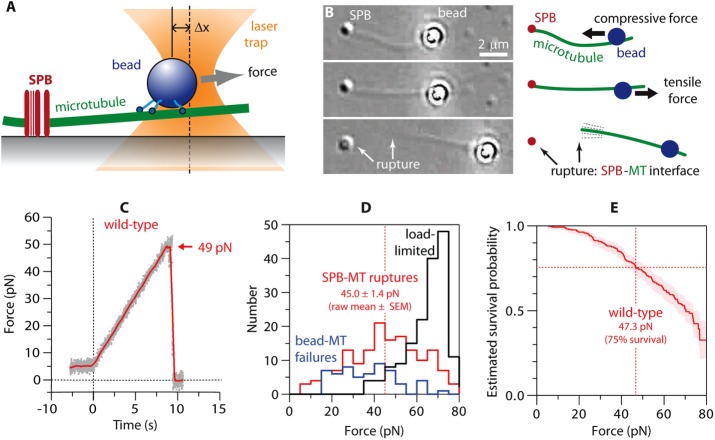
Measuring the strength of individual SPB–microtubule attachments. (A) Beads decorated with kinesins (in the absence of ATP, i.e., in “rigor”) form strong linkages to the microtubules, allowing application of high forces to individual SPB–microtubule attachments using a laser trap. (B) The laser trap (not visible) applies force to a bead linked to an SPB-attached microtubule (MT). Top, initially, the trap pushes the bead toward the SPB, applying compressive force to the SPB-MT interface and causing the MT to buckle. Middle, the direction of force is reversed to apply tensile force to the SPB-MT attachment. Bottom, after the SPB-MT interface ruptures (arrows), the filament remains linked to and pivots around the trapped bead. The SPB appears slightly different in this image due to a slight change in microscope focus. See also Supplemental Movie S2. (C) Tensile force vs. time (force ramp, 5 pN/s) applied to a microtubule attached to a wild-type SPB. Grey dots show raw data. Red trace shows the same data after smoothing (500-ms sliding boxcar average). Dashed vertical line marks the start of the force ramp. Arrow marks rupture at the SPB–microtubule (SPB-MT) interface. (D) Histograms showing outcomes from a series of force ramp experiments using wild-type SPBs. Vertical dashed line marks raw the mean rupture force, computed from only the subset of trials ending with SPB–microtubule rupture. (E) Survival probability as a function of force for microtubule attachments to wild-type SPBs, estimated using Kaplan–Meier analysis. Shaded area shows 95% confidence intervals. Dashed vertical line marks the estimated force at 75% survival.
The tension imposed by the laser trap in our SPB assay is transmitted through the bead–microtubule interface, through the microtubule itself, through the SPB, and into the glass coverslip. To rupture the SPB–microtubule interface, all of the other molecular interfaces along the path of force transmission must remain intact. Fortunately, the SPBs adhered very strongly to and never released from the coverslip. However, the bead–microtubule interface was not always strong enough to remain intact during the force ramp. Consequently a fraction of trials ended in failure at the bead–microtubule interface rather than at the SPB–microtubule interface. A second challenge was that SPB strengths often exceeded the load limit of our instrument (∼65 pN), such that a fraction of trials ended without any rupture. Both alternative outcomes imposed a downward bias on our measurements of SPB–microtubule rupture forces, tending to shift the distributions of rupture force toward lower values.
In experiments with wild-type SPBs, rupture occurred at the SPB–microtubule interface in 41% of trials (156 of 376 trials). The raw mean rupture force, computed from only the subset of trials ending with SPB–microtubule rupture, was 45.0 ± 1.4 pN (mean ± SEM; N = 156; Figure 2D). Failure at the bead–microtubule interface accounted for 15% of the trials (56 of 376) and occurred on average at 36.9 ± 1.8 pN (raw mean ± SEM; N = 56). In the remaining 44% of trials (164 of 376), the trap load-limit was reached at 65.2 ± 0.7 pN (raw mean ± SEM; N = 164), without any rupture or failure. These trials during which the SPB–microtubule interface outlived the bead–microtubule interface or when it survived up to the load limit are analogous to the common situation in clinical trials when a patient drops out of the study before the outcome of interest has occurred (e.g., before death). Clinical data from such patients are “censored” when they are lost from the study, but their survival up to the time of dropout nevertheless provides useful information about the efficacy of the treatment. Similarly, in our experiments, failures at the bead–microtubule interface and trials in which the load limit was reached provide censored data indicating that an SPB–microtubule attachment survived up to the point of censoring. Thus we used all of the data, including censored data, to estimate the true (unbiased) survival probability as a function of force by applying Kaplan–Meier analysis (Kaplan and Meier, 1958), a statistical method used routinely in clinical trials (Rich et al., 2010). This method estimates survival from the number of SPB–microtubule ruptures at each level of force, normalized by the number still “at risk” of rupturing at that force (including those that are later censored; see Materials and Methods). Based on this analysis, estimated force at 75% survival for wild-type SPB attachments was 47.3 pN (95% confidence interval, 42.2–52.5 pN; Figure 2E and Table 1).
TABLE 1:
Summary of rupture strengths for microtubule attachments to wild-type and mutant SPBs and to kinetochores.
| Attachment type | Estimated force at 75% survivala (pN) | Confidence interval, lower bounda (pN) | Confidence interval, upper bounda (pN) | Number of rupture eventsb | Number of events censored by failure at bead | Number of events censored by load limit | p value compared with wild-type SPBs (log-rank test) |
|---|---|---|---|---|---|---|---|
| Wild-type SPBs | 47.3 | 42.2 | 52.5 | 156 | 56 | 164 | — |
| Spc72-AID SPBs | 50.3 | 41.0 | 60.0 | 42 | 47 | 51 | 0.95 |
| Spc110-407 SPBs | 32.0 | 24.5 | 46.1 | 47 | 78 | 31 | 0.041 |
| Kinetochores | 11.6 | 8.7 | 15.5 | 30 | 0 | 0 | <0.0001 |
Forces at 75% survival and their 95% confidence intervals were estimated by Kaplan–Meier analysis.
All individual force values for ruptures at the SPB–microtubule interface, failures at the bead–microtubule interface, and events that reached the load limit of the trap are given in Supplemental Data File 1.
Removal of the cytoplasmic face does not affect the measured strengths
During closed mitosis in yeast, the SPB nucleates microtubules from both its nuclear and cytoplasmic faces (Jaspersen and Winey, 2004; Kilmartin, 2014). The nuclear face nucleates ∼20 microtubules, whereas the cytoplasmic face nucleates only two or three (Winey and O’Toole, 2001). Because there are many more nuclear microtubules in vivo, we expected that most of our in vitro SPB-nucleated microtubules probably emanated from the nuclear face. To differentiate between nuclear and cytoplasmic attachments, we purified SPBs lacking the cytoplasmic face. In yeast, Spc72 links the γ-tubulin small complex to the cytoplasmic face (Knop and Schiebel, 1998; Souès and Adams, 1998; Usui et al., 2003). Thus we ablated the cytoplasmic face by depleting Spc72 with an auxin-inducible degron (Spc72-AID) before SPB purification (Nishimura et al., 2009). More than 98% of auxin-treated cells were multibudded and contained multiple SPBs (Supplemental Figure S1B), as expected for cells lacking Spc72, which have a multinuclear phenotype arising from failure to move the daughter nucleus into the bud (Chen et al., 1998; Souès and Adams, 1998). This observation confirms that Spc72 was efficiently degraded.
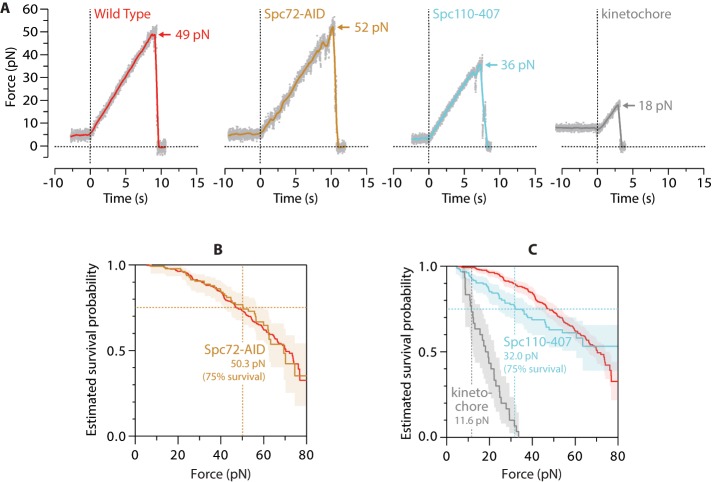
When microtubule attachment strengths to Spc72-depleted SPBs were measured by our laser trap assay, neither the estimated force at 75% survival, 50.3 pN (95% confidence interval, 41.0–60.0 pN) nor the estimated Kaplan–Meier survival curve differed significantly from wild type (Figure 3, A and B; Table 1 summarizes all measurement statistics and reports p values from log-rank tests; Bland and Altman, 2004). These observations are consistent with our hypothesis that cytoplasmic microtubules made little or no contribution. Formally, they are also consistent with an alternative scenario in which our experiments measured both cytoplasmic and nuclear attachments and both attachment types are of identical strength. Such a scenario, however, seems unlikely, given the systematic weakening caused by a mutation on the nuclear side, described later.
FIGURE 3:
Removal of calmodulin from the nuclear-side tether weakens SPB–microtubule attachments. (A) Individual rupture force traces for wild-type SPBs, Spc72-AID SPBs, Spc110-407 SPBs, and kinetochores. Gray dots show raw data. Colored traces show the same data after smoothing (500-ms sliding boxcar average). Dashed vertical lines mark start of the force ramp. Arrows mark ruptures at SPB–microtubule or kinetochore–microtubule interface. (B) Estimated survival probability as a function of force for microtubule attachments to wild-type and Spc72-AID SPBs (red and yellow curves, respectively). Shaded area shows 95% confidence interval for the Spc72-AID curve. Dashed vertical line marks the estimated force at 75% survival for Spc72-AID. The curve for Spc72-AID is not significantly different from wild type, indicating that ablation of the cytoplasmic face of the SPB does not affect the measured strengths. (C) Estimated survival probability as a function of force for indicated attachments. Shaded areas show 95% confidence intervals. Dashed vertical lines mark estimated forces at 75% survival for kinetochores and Spc110-407 SPBs (gray and blue, respectively). Survival for Spc110-407 is reduced significantly compared with wild type. Measurement statistics and p values are summarized in Table 1. Wild-type data in A–C are recopied from Figure 2, C and E, for comparison.
Calmodulin is required for strong SPB–microtubule attachments
In vivo, Spc110 tethers microtubule minus ends to the nuclear face of the SPB, spanning the distance from the γ-tubulin small complex to the SPB core (specifically, to the “central plaque”; Kilmartin et al., 1993). Spc110 binds the SPB core via a C-terminal domain that also contains a calmodulin-binding site (Geiser et al., 1993; Stirling et al., 1994). Immuno–electron microscopy and fluorescence resonance energy transfer analysis confirm that calmodulin resides within the SPB core, very close to the C-terminus of Spc110 and also near other core proteins (e.g., Spc29 and Spc42; Sundberg et al., 1996; Muller et al., 2005). Temperature-sensitive mutations in either calmodulin or in the calmodulin-binding site of Spc110 cause misassembly of Spc110 away from the SPB (Sundberg et al., 1996). The function of calmodulin in the SPB is unknown, but given its localization at the core and the phenotype of the temperature-sensitive mutants, we hypothesized that calmodulin might contribute to the mechanical integrity of the SPB. To test this idea directly, we took advantage of a suppressor mutant allele of SPC110 that overcomes the requirement for calmodulin at the SPB. The allele, SPC110-407, encodes a C-terminal truncation that removes the calmodulin-binding site and fails to localize calmodulin to the SPB in vivo (Geiser et al., 1993).
We purified calmodulin-depleted SPBs from cells carrying SPC110-407 in place of the wild-type gene and tested them in our rupture strength assay. The Spc110-407 poles attached more weakly to microtubules than wild-type SPBs. Their estimated force at 75% survival, 32.0 pN (95% confidence interval, 24.5–46.1 pN), and their estimated Kaplan–Meier survival curve were significantly lower than those for wild-type poles (Figure 3, A and C, and Table 1). These results show that calmodulin and the C-terminal calmodulin-binding domain of Spc110 contribute to the strength of SPB–microtubule attachments.
Wild-type SPBs withstand forces much higher than kinetochores
In the context of the mitotic spindle, a tug-of-war can occur between kinetochores and spindle poles. Tension applied by the kinetochores to microtubule plus ends (Chacón et al., 2014) is transmitted to their minus ends tethered at the poles. The kinetochore-attached plus ends are dynamic, continuously assembling or disassembling, and eukaryotic cells possess efficient mechanisms for rapidly correcting and reestablishing these dynamic attachments, making ruptures at the kinetochore–microtubule interface repairable (Nicklas and Koch, 1969). In contrast, microtubule minus ends are stably anchored and nondynamic at yeast SPBs (Mallavarapu et al., 1999; Maddox et al., 2000; Khodjakov et al., 2004). Their dissociation from the nuclear face correlates with delamination of the SPB, spindle collapse, and irreversible cell cycle arrest (Yoder et al., 2005). Thus it might be crucial that SPBs can normally sustain all tensile forces exerted on them by kinetochores via microtubules, such that SPBs always win the tug-of-war (Figure 4A). Indeed, the strengths we measured for minus-end attachments to SPBs were higher than those measured previously for attachments between kinetochores and growing plus ends (Akiyoshi et al., 2010; Sarangapani et al., 2014). However, this difference is probably exaggerated by the 20-fold higher loading rate used in our SPB assays (5 vs. 0.25 pN/s for kinetochores), given that faster loading rates are well known to increase rupture forces for intermolecular bonds (Merkel et al., 1999). We therefore remeasured rupture strengths for kinetochore attachments to microtubule plus ends at the same loading rate we used for SPBs. Under identical loading rates, the kinetochore attachments remained far weaker than SPB attachments. Their estimated force at 75% survival, 11.6 pN (95% confidence interval, 8.7–15.5 pN), was fourfold lower than the corresponding value for wild-type SPBs, and their estimated Kaplan–Meier survival curve was very significantly lower as well (Figure 3, A and C, and Table 1).
FIGURE 4:
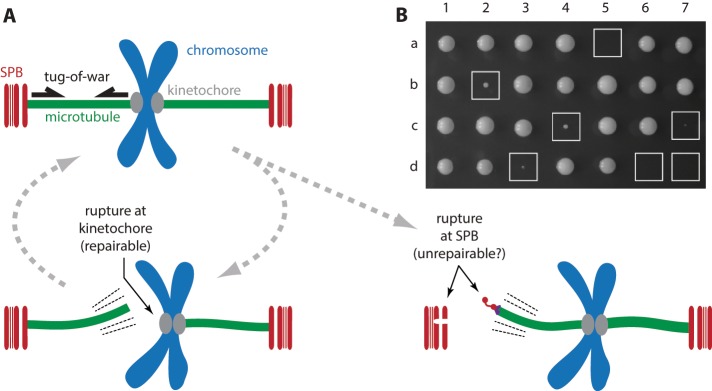
Removal of calmodulin from nuclear-side tether sensitizes cells to increased SPB stress. (A) Tug-of-war model, kinetochores vs. SPBs. Top left, after chromosome biorientation, tension generated at kinetochore-attached microtubule plus ends is transmitted to SPB-attached minus ends. Bottom left, strong minus-end anchorage normally prevents detachment at the SPB, such that ruptures, if they occur, happen at the kinetochore–microtubule interface, which is weaker but readily repairable. Bottom right, altering the relative strengths of SPB and kinetochore attachments (e.g., by combining SPC110-407 and DAM1-765) can lead to ruptures at minus ends, possibly causing SPB delamination, cell cycle arrest, and lethality. (B) Tetrad analysis confirms that the mutant SPB allele SPC110-407 is lethal when combined with a mutant kinetochore allele, DAM1-765, which was previously shown to increase mechanical stresses on SPBs (Shimogawa et al., 2006). Each column shows the four colonies derived from a single tetrad produced by mating a haploid SPC110-407 strain with a haploid DAM1-765 strain and then inducing meiosis (sporulation). Boxed colonies, expressing both mutant alleles, exhibited slow or no growth.
The much higher strength of SPB– versus kinetochore–microtubule attachments may be explained by structural and kinetic differences between these two types of interfaces. All of the terminal α-tubulins at an SPB-attached microtubule minus end are stably bound by γ-tubulins, which associate in pairs with two accessory proteins, Spc97 and Spc98 (GCP2 and GCP3), to form γ-tubulin small complexes (Knop and Schiebel, 1997; Oegema et al., 1999; Kollman et al., 2015). The γ-tubulin small complexes, in turn, are stably tethered into the SPB core by dimers of Spc110 (or its equivalent, Spc72, on the cytoplasmic face; Knop and Schiebel, 1997, 1998; Sundberg and Davis, 1997; Nguyen et al., 1998; Souès and Adams, 1998). Thus tensile loads on the SPB are probably distributed across at least 13 high-affinity protein–protein bonds. Yeast kinetochores carry a similarly large number of microtubule-binding elements, including 8–20 Ndc80 complexes and 16 to ∼40 Dam1 complexes (Joglekar et al., 2006; Lawrimore et al., 2011). However, the microtubule binders of the kinetochore must dynamically bind and unbind or slide along the filament to maintain attachment during tip assembly and disassembly. Tensile load on the kinetochore might therefore be shared by only a subset of elements that happen to be tightly bound at any given moment.
Removal of calmodulin from SPBs sensitizes cells to increased SPB stress
Although removal of the calmodulin-binding site from Spc110 weakened the in vitro SPB–microtubule attachments compared with wild-type SPBs, the calmodulin-free Spc110-407 poles were still stronger than kinetochores (Figure 3C and Table 1). In vivo, these mutant poles support normal cell growth rates (Geiser et al., 1993), so we expected that their strength in vitro would exceed that of kinetochores. We hypothesized, however, that under conditions of increased SPB stress, the reduced strength of Spc110-407 SPBs might no longer suffice, and cell growth might be compromised. To test this idea, we crossed SPC110-407 cells with cells carrying a mutant kinetochore allele, DAM1-765, that was previously shown to increase mechanical stresses on SPBs (Shimogawa et al., 2006). Cells carrying DAM1-765 alone are viable, but we found that the allele is lethal when combined with SPC110-407 (Figure 4B). DAM1-765 is also synthetic lethal in combination with another SPC110 mutant allele, spc110-226 (Shimogawa et al., 2006), which sensitizes cells to delamination of their SPBs at high growth temperatures (Yoder et al., 2005). The finding that SPC110-407, like spc110-226, is synthetic lethal in combination with DAM1-765 suggests that SPC110-407 similarly compromises SPB strength in vivo, consistent with the weakening it caused in our in vitro experiments.
Conclusion
In summary, our new in vitro assay has allowed a direct assessment of the strength of attachment between microtubule minus ends and yeast SPBs, which are functionally equivalent to the centrosomes of other eukaryotes. The strength of SPB–microtubule attachment is very high, exceeding the strength of attachment between kinetochores and growing microtubule plus ends by approximately fourfold. We suggest that this strong anchorage to SPBs might be important for spindle integrity in mitotic cells so that tensile forces generated at kinetochores do not cause microtubule detachment and delamination at SPBs. Quantitative comparison between wild-type and mutant SPBs indicates that microtubule attachment strength depends on the tethering molecule, Spc110, and calmodulin. The dependence of attachment strength on calmodulin implies that this molecule helps reinforce the anchorage of Spc110 into the SPB core and predicts that, under conditions of high SPB stress, loss of calmodulin sensitizes cells to SPB delamination. Given the homology that Spc110 shares with pericentrin (kendrin), which also binds calmodulin, we speculate that these mammalian proteins might also contribute mechanical strength to minus-end attachments at mammalian centrosomes. We anticipate that our in vitro mechanical approach to studying SPB–microtubule attachments will allow further dissection of the molecular basis for their very high strength. Our work should also guide efforts to study the centrosomes of other eukaryotes and facilitate direct tests for whether forces control phosphoregulation or other signaling events at SPBs.
MATERIALS AND METHODS
Strains, plasmids, and media
All of the strains used in this study were derived from W303. In BGY72–8A (Fong et al., 2016), (MAT a), Spc97 was C-terminally TAP-tagged by PCR amplification of the TAP-kanMX cassette from the plasmid TAP-2xPA using primers that shared homology with the flanking sequences of the SPC97 stop codon. Spc42 was C-terminally tagged with mCherry by PCR amplification of the mCherry-hphMX3 cassette from pBS35 (a gift from the Yeast Resource Center, University of Washington, Seattle, WA) with homology flanking the SPC42 stop codon.
For the Spc72-AID strain, the auxin degron IAA7 was PCR amplified from pSB2065 (a gift from Sue Biggins, Fred Hutchinson Cancer Research Center, Seattle, WA) with primers that shared homology with the flanking sequences of the SPC72 stop codon. The diploid was sporulated and dissected, and haploids were selected for pADH1-OsTIR1-9myc::URA3, SPC42-mCherry::hphMX, SPC72-AID::kanMX, and SPC97-TAP::kanMX, resulting in strain KFY329-24B.
BGY72-8A was crossed with a haploid containing SPC110-407. The diploids were sporulated and dissected, and haploids were selected for SPC42-mCherry::hphMX and SPC97-TAP::kanMX. The presence of Spc110–407 was determined by TCA precipitation and Western blot analysis, resulting in strain KFY304-2D.
A strain expressing SPC110-407 was crossed to a strain expressing DAM1-765. Two-thirds of the SPC110-407 DAM1-765 spores were dead, and the remaining one-third showed slowed growth, indicating synthetic lethality between SPC110-407 and DAM1-765. Yeast extract peptone dextrose (YPD) medium is as described in Burke et al. (2000).
Growth of yeast strains
Wild type and SPC110-407 yeast strains were grown in YPD and harvested at 150 Klett units. Spc72-AID cells were grown to 100 Klett units. Auxin (indole-3-acetic acid) in dimethyl sulfoxide was then added to a final concentration of 1 mM. The cells were then grown for an additional 3 h with auxin, to deplete Spc72, before harvesting. More than 98% of these Spc72-depleted cells were multibudded with multipolar spindles, as determined by fluorescence microscopy (Supplemental Figure S1B).
Yeast cells were harvested and lysed as previously described (Fong et al., 2016). Yeast cells were pelleted and washed with distilled H2O twice before resuspension in buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES] buffer, pH 7.4, 1.2% polyvinylpyrrolidone [average molecular weight 40,000], 1 mM dithiothreitol [DTT], 1 mM phenylmethanesulfonyl fluoride [PMSF], 4 µg/ml aprotinin, 4 µg/ml chymostatin, 4 µg/ml leupeptin, 4 µg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate). Cells were pelleted again, and the cell paste was flash frozen in liquid nitrogen. Cell pellets were lysed by cryogrinding in a PM100 ball mill grinder (Retsch).
TAP purification and velocity sedimentation
Yeast SPBs were purified by a TAP-tag on Spc97 as previously described (Fong et al., 2016). Lysed cells were resuspended in extraction buffer (20 mM HEPES buffer, pH 7.4, 0.5% Triton X-100, 2 mM MgCl2, 100 µM GTP, 1 mM ATP, 1 mM DTT, 1 mM PMSF, 4 µg/ml aprotinin, 4 µg/ml chymostatin, 4 µg/ml leupeptin, 4 µg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 5% glycerol) with 300 mM NaCl and homogenized. The lysate was cleared at 2000 × g for 10 min at 4°C. Dynabeads conjugated to rabbit immunoglobulin G (according to manufacturer’s protocols) were added to cleared lysate and incubated for 30 min at 4°C. The Dynabeads were washed three times with extraction buffer with 200 mM NaCl. To elute the spindle pole bodies, the Dynabeads were resuspended in TEV cleavage buffer (40 mM HEPES buffer, pH 7.4, 200 mM NaCl, 2 mM MgCl2, 1 mM GTP, 1 mM ATP, 1 mM EDTA, pH 8, 1 mM DTT, 5% glycerol) and incubated with 1 μg of TEV for 2 h at 4°C.
Sucrose gradients were generated by allowing five steps of sucrose solutions (200 µl each of 10, 20, 30, and 40% and 2.5 M sucrose in 10 mM Bis-Tris, pH 6.5, 0.1 mM MgCl2) to equilibrate at 4°C for 2 h. The TEV eluate was then applied to the sucrose gradient and spun at 50,000 rpm for 5 h at 4°C in a TLS55 rotor (Beckman Coulter). Fractions (90 µl) were removed from the top of the gradient with wide-bore tips. The presence of SPBs was determined by Western blot analysis, probing for Spc110 and Spc97 (Supplemental Figure S1A).
Laser trap instrument
The laser trap was described previously (Akiyoshi et al., 2010; Franck et al., 2010; Sarangapani et al., 2014). Position sensor response was mapped using the piezo stage to raster-scan a stuck bead through the beam, and trap stiffness was calibrated along the two principal axes using the drag force, equipartition, and power spectrum methods. Force feedback was implemented with custom LabView software. During rupture force measurements, bead-trap separation was sampled at 40 kHz, and stage position was updated at 50 Hz to maintain the desired tension (force-clamp assay) or ramp rate (force-ramp assay). Bead and stage position data were decimated to 200 Hz before being stored to disk.
Preparation of kinesin-coated beads
To minimize the bias caused by failures at the bead–microtubule interface, we tested several bead–microtubule linkage strategies, including biotinylated tubulin with streptavidin-coated beads and beads coated with anti-tubulin antibodies. Surprisingly, even though individual kinesin–microtubule bonds are weaker than streptavidin–biotin bonds at the single-molecule level (Merkel et al., 1999; Uemura and Ishiwata, 2003), the streptavidin–biotin linkages proved to be weaker in our experiments, which used beads coated densely with the proteins. This observation suggests that at high density on the beads, many kinesin molecules shared the load effectively, whereas relatively fewer streptavidins could do so. We speculate that the more elongated and flexible structure of the kinesins allowed their microtubule-binding heads to project farther from the bead surface than streptavidin, thereby facilitating more interactions with the microtubule. Anti-tubulin antibodies also proved to be weaker, so we performed all of the laser trap assays reported here using kinesin-coated beads.
An N-terminal derivative of the Drosophila melanogaster kinesin heavy chain, DmK401, was prepared as previously described (Asbury et al., 2003). DmK401 consisted of a homodimer of the N-terminal 401 amino acids of the kinesin heavy chain with a hexahistidine (6H) tag on the C-terminus. DmK401 was expressed in BL21 Star (DE3) cells (Invitrogen). We grew 250-ml cultures at 20°C for 2 h after 1 mM isopropyl-β-d-thiogalactoside induction. Rifampicin was added to a final concentration of 200 μM, and cultures were grown overnight at 20°C. Cells were pelleted and resuspended in an equivalent volume of lysis buffer (250 mM potassium phosphate, pH 7.6, 10 mM imidazole, pH 7.0, 1 mM β-mercaptoethanol, 4 mM MgCl2, Complete protease inhibitors [Roche]), lysed with a French press, and clarified by ultracentrifugation at 50,000 rpm for 40 min at 4°C. The clarified lysate was mixed with an equivalent volume of glycerol and stored at −20°C.
Streptavidin-coated polystyrene beads (0.44 μm in diameter; Spherotech) were functionalized with biotinylated anti-His5 antibodies (Qiagen) and stored with continuous rotation at 4°C in BRB80 (80 mM 1,4-piperazinediethanesulfonic acid [PIPES], 1 mM MgCl2, and 1 mM ethylene glycol tetraacetic acid [EGTA], pH 6.9) supplemented with 8 mg/ml bovine serum albumin [BSA] for up to 3 mo. Before each experiment, beads were decorated with kinesin by incubating 6 pM anti-His5 beads with 20 μl of cleared lysate from the kinesin expression cells for 1 h at 4°C diluted in assay buffer (BRB80; concentration as before, 5 mg/ml BSA, 11.5 μM Taxol, and 1 mM DTT).
Chamber preparation for laser trap experiment
Flow chambers (∼10-µl volume) were made using glass slides, double-stick tape, and KOH-cleaned coverslips. Briefly, two lengths of double-stick tape were placed across a microscope slide to create a channel. A KOH-cleaned coverslip was placed over the channel and sealed with pressure. It was then functionalized in the following manner. An aliquot of purified SPBs was diluted with 5× BRB80 (400 mM PIPES, 5 mM MgCl2, and 5 mM EGTA), 40 mg/ml BSA, and 2.67 M KCl to a final concentration of 1× BRB80, 8 mg/ml BSA and 500 mM KCl. The diluted SPBs were introduced into the flow chamber and allowed to nonspecifically adhere to the coverslip for 30 min. The SPBs were washed thoroughly by flowing in BRB80. Bovine brain tubulin, purified as previously described (Castoldi and Popov, 2003), was cleared of aggregates by ultracentrifugation at 90,000 rpm for 10 min at 4°C in a TLA100 rotor (Beckman Coulter). The tubulin polymerization buffer (1× BRB80, 1 mg/ml κ-casein, 5 mg/ml BSA, 2 μM Taxol, 1 mM GTP, 1 mM DTT, and 20 μM cleared tubulin) was added to the flow cell, and microtubules were allowed to nucleate at room temperature for several minutes. Free tubulin was washed out of the chamber with wash buffer (1× BRB80, 1 mg/ml κ-casein, 5 mg/ml BSA, 11.5 μM Taxol, and 1 mM DTT). Kinesin-coated beads (described earlier) were then flowed in along with an oxygen-scavenging system (500 µg/ml glucose oxidase, 60 µg/ml catalase, and 25 mM glucose). The edges of the flow chamber were sealed to prevent evaporation. All laser trap experiments were performed in a temperature-controlled room maintained at 23°C.
For kinetochore rupture force experiments, mitotic kinetochore particles were isolated from budding yeast by affinity purifying Dsn1–6His–3Flag protein as previously described (Akiyoshi et al., 2010) and linked to anti-His5 antibody–functionalized polystyrene beads as previously described (Akiyoshi et al., 2010; Sarangapani et al., 2013, 2014). Briefly, before each experiment, beads were decorated with kinetochore particles by incubating 6 pM anti-His5 beads for 60 min at 4°C with purified kinetochore material, corresponding to Dsn1-His-Flag concentrations of 6 nM. Finally, kinetochore particle–coated beads were introduced at an eightfold dilution from the incubation mix in a solution of growth buffer containing 1.5 mg/ml purified bovine brain tubulin and an oxygen-scavenging system as described.
Rupture force assay
Spindle pole body puncta were identified using the DIC imaging module (part of the laser trap setup), and those that had single microtubules emanating from them were specifically chosen for pulling experiments. Many of these microtubules had prebound beads (i.e., beads already associated with the SPB-attached microtubule), but if no bead was bound to the microtubule, free beads from solution were trapped and allowed to bind to these microtubules. The beads were initially subjected to a preload force of ∼5 pN. After a brief preload period, the laser trap was programmed to ramp the force at a constant rate of 5 pN/s until the microtubule ruptured from the SPB, the bead broke away from the microtubule, or the load limit of the trap was reached.
For kinetochore rupture force experiments, the laser trap was used to place individual free beads close to the ends of growing microtubules to allow binding. On binding, the attachments were preloaded with a constant force of ∼5 pN. After a brief preload period during which we verified that the beads were moving at a rate consistent with that of microtubule growth, the laser trap was programmed to ramp the force at a constant rate (5 pN/s) until the kinetochore–microtubule attachment ruptured.
Survival curve analysis
The Kaplan–Meier survival curve was estimated at each force associated with an SPB–microtubule rupture as
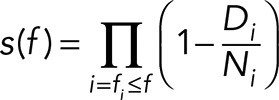 |
where ∏ denotes a product series, Di represents the number of SPB–microtubule attachments that ruptured at force fi, and Ni represents the number of attachments that remained “at risk” of rupturing at force fi (excluding those that were censored at lower forces). An important assumption of this analysis is that SPB–microtubule attachments censored at any particular force (i.e., those that fail at the bead–microtubule interface or reach the load limit) had the same risk of rupture as uncensored attachments.
To calculate the p values shown in Table 1, the Kaplan–Meier survival curves were compared using the log-rank test (Bland and Altman, 2004), a standard method for determining the degree of statistical significance in clinical trials. We also computed the estimated force at 75% survival for each SPB type. Statistical analysis was performed using the Stata data analysis package. All the individual force values for ruptures at the SPB–microtubule interface, failures at the bead–microtubule interface, and events that reached the load limit of the trap are reported in Supplemental Data File 1.
Supplementary Material
Acknowledgments
This work was supported by a National Research Service Award Fellowship to E.C.Y. (F32GM109493), a fellowship from the Raymond and Beverly Sackler Scholars Program at the University of Washington to A.L., a Packard Fellowship to C.L.A. (2006–30521), and National Institutes of Health grants to C.L.A. (P01GM105537) and T.N.D. (P01GM105537).
Abbreviations used:
- AID
auxin-inducible degron
- SPB
spindle pole body
- TAP
tandem affinity purification
- VE-DIC
video-enhanced differential interference contrast.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0034) on March 22, 2017.
REFERENCES
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell R, Edelmaier C, Sweezy-Schindler O, Lamson A, Gergely ZR, O’Toole E, Crapo A, Hough LE, McIntosh JR, Glaser MA, et al. Physical determinants of bipolar mitotic spindle assembly and stability in fission yeast. Sci Adv. 2017;3:e1601603. doi: 10.1126/sciadv.1601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG. The logrank test. BMJ. 2004;328:1073. doi: 10.1136/bmj.328.7447.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DJ, Dawson D, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 2000.
- Byers B, Goetsch L. Behavior of spindles and spindles plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Cane S, Ye AA, Luks-Morgan SJ, Maresca TJ. Elevated polar ejection forces stabilize kinetochore-microtubule attachments. J Cell Biol. 2013;200:203–218. doi: 10.1083/jcb.201211119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- Chacón JM, Mukherjee S, Schuster BM, Clarke DJ, Gardner MK. Pericentromeric tension is self-regulated by spindle structure in metaphase. J Cell Biol. 2014;205:313–324. doi: 10.1083/jcb.201312024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Davis TN. The centrosomal proteins pericentrin and kendrin are encoded by alternatively spliced products of one gene. Genomics. 2003;82:401–405. doi: 10.1016/s0888-7543(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Flory MR, Moser MJ, Monnat RJ, Davis TN. Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc Natl Acad Sci USA. 2000;97:5919–5923. doi: 10.1073/pnas.97.11.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KK, Graczyk B, Davis TN. Purification of fluorescently labeled saccharomyces cerevisiae spindle pole bodies. Methods Mol Biol. 2016;1413:189–195. doi: 10.1007/978-1-4939-3542-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Direct physical study of kinetochore–microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51:242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina I, Nandi A, Delivani P, Chacón MR, Klemm AH, Ramunno-Johnson D, Krull A, Lindner B, Pavin N, Tolić-Nørrelykke IM. Pivoting of microtubules around the spindle pole accelerates kinetochore capture. Nat Cell Biol. 2013;15:82–87. doi: 10.1038/ncb2640. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast: role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. Lessons from yeast: the spindle pole body and the centrosome. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130456. doi: 10.1098/rstb.2013.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcription is cell-cycle regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez JJ, Sali A, Kilmartin J, Davis TN, et al. Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat Struct Mol Biol. 2015;22:132–137. doi: 10.1038/nsmb.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Roth S, Dogterom M. End-on microtubule-dynein interactions and pulling-based positioning of microtubule organizing centers. Cell Cycle. 2012;11:3750–3757. doi: 10.4161/cc.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Neuner A, Schiebel E. Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 2015;25:296–307. doi: 10.1016/j.tcb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Lin T, Neuner A, Schlosser YT, Scharf AN, Weber L, Schiebel E. Cell-cycle dependent phosphorylation of yeast pericentrin regulates γ-TuSC-mediated microtubule nucleation. Elife. 2014;3:1–29. doi: 10.7554/eLife.02208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lampson MA. Regulation of kinetochore-microtubule attachments by Aurora B kinase. Biochem Soc Trans. 2009;37:976–980. doi: 10.1042/BST0370976. [DOI] [PubMed] [Google Scholar]
- Lyon AS, Morin G, Moritz M, Yabut KCB, Vojnar T, Zelter A, Muller E, Davis TN, Agard DA. Higher-order oligomerization of Spc110p drives γ-tubulin ring complex assembly. Mol Biol Cell. 2016;27:2245–2258. doi: 10.1091/mbc.E16-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen) J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O’Toole ET, Giddings TH, Sundin BA, Davis TN. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Vinh DBN, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast γ-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MP, Höök P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun. 2015;6:7206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JT, Neely JG, Paniello RC, Voelker CCJ, Nussenbaum B, Wang EW. A practical guide to understanding Kaplan-Meier curves. Otolaryngol Head Neck Surg. 2010;143:331–336. doi: 10.1016/j.otohns.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, Asbury CL. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc Natl Acad Sci USA. 2013;110:7282–7287. doi: 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Asbury CL. Catch and release: how do kinetochores hook the right microtubules during mitosis. Trends Genet. 2014;30:150–159. doi: 10.1016/j.tig.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Duro E, Deng Y, Alves FL, Ye Q, Opoku KN, Ceto S, Rappsilber J, Corbett KD, Biggins S, et al. Sister kinetochores are mechanically fused during meiosis I in yeast. Science. 2014;346:248–251. doi: 10.1126/science.1256729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Forth S, Kapoor TM. Measuring pushing and braking forces generated by ensembles of kinesin-5 crosslinking two microtubules. Dev Cell. 2015;34:669–681. doi: 10.1016/j.devcel.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR 3rd, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souès S, Adams IR. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J Cell Sci. 1998;111:2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stirling DA, Welch KA, Stark MJ. Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO J. 1994;13:4329–4342. doi: 10.1002/j.1460-2075.1994.tb06753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Davis TN. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body compenents. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat Struct Biol. 2003;10:308–311. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- Umbreit NT, Davis TN. Mitosis puts sisters in a strained relationship: force generation at the kinetochore. Exp Cell Res. 2012;318:1361–1366. doi: 10.1016/j.yexcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Maekawa H, Pereira G, Schiebel E. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 2003;22:4779–4793. doi: 10.1093/emboj/cdg459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RGHP, Tanenbaum ME, Medema RH. Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep. 2014;8:948–956. doi: 10.1016/j.celrep.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu T, Shi L, Zhang L, Zheng W, Qu JY, Niu R, Qi RZ. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem. 2010;285:22658–22665. doi: 10.1074/jbc.M110.105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O’Toole ET. The spindle cycle in budding yeast. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci. 2014;369, pii: 20130459 doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Viscardi V, Mahamid J, Ochoa SD, Bunkenborg J, Widlund PO, Pozniakovsky A, Zanin E, Bahmanyar S, et al. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science. 2015;348:808–812. doi: 10.1126/science.aaa3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder TJ, McElwain MA, Francis SE, Bagley J, Muller EG, Pak B, O’Toole ET, Winey M, Davis TN. Analysis of a spindle pole body mutant reveals a defect in biorientation and illuminates spindle forces. Mol Biol Cell. 2005;16:141–152. doi: 10.1091/mbc.E04-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.