Abstract
The nuclear paraspeckle assembly transcript 1 (NEAT1) is a long non-coding RNA. Many studies have reported that NEAT1 plays critical oncogenic roles and facilitates tumorigenesis of various human cancers. High NEAT1 expression is associated with a poor prognosis in cancer patients. This meta-analysis was conducted to assess the association between NEAT1 levels and survival times of cancer patients. Overall survival (OS) was the primary endpoint. Thirteen publications with 1,496 cancer patients from 5 databases (PubMed, EMBASE, Cochrane Library, Wiley Online Library, and Medline) met the criteria for this meta-analysis. Results of the analysis showed that NEAT1 expression in human cancer was significantly associated with OS (hazard ratio [HR]=1.53, 95% confidence interval [CI]: 1.39–1.68), including digestive system tumor (HR=1.54, 95% CI: 1.37–1.73) and respiratory carcinomas (HR=1.44, 95% CI: 1.11–1.85). The results also indicated that NEAT1 expression was highly associated with tumor size (>3 cm vs. ≤3 cm; odds ratio [OR]=2.51, 95% CI: 1.27–4.99; p=0.009), TNM stage (III+IV vs. I+II; OR=4.17, 95% CI: 2.42–7.18; p=0.00001), and distant metastasis (OR=2.73, 95% CI: 1.28–5.79; p=0.01). However, there was no significant association with differentiation (poor vs. well + moderate, OR=1.45, 95% CI: 0.72–2.91) and lymph node metastasis (OR=1.39, 95% CI: 0.54–3.60). This meta-analysis showed that NEAT1 expression may be a useful biomarker for predicting a poor prognosis in patients with cancer.
Keywords: NEAT1, cancer, OS, meta-analysis, poor prognosis
INTRODUCTION
Long non-coding RNA (LncRNA) is a class of non-coding RNA, and has more than 200 nucleotides [1]. Initially, LncRNA was considered as spurious transcriptional noise without biological functions because of its lack of protein-coding function [2]. However, later studies indicated that LncRNA plays an important role in pathophysiological processes such as cancers [3, 4]. In recent years, few LncRNAs, such as AFAPI-ASI, MALAT-1, and UCA1 have been confirmed to play a significant role in cancer progression [5–8]. Therefore, LncRNAs might have complex and extensive functions in carcinogenesis and progression of human cancers [9].
The nuclear paraspeckle assembly transcript 1 (NEAT1) is a newly identified nuclear-restricted LncRNA and is located on chromosome 11 (11q13.1). It is an essential component of nuclear paraspeckles [10, 11], and has been confirmed to overexpress in many human cancers, including prostate, lung, and breast cancers [12–14]. High NEAT1 expression in cancerous tissues was reported to be associated with prognosis and overall survival (OS) in several cancers. However, the effect of NEAT1 on the outcome of cancer patients has been controversial. No meta-analysis has been conducted to date on the correlation of NEAT1 with the survival of cancer patients. NEAT1 might act as a potential diagnostic biomarker and prognostic factor. This meta-analysis is the first to explore the correlation between NEAT1 expression and prognosis of cancer patients.
RESULTS
Literature search and description of studies
As shown on Figure 1, 163 studies were found in the 5 databases (PubMed, EMBASE, Cochrane Library, Wiley Online Library and Medline). We reviewed the titles and abstracts, and 49 irrelevant studies and duplicates were excluded. Of the remaining 99 studies, 86 were eliminated because they used different statistics methods, were animal experiments, or were not in English. After data extraction, 13 studies [14–26], all from China, were selected for the final meta-analysis with a total of 1,496 cancer patients. The main characteristics are summarized in Table 1. The 13 studies covered 9 different types of cancer, including ovarian cancer (n=2), colorectal cancer (n=3), non-small cell lung cancer (NSCLC; n=2), gastric cancer (n=1), hepatocellular carcinoma (n=1), glioma (n=1), nasopharyngeal carcinoma (NPC; n=1), pancreatic cancer (PC; n=1) and esophageal squamous cell carcinoma (n=1). Among the 13 studies, 12 involved tissue collection, and 1 involved a whole blood analysis. All of the detection methods used quantitative polymerase chain reaction.
Figure 1. Flow chart of studies selection procedure.
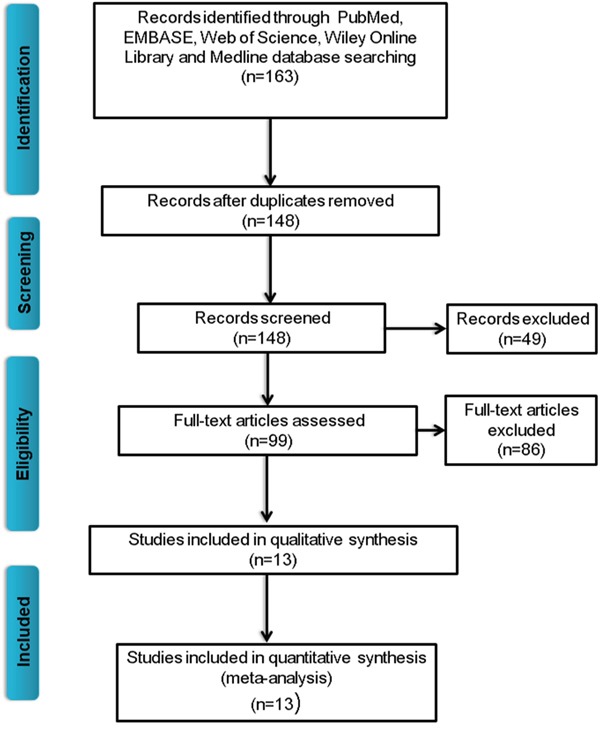
Table 1. Characteristics of studies included in this meta-analysis.
| Author | Year | Country | Sample size | Sample type | Cancer type | Tumor size (cm) | TNM stage | Follow-up | Method | Outcome | HR statistics | Variance | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤3VS>3 | I/II Vs III/IV | (Month) | Analysis | ||||||||||
| He | 2015 | China | 94 | Tissue | Glioma | 30/64 | 23/71 | >50 | qRT-PCR | OS | Reported | Univariate | 8 |
| Guo | 2015 | China | 95 | Tissue | HCC | NA | 22/73 | >60 | qRT-PCR | OS | Survival curve | Univariate | 7 |
| Fu | 2016 | China | 140 | Tissue | GC | NA | 63/77 | 96 | qRT-PCR | OS | Reported | Multivariate | 8 |
| Sun | 2016 | China | 96 | Tissue | NSCLC | 41/55 | 28/68 | 41 | qRT-PCR | OS | Survival curve | Univariate | 8 |
| Li | 2015 | China | 239 | Tissue | CC | 82/157 | 92/147 | >60 | qRT-PCR | OS, DFS | Reported | Univariate | 8 |
| Wu | 2015 | China | 191 | Whole blood | CC | NA | 26/165 | 80 | qRT-PCR | OS | Reported | Multivariate | 8 |
| Chen | 2015 | China | 96 | Tissue | ESCC | NA | 35/61 | >60 | qRT-PCR | OS | Reported | Multivariate | 8 |
| Lu | 2015 | China | 71 | Tissue | NPC | NA | 36/35 | >40 | qRT-PCR | OS | Reported | Multivariate | 8 |
| Aderiaens | 2016 | Belgium | 58 | Tissue | OC | NA | NA | >60 | qRT-PCR | OS | Reported | Univariate | 8 |
| Huang | 2016 | China | 86 | Tissue | PC | NA | 56/32 | >50 | qRT-PCR | OS | Survival curve | Univariate | 7 |
| Peng | 2016 | China | 56 | Tissue | CC | NA | NA | 60 | qRT-PCR | OS | Survival curve | Multivariate | 7 |
| Chen | 2016 | China | 149 | Tissue | OC | NA | 53/96 | >60 | qRT-PCR | OS | Reported | Multivariate | 8 |
| Pan | 2015 | China | 125 | Tissue | NSCLC | 60/65 | 54/71 | >40 | qRT-PCR | OS | Survival curve | Univariate | 8 |
HCC: hepatocellular carcinoma; GC: gastric cancer; NSCLC: non-small cell lung cancer; CC: colorectal cancer; NPC: nasopharyngeal carcinoma; OC: ovarian cancer; PC: pancreatic cancer; ESCC: esophageal squamous cell carcinoma; NA: not available.
Correlation of high NEAT1 expression with OS in human cancer
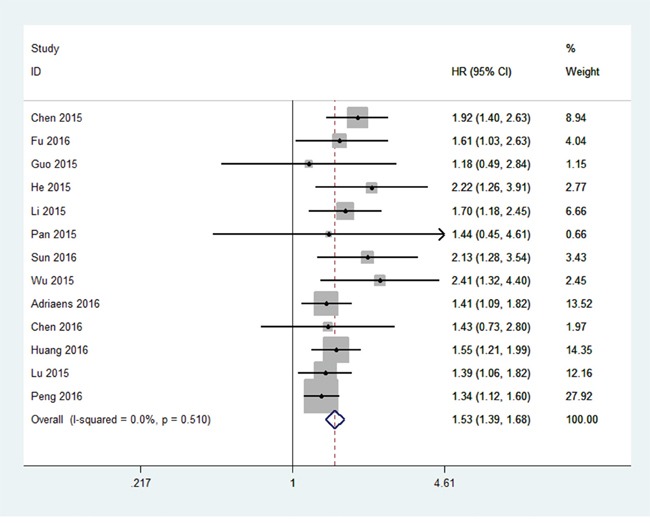
All 13 studies showed OS according to NEAT1 expression levels among the 1,496 patients. The pooled hazard ratio (HR) was 1.53 (95% confidence interval [CI]: 1.39–1.68) for the high NEAT1 expression group versus low expression group (p<0.00001; Figure 2). There was no significant heterogeneity (I2=0%, PQ=0.51), and the fixed-effects model was chosen to estimate the pooled HR. This suggested that high expression of NEAT1 was a predicator of poor prognosis among human cancers.
Figure 2. Forest plot of HR for NEAT1 high expression and overall survival.
Correlation of NEAT1 expression with clinicopathological parameters and cancer type
In this meta-analysis, correlation of NEAT1 expression with digestive system tumor and respiratory carcinomas and clinicopathological parameters are illustrated in Figure 3 and Table 2. High NEAT1 expression correlated with poor prognosis of digestive system tumor (HR=1.54, 95% CI: 1.37–1.73; Figure 3A) and Respiratory carcinomas (HR=1.44, 95% CI: 1.11–1.85; Figure 3B) patients. Furthermore, we found correlation of NEAT1 expression with more advanced tumor size (>3 cm vs. ≤3 cm; odds ratio [OR]=2.51, 95% CI: 1.27–4.99; p=0.009; Figure 4A), distant metastasis (M1 vs. M0; OR=2.72, 95% CI: 1.28–5.79; p=0.010; Figure 4B), and TNM stage (III+IV vs. I+II; OR=4.17, 95% CI: 2.42–7.18; p=0.00001; Figure 4C). However, NEAT1 expression was not significantly associated with differentiation (poor vs. well + moderate, OR=1.45, 95% CI: 0.72–2.91; p=0.30) and lymph node metastasis (yes vs. no, OR=1.39, 95% CI: 0.54–3.60, p=0.5) (Table 2).
Figure 3. Forest plot of NEAT1 expression with OS in digestive system tumor and respiratory carcinomas patients.
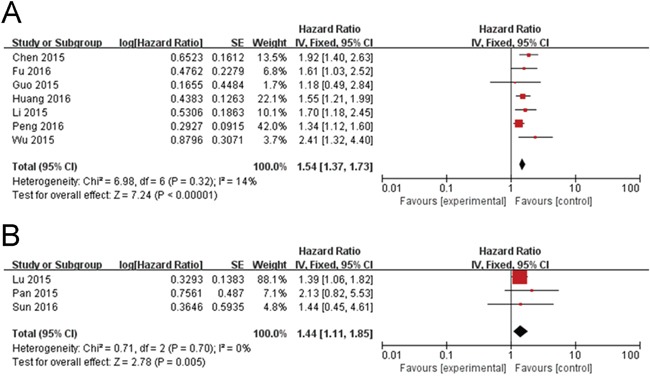
(A) Digestive system tumor and (B) respiratory carcinomas.
Table 2. Results of the association between NEAT1 and clinicopathological parameters.
| Outcome | Studies (n) | OR (HR) | 95%CI | P Value | Model | Heterogeneity |
|---|---|---|---|---|---|---|
| Chi2, I2, P Value | ||||||
| Tumor size (>3 cm vs. ≤3 cm) | 3 | 2.51 | 1.27-4.99 | 0.009 | Random | 4.60, 57%, 0.10 |
| TNM stage (III+IV vs. I+II) | 5 | 4.17 | 2.42-7.18 | 0.00001 | Random | 8.46, 53%, 0.08 |
| Differentiation (poor vs. well + moderate) | 4 | 1.45 | 0.72-2.91 | 0.3 | Random | 11.97, 75%, 0.007 |
| Lymph node metastasis (Yes vs. No) | 5 | 1.39 | 0.54-3.60 | 0.5 | Random | 29.01, 86%, 0.0001 |
| Distant metastasis (M1 vs. M0) | 6 | 2.72 | 1.28-5.79 | 0.01 | Random | 17.75, 72%, 0.003 |
| Digestive system tumor | 7 | 1.54 | 1.37-1.73 | 0.00001 | Fixed | 6.98, 14%, 0.32 |
| Respiratory carcinomas | 3 | 1.44 | 1.11-1.85 | 0.005 | Fixed | 0.71, 0%, 0.70 |
Figure 4. Forest plot of NEAT1 expression and OR for clinicopathological features.
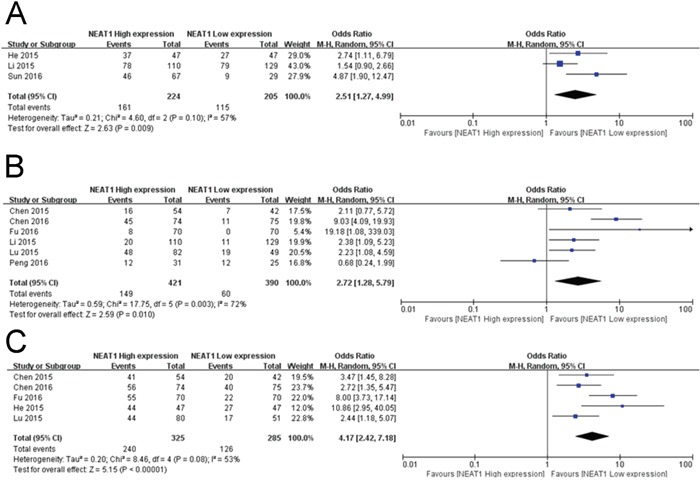
The investigated clinicopathological parameters are (A) tumor size, (B) DM and (C) TNM stage.
NEAT1 expression in other cancer types
In order to understand the expression level of NEAT1 in other cancer types, we used two public cancer database The Cancer Genome Atlas (TCGA) and Oncomine (https://www.oncomine.org) to analyze the expression of NEAT1 in 15 cancer types. The results indicated that the expression of NEAT1 was higher in the tumor tissues than the corresponding normal issues (Figure 5A and 5B).
Figure 5. The expression level of NEAT1 analyzed by cancer public database.
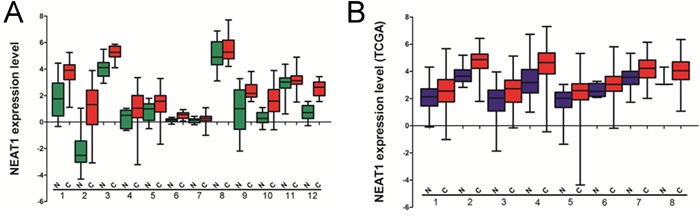
(A) The expression of NEAT1 analyzed by Oncomine. 1, prostate cancer; 2, myeloma; 3, breast cancer; 4, lung cancer; 5, gastric cancer; 6, adrenal cancer; 7, colon cancer; 8, liver cancer; 9, renal cancer; 10, lymphoma; 11, pancreas cancer; 12, leukemia. (B) The expression of NEAT1 analyzed by TCGA database.1, head and neck squamous cancer; 2, kidney cancer; 3, hepatocellular carcinoma; 4, prostate cancer; 5, stomach adenocarcinoma; 6, uterine corpus endometrioid carcinoma; 7, bladder urothelial carcinoma; 8, cervical squamous cell carcinoma and endocervical adenocarcinoma.
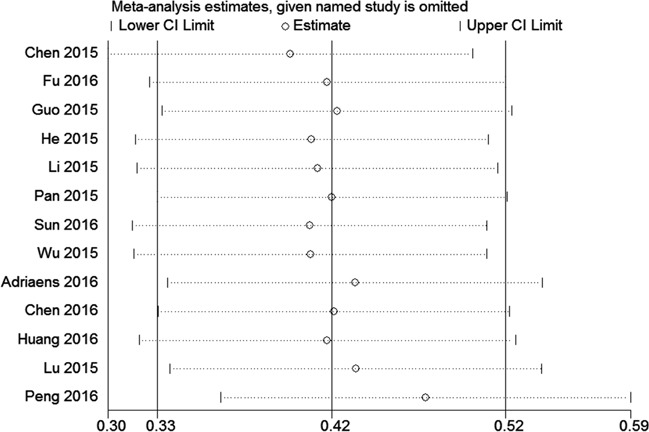
Sensitivity analysis
A sensitivity analysis was conducted to analyze the association between NEAT1 and OS by deleting one study at a time from the pooled analysis to examine the influence of the removed data set to the overall HR. The result was not influenced by the exclusion of each study, suggesting that the result of the synthetic analysis was robust (Figure 6).
Figure 6. Sensitivity analysis of the effect of the individual study on the pooled HRs for the correlation between NEAT1 expression and overall survival (OS).
Publication bias
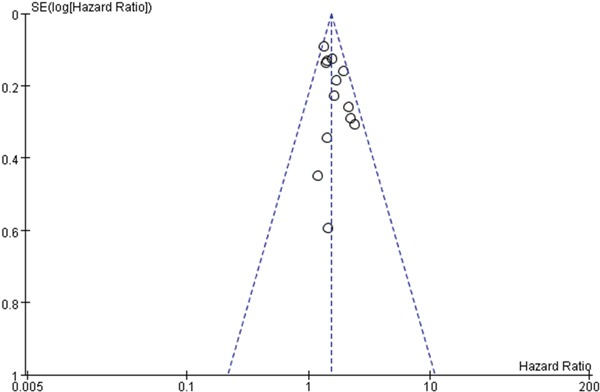
To evaluate the potential for publication bias in this meta-analysis, funnel plots were made (Figure 7). No significant publication bias was observed across studies.
Figure 7. Funnel plot was used to evaluate publication bias on OS.
DISCUSSION
This meta-analysis is the first to evaluate the association between NEAT1 levels and cancer prognosis. The results indicated that high NEAT1 expression was significantly associated with shorter OS times in cancer patients. Subgroup analyses showed that NEAT1 levels were significantly associated with tumor size, TNM stage, and distant metastasis. Yang et al.[27] reported that high expression level of NEAT1 was significantly associated with shorter overall survival in cancer patients, which is similar to our results. However, we also found that the expression of NEAT1 was higher in respiratory carcinomas and correlated with poor prognosis of respiratory carcinomas patients. Xiong et al.[28] used 57 microarrays and other RNA-seq datasets to analyzed correlation of NEAT1 expression with digestive system tumor, and arrived at the same conclusion with us. These results also shown the reliability of our conclusion.
Recently, many LncRNAs have been reported to function as oncogenes or tumor suppressor genes, including PVT1 [29], MALAT1 [30], and HOTAIR [31]. Choudhry et al. reported that NEAT1 is a direct transcription target of HIF-2 in many breast cancer lines and solid tumors. NEAT1 is an essential structural component of paraspeckles, and the hypoxic induction of NEAT1 induces paraspeckle formation in a manner that is dependent upon both NEAT1 and HIF-2. This then leads to accelerated cellular proliferation, improved clonogenic survival, and reduced apoptosis [14]. You et al. reported that NEAT1 is a target of miR-449a and is involved in cell growth and apoptosis of lung cancer cell lines [12]. Yoon et al. reported that NEAT1 can be modulated by RNA-binding protein AUF1, thus affecting the organization of nuclear paraspeckles [32]. Blume et al. reported that NEAT1 is also an important element of the p53-dependent DNA damage response machinery in chronic lymphocytic leukemia [33]. Lastly, Chakravarty et al. reported that NEAT1, as an estrogen receptor alpha specific LncRNA, drives oncogenic growth by altering the epigenetic landscape of target gene promoters [13]. Previous studies revealed that NEAT1 overexpression could influence cell apoptosis and migration [34, 35]. On the contrary, silencing of NEAT1 expression by small interfering RNA could suppress cell apoptosis and metastasis. Therefore, NEAT1 expression level may be an indicator of the intrinsic characteristics of cancer progression. Furthermore, the relationship between NEAT1 expression and a variety of cancer patients’ clinicopathological parameters were reported in previous studies [14–21]. These data might explain why high levels of NEAT1 were significantly associated with shorter OS in cancer patients in this meta-analysis. However, the detailed mechanisms of why NEAT1 was associated with oncologic outcome in cancer were unclear. Wu et al. reported that a 5-LncRNA signature was detected in clear cell renal cell carcinoma patients and normal controls, and further study indicated that this 5-LncRNA signature appears to provide a promising biomarker for the detection of clear cell renal cell carcinoma [36]. Therefore, NEAT1 might also be a tumor marker for predicting tumorigenesis and cancer progression. Although the results revealed the relationship between NEAT1 and OS and patients’ clinicopathological parameters, there are some limitations of this meta-analysis: the number of studies and cancer patients were limited. Most of the studies were conducted in China; therefore, differences may occur between ethnic groups. Additionally, there were 5 studies in which HR could not directly be calculated, thus they may not have provided the most accurate HR estimate. Our results should be confirmed in further studies.
In all, our results indicated that high NEAT1 expression may be a risk factor for shorter OS and a useful biomarker to predict poorer prognosis in human cancers. However, to reinforce the findings, better standardized methods with large sample sizes are needed to further confirm the association between NEAT1 and clinical outcomes of cancers in various ethnic populations.
MATERIALS AND METHODS
Literature search strategies
PubMed, EMBASE, Cochrane Library, Wiley Online Library, and Medline were searched, and articles published from January 1, 1996 to February 10, 2017 were considered. The search strategies were the following: “long non-coding RNA NEAT1” or “lncRNA NEAT1” or “NEAT1” or “nuclear paraspeckle assembly transcript 1” and “Cancer” or “Tumor” or “Neoplasia” and “prognos*” or “surviv*” or “outcome” or “mortality” or “predict.” Reference lists of relevant articles and review papers were also searched manually to identify potential studies.
Inclusion and exclusion criteria
Inclusion criteria were: (1) Studies were written in English, (2) the level of NEAT1 expression was examined in cancer tissues, (3) investigation of the correlation between NEAT1 expression level and survival outcome (OS or progression-free survival), and (4) HR and its 95% CI for survival time were reported or could be calculated from the reported data. Exclusion criteria were: (1) animal studies, case reports, meta-analyses, and review articles and (2) papers lacking all raw data or inability to calculate its HR, 95% CI, and p values.
Data extraction and study quality assessment
All data information of eligible studies included the first author's name, publication year, study regions, sample size, cancer type, tumor size, TNM stage, and method of NEAT1 testing. The HR, corresponding 95% CI, and HR statistics for outcome (OS and progression-free survival [PFS]) were calculated independently.
Study quality was assessed using the Newcastle–Ottawa quality assessment scale (NOS). The NOS score items included selection, outcome, and comparability, and ranged between 0 and 9.
Statistical analysis
The extracted data of this meta-analysis were analyzed using Review Manager 5.3 software (Cochrane network). The HR and its 95% CI were used to evaluate the strength of the association between NEAT1 and OS. If they were reported in a study, HR with 95% CI was extracted directly. If not, the data were extracted from Kaplan–Meier curve using Engauge Digitizer version 4.1 (http://digitizer.sourceforge.net/) [37]. The chi-square-based Q test and I2 statistics were used to determine the heterogeneity [38], and I2 >50% and a P-value for Q test <0.05 were considered significant heterogeneity. Conversely, the I2<50% and a P-value for Q test >0.05 were considered as having no heterogeneity. If there was heterogeneity in the included studies, we chose the random-effects model. The fixed-effects model was chosen when no significant heterogeneity was observed [39]. Sensitivityanalyses were carried out using Stata 12.0 (Stata Corporation, College Station, TX, USA).
Footnotes
CONFLICTS OF INTEREST
All authors declare that they have no conflicts.
FUNDING
This work were funded by the National Natural Science Foundation of China (Grant no. 81271795), Natural Science Foundation of Jiangsu Province, China (Grant no. BK20161345) and Key R and D programs - social development of Zhenjiang, Jiangsu, China (Grant no. SH2015080).
REFERENCES
- 1.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 4.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, Suzuki M, Yu Y, Chen W, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You J, Zhang Y, Liu B, Li Y, Fang N, Zu L, Li X, Zhou Q. MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer. 2014;51:e77–e81. doi: 10.4103/0019-509X.154055. [DOI] [PubMed] [Google Scholar]
- 13.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, Mole DR. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He C, Jiang B, Ma J, Li Q. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS. 2016;124:169–174. doi: 10.1111/apm.12480. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–5402. [PMC free article] [PubMed] [Google Scholar]
- 17.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PubMed] [Google Scholar]
- 18.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, Zhao Q, Li J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6:27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z, Xu Y. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Kong J, Ma Z, Gao S, Feng X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5:2808–2815. [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20:3373–3377. [PubMed] [Google Scholar]
- 23.Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, Zhang K, Zeng D, Liao R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 24.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E, Lapouge G, Beck B, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 25.Huang B, Liu C, Wu Q, Zhang J, Min Q, Sheng T, Wang X, Zou Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828–834. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 26.Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of Akt signaling. Pathol Oncol Res. 2016 doi: 10.1007/s12253-016-0172-4. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Li Z, Li Y, Xu R, Wang Y, Tian Y, Chen W. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017;8:2672–2680. doi: 10.18632/oncotarget.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong DD, Feng ZB, Cen WL, Zeng JJ, Liang L, Tang RX, Gan XN, Liang HW, Li ZY, Chen G. The clinical value of lncRNA NEAT1 in digestive system malignancies: a comprehensive investigation based on 57 microarray and RNA-seq datasets. Oncotarget. 2017;8:17665–17683. doi: 10.18632/oncotarget.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu X, Zhan Y, Liu L, Lin J, Zhou Q, Xu W, Zhao G, Cai Z, Huang W. Tetracycline-inducible shRNA targeting long non-coding RNA PVT1 inhibits cell growth and induces apoptosis in bladder cancer cells. Oncotarget. 2015;6:41194–41203. doi: 10.18632/oncotarget.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH, Chang JG, Liu TC. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, Yang X, Kang MJ, Wood WR, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blume CJ, Hotz-Wagenblatt A, Hullein J, Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A, Benner A, Dietrich S, Oakes CC, Dreger P, et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29:2015–2023. doi: 10.1038/leu.2015.119. [DOI] [PubMed] [Google Scholar]
- 34.XianGuo C, ZongYao H, Jun Z, Song F, GuangYue L, LiGang Z, KaiPing Z, YangYang Z, ChaoZhao L. Promoting progression and clinicopathological significance of NEAT1 over-expression in bladder cancer. Oncotarget. 2016 Jun;15 doi: 10.18632/oncotarget.10084. [Epub ahead of print] [DOI] [Google Scholar]
- 35.Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, Su X, Peng L, Jiao B. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10:11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, Wang YQ, Weng WW, Zhang QY, Yang XQ, Gan HL, Yang YS, Zhang PP, Sun MH, Xu MD, Wang CF. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 39.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]


