Abstract
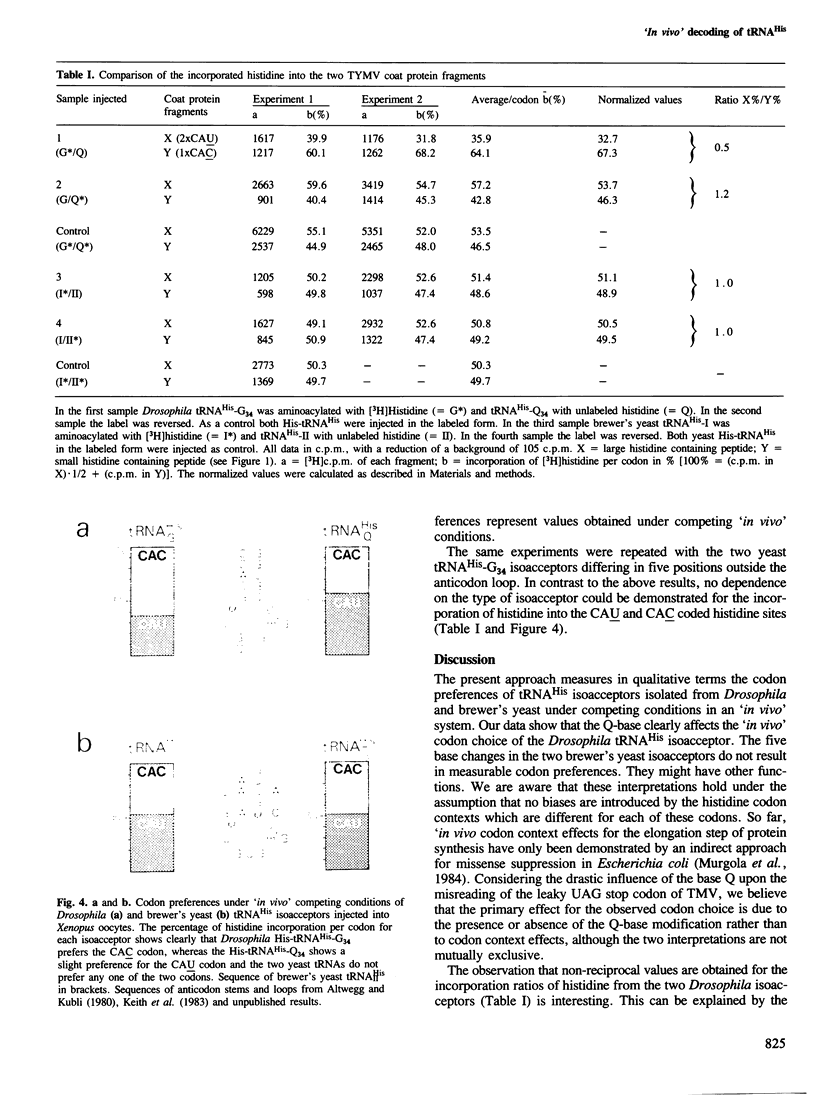
The 'in vivo' decoding properties of four tRNAHis isoacceptors, two from Drosophila melanogaster and two from brewer's yeast, were studied after their microinjection, along with turnip yellow mosaic virus (TYMV) coat protein mRNA, into Xenopus laevis oocytes. The two Drosophila isoacceptors are identical besides containing either a guanosine (G) or the hypermodified nucleoside queuosine (Q) in the wobble position. The brewer's yeast isoacceptors differ by four bases in the anticodon stem, and by one base in the amino acceptor stem. Our results show that, under competing 'in vivo' conditions, the Drosophila tRNAHis with the anticodon GUG clearly prefers the histidine codon CAC to the codon CAU, whereas little preference is observed for the tRNAHis with the anticodon QUG for the codon CAU, and no preference for either codon by the two yeast isoacceptors. Hence, it can be concluded that the presence of the Q-base clearly affects the choice of the codon. This is the first demonstration of an 'in vivo' codon preference by tRNA isoacceptors differing in the modification of the wobble base during the elongation step of protein synthesis. These results imply that one function of the Q-base is at the translational level.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altwegg M., Kubli E. The nucleotide sequence of histidine tRNA gamma of Drosophila melanogaster. Nucleic Acids Res. 1980 Aug 11;8(15):3259–3262. doi: 10.1093/nar/8.15.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M., Kubli E., Kohli J., de Henau S., Grosjean H. Nonsense suppression in eukaryotes: the use of the Xenopus oocyte as an in vivo assay system. Nucleic Acids Res. 1980 Nov 25;8(22):5169–5178. doi: 10.1093/nar/8.22.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G. W., Dyke K. V. Scintillation counting: a comparison of the counting efficiencies of serveral aqueous solubilizers. Anal Biochem. 1973 Aug;54(2):624–627. doi: 10.1016/0003-2697(73)90398-9. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Serra V. Reiterated transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):391–410. doi: 10.1016/0022-2836(73)90013-2. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigliatti T. A., White B. N., Tener G. M., Kaufman T. C., Holden J. J., Suzuki D. T. Studies on the transfer RNA genes of Drosophila. Cold Spring Harb Symp Quant Biol. 1974;38:461–474. doi: 10.1101/sqb.1974.038.01.050. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Guilley H., Briand J. P. Nucleotide sequence of turnip yellow mosaic virus coat protein mRNA. Cell. 1978 Sep;15(1):113–122. doi: 10.1016/0092-8674(78)90087-9. [DOI] [PubMed] [Google Scholar]
- Hansen B. S., Vaughan M. H., Wang L. Reversible inhibition by histidinol of protein synthesis in human cells at the activation of histidine. J Biol Chem. 1972 Jun 25;247(12):3854–3857. [PubMed] [Google Scholar]
- Hatfield D., Varricchio F., Rice M., Forget B. G. The aminoacyl-tRNA population of human reticulocytes. J Biol Chem. 1982 Mar 25;257(6):3183–3188. [PubMed] [Google Scholar]
- Hosbach H. A., Kubli E. Transfer RNA in aging Drosophila: II. Isoacceptor patterns. Mech Ageing Dev. 1979 Apr;10(1-2):141–149. doi: 10.1016/0047-6374(79)90077-0. [DOI] [PubMed] [Google Scholar]
- Kasai H., Kuchino Y., Nihei K., Nishimura S. Distribution of the modified nucleoside Q and its derivatives in animal and plant transfer RNA's. Nucleic Acids Res. 1975 Oct;2(10):1931–1939. doi: 10.1093/nar/2.10.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Gangloff J., Dirheimer G. Conditions optimales d'aminoacylation du tRNA Asp et du tRNA Trp de levure. Biochimie. 1971;53(5):661–669. doi: 10.1016/s0300-9084(71)80022-6. [DOI] [PubMed] [Google Scholar]
- Keith G., Pixa G., Fix C., Dirheimer G. Primary structure of three tRNAs from brewer's yeast: tRNAPro2, tRNAHis1 and tRNAHis2. Biochimie. 1983 Nov-Dec;65(11-12):661–672. doi: 10.1016/s0300-9084(84)80030-9. [DOI] [PubMed] [Google Scholar]
- Kubli E., Schmidt T. The localization of tRNA4Glu genes from Drosophila melanogaster by "in situ" hybridization. Nucleic Acids Res. 1978 May;5(5):1465–1478. doi: 10.1093/nar/5.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T., Hijazi K. A. Codon context effects in missense suppression. J Mol Biol. 1984 May 5;175(1):19–27. doi: 10.1016/0022-2836(84)90442-x. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Structure, biosynthesis, and function of queuosine in transfer RNA. Prog Nucleic Acid Res Mol Biol. 1983;28:49–73. doi: 10.1016/s0079-6603(08)60082-3. [DOI] [PubMed] [Google Scholar]
- Noguchi S., Nishimura Y., Hirota Y., Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J Biol Chem. 1982 Jun 10;257(11):6544–6550. [PubMed] [Google Scholar]
- Olsen C. E., Penhoet E. E. Chromatographic and functional comparison of human placenta and HeLa cell tyrosine transfer ribonucleic acids. Biochemistry. 1976 Oct 19;15(21):4649–4654. doi: 10.1021/bi00666a016. [DOI] [PubMed] [Google Scholar]
- Smith D. W., McNamara A. L. The effect of the Q base modification on the usage of tRNAHis in globin synthesis. Biochem Biophys Res Commun. 1982 Feb 26;104(4):1459–1463. doi: 10.1016/0006-291x(82)91414-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- White B. N., Tener G. M. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. J Mol Biol. 1973 Mar 15;74(4):635–651. doi: 10.1016/0022-2836(73)90054-5. [DOI] [PubMed] [Google Scholar]
- Yarus M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science. 1982 Nov 12;218(4573):646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]