Abstract

Triosephosphate isomerase (TIM) is a proficient catalyst of the reversible isomerization of dihydroxyacetone phosphate (DHAP) to d-glyceraldehyde phosphate (GAP), via general base catalysis by E165. Historically, this enzyme has been an extremely important model system for understanding the fundamentals of biological catalysis. TIM is activated through an energetically demanding conformational change, which helps position the side chains of two key hydrophobic residues (I170 and L230), over the carboxylate side chain of E165. This is critical both for creating a hydrophobic pocket for the catalytic base and for maintaining correct active site architecture. Truncation of these residues to alanine causes significant falloffs in TIM’s catalytic activity, but experiments have failed to provide a full description of the action of this clamp in promoting substrate deprotonation. We perform here detailed empirical valence bond calculations of the TIM-catalyzed deprotonation of DHAP and GAP by both wild-type TIM and its I170A, L230A, and I170A/L230A mutants, obtaining exceptional quantitative agreement with experiment. Our calculations provide a linear free energy relationship, with slope 0.8, between the activation barriers and Gibbs free energies for these TIM-catalyzed reactions. We conclude that these clamping side chains minimize the Gibbs free energy for substrate deprotonation, and that the effects on reaction driving force are largely expressed at the transition state for proton transfer. Our combined analysis of previous experimental and current computational results allows us to provide an overview of the breakdown of ground-state and transition state effects in enzyme catalysis in unprecedented detail, providing a molecular description of the operation of a hydrophobic clamp in triosephosphate isomerase.
Introduction
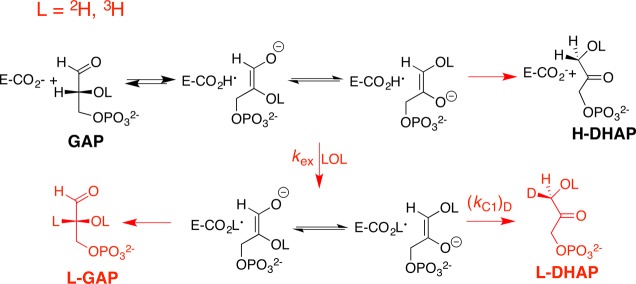
Triosephosphate isomerase (TIM) catalyzes the reversible isomerization of dihydroxyacetone phosphate (DHAP) to d-glyceraldehyde phosphate (GAP, Scheme 1) through a pair of enediolate phosphate reaction intermediates.1−4 TIM is an abundant and readily isolatable glycolytic enzyme, and has been the target of enlightening mechanistic studies for more than 50 years.5 The rate acceleration for TIM is sufficient to obtain partially diffusion-limited turnover of substrate.6,7 The enzyme’s catalytic proficiency, which is related to the burden borne in deprotonating its carbon acid substrate DHAP of pKa = 18,8 is greater than that for ketosteroid isomerase, which has the smaller burden of deprotonation of carbon acid substrate of pKa = 13,9 but less than for proline racemase-catalyzed deprotonation of the weakly acidic α-amino carbon of proline (pKa ≈ 29).10
Scheme 1.
The kinetic parameters and products from TIM-catalyzed isomerization in HOH,11 DOD,12,13 and HOT14 document the formation of enediolate reaction intermediates,12,14 and provide the relative rates for partitioning of this intermediate between the reaction pathways shown in Scheme 1, but do not provide specific insight into the chemical events at the enzyme active site. The chemical reaction mechanism for TIM was inferred from the placement of catalytic side chains relative to the substrate, as determined by X-ray crystallography (Figure 1).1,2,15−18 The roles of these side chains in catalysis were confirmed in studies on the effect of site-directed mutations on enzyme activity, which were largely completed by the late 1990s.1,2,4
Figure 1.
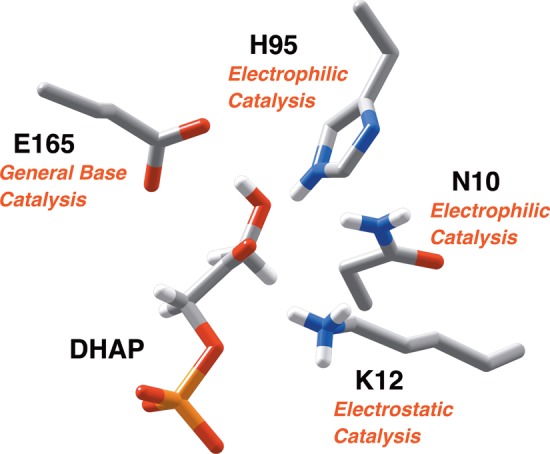
A representation of the catalytic side chains at the active site of TIM (PDB ID: 1NEY), using the numbering of the residues from the wild-type TIM from Saccharomyces cerevisiae.15,19,20 The substrate dihydroxyacetone phosphate (DHAP) is deprotonated by the carboxylate side chain of E165.21,22 The neutral imidazole side chain of H95 is positioned to stabilize negative charge that develops at O-1 or O-2 of the isomeric enediolate phosphate reaction intermediates,23,24 and the amide side chain of N10 is positioned to stabilize negative charge that develops at O-1.25 The alkyl ammonium cation side chain of K12 interacts with both the phosphodianion and the negatively charged oxygen of an enediolate phosphate reaction intermediate.26−29
Richard and Amyes initiated experimental studies to close the gap between the prejudice that the mechanism of action for TIM is well understood, and the lack of clarity about the origin of the enzyme’s catalytic rate acceleration.3,30 Specifically, the tethering of amino acid side chains at the positions appropriate for their catalytic functions is alone unlikely to produce robust catalytic activity, and so it was proposed that these side chains will in “some sense be activated for catalysis at the enzyme active site compared to water”3 (i.e., the corresponding nonenzymatic base-catalyzed deprotonation of the substrate in aqueous solution). This model, which is conceptually similar to the electrostatic preorganization model,31,32 was confirmed by the demonstration that interactions between the protein catalyst and phosphite dianion activate the side chains at the active site for catalysis of deprotonation of the truncated substrate glycolaldehyde.33,34
The binding energy of the phosphodianion of substrates DHAP and GAP is utilized to drive a conformational change of TIM that converts an inactive open protein to an active closed form.16,35 Two important components of this activating conformational change are the placement of the conserved hydrophobic side chains of I170 and L230 on opposite faces of the carboxylate side chain of E165 and the extrusion of water molecules, which solvate this side chain, from the active site to bulk solvent.36−39 The observation that the modest I170V mutation causes a genetic disease in humans suggests a critical role for I170 in catalysis.40,41 The importance of these side chains was confirmed by the demonstration that the I170A, L230A, and I170A/L230A mutations of TIM from Trypanosomes result in significant falloffs in the kinetic parameters for the TIM-catalyzed reactions of the whole substrates DHAP and GAP, and for phosphite dianion activation of the TIM-catalyzed reactions of the truncated substrate glycolaldehyde.42−44
These results show that I170 and L230 are linchpins in the active site architecture of TIM, but do not provide a description of their role in promoting substrate deprotonation. This folds into the deeper question of why the active site architecture of TIM has been highly conserved during evolution.4,45,46 The requirement of the hydrophobic side chains of I170 and L230 for optimal catalytic activity42−44 may reflect either a specific role for the side chains in effecting an increase in the ground-state basicity of the carboxylate of E165,38,47 or an integral role as building blocks for this highly evolved enzyme active site, where there are optimal stabilizing electrostatic interactions between the deprotonation transition state and polar protein side chains.48,49 In the second case, the main effect of side chain truncation would be to perturb the protein structure and reduce these optimal stabilizing electrostatic interactions.
We report here the results of empirical valence bond (EVB) calculations to simulate the action of clamping side chains of I170 and L230 in catalysis by TIM, using base-catalyzed substrate deprotonation by propionate anion as a model for the nonenzymatic reaction, and the X-ray crystal structure of TIM from Saccharomyces cerevisiae (yTIM) as a model for the enzymatic reaction.15 We report that the activation barrier for the wild-type TIM-catalyzed reaction determined by calculations is in good agreement with the activation barrier determined by experiment. In addition, the calculated effect of the I170A, L230A, and I170A/L230A mutations on the activation barrier (ΔG⧧) for the wild-type TIM-catalyzed deprotonation of GAP and DHAP likewise show exceptionally good agreement with the experiment. Combining our current simulation data with detailed analysis of previous experimental results shows that the reduction in the Gibbs free energy (ΔG°) for wild-type as compared to mutant TIMs reflects (i) a small destabilization of the Michaelis complex to wild-type TIM, from replacement of solvating waters for the E165 carboxylate by the hydrophobic side chains of I170 and L230; and (ii) the larger effect of disrupting the precise placement of side chains at the enzyme active site, which results in the weakening of stabilizing interactions between the protein and transition state.
Methodology
We have used the empirical valence bond approach50,51 to model the TIM-catalyzed deprotonation of DHAP and GAP, following our recent computational study of both wild-type and mutant TIM as a prototype system for computational enzyme design.52 The system and simulation setup are largely as described in our previous work.52 These simulations are described in detail in the Supporting Information.
All simulations in this work were performed using the 1.2 Å resolution structure of TIM in complex with DHAP (PDB ID: 1NEY).19,20,53 This structure was prepared for simulations as described in ref (52) and in the Supporting Information. The I170A, L230A, and I170A/L230A mutants were obtained through simple truncation of the relevant side chains. We note that, for simplicity, we use the residue numbering from the wild-type TIM from Saccharomyces cerevisiae throughout this work. In addition, it was demonstrated in the crystal structures of the corresponding variants from Trypanosoma brucei brucei (TbbTIM) that the only impact of these truncations relative to the wild-type structure is the introduction of additional water molecules in the place of the truncated side chains (see ref (44)). GAP was manually placed in the active site using the DHAP conformation in the crystal structure as a template (i.e., by overlaying it on the DHAP molecule in the crystal structure), leading to eight independent starting structures for our simulations of the deprotonation reactions catalyzed by wild-type and mutant TIM (i.e., four variants and two substrates per variant). Each system was then solvated in a water droplet of 20 Å radius of TIP3P water molecules,54 centered on the C1 atom of bound DHAP and GAP, with all protein atoms outside this droplet restrained to their crystallographic positions to avoid system instabilities during the simulation. The protonation states of all ionizable side chains within the inner 85% of the simulation sphere as well as the protonation patterns of all histidine side chains were assigned on the basis of standard pKa values in aqueous solution and by visual inspection, and these were then verified using PROPKA 3.155,56 and the MolProbity server.57 All residues that were ionized in these simulations are listed in Table S1. All other ionizable residues fell in the restrained region of the simulation, and were thus kept in their neutral states to avoid having charges near the surface of or outside the water droplet in the simulations. All simulations in this work were performed using the OPLS-AA force field.58 OPLS-AA compatible parameters for DHAP and GAP were obtained as described in the Supporting Information, and all nonstandard parameters used can be found in the Supporting Information of either ref (52) (DHAP) or this work (GAP).
Molecular Dynamics Simulations and Empirical Valence Bond Calculations
All molecular dynamics (MD) and EVB simulations were performed using the Q simulation package,59 with a 1 fs time step. Each system was gradually heated from 0.01 to 300 K over the course of 140 ps simulation time, while gradually dropping an initial harmonic restraint of 200 kcal mol–1 Å–2 on all heavy atoms to only 0.5 kcal mol–1 Å–2 on the reacting atoms (i.e., the substrate and the E165 side chain). This weak restraint was retained for all subsequent simulations. Once the temperature reached 300 K, we performed additional MD simulations to equilibrate the system for subsequent EVB simulations. Specifically, for each system, we performed three initial 40 ns MD equilibrations at 300 K with different random seeds to assign the initial velocities, resulting in 120 ns equilibration time per system, and 960 ns total equilibration time over all systems. At the end of each individual 40 ns equilibration, we then performed a further 10 short (110 ps) MD simulations with new random seeds, to generate 30 independent starting points (from three initial equilibration runs) for our subsequent EVB simulations. Additionally, for computational efficiency, all initial equilibrations were performed at the transition state as in our previous work,52 as this allowed us to simultaneously propagate EVB trajectories downhill to either reactant or product, thus greatly accelerating the simulations. The RMSD values of all backbone atoms in each system, averaged over the three different replicas, are shown in Figures S1 and S2, demonstrating the convergence of our calculations.
The subsequent EVB simulations were performed using the standard EVB free energy perturbation/umbrella sampling (EVB-FEP/US) procedure described in detail elsewhere,50,51,60 and the successful applications of EVB to TIM have been demonstrated both by us52 and by others.61 The protocol used here is modeled on that used in our previous work,52 and the valence bond states used to describe the reaction are shown in Figure 2. Each EVB trajectory was generated using 51 EVB mapping frames of 200 ps of simulation time per frame, sampling over 30 starting conformations per system as described above, leading to a total of 10.2 ns of simulation time per individual EVB trajectory, 306 ns of simulation time per system (over 30 trajectories), and 2.448 μs of total simulation time over all eight systems considered in this work. A more detailed description of the EVB calibration and parametrization procedures, and the analysis software used, can be found in the Supporting Information, where we have also presented all EVB parameters used to describe the TIM-catalyzed deprotonation of GAP. The corresponding DHAP parameters have already been provided in the Supporting Information of ref (52).
Figure 2.
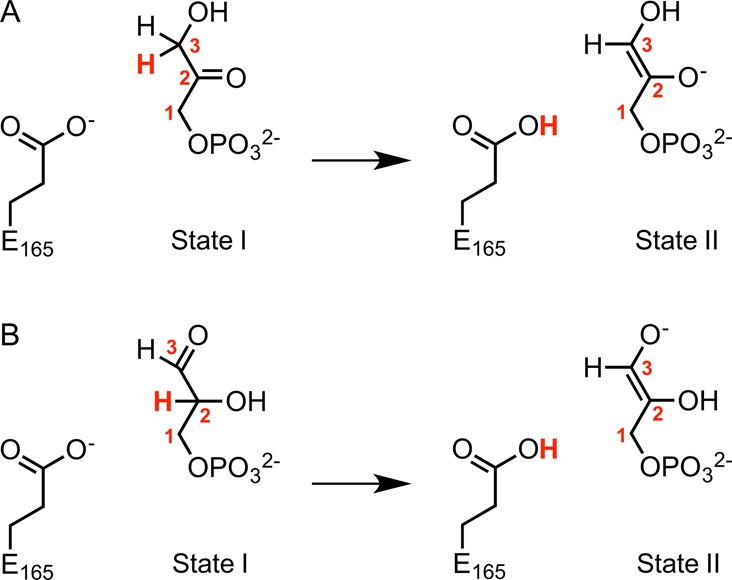
Valence bond states used in this work to describe the TIM-catalyzed deprotonation of the enzyme-bound substrates (A) DHAP and (B) GAP. State I and State II correspond to complexes to the reactant and to the enediolate phosphate intermediate, respectively. In the case of the corresponding uncatalyzed reaction, the carboxylate side chain of E165 was modeled using a propionate anion. The numbering of the carbon atoms of the two substrates is shown, and the transferred proton is highlighted in bold red text.
Results
The valence-bond (VB) states used in this work to describe deprotonation of the acidic carbon of DHAP and GAP by propionate anion to form the enediolate phosphate reaction intermediates are shown in Figure 2. The energetics of the general base-catalyzed reaction in aqueous solution were calibrated as described in the Methodology to give a set of reaction parameters that were used to model the reaction at the active sites for wild-type and mutant TIMs. Table 1 reports the calculated activation barriers (ΔG⧧) and Gibbs (ΔG°) free energies to formation of the enediolate phosphate from the deprotonation of enzyme-bound DHAP and GAP by either propionate anion in water or by the carboxylate side chain of E165 at wild-type, I170A, L230A, and I170A/L230A mutant TIMs. The values for the activation free energies (ΔG⧧) determined by experiment are reported in Table S2, and are used in the comparison of the effects of site-directed mutations on the relative calculated (ΔΔG⧧calc) and experimental (ΔΔG⧧exp) activation barriers reported in Table 1.
Table 1. Activation (ΔG⧧) and Gibbs Free Energies (ΔG°) for the Deprotonation of DHAP and GAP by Wild-Type and Mutant Forms of TIM, To Form Enediolate Phosphate Reaction Intermediatesa.
| substrate | catalyst | ΔGcalc⧧b | ΔGcalc⧧ – ΔGexpc | ΔGTIM⧧ – ΔGnond | ΔGWT⧧ – ΔGmute | ΔGcalc°f | ΔGTIM⧧ – ΔGnong |
|---|---|---|---|---|---|---|---|
| DHAP | CH3CH2CO2– in water | 25.2 ± 0.2 | 18.9 ± 0.2 | ||||
| WT-TIM | 14.5 ± 1.4 | 0.4 | –10.7 | 5.6 ± 1.8 | –13.3 | ||
| I170A | 16.3 ± 1.5 | 0.5 | –8.9 | –1.8 | 7.6 ± 1.4 | –11.3 | |
| L230A | 16.7 ± 0.8 | 0.1 | –8.5 | –2.2 | 8.6 ± 0.8 | –10.3 | |
| I170A/L230A | 18.5 ± 1.0 | 1.1 | –6.7 | –4.0 | 11.0 ± 1.3 | –7.9 | |
| GAP | CH3CH2CO2– in water | 24.1 ± 0.2 | 16.1 ± 0.2 | ||||
| WT-TIM | 12.9 ± 0.8 | 0.0 | –11.2 | 2.5 ± 0.9 | –13.6 | ||
| I170A | 16.2 ± 1.7 | 0.2 | –7.9 | –3.3 | 5.7 ± 1.9 | –10.4 | |
| L230A | 14.9 ± 0.8 | 0.7 | –9.2 | –2.0 | 3.1 ± 1.0 | –13.0 | |
| I170A/L230A | 16.5 ± 1.4 | 0.2 | –7.6 | –3.6 | 5.4 ± 1.8 | –10.7 |
All energies are shown in kcal mol–1.
Activation barrier for proton transfer from the carbon acid substrate to a carboxylate base, for reactions in water or at the active site of yTIM. The calculated energies and standard deviations are obtained as the average of 30 independent EVB trajectories/system (Methodology).
Difference between the calculated activation barriers for proton transfer and the barriers determined by experiment reported in Table S2.
Difference between the activation barriers for proton transfer at TIM and in aqueous solution.
Difference between the activation barriers for proton transfer at wild-type and the specified mutant TIM.
Calculated change in Gibbs free energy for proton transfer from the carbon acid substrate to a carboxylate base in water or at the active site of yTIM.
The difference in ΔGcalc° for proton transfer at TIM (GTIM) and in aqueous solution (Gnon°).
Representative geometries of the key stationary points for the Michaelis complex (MC), the transition state (TS), and the enediolate phosphate intermediate (IS) obtained from EVB simulations of the deprotonation of DHAP and GAP by wild-type TIM are provided in Figure S3. Table S3 reports the average values of the distances, from EVB simulations, separating the donor (D) and acceptor (A) atoms from the transferred proton (H), the average D–H distance, and the D–H–A bond angles, at these stationary points. The corresponding distances separating the side chains of H95 and K12 from substrate atoms that lie within hydrogen-bonding distance of these side chains are shown in Table S4.
We have calculated for each reacting state the number of water molecules that lie within 4 Å of at least one carboxylate oxygen of E165 during the initial equilibration runs for each system, as in our recent study of serum paraoxonase 1.62Figure 3 and Tables S5 and S6 show the average of the water counts at the transition states from our EVB simulations for the deprotonation of DHAP and GAP by wild-type and mutant forms of TIM, as well as the corresponding average number of hydrogen-bond interactions between these water molecules and E165, calculated using VMD.63 Finally, Figure 3 shows the average electrostatic contribution from E165–water interactions to the calculated activation free energies. These were extracted from the EVB trajectories by using the linear response approximation64,65 for the data presented in Table S7.
Figure 3.
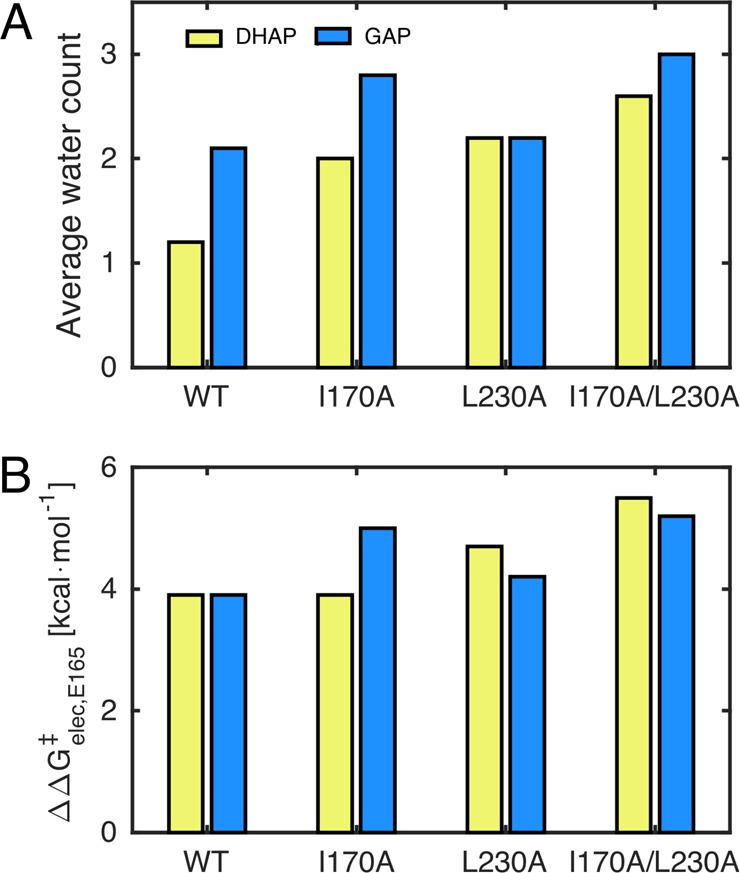
Comparison of parameters for reactions of DHAP (yellow) and GAP (blue). (A) The average number of water molecules within 4 Å of the E165 side chain at the transition state. (B) The electrostatic contribution from E165–solvent interactions to the total calculated activation free energy. The raw data for this figure, a description of how these data were obtained, and the average number of hydrogen bonds between the water molecules shown in panel (A) and the E165 side chain are presented in Tables S5–S7 and the main text.
We have also calculated an estimate for the total electrostatic contribution to the calculated activation energies for the deprotonation of DHAP and GAP by wild-type and mutant TIM, and an estimate for the breakdown of the total interaction into contributions from protein–solvent and solute–solvent interactions. These data were obtained by applying the linear response approximation64,65 to the calculated EVB trajectories, as was done for other systems in our previous work,62 but that were in the present case scaled for simplicitly using an assumed dielectric constant of 4, which is the value most commonly used in simulations to approximate for a hydrophobic active site (see, e.g., refs (66) and (67)). These data are shown visually in Figure 4, with the corresponding raw data being shown in Table S7. Finally, Figure 5 shows the corresponding breakdown of the contribution of individual amino acids to the calculated electrostatic contribution from the protein to the deprotonation of DHAP and GAP in wild-type and mutant forms of TIM, respectively, and the corresponding raw data can be found in Table S8. From these data, it can be seen that while the absolute electrostatic contributions for each system are significant, the effect of the I170A, L230A, and I170A/L230A mutations on the relative electrostatic contributions from each residue (as compared to those observed in the wild-type enzyme) is negligible.
Figure 4.
Comparison of (A) the experimental activation free energies for the deprotonation of DHAP (yellow) and GAP (blue), (B) the total electrostatic contribution from both protein and solvent to the calculated activation free energies (ΔG⧧), as well as the individual contributions from (C) protein and (D) solvent, scaled assuming a dielectric constant of 4 for the active site. For the corresponding raw data, see Table S7.
Figure 5.

Electrostatic contributions of individual residues to the calculated activation free energies (ΔΔG⧧elec) for the TIM-catalyzed deprotonation of DHAP (A,B) and GAP (A,C) by (A) wild-type TIM and (B,C) the TIM variants studied in this work. Note that, for clarity, only residues with contributions >0.2 kcal mol–1 are shown in this figure. All values were obtained by applying the linear response approximation64,65 to the calculated EVB trajectories, as in our previous works,62,68−70 and scaled assuming a dielectric constant of 4 for the active site. The corresponding raw data for this figure are shown in Table S8.
Finally, the “flexibility” in protein structure during our simulations for reactions catalyzed by wild-type and mutant forms of TIM was evaluated by examining the root-mean-square fluctuations (RMSF) of the backbone α-amino acid carbons during our initial molecular dynamics simulations of the transition state complexes of wild-type and mutant forms of TIM in complex with DHAP and GAP (Figures 6 and S4, respectively). The first 10 ns of every 40 ns simulation was discarded from the analysis as equilibration, and 3 replicates were performed for each system, leading to a total of 90 ns of simulation time per system. The top panel in Figure 6 shows the calculated RMSF of the backbone α-amino acid carbons at the transition state for the wild-type TIM-catalyzed deprotonation of DHAP for a single enzyme subunit. The next three panels at the bottom half of Figure 6 show the effect of I170A, L230A, and I170A/L230A mutations on the RMSF determined for the wild-type TIM-catalyzed reactions of DHAP. The corresponding data for the transition state complexes for the TIM-catalyzed deprotonation of GAP are shown in Figure S4.
Figure 6.
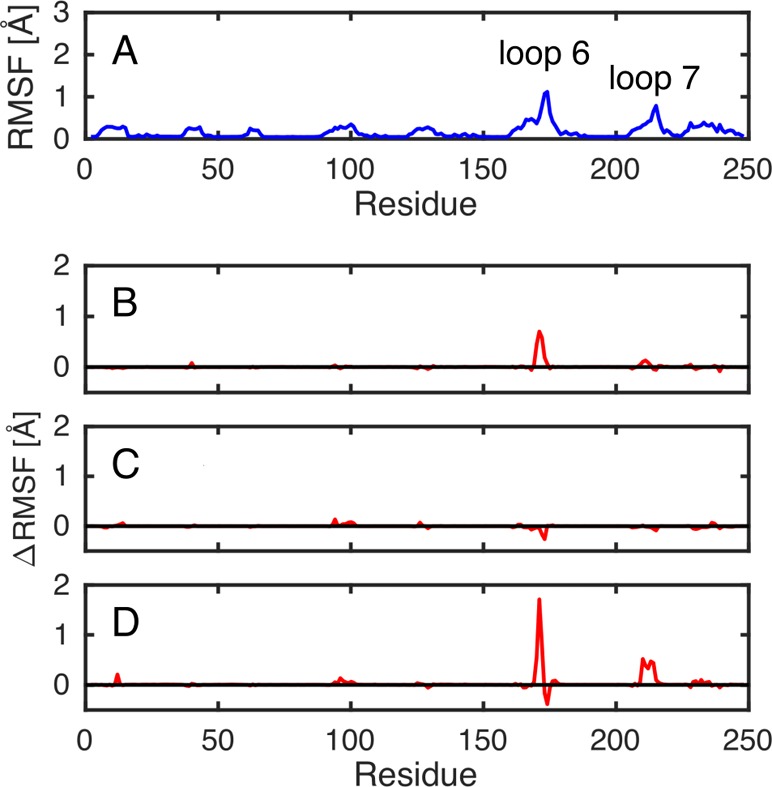
Root-mean-square fluctuations (RMSF) of the backbone α-amino acid carbons at the transition state complexes for the TIM-catalyzed deprotonation of DHAP determined for a single enzyme subunit. (A) The total fluctuations observed for the wild-type enzyme. (B–D) The difference between the fluctuations of wild-type TIM and the specified enzyme variant (where (B), (C), and (D) denote the I170A, L230A, and I170A/L230A variants, respectively). Data were collected every 5 ps from three individual 40 ns molecular dynamics trajectories for each system, and represent a total of 90 ns simulation time per system (the first 10 ns of each trajectory was discarded as equilibration; see the description in the main text). The corresponding plots of data for TIM-catalyzed deprotonation of GAP at the transition state are given in Figure S4.
Discussion
Previous computational studies to model catalysis by TIM have pushed at the frontiers of computational and mechanistic enzymology, while providing insight into the mechanism of this enzyme-catalyzed proton transfer reaction.52,61,71−82 Our present work has focused on using EVB methods to model catalysis by wild-type TIM. This provides a starting point for computational studies that address issues about the role of the highly conserved hydrophobic side chains of I170 and L230 at an enzyme active site that contains the more intensively studied polar side chains, which facilitate the proton transfer reactions at the enzyme-bound substrate (Figure 1).
The activation barrier for TIM-catalyzed isomerization in water is composed mainly of the thermodynamic barrier (corresponding to the Gibbs free energy) to deprotonation of enzyme-bound substrate to form the enediolate phosphate reaction intermediate (Scheme 1).83 We have therefore focused our computational effort on modeling the large barrier for this step, but not the small barriers for the rapid proton transfer reactions that connect the two enediolate phosphate intermediates (Scheme 1).
The activation barriers, ΔGexp⧧, calculated from the experimental values of kcat for isomerization of triosephosphates catalyzed by wild-type and mutant TIMs from Trypanosoma brucei brucei (TbbTIM), are compared in Table 1 with the computed ΔGcalc for deprotonation catalyzed by TIM from yeast, which are obtained by in silico mutation of an atomic-level 1.2 Å resolution crystal structure of the enzyme·DHAP complex.15 We note that there are no large differences in the kinetic parameters for isomerization of the whole substrates and substrate pieces catalyzed by TIM from Tbb,46 yeast,84 and rabbit muscle,33 due to the high structural homology of the active sites of TIM from different organisms.4,44,46
Wild-Type TIM
There is excellent agreement between the experimental activation barriers ΔG⧧ of 14.1 and 12.9 kcal mol–1 for the TIM-catalyzed deprotonation of DHAP and GAP, respectively (Scheme 1), and the activation barriers of 14.5 and 12.9 kcal mol–1 calculated using the EVB approach. This corresponds to 10.7 and 11.2 kcal mol–1 reductions, respectively, in the activation barriers ΔGcalc⧧ for propionate anion-catalyzed deprotonation of DHAP or GAP upon substrate binding to TIM. These reductions in ΔG⧧ are about 80% of the respective 13.3 and 13.6 kcal mol–1 decreases in the Gibbs free energy, ΔGcalc, for substrate deprotonation (Scheme 1), so that about 80% of the change in the reaction driving force is expressed as a change in the stability of the transition state for proton transfer.
This reduction in the activation barrier for proton transfer at TIM as compared to an aqueous solvent is achieved through either the selective stabilization of the transition state, which brings its energy closer to that for the reactant, or the selective destabilization of the reactant, which brings its energy closer to that of the transition state. EVB and other approaches to model enzyme reaction mechanisms typically do not consider the substrate binding step, and therefore cannot be used to directly evaluate the binding energy of DHAP and GAP that is utilized to destabilize the Michaelis complex, either by inducing strain into the substrate, or to drive a thermodynamically unfavorable conformational change at the enzyme.85,86 The EVB approach provides the difference between the electrostatic interactions of the protein catalyst TIM with the reactant and with the transition state complexes, which are assumed to be the underlying cause for transition state stabilization. These were extracted from the calculated EVB trajectories using the linear response approximation,62,64,65,68−70 which can provide an estimate of the contribution of electrostatic interactions between catalytic side chains/solvent molecules and the reacting atoms to the enzymatic rate acceleration.
Figures 4 and 5 show the electrostatic contributions from protein and solvent and the corresponding breakdown of the contributions from individual amino acid side chains (>0.2 kcal mol–1) to the calculated activation free energies (ΔG⧧) for the formation of the enediolate phosphate intermediate, extracted from the EVB calculations. The sum of the electrostatic stabilization provided by the protein to the transition states for the deprotonation of DHAP and GAP is −6.1 and −4.9 kcal mol–1, respectively (see Figure 4 and Table S7). This shows that stabilizing electrostatic interactions make an important, and perhaps the primary, contribution to the corresponding computed 11.9 and 11.3 kcal mol–1 reductions in the activation barriers for proton transfer in the enzyme active site as compared to that in water.
Figure 5 provides a qualitative description of the effect of protein-side chain interactions on transition state and intermediate stability, which is fully consistent with observations from earlier experimental studies on the mechanism of the action of TIM. In particular, we note that:
(1) The substrate for TIM is locked in a protein cage,37,39 with the phosphodianion occluded from interaction with solvent water, and ion-paired to the surface alkyl ammonium side chain of K12. This side chain interaction is estimated to provide 3.9 and 2.7 kcal mol–1 stabilization, respectively, of the transition states for TIM-catalyzed deprotonation of DHAP and GAP. The ion pair between the buried side chain anion of E97 and K12 immobilizes the cationic side chain at the protein surface.26−29 The electrostatic interactions of the side chain anion with the transition state are estimated to be destabilizing. However, the sum of the net interactions of the K12·E97 ion pair with the transition state is still stabilizing by 2.6 and 1.9 kcal mol–1, respectively, at the transition state for the TIM-catalyzed deprotonation of DHAP and GAP, making this the largest individual contribution to electrostatic stabilization from the protein. For comparison, the K12G mutation of yTIM results in a 7.8 kcal mol–1 decrease in the activation barrier to kcat/Km, for the second-order reaction of free TIM and substrate, but roughly one-half of this interaction is expressed at the Michaelis complex.28
(2) The neutral imidazole side chain of H95 interacts with the carbonyl oxygen of DHAP and GAP (Figure 2).23,24 The small 0.7 and 0.9 kcal mol–1 stabilizing interactions between this side chain and the endiolate phosphate-like transition state for TIM-catalyzed deprotonation of DHAP and GAP, respectively, emphasize the relatively minor role of electrophilic catalysis by this neutral side chain in TIM-catalyzed isomerization, as compared to that of the cationic side chain of K12.3,23,26,28
In summary, early discussions of the mechanism of action of TIM were focused on the contribution of Brønsted acid catalysis at the carbonyl group to the enzymatic rate acceleration.23,24,87,88 By contrast, the results of recent experimental studies emphasize the importance of electrostatic stabilization of the charged enolate transition state for the fully stepwise deprotonation of carbon.3,28 This viewpoint is consistent with the results of the EVB calculations reported in this work.
There is a large activation barrier of ΔGcalc⧧ = 25.2 kcal mol–1 and a Gibbs free energy of ΔGcalc = 18.9 kcal mol–1 to deprotonation of DHAP by propioniate anion in water to form an endiolate phosphate. John Gerlt and Paul Gassman proposed that enzymatic catalysis of deprotonation of carbon is due to a reduction in ΔGcalc° for formation of unstable enolates, which is largely expressed as a reduction in ΔGcalc from stabilization of “late” reaction transition states.89 This proposal is supported by data from Table 1, which show that 80% of 13.3 kcal mol–1 reduction in ΔGcalc° for deprotonation of DHAP at TIM is expressed as a 10.9 kcal mol–1 reduction in the activation barrier ΔGcalc. Similarly, 80% of 13.6 kcal mol–1 reduction in ΔGcalc° for deprotonation of GAP at TIM is expressed as an 11.2 kcal mol–1 reduction in the activation barrier ΔGcalc. We conclude that the position of transition states for these TIM-catalyzed reactions is close to the product enediolate phosphate, because the two species show stabilizing interactions similar to those of the protein catalyst.
I170A and L230A Mutants of TIM
Figures 1 and 7 show representations of the side chains at the tightly packed active site of TIM. The I170A and L230A mutations of side chains that lie close to the carboxylate group of E165 result in increases in the activation barriers for deprotonation of TIM-bound substrates,42,43 which we have modeled in this work. The average absolute difference between the calculated and experimental activation barriers, (ΔΔG⧧calc – ΔΔG⧧exp) = 0.4 kcal mol–1, is close to the uncertainty of ca. 0.2 kcal mol–1 in the experimental barriers, and our calculations also capture the fact that the experimentally observed order of the I170A and L230A mutations in terms of increasing activation free energy (ΔΔG⧧mut – ΔΔG⧧WT) is reversed between the two substrates. We conclude, therefore, that these EVB calculations do an excellent job in reproducing the effects of I170A and L230A mutations on enzyme activity determined by experiment.
Figure 7.
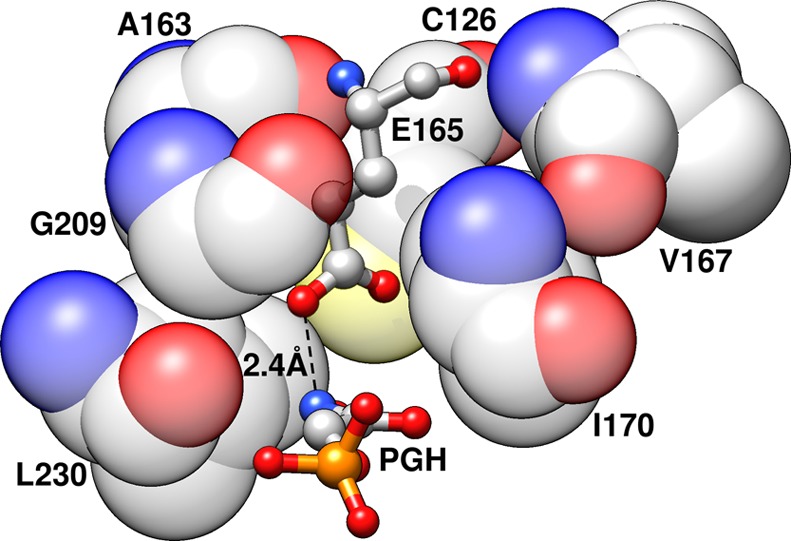
A representation of clamping of the basic side chain of E165 by the hydrophobic side chains of I170 and L230 at the active site of wild-type TbbTIM (PDB ID: 1TRD)19,20,90 in complex with the intermediate analogue phosphoglycolhydroxamate (PGH). The I170 side chain is, in turn, clamped by the side chains of V167 and (the CH2 groups of the side chain of) E165. For consistency, the figure has been annotated using the numbering for the wild-type yTIM.
Figure 8 shows linear free energy relationships between the calculated activation barriers ΔGcalc⧧ and the free energy changes ΔGcalc (●) for wild-type and mutant TIM-catalyzed deprotonation of DHAP and GAP to form the enediolate phosphate intermediates, as well as the corresponding activation barriers observed in experiment (+). The slopes of 0.80 and 0.76 for the relationships for TIM-catalyzed reactions of DHAP and GAP, respectively (Figure 8), are similar to the slopes of the two-point correlations for the uncatalyzed and wild-type TIM-catalyzed reactions discussed above. These results show that the side chains of I170A and L230A function to reduce the Gibbs free energy for TIM-catalyzed deprotonation, and that a large fraction of this effect is expressed as stabilization of an enediolate phosphate-like transition state.
Figure 8.
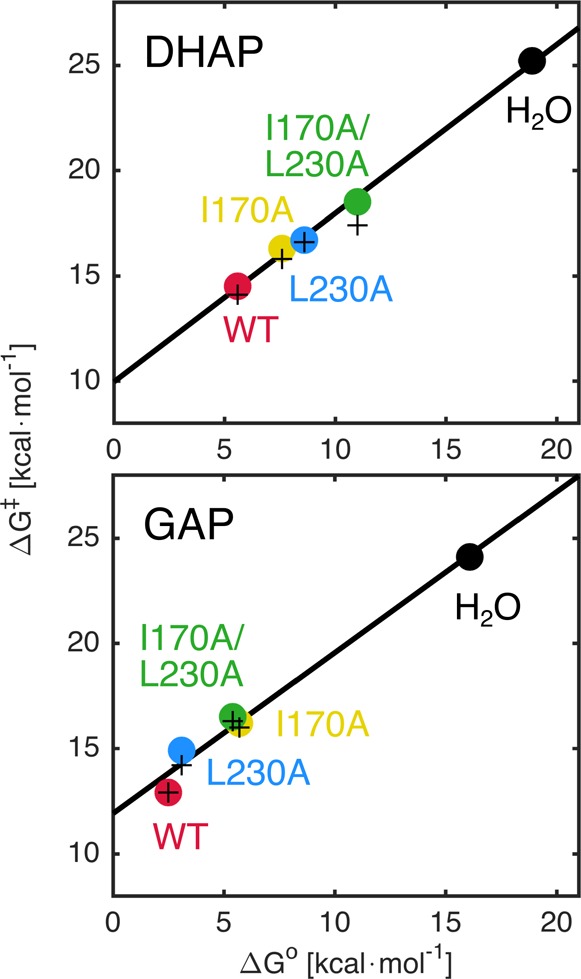
Linear free energy relationships between the activation barriers, ΔG⧧, and the Gibbs free energy, ΔG°, for the deprotonation of DHAP and GAP catalyzed by wild-type and mutant forms of TIM, using values of ΔG° determined in this work by EVB calculations, and values of ΔG⧧ determined either by EVB calculations (●) or by experiment (+). The correlation coefficients are 0.9921 and 0.9987 for DHAP, and 0.9909 and 0.9898 for GAP (calculated and experimental values, respectively), calculated using linear regression analysis. The corresponding calculated and experimental data are summarized in Tables 1 and S2, respectively.
The results from EVB calculations show that the positions of the transition states for TIM-catalyzed deprotonation of bound substrates lie closer to the intermediate than to the Michaelis complex (see Figure S5). One consequence of a late transition state for TIM-catalyzed deprotonation of bound substrate is that the effect of substitutions on the reaction driving force ΔG° is strongly expressed as changes in the activation barrier ΔG⧧ for substrate deprotonation. Consequently, the effects of ΔG⧧ can be rationalized by a consideration of the effects on ΔG° for substrate deprotonation. This situation plays to the strengths of the EVB method in modeling reactions that proceed through a high energy polar intermediate, because: (i) It is easier to model computationally the effects of changing enzyme structure on the stability of a fully formed intermediate, than the effects on the stability of a metastable transition state. (ii) These effects on the stability of the highly charged enediolate phosphate trianion reaction intermediate should be dominated by the changes in the electrostatic interactions between the protein catalyst and the intermediate that the EVB method is particularly adept at modeling.51
Complex Interpretations
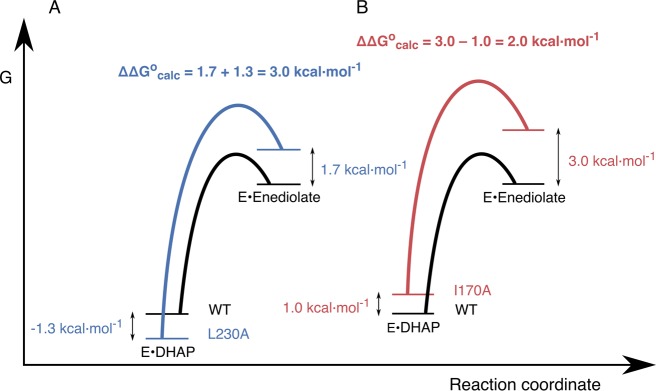
The I170A, L230A, and I170A/L230A mutations each result in increases in the activation barrier to ΔG°to formation of the enediolate phosphate. This change in reaction barrier may be due either to the destabilization of the enediolate reaction intermediate (ΔΔGI°) or to the stabilization of the ground-state Michaelis complex [−(ΔΔGR)]. In the first case, the change in ΔG° may reflect a reorganization of the active site that results in the destabilization of the reaction intermediate. In the second case, this would reflect the elimination of interactions from the deleted side chains that destabilize the Michaelis complex relative to the enediolate intermediate and result in an increase in the barrier ΔGo for deprotonation of bound substrate to form this intermediate.91
As shown in Figure 8, we obtain a linear relationship between the calculated activation free energies (ΔG⧧) and the corresponding Gibbs free energies (ΔG°) for the deprotonation of DHAP and GAP by wild-type and mutant TIM. The calculated total effect of the I170A, L230A, or I170A/L230A mutations on the Gibbs free energy, ΔG°, for the formation of the enediolate phosphate intermediate, is the sum of the effect on the stability of the Michaelis complex (−ΔΔGR°), and the effect on the stability of the corresponding reaction intermediate (ΔΔGI). That is, ΔΔG° = −ΔΔGR° + ΔΔGI. The effect of these mutations on the stability of the Michaelis complex (ΔΔGR°) can been determined from the change in the experimental value of Km for the TIM-catalyzed deprotonation of DHAP and GAP (Table 2). The overall Gibbs free energy, ΔΔG°, for the deprotonation of the substrate is provided by our calculations (Table 1). From these data, we can therefore obtain the values of ΔΔGI = ΔΔG° + ΔΔGR° (Table 2) and provide graphical representations in Figure 9A and B of data for deprotonation of DHAP bound to the L230A and the I170A mutant enzymes, respectively.
Table 2. Effect of Mutations of TIM on the Relative Activation Barriers (ΔΔG⧧), Reactant Stability (ΔΔGR°), Intermediate Stability (ΔΔGI), and on the Total Change in Gibbs Free Energy for Conversion of the Michaelis Complex to the Intermediate (ΔΔG°)a.
| variant | substrate | ΔΔG⧧b,f | ΔΔG°c,f | ΔΔGR°d | ΔΔGI°e |
|---|---|---|---|---|---|
| I170A | DHAP | 1.8 | 2.0 | 1.0 | 3.0 |
| GAP | 3.3 | 3.2 | –0.3 | 2.9 | |
| L230A | DHAP | 2.2 | 3.0 | –1.3 | 1.7 |
| GAP | 2.0 | 0.6 | –0.3 | 0.3 | |
| I170A/L230A | DHAP | 4.0 | 5.4 | –1.4 | 4.0 |
| GAP | 3.6 | 2.9 | –1.6 | 1.3 |
All energies are shown in kcal mol–1.
The difference in the calculated activation barriers (ΔGWT⧧ – ΔGmut) for the wild-type and mutant yTIM (Table 1).
The difference in the calculated free energy barriers for substrate deprotonation (ΔGWT° – ΔGmut) for the wild-type and mutant yTIM (Table 1).
Estimated from the ratio of the values for the experimental values for Km determined the wild-type and mutant yTIM.43
Estimated as the difference between the value of ΔΔG° for the overall effect of the mutation on the reaction driving force and ΔΔGR°: ΔΔGI = (ΔΔG° + ΔΔGR°).
The calculated energies are averages over 30 independent EVB trajectories for each enzyme, as described in the Methodology.
Figure 9.
Free energy profiles for wild-type and mutant TIM-catalyzed deprotonation of enzyme-bound DHAP, where the energy of the Michaelis complex is shown relative to the energy for unliganded TIM and DHAP. (A) Profiles for the reactions catalyzed by wild-type TIM and the L230A mutant. (B) Profiles for the reactions catalyzed by wild-type TIM and the I170A mutant. The values of ΔΔGR°, ΔΔGI, and ΔΔG° = ΔG°mut – ΔG°WT from Table 2 are −1.3, 1.7, and 3.0 kcal mol–1 for the L230A mutant, and 1.0, 3.0, and 2.0 kcal mol–1 for the I170A mutants, respectively.
Figure 9 provides insight into the roles of the hydrophobic side chains in TIM-catalyzed deprotonation of substrate. The relative values of kcat for isomerization catalyzed by L230A and I170A mutants show that there is a 1.0 kcal mol–1 larger destabilization of the transition state for deprotonation of DHAP catalyzed by the L230A mutant as compared to the I170A mutant. This implies that the hydrophobic side chain of L230 at wild-type TIM provides a 1.0 kcal mol–1 stronger stabilization of the enediolate intermediate relative to DHAP than the side chain of I170. However, the I170A and L230A substitutions result in a 1.0 kcal mol–1 destabilization and 1.3 kcal mol–1 stabilization, respectively, of the Michaelis complexes to DHAP (Figure 9A and B).43 This corresponds to a 2.3 kcal mol–1 change in the relative energies of the Michaelis complexes, which is larger than the 1.0 kcal mol–1 difference in the effects of these mutations on ΔG° for substrate ionization. A correction for these changes in ground-state energies shows that the presence of the hydrophobic side chain from I170, as compared to L230, in fact, provides a 1.3 kcal mol–1 stronger stabilization of the enediolate intermediate relative to DHAP.
There is a 1.3 kcal mol–1 larger stabilization of the Michaelis complex to GAP as compared to DHAP for the I170A mutant (ΔΔGR°, Table 2), which is reflected in the 1.2 kcal mol–1 larger barrier ΔG° for deprotonation of DHAP as compared to GAP to form the enediolate phosphate. By contrast, the L230A mutation provides a 1.0 kcal mol–1 larger stabilization of the Michaelis complex to DHAP as compared to GAP that favors deprotonation of GAP relative to DHAP. These results show that the I170A and L230A mutations provide preferential stabilization of enzyme complexes to GAP and DHAP, respectively, and suggest that these differences in the stabilizing binding interactions are lost upon substrate deprotonation to form the similarly structured enediolate reaction intermediates (Scheme 1).
The results of the present calculations, summarized by Figure 9, emphasize the importance of I170 and L230 in ensuring optimal stabilizing interactions between TIM and transition state for substrate deprotonation.92 The estimated electrostatic contributions from both protein and solvent to the calculated activation free energies for the deprotonation of DHAP and GAP are shown in Figures 4 and 5. These electrostatic interactions are weakened by the I170A, L230A, and I170A/L230A mutations, where the changes in the total electrostatic interactions are the sum of many small changes in individual interaction, including the electrostatic contributions from N10, K12, H95, S96, and E129 shown in Tables S7 and S8. These results support the notion that the placement of I170 and L230 at the tightly packed active site of TIM is critical to the observation of optimal electrostatic stabilization of the transition state for substrate deprotonation. We conclude that the replacement of these bulky side chains by the methyl group of alanine is accompanied by a repositioning of the catalytic side chains, and a reduction in the optimal electrostatic stabilization of the transition state for substrate deprotonation.
Ground-State Effects
The ground-state effects of the I170A and L230A mutations are too complex to be modeled in full by these calculations. However, our results provide insight into their contribution to the observed effects of I170A and L230A mutations on enzyme activity. The X-ray crystal structures for unliganded wild-type, I170A, L230A, and I170A/L230A TbbTIM and for the corresponding complexes to phosphoglycolate (PGA), at resolutions ranging from 1.7 to 2.3 Å, show that the mutations cause no significant changes in the structure of the complexes to PGA, but that each deleted hydrophobic side chain at these complexes is replaced by a single water molecule.44 The results of our simulations (Figure 3) are in agreement with X-ray structural data. These simulations show that I170A, L230A, and I170A/L230A mutations each result in an increase in the average number of water molecules within 4 Å of the E165 side chain at the transition state for these reactions (Figure 3A), and in the corresponding average number of hydrogen bonds that these water molecules form to the carboxylate side chain of E165 (Table S6). We note also that the additional water molecules introduced in our simulations are located in positions similar to those observed in ref (44).
Figure 3B and Table S6 suggest that the 1.3 kcal mol–1 stabilization of the Michaelis complex at the L230A mutant is due to the stabilizing interactions between water and the side chain of E165. However, a consideration of the effect of these mutations on the ground-state solvation of I170 cannot rationalize the opposite 1.0 kcal mol–1 destabilization of the Michaelis complex observed for the I170A mutant TIM. Insight into the explanation for the different effects of the I170 and L230 mutations on Km is obtained from a comparison of the effect of these mutations on the mobility of amino acid side chains at the Michaelis complex. We have examined the root-mean-square fluctuations (RMSF) of individual residues at the transition states complexes of the wild-type and mutant TIMs (Figures 6 and S4). The RMSF plots (Figures 6 and S4) show both the absolute values for the simulations of wild-type TIM in complex with DHAP and GAP, as well as the difference between the RMSF (ΔRMSF) for wild-type and the I170A, L230A, or I170A/L230A mutant forms of yTIM (Figure 10).
Figure 10.
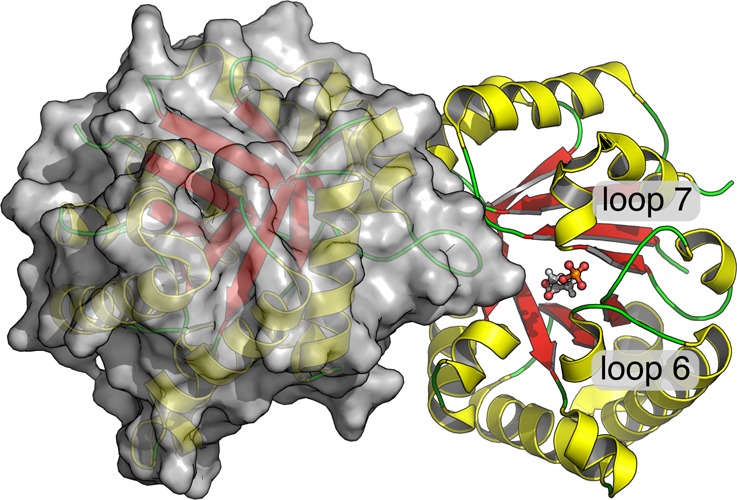
An overview of the quarternary structure of TIM, highlighting the positions of the bound substrate (DHAP) in chain A, as well as of loops 6 and 7 (PDB ID: 1NEY).15,19,20
The increase in the RMSF for the I170A mutant is consistent with an increase in the flexibility of loop 6, which suggests that the mutation results in a weakening of the hydrophobic interaction between the side chains of I170 and V167 (Figure 7). We propose that the different effects of the I170A and L230A mutations on Km for substrate binding reflect the different effects of these mutations on the relative stability of the open and closed forms of TIM for mutations at flexible loop 6 and at a site distant from loop 6. The Michaelis complex of substrate bound to the closed form of each mutant is stabilized by interactions between the additional water(s) and the carboxylate side chain of E165. This is the dominant effect that controls the decrease in Km for the L230A mutant. By contrast, the effect of the I170A mutation is controlled by the destabilization of the closed form of the TIM from the loss of the hydrophobic interactions between the protein (V176) and the excised side chain (Figure 7).
The L230A mutation results in surprising (15–20)-fold increases in the second-order rate constant for TIM-catalyzed deprotonation of the truncated substrate glycolaldehyde and in the affinity of phosphite dianion in activation of the TIM-catalyzed deprotonation of glycolaldehyde.42,43 We attribute these changes to ground-state effects of the L230A mutation, which result in increases in the fractional concentration of TIM present in a high energy active form.42,43 We are, however, unable to use the results of the present calculations to provide insight into the origin of these proposed ground-state effects.
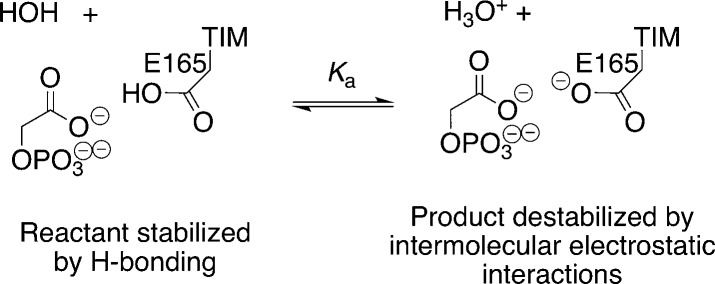
The pKa of the Carboxylic Acid Side Chain of E165
The I170A mutation of TbbTIM is accompanied by a decrease in the pKa for deprotonation of the hydrogen-bonded complex between the PGA trianion and the carboxylic acid side chain of E165 (Scheme 2), from pKa > 10 for wild-type TIM to pKa = 7.8.47 It was proposed that this result supports the conclusion that “the hydrophobic side chain of Ile170 plays a critical role in effecting the large increase in the basicity of the catalytic base upon the binding of substrate or inhibitors.”47 This conclusion includes the implicit assumption that the I170A mutation results in the same decrease in pKa’s of the carboxylic acid side chain of E165 at the TIM·substrate and at the TIM·PGA complexes.
Scheme 2.
We note the following complications, and suggest that this assumption is inappropriate:
(1) The proposed ground-state effect of the I170A mutation on the basicity of E165 at the TIM·substrate complex is presumably due to an increase in the stabilizing solvation of the side chain anion at the mutant enzyme that would result in a decrease in Km. We find instead that this mutation results in an increase in Km.44
(2) The effect of the I170A mutation on the pKa of the binary complex to PGA represents, in part, the combined effects of the mutation on the stability of the enzyme–inhibitor hydrogen bond, and on destabilizing electrostatic interactions between the inhibitor trianion and the carboxylate anion at the deprotonated enzyme (Scheme 2). There should be a smaller effect of this mutation on the pKa of the binary complex to substrate dianion, because of the weaker destabilizing electrostatic interactions between the carboxylate anion with the substrate dianion, and the lack of a significant hydrogen-bonding interaction between the carboxylic acid and substrate.
(3) Proton transfer from substrate to TIM to form the enediolate trianion, whose charge is similar to that for PGA, should result in a significant increase in the pKa for the carboxylic acid side chain of E165. We propose that (i) the pKa of this side chain at the complex to the enediolate intermediate is similar to the high pKa determined for the complex to PGA (Scheme 2).47 (ii) An important role of I170 is to optimize the increase in side chain basicity that accompanies substrate deprotonation.93 (iii) The I170A mutation has the effect of weakening the destabilizing electrostatic interactions of the carboxylate anion with the intermediate trianion. (iv) A large fraction of this increase in the pKa of the side chain at the complex to the substrate dianion, as compared to the enediolate trianion, is expressed at a late transition state for substrate deprotonation (Figure 8).93
Broader Issues
The large variations in the amino acid sequences of TIMs from organisms that span the known course of evolution contrast sharply with the small changes in the positions of the amino acid side chains at enzyme active sites, which are nearly superimposable for TIMs from different organisms.39,46 This suggests that the optimal, or “perfect”, structure for the active site of TIM may have been achieved relatively early in evolution,6,94 and that this structure has been conserved with the drifting in the sequence of amino acids at other regions of the protein. We expect that positions of the side chains that participate directly in the chemistry of the enzyme-catalyzed proton transfer reaction such as K12, H95, and E165 should be conserved to maintain their optimal stabilizing interactions with the transition state. Our investigations of I170 and L230 have focused on developing an understanding of the imperatives for the high conservation in the position of these conserved active site side chains.
The evolution of the catalytic efficiency of TIM and other enzymes is driven by two imperatives: (1) the requirement for optimal stabilizing interactions between the protein catalyst and the enzyme-bound transition state; and (2) the requirement to defer the expression of these optimal interactions from the Michaelis complex to the transition state, to avoid irreversible substrate binding.85,86,91 The latter may be accomplished through the utilization of substrate binding energy to introduce destabilizing interactions into the protein and/or substrate, which are relieved at the transition state for the catalyzed reaction.49,91,95
A comparison of the X-ray crystal structures for wild-type, I170A, L230A, and I170A/L230A mutants of TIM shows that the effects of these mutations on the structure of the unliganded protein and the complex to PGA are minimal, except that the excised amino acid side chains are replaced by molecules of water at the PGA complex.44 The effects on the structure of the binary enzyme–inhibitor complex suggest that the hydrophobic side chains function to enhance the ground-state basicity of E165. These appearances are deceiving. The experimental and computational results to model the effect of these mutations on the wild-type TIM-catalyzed reaction provide support for the existence of a modest ca. 1.3 kcal mol–1 effect of the L230A mutation of the stability of ground-state Michaelis complex (Figure 9). By contrast, our results provide strong evidence that these hydrophobic side chains are building blocks at a complex active site architecture, which provides for optimal stabilizing electrostatic interactions between the protein catalyst and the transition state for TIM-catalyzed substrate deprotonation.92
The phosphodianion binding energy of the substrates for TIM, orotidine 5′-monophosphate decarboxylase (OMPDC),96,97 glycerol 3-phosphate dehydrogenase (GPDH),98,99 and other enzymes100,101 is utilized to drive a large activating enzyme conformational change.85 By contrast, experimental results on TIM discussed here and on OMPDC85,102 provide only limited support for the proposal that the phosphodianion binding energy is utilized to introduce steric or electrostatic stress into the substrate. We have proposed that the ligand phosphodianion binding energy is utilized to pay the significant entropic and enthalpic price for the conversion of floppy, well-solvated open enzymes to tight catalytically active desolvated substrate cages. This provides a general, and perhaps prevailing, mechanism for avoiding irreversible substrate binding by deferring the expression of large intrinsic substrate binding energies from the Michaelis complex to the enzymic transition state.
Acknowledgments
This work was funded by the Swedish Research Council (VR, Grant 2015-04928), the National Institutes of Health (GM03597 and GM116921), and “Strategischer Forschungsfonds” of Heinrich Heine University Düsseldorf (F2014/730-11). D.P. is financially supported by the Jürgen Manchot Foundation, and Y.S.K. is financially supported by the Sven and Lilly Lawski Foundation for Natural Sciences Research. We would also like to thank the Swedish National Allocations Committee (SNAC) for the generous allocation of computer time, through SNAC projects SNIC 2015/16-12 and 2016/1-293. All calculations were performed on the Triolith cluster at the National Supercomputing Center at Linköping University, and the Hebbe cluster at the C3SE center at the Chalmers University of Technology.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b05576.
Extended simulation details, supplementary figures and tables outlining key geometric parameters and protein flexibility in our simulations, and full set of EVB parameters used to simulate the deprotonation of GAP (the corresponding DHAP parameters have been published previously) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Knowles J. R. Phil. Trans. R. Soc., B 1991, 332, 115–121. 10.1098/rstb.1991.0039. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. Nature 1991, 350, 121–124. 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- Richard J. P. Biochemistry 2012, 51, 2652–2661. 10.1021/bi300195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K. Cell. Mol. Life Sci. 2010, 67, 3961–3982. 10.1007/s00018-010-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. V.; Rose I. A. J. Biol. Chem. 1959, 234, 1007–10. [PubMed] [Google Scholar]
- Knowles J. R.; Albery W. J. Acc. Chem. Res. 1977, 10, 105–111. 10.1021/ar50112a001. [DOI] [Google Scholar]
- Blacklow S. C.; Raines R. T.; Lim W. A.; Zamore P. D.; Knowles J. R. Biochemistry 1988, 27, 1158–1165. 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- Richard J. P.; Amyes T. L. Curr. Opin. Chem. Biol. 2001, 5, 626–633. 10.1016/S1367-5931(01)00258-7. [DOI] [PubMed] [Google Scholar]
- Hawkinson D. C.; Eames T. C. M.; Pollack R. M. Biochemistry 1991, 30, 10849–10858. 10.1021/bi00109a007. [DOI] [PubMed] [Google Scholar]
- Williams G.; Maziarz E. P.; Amyes T. L.; Wood T. D.; Richard J. P. Biochemistry 2003, 42, 8354–8361. 10.1021/bi0345992. [DOI] [PubMed] [Google Scholar]
- Putman S. J.; Coulson A. F. W.; Farley I. R. T.; Riddleston B.; Knowles J. R. Biochem. J. 1972, 129, 301–310. 10.1042/bj1290301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue A. C.; Amyes T. L.; Richard J. P. Biochemistry 2005, 44, 2610–2621. 10.1021/bi047954c. [DOI] [PubMed] [Google Scholar]
- O’Donoghue A. C.; Amyes T. L.; Richard J. P. Biochemistry 2005, 44, 2622–2631. 10.1021/bi047953k. [DOI] [PubMed] [Google Scholar]
- Herlihy J. M.; Maister S. G.; Albery W. J.; Knowles J. R. Biochemistry 1976, 15, 5601–5607. 10.1021/bi00670a026. [DOI] [PubMed] [Google Scholar]
- Jogl G.; Rozovsky S.; McDermott A. E.; Tong L. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 50–55. 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolis E.; Petsko G. A. Biochemistry 1990, 29, 6619–6625. 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- Alber T.; Banner D. W.; Bloomer A. C.; Petsko G. A.; Phillips D.; Rivers P. S.; Wilson I. A. Phil. Trans. R. Soc., B 1981, 293, 159–171. 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- Banner D. W.; Bloomer A. C.; Petsko G. A.; Phillips D. C.; Pogson C. I.; Wilson I. A.; Corran P. H.; Furth A. J.; Milman J. D.; Offord R. E.; Priddle J. D.; Waley S. G. Nature 1975, 255, 609–614. 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Berman H.; Henrick K.; Nakamura H. Nat. Struct. Biol. 2003, 10, 980–980. 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. Nucleic Acids Res. 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines R. T.; Sutton E. L.; Straus D. R.; Gilbert W.; Knowles J. R. Biochemistry 1986, 25, 7142–7154. 10.1021/bi00370a057. [DOI] [PubMed] [Google Scholar]
- Komives E. A.; Lougheed J. C.; Liu K.; Sugio S.; Zhang Z.; Petsko G. A.; Ringe D. Biochemistry 1995, 34, 13612–13621. 10.1021/bi00041a041. [DOI] [PubMed] [Google Scholar]
- Lodi P. J.; Knowles J. R. Biochemistry 1991, 30, 6948–6956. 10.1021/bi00242a020. [DOI] [PubMed] [Google Scholar]
- Komives E. A.; Chang L. C.; Lolis E.; Tilton R. F.; Petsko G. A.; Knowles J. R. Biochemistry 1991, 30, 3011–3019. 10.1021/bi00226a005. [DOI] [PubMed] [Google Scholar]
- Kursula I.; Partanen S.; Lambeir A. M.; Antonov D. M.; Augustyns K.; Wierenga R. K. Eur. J. Biochem. 2001, 268, 5189–5196. 10.1046/j.0014-2956.2001.02452.x. [DOI] [PubMed] [Google Scholar]
- Lodi P. J.; Chang L. C.; Knowles J. R.; Komives E. A. Biochemistry 1994, 33, 2809–2814. 10.1021/bi00176a009. [DOI] [PubMed] [Google Scholar]
- Joseph-McCarthy D.; Lolis E.; Komives E. A.; Petsko G. A. Biochemistry 1994, 33, 2815–2823. 10.1021/bi00176a010. [DOI] [PubMed] [Google Scholar]
- Go M. K.; Koudelka A.; Amyes T. L.; Richard J. P. Biochemistry 2010, 49, 5377–5389. 10.1021/bi100538b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go M. K.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2010, 132, 13525–13532. 10.1021/ja106104h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; O’Donoghue A. C.; Richard J. P. J. Am. Chem. Soc. 2001, 123, 11325–11326. 10.1021/ja016754a. [DOI] [PubMed] [Google Scholar]
- Warshel A. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 5250–5254. 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A. J. Biol. Chem. 1998, 273, 27035–27038. 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- Amyes T. L.; Richard J. P. Biochemistry 2007, 46, 5841–5854. 10.1021/bi700409b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go M. K.; Amyes T. L.; Richard J. P. Biochemistry 2009, 48, 5769–5778. 10.1021/bi900636c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompliano D. L.; Peyman A.; Knowles J. R. Biochemistry 1990, 29, 3186–94. 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- Kursula I.; Wierenga R. K. J. Biol. Chem. 2003, 278, 9544–9551. 10.1074/jbc.M211389200. [DOI] [PubMed] [Google Scholar]
- Richard J. P.; Amyes T. L.; Goryanova B.; Zhai X. Curr. Opin. Chem. Biol. 2014, 21, 1–10. 10.1016/j.cbpa.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabanan M. M.; Amyes T. L.; Richard J. P. Curr. Opin. Struct. Biol. 2010, 20, 702–710. 10.1016/j.sbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P.; Zhai X.; Malabanan M. M. Bioorg. Chem. 2014, 57, 206–212. 10.1016/j.bioorg.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R.; Lalloz M. R.; Bellingham A. J.; Layton D. M. Hum. Mutat. 1997, 10, 290–294. . [DOI] [PubMed] [Google Scholar]
- Roland B. P.; Amrich C. G.; Kammerer C. J.; Stuchul K. A.; Larsen S. B.; Rode S.; Aslam A. A.; Heroux A.; Wetzel R.; vanDemark A. P.; Palladino M. J. Biochim. Biophys. Acta, Mol. Basis Dis. 2015, 1852, 61–69. 10.1016/j.bbadis.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabanan M. M.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2011, 133, 16428–16431. 10.1021/ja208019p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabanan M. M.; Koudelka A. P.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2012, 134, 10286–10298. 10.1021/ja303695u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J. P.; Amyes T. L.; Malabanan M. M.; Zhai X.; Kim K. J.; Reinhardt C. J.; Wierenga R. K.; Drake E. J.; Gulick A. M. Biochemistry 2016, 55, 3036–3047. 10.1021/acs.biochem.6b00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. J.; Durani V.; Magliery T. J. J. Mol. Biol. 2011, 413, 195–208. 10.1016/j.jmb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Malabanan M. M.; Go M. K.; Amyes T. L.; Richard J. P. Biochemistry 2011, 50, 5767–5779. 10.1021/bi2005416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabanan M. M.; Nitsch-Velasquez L.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2013, 135, 5978–5981. 10.1021/ja401504w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A.; Sharma P. K.; Kato M.; Parson W. W. Biochim. Biophys. Acta, Proteins Proteomics 2006, 1764, 1647–1676. 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Warshel A.; Strajbl M.; Villa J.; Florian J. Biochemistry 2000, 39, 14728–14738. 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- Warshel A.; Weiss R. M. J. Am. Chem. Soc. 1980, 102, 6218–6226. 10.1021/ja00540a008. [DOI] [Google Scholar]
- Kamerlin S. C. L.; Warshel A. WIREs Comput. Mol. Sci. 2011, 1, 30–45. 10.1002/wcms.10. [DOI] [Google Scholar]
- Amrein B. A.; Steffen-Munsberg F.; Szeler I.; Purg M.; Kulkarni Y.; Kamerlin S. C. L. IUCrJ 2017, 4, 50–64. 10.1107/S2052252516018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogl G.; Rozovsky S.; McDermott A. E.; Tong L. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (1), 50–55. 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Olsson M. H. M.; Chresten R.; Sondergaard M. R.; Jensen J. H. J. Chem. Theory Comput. 2011, 7, 525–527. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Sondergaard C. R.; Olsson M. H. M.; Rostkowski M.; Jensen J. H. J. Chem. Theory Comput. 2011, 7, 2284–2295. 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. 3rd; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Maxwell D. S.; Tirado-Rives J. J. Am. Chem. Soc. 1996, 118, 11225–11236. 10.1021/ja9621760. [DOI] [Google Scholar]
- Marelius J.; Kolmodin K.; Feierberg I.; Åqvist J. J. Mol. Graphics Modell. 1998, 16, 213–225. 10.1016/S1093-3263(98)80006-5. [DOI] [PubMed] [Google Scholar]
- Hwang J. K.; King G.; Creighton S.; Warshel A. J. Am. Chem. Soc. 1988, 110, 5297–5311. 10.1021/ja00224a011. [DOI] [Google Scholar]
- Åqvist J.; Fothergill M. J. Biol. Chem. 1996, 271, 10010–10016. 10.1074/jbc.271.17.10010. [DOI] [PubMed] [Google Scholar]
- Blaha-Nelson D.; Krüger D.; Szeler K.; Ben-David M.; Kamerlin S. C. L. J. Am. Chem. Soc. 2017, 139, 1155–1167. 10.1021/jacs.6b10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Lee F. S.; Chu Z. T.; Bolger M. B.; Warshel A. Protein Eng., Des. Sel. 1992, 5, 215–228. 10.1093/protein/5.3.215. [DOI] [PubMed] [Google Scholar]
- Muegge I.; Tao H.; Warshel A. Protein Eng., Des. Sel. 1997, 10, 1363–1372. 10.1093/protein/10.12.1363. [DOI] [PubMed] [Google Scholar]
- Gilson M. K.; Honig B. Proteins: Struct., Funct., Genet. 2004, 4, 7–18. 10.1002/prot.340040104. [DOI] [Google Scholar]
- Li L.; Li C.; Zhang Z.; Alexov E. J. Chem. Theory Comput. 2013, 9, 2126–2136. 10.1021/ct400065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein B. A.; Bauer P.; Duarte F.; Janfalk Carlsson Å.; Naworyta A.; Mowbray S. L.; Widersten M.; Kamerlin S. C. L. ACS Catal. 2015, 5, 5702–5713. 10.1021/acscatal.5b01639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrozo A.; Duarte F.; Bauer P.; Carvalho A. T. P.; Kamerlin S. C. L. J. Am. Chem. Soc. 2015, 137, 9061–9076. 10.1021/jacs.5b03945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David M.; Sussman J. L.; Maxwell C. I.; Szeler K.; Kamerlin S. C. L.; Tawfik D. S. J. Mol. Biol. 2015, 427, 1359–1374. 10.1016/j.jmb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Alagona G.; Desmeules P.; Ghio C.; Kollman P. A. J. Am. Chem. Soc. 1984, 106, 3623–3632. 10.1021/ja00324a035. [DOI] [Google Scholar]
- Alagona G.; Ghio C.; Kollman P. A. J. Mol. Biol. 1986, 191, 23–27. 10.1016/0022-2836(86)90419-5. [DOI] [PubMed] [Google Scholar]
- Brown F. K.; Kollman P. A. J. Mol. Biol. 1987, 198, 533–546. 10.1016/0022-2836(87)90298-1. [DOI] [PubMed] [Google Scholar]
- Joseph D.; Petsko G. A.; Karplus M. Science 1990, 249, 1425–1428. 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- Bash P. A.; Field M. J.; Davenport R. C.; Petsko G. A.; Ringe D.; Karplus M. Biochemistry 1991, 30, 5826–5832. 10.1021/bi00238a003. [DOI] [PubMed] [Google Scholar]
- Karplus M.; Evanseck J. D.; Joseph D.; Bash P. A.; Field M. J. Faraday Discuss. 1992, 93, 239–248. 10.1039/fd9929300239. [DOI] [PubMed] [Google Scholar]
- Neria E.; Karplus M. Chem. Phys. Lett. 1997, 267, 23–30. 10.1016/S0009-2614(97)00068-7. [DOI] [Google Scholar]
- Cui Q.; Karplus M. J. Am. Chem. Soc. 2001, 123, 2284–2290. 10.1021/ja002886c. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Karplus M. J. Am. Chem. Soc. 2002, 124, 3093–3124. 10.1021/ja0118439. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Karplus M. J. Phys. Chem. B 2002, 106, 7927–7947. 10.1021/jp0205057. [DOI] [Google Scholar]
- Alagona G.; Ghio C.; Kollman P. A. J. Comput. Chem. 2003, 24, 46–56. 10.1002/jcc.10153. [DOI] [PubMed] [Google Scholar]
- Guallar V.; Jacobson M.; McDermott A.; Friesner R. A. J. Mol. Biol. 2004, 337, 227–239. 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Richard J. P. J. Am. Chem. Soc. 1984, 106, 4926–4936. 10.1021/ja00329a050. [DOI] [Google Scholar]
- Zhai X.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2015, 137, 15185–15197. 10.1021/jacs.5b09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Malabanan M. M.; Zhai X.; Reyes A. C.; Richard J. P. Protein Eng., Des. Sel. 2017, 30, 159–168. 10.1093/protein/gzw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyes T. L.; Richard J. P. Biochemistry 2013, 52, 2021–2035. 10.1021/bi301491r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Gassman P. G. Biochemistry 1993, 32, 11943–11952. 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A.; Kozarich J. W.; Kenyon G. L.; Gassman P. G. J. Am. Chem. Soc. 1991, 113, 9667–9669. 10.1021/ja00025a039. [DOI] [Google Scholar]
- Gerlt J. A.; Gassman P. G. J. Am. Chem. Soc. 1993, 115, 11552–11568. 10.1021/ja00077a062. [DOI] [Google Scholar]
- Noble M. E. M.; Zeelen J. P.; Wierenga R. K. Proteins: Struct., Funct., Genet. 1993, 16, 311–326. 10.1002/prot.340160402. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. Adv. Enzymol. Relat. Areas Mol. Biol. 2006, 43, 219–410. 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Warshel A. J. Biol. Chem. 1998, 273, 27035–27038. 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- Richard J. P. Biochemistry 1998, 37, 4305–4309. 10.1021/bi972655r. [DOI] [PubMed] [Google Scholar]
- Albery W. J.; Knowles J. R. Biochemistry 1976, 15, 5631–5640. 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- Wu N.; Mo Y.; Gao J.; Pai E. F. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 2017–2022. 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman L. M.; Amyes T. L.; Goryanova B.; Gerlt J. A.; Richard J. P. J. Am. Chem. Soc. 2014, 136, 10156–10165. 10.1021/ja505037v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryanova B.; Goldman L. M.; Amyes T. L.; Gerlt J. A.; Richard J. P. Biochemistry 2013, 52, 7500–7511. 10.1021/bi401117y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. C.; Zhai X.; Morgan K. T.; Reinhardt C. J.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2015, 137, 1372–1382. 10.1021/ja5123842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. C.; Amyes T. L.; Richard J. P. J. Am. Chem. Soc. 2016, 138, 14526–14529. 10.1021/jacs.6b07028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodar S. A.; Allen C. L.; Gulick A. M.; Murkin A. S. J. Am. Chem. Soc. 2015, 137, 2748–2756. 10.1021/ja512911f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray W. J. Jr.; Long J. W.; Owens J. D. Biochemistry 1976, 15, 4006–4017. 10.1021/bi00663a015. [DOI] [PubMed] [Google Scholar]
- Chan K. K.; Wood B. M.; Fedorov A. A.; Fedorov E. V.; Imker H. J.; Amyes T. L.; Richard J. P.; Almo S. C.; Gerlt J. A. Biochemistry 2009, 48, 5518–5531. 10.1021/bi900623r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.