Summary
Dendritic cells (DCs) in mesenteric lymph nodes (MLNs) induce Foxp3+ regulatory T cells to regulate immune responses to beneficial or non‐harmful agents in the intestine, such as commensal bacteria and foods. Several studies in MLN DCs have revealed that the CD103+ DC subset highly induces regulatory T cells, and another study has reported that MLN DCs from programmed death ligand 1 (PD‐L1) ‐deficient mice could not induce regulatory T cells. Hence, the present study investigated the expression of these molecules on MLN CD11c+ cells. Four distinct subsets expressing CD103 and/or PD‐L1 were identified, namely CD11b+ CD103+ PD‐L1High, CD11b− CD103+ PD‐L1High, CD11b− CD103+ PD‐L1Low and CD11b+ CD103− PD‐L1Int. Among them, the CD11b− CD103+ PD‐L1High DC subset highly induced Foxp3+ T cells. This subset expressed Aldh1a2 and Itgb8 genes, which are involved in retinoic acid metabolism and transforming growth factor‐β (TGF‐β) activation, respectively. Exogenous TGF‐β supplementation equalized the level of Foxp3+ T‐cell induction by the four subsets whereas retinoic acid did not, which suggests that high ability to activate TGF‐β is determinant for the high Foxp3+ T‐cell induction by CD11b− CD103+ PD‐L1High DC subset. Finally, this subset exhibited a migratory DC phenotype and could take up and present orally administered antigens. Collectively, the MLN CD11b− CD103+ PD‐L1High DC subset probably takes up luminal antigens in the intestine, migrates to MLNs, and highly induces regulatory T cells through TGF‐β activation.
Keywords: dendritic cells, intestinal immunity, mesenteric lymph nodes, oral tolerance, regulatory T cells
Abbreviations
- APC
allophycocyanin
- CFSE
5(6)‐Carboxyfluorescein N‐hydroxysuccinimidyl ester
- DC
dendritic cell
- FCS
fetal calf serum
- IDO
indoleamine 2,3‐dioxygenase
- Int.
intermediate
- MLN
mesenteric lymph node
- OVA
ovalbumin
- OVAp
ovalbumin peptide
- PD‐L1
programmed death ligand 1
- PE
phycoerythrin
- pTreg cell
peripheral regulatory T cell
- RA
retinoic acid
- TGF
transforming growth factor
- Treg cell
regulatory T cell
- tTreg cell
thymus‐derived regulatory T cell
Introduction
The intestinal immune system should be precisely regulated to prevent destructive immune responses toward antigens derived from beneficial or non‐harmful substances and organisms, such as foods and commensal bacteria. Otherwise, excessive immune responses could lead to, for example, food allergy and inflammatory diseases. The intestinal immune system is set to suppress immune responses to orally administered antigens in the steady state. Further, oral administration of an antigen can suppress immune responses to subsequent systemic challenge of the antigen, referred to as oral tolerance.1, 2, 3, 4 Hence, elucidation of the mechanism underlying oral tolerance can contribute to developing preventive and therapeutic strategies for food allergy and inflammatory diseases in the intestine. Oral tolerance is probably a useful way to treat systemic diseases that have immunological pathology, such as autoimmune diseases.
Antigen‐specific immune suppression is largely accomplished by inducing regulatory T (Treg) cells that suppress immune responses to specific antigens.5, 6, 7, 8, 9 Treg cells are classified into two types based on their origins, i.e. thymus‐derived Treg (tTreg) cells and peripherally derived Treg (pTreg) cells.10 Among them, pTreg cells play a pivotal role in inducing oral tolerance.11 Indeed, a large proportion of the pTreg cells in the small intestine are dependent on food‐derived antigens.12 Oral tolerance is induced mainly in mesenteric lymph nodes (MLNs), considering surgical removal of MLNs in mice abrogates induction of oral tolerance.13 Hence, the MLN is probably a main site for the induction of pTreg cells specific to orally administered antigens, which are required for oral tolerance. Further, these pTreg cells should migrate to the intestine to induce oral tolerance.14, 15 T‐cell homing to the small intestine is mediated by CCR9 and integrin α 4 β 7.16, 17
In the intestine, CD103+ dendritic cells (DCs) play a crucial role in tolerogenic responses including Treg cell induction.18 Many studies have revealed that MLN CD11b+ CD103+ and CD11b− CD103+ DCs highly induce Foxp3+ Treg cells through several factors. Treg cells are induced by transforming growth factor‐β (TGF‐β), and this induction is enhanced by retinoic acid (RA), i.e. RA enhances Treg cell induction only in the presence of TGF‐β.19, 20, 21 MLN CD103+ DCs highly express RALDH2 enzyme, which metabolizes retinal to RA.19, 22, 23 In addition to Treg cell induction, RA also induces gut‐homing receptors, CCR9 and integrin α 4 β 7, on T cells.24 Hence, CD103+ DCs, which highly express RALDH2, can induce gut‐homing Treg cells.19, 25 MLN CD103+ DCs also highly express TGF‐β.19 The TGF‐β is secreted as a latent form and needs to be cleaved into the active form. The intestinal CD103+ DCs further mediate this activation process through integrin α v β 8.26, 27, 28, 29 Besides, the intestinal CD103+ DCs induce Treg cells and subsequent oral tolerance through indoleamine 2,3‐dioxygenase (IDO) activity, which mediates tryptophan catabolism.30
To induce Treg cells specific to orally administered antigens in MLNs, DCs should obtain the antigens in the intestine and then migrate to MLNs. CD103+ DCs capture luminal antigens directly by extending their dendrites to the lumen and indirectly through goblet cells.31, 32 In addition, they can receive the antigens from CD103− CX3CR1+ macrophages, which can take up antigens directly from the lumen, in the intestinal lamina propria.33, 34 Lamina propria CD103+ DCs can migrate to MLNs in a CCR7‐dependent manner.35, 36, 37 Hence, CD103+ DCs capture luminal antigens, migrate to MLNs, and present the antigens to T cells, which results in induction of Treg cells specific to orally administered antigens.
Besides the many studies on CD103+ DCs mentioned above, another study has reported that programmed death ligand 1 (PD‐L1) and PD‐L2, co‐stimulatory molecules expressed on DCs, are important for Treg cell induction by MLN DCs.38 MLN DCs from PD‐L1‐ or PD‐L2‐deficient mice cannot induce Treg cells. However, the relationship between CD103, PD‐L1 and PD‐L2 molecules on MLN DCs remained unclear. In the present study, we investigated CD103, PD‐L1 and PD‐L2 expression on MLN CD11c+ cells and identified four subsets expressing CD103 and/or PD‐L1 based on CD11b, CD103 and PD‐L1. Among the subsets, CD11b− CD103+ PD‐L1High DCs highly induced Foxp3+ Treg cells. This Treg cell induction was probably dependent on TGF‐β activation through integrin α v β 8. Further, the CD11b− CD103+ PD‐L1High DCs highly expressed CCR7 and obtained orally administered antigens, so this DC subset is migratory. These results revealed that CD11b− CD103+ PD‐L1High DCs capture luminal antigens in the intestine, migrate to MLNs, and induce Treg cells through TGF‐β activation. This newly characterized DC subset may be important for oral tolerance induction and has implications as a target for therapeutic manipulation using oral tolerance.
Materials and methods
Mice
BALB/c mice (CLEA Japan, Tokyo, Japan) and DO11.10 mice39 were used at 7–20 weeks old. In some experiments, BALB/c mice were fed water containing ovalbumin (OVA; Wako, Osaka, Japan) (200 mg/ml) ad libitum for 3 days before cell isolation. All experiments were approved by the Animal Use Committee of the Faculty of Agriculture at the University of Tokyo and were performed in accordance with The University of Tokyo guidelines for animal care and use.
Media and reagents
RPMI media and 10% fetal calf serum (FCS)‐RPMI media were prepared as described previously.40 For flow cytometry, anti‐α 4 β 7‐allophycocyanin (APC) (DATK32), anti‐CCR7‐APC (4B12), anti‐CCR9‐FITC (CW‐1.2), anti‐CD4‐APC (GK1.5), anti‐CD4‐biotin (GK1.5), anti‐CD11b‐FITC (M1/70), anti‐CD11c‐APC (N418), anti‐CD11c‐APC/Cy7 (N418), purified anti‐CD16/32 (93), anti‐CD64‐APC (X54‐5/7.1), anti‐CD80‐biotin (16‐10A1), anti‐CD86‐biotin (GL‐1), anti‐CD103‐biotin (2E7), anti‐CD172a‐FITC (P84), anti‐F4/80‐biotin (BM8), anti‐PD‐L1 (10F.9G2), anti‐PD‐L1‐phycoerythrin (PE) (10F.9G2), anti‐XCR1‐FITC (ZET), streptavidin‐Peridinin chlorophyll protein, streptavidin‐PE/Cy7, and streptavidin‐APC were purchased from BioLegend (San Diego, CA); anti‐CD103‐FITC (2E7), anti‐Foxp3‐PE (FJK‐16s), anti‐PD‐L2 (TY25), and anti‐PD‐L2‐biotin (TY25) were purchased from eBioscience (San Diego, CA); anti‐CD8α‐PE/Cy7 (53‐6.7) and anti‐CD11b‐APC (M1/70) were purchased from TONBO biosciences (San Diego, CA); anti‐I‐A/I‐E‐PE (M5/114.15.2) and streptavidin‐PE/Cy5 were purchased from BD Bioscience (San Jose, CA). OVA323‐339 peptide (OVAp; ISQAVHAAHAEINEAGR) was purchased from Operon Biotechnologies (Tokyo, Japan). DMSO was purchased from Sigma (St Louis, MO). LE540 (Wako) and all‐trans‐RA (Wako) were stored at 10 mm in DMSO, and human TGF‐β 1 (hTGF‐β; R&D Systems, Minneapolis, MN) was stored at 4 μg/ml in PBS. These stocks were diluted by media for use. 5(6)‐Carboxyfluorescein N‐hydroxysuccinimidyl ester (CFSE) was purchased from Molecular Probes (Eugene, OR).
Cell isolation
The MLNs were removed and incubated in 10% FCS‐RPMI containing 0·5 mg/ml collagenase (Wako, Osaka, Japan) and 10 μg/ml DNase I (Roche, Basel, Switzerland), and a single‐cell suspension was obtained. The cell suspension was filtered with a nylon mesh and was washed with RPMI. From the obtained cells, CD11c+ cells were separated using MACS system (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instructions. Hereafter, we describe cells separated once and twice by MACS as enriched CD11c+ cells and purified CD11c+ cells, respectively.
Subsets of MLN CD11c+ cells were sorted by FACS Vantage (BD Bioscience, San Jose, CA) or FACS Aria II (BD Bioscience). FACS Vantage was used to sort subsets by CD11b, CD103 and PD‐L1 expression from purified CD11c+ cells whereas FACS Aria II was used to sort subsets by CD11b, CD11c, CD103 and PD‐L1 (or CD8α) expression from enriched CD11c+ cells. PBS containing 3% FCS was used as staining and washing buffer. Detailed staining protocol is described below (see Flow cytometry).
For preparing splenic cells, spleens were mashed and were filtered with nylon mesh. From the obtained cells, CD4+ cells were separated using MACS system according to the manufacturer's instructions. For CFSE‐labelling, the CD4+ cells were incubated in PBS containing CFSE for 6–7 min at room temperature, and then, the cells were washed twice.
Flow cytometry
Surface molecules on cells were stained with fluorescently labelled antibodies after Fc receptor block by anti‐CD16/32 antibody. When biotinylated antibodies were used in the staining, secondary staining was performed using streptavidin‐conjugated fluorescent reagents after the primary staining antibodies were washed out. In the case of dead cell‐staining by propidium iodide, propidium iodide (2 μg/ml; Sigma) was added to samples after staining by antibodies and was washed out immediately. Intracellular Foxp3 was stained using Foxp3 staining buffer set (eBioscience) according to the manufacturer's instructions. Fluorescence levels were measured by FACS Verse (BD Bioscience) and data were analysed using flowjo software (Tree Star, Ashland, OR).
Cell culture
For Treg cell induction assay, MLN DC subsets from BALB/c mice (5 × 104 cells/ml) and splenic CD4+ cells from DO11.10 mice (5 × 105 cells/ml) were co‐cultured in the presence of OVAp (10 nm). In some experiments, reagents, such as anti‐PD‐L1 antibody, anti‐PD‐L2 antibody, their isotype control antibodies, LE540, RA, DMSO and hTGF‐β, were added. These cells and reagents were cultured in 200 μL of 10% FCS‐RPMI in 96‐well round‐bottom plates for 3·5 days in a 5% CO2 humidified atmosphere at 37°.
For antigen uptake assay, MLN DC subsets from OVA‐fed or normal BALB/c mice (1 × 105 cells/ml) and CFSE‐labelled splenic CD4+ cells from DO11.10 mice (1 × 106 cells/ml) were co‐cultured in the presence or absence of OVA protein (100 ng/ml) in 200 μl of 10% FCS‐RPMI in 96‐well flat‐bottom plates for 3 days in a 5% CO2 humidified atmosphere at 37°.
Quantitative PCR
Total RNA was extracted from cells using QIAShredder (QIAGEN, Hilden, Germany) and RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. From the total RNA, single‐stranded cDNA was synthesized using Superscript II reverse transcriptase and Oligo dT primers (Invitrogen, Carlsbad, CA). Subsequently, real‐time PCR was performed using Light Cycler (Roche) with SYBR Green PCR kit (QIAGEN) and subsequent primers – Hprt forward: 5′‐GAAGAGACTGGGGATCACTC‐3′, reverse: 5′‐CATGCCATCTTCCATATTGT‐3′; Aldh1a2 forward: 5′‐GACTTGTAGCAGCTGTCTTCACT‐3′, reverse: 5′‐TCACCCATTTCTCTCCCATTTCC‐3′; Tgfb1 forward: 5′‐ATTGAGGGCTTGTTGAGATG‐3′, reverse: 5′‐GACTGGCGAGCCTTAGTTTG‐3′; Ido1 forward: 5′‐TCCAGTGCAGTAGAGCGTTCA‐3′, reverse: 5′‐GAAAAACGTGTCTGGGTCCA‐3′; Itgav forward: 5′‐GAGGGAGATGTTCACACTTTG‐3′, reverse: 5′‐AGCAGGGATTTCACGTCAG‐3′; Itgb8 forward: 5′‐TGTACTGATCCCAGAAGCATTG‐3′, reverse: 5′‐TGGGCCAGATAAACATTCTGAT‐3′; Ccr7 forward: 5′‐GTGTGCTTCTGCCAAGATGA‐3′, reverse: 5′‐CCACGAAGCAGATGACAGAA‐3′. Relative gene expression was calculated as described previously except that target gene expression was normalized to Hprt gene expression as an internal control.40
Results
Mesenteric lymph node CD11c+ cells contain four subsets expressing CD103 and/or PD‐L1
Previous studies revealed that MLN CD103+ DCs highly induce Treg cells.18 Meanwhile, another study reported that MLN DCs from PD‐L1−/− cannot induce Treg cells.38 Hence, we examined CD103 and PD‐L1 expression on MLN CD11c+ cells. MLN CD11c+ cells contained CD11b+ CD103+, CD11b− CD103+, CD11b+ CD103− and CD11b− CD103− subsets (Fig. 1a). Among them, we found that the CD11b− CD103+ subset was further classified into two subsets based on PD‐L1 expression, namely PD‐L1High and PD‐L1Low subsets (Fig. 1b). Hence, MLN CD11c+ cells include four subsets expressing CD103 and/or PD‐L1, including CD11b+ CD103+ PD‐L1High, CD11b− CD103+ PD‐L1High, CD11b− CD103+ PD‐L1Low and CD11b+ CD103− PD‐L1Intermediate (Int) subsets.
Figure 1.
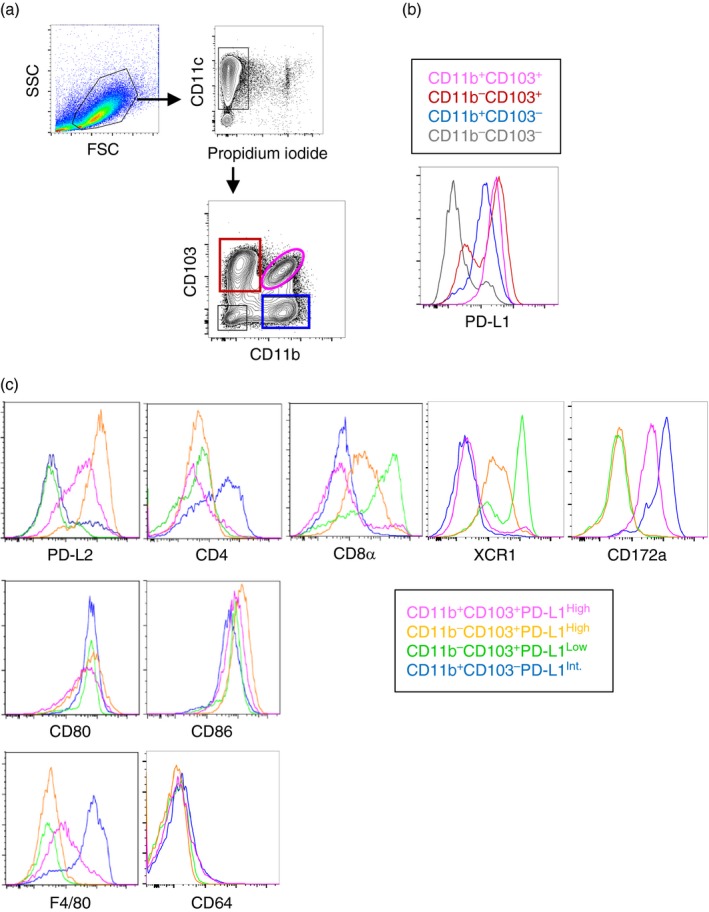
Mesenteric lymph node (MLN) CD11c+ cells are classified into four subsets based on CD11b, CD103 and programmed death ligand 1 (PD‐L1) expression. Enriched MLN CD11c+ cells were analysed by flow cytometry. (a) CD11b and CD103 expression on live (propidium iodide–) CD11c+ cells was analysed. (b) PD‐L1 expression on the four subsets in (a) was analysed. (c) Cell surface molecules on the four subsets expressing CD103 and/or PD‐L1 in (b) were analysed. The results are representatives of three independent experiments.
We further characterized four subsets: namely CD11b+ CD103+ PD‐L1High, CD11b− CD103+ PD‐L1High, CD11b− CD103+ PD‐L1Low and CD11b+ CD103−PD‐L1Int. Results are shown in Fig. 1(c) and Table 1. PD‐L2, another molecule required for Treg cell induction, was expressed on CD11b+ CD103+ PD‐L1High and CD11b− CD103+ PD‐L1High subsets whereas the other two subsets did not express PD‐L2. CD4 and CD8α were also differently expressed among the subsets whereas co‐stimulatory molecules, CD80 and CD86, were equally expressed. Recent studies have revealed that DCs can be classified into subsets based on XCR1 and CD172a expression.41, 42, 43, 44, 45, 46 Consistent with the previous studies, CD11b− CD103+ DCs including PD‐L1High and PD‐L1Low subsets expressed XCR1 but not CD172a whereas CD11b+ CD103+ and CD11b+ CD103− DCs expressed CD172a but not XCR1. The CD11b− CD103+ PD‐L1High subset expressed low XCR1, whereas the CD11b− CD103+ PD‐L1Low subset expressed high XCR1. The CD11b+ CD103− PD‐L1Int subset highly expressed F4/80, which suggested that this subset contained macrophages. However, none of the subsets, including this CD11b+ CD103− PD‐L1Int subset, expressed a macrophage‐specific marker, CD64, consistently with a previous study.47 Hence, we concluded that these four CD11c+ cell subsets are classified as DC subsets.
Table 1.
Phenotype of mesenteric lymph node CD11c+ cell subsets
| Subset 1 | Subset 2 | Subset 3 | Subset 4 | |
|---|---|---|---|---|
| CD11b | + | − | − | + |
| CD103 | + | + | + | − |
| PD‐L1 | High | High | Low | Int. |
| PD‐L2 | Int. | High | − | − |
| CD4 | − | − | − | +/− |
| CD8α | − | Int. | High | − |
| XCR1 | − | Low | High | − |
| CD172a | Int. | − | − | High |
| F4/80 | Int. | − | − | High |
| CD64 | − | − | − | − |
Phenotypes of mesenteric lymph node CD11c+ cells were analysed by flow cytometry. Representative plots are shown in Fig. 1.
CD11b− CD103+ PD‐L1High DC subset highly induces Treg cells
Next, we compared Treg cell induction by the four MLN CD11c+ cell subsets. The subsets were sorted and co‐cultured with OVA‐specific T cells in the presence of OVAp. To avoid blocking PD‐L1 by anti‐PD‐L1 antibodies in the sorting process, the subsets were sorted using CD8α instead of PD‐L1. Hence, CD11b+ CD103+ CD8α −, CD11b− CD103+ CD8α Int, CD11b− CD103+ CD8α High and CD11b+ CD103− CD8α − subsets were sorted. These subsets correspond to CD11b+ CD103+ PD‐L1High, CD11b− CD103+ PD‐L1High, CD11b−CD103+ PD‐L1Low and CD11b+ CD103− PD‐L1Int subsets, respectively (see Fig. 1c and Table 1). Among those subsets, CD11b− CD103+ CD8α Int (PD‐L1High) DCs induced a higher proportion of Foxp3+ T cells than the other subsets (Fig. 2a,b). Hence, we found that MLN CD11b−CD103+ PD‐L1High DCs highly induce Treg cells.
Figure 2.
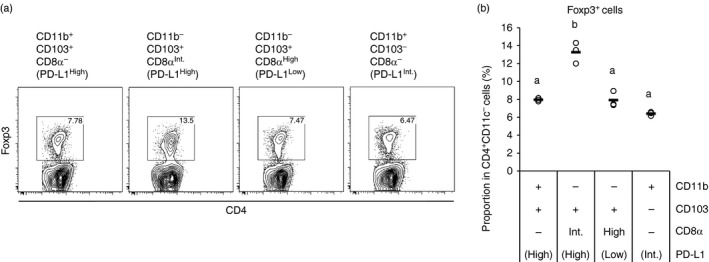
CD11b− CD103+ PD‐L1High dendritic cells (DCs) highly induce regulatory T (Treg) cells. DO11.10 CD4+ T cells (5 × 105 cells/ml) were co‐cultured with the indicated CD11c+ cell subsets (5 × 104 cells/ml) in the presence of ovalbumin peptide (OVAp; 10 nm). After 3·5 days, the cells in each well were collected and were analysed by flow cytometry. Two independent experiments were performed. (a) CD4 and Foxp3 expression on CD4+ CD11c− cells are shown. Data are from a representative well of each sample. (b) Proportions of Foxp3+ cells in CD4+ CD11c− cells [gates shown in (a)] were analysed. The plot shows representative data from one experiment (n = 3). Circles and horizontal bars indicate data from one well and mean of results from three wells, respectively. Statistical analysis was performed by Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05).
TGF‐β activation is critical for Treg cell induction by CD11b− CD103+ PD‐L1High DC subset
To investigate the mechanism involved in the highest Treg cell induction by the CD11b− CD103+ PD‐L1High subset, we examined the involvement of PD‐L1 and PD‐L2 by blocking these molecules using antibodies in the culture system. As a result, blocking of neither PD‐L1 nor PD‐L2 had any effect on the Treg cell induction by the subsets (Fig. 3a–d). Further, simultaneous blocking of these molecules had no effect (Fig. 3e,f). Hence, these molecules are not critical for Treg cell induction by the MLN DC subset, although it has been reported that PD‐L1‐ or PD‐L2‐deficient MLN DCs cannot induce Treg cells.38 This suggests that these molecules may be involved indirectly in Treg cell induction of MLN DCs.
Figure 3.
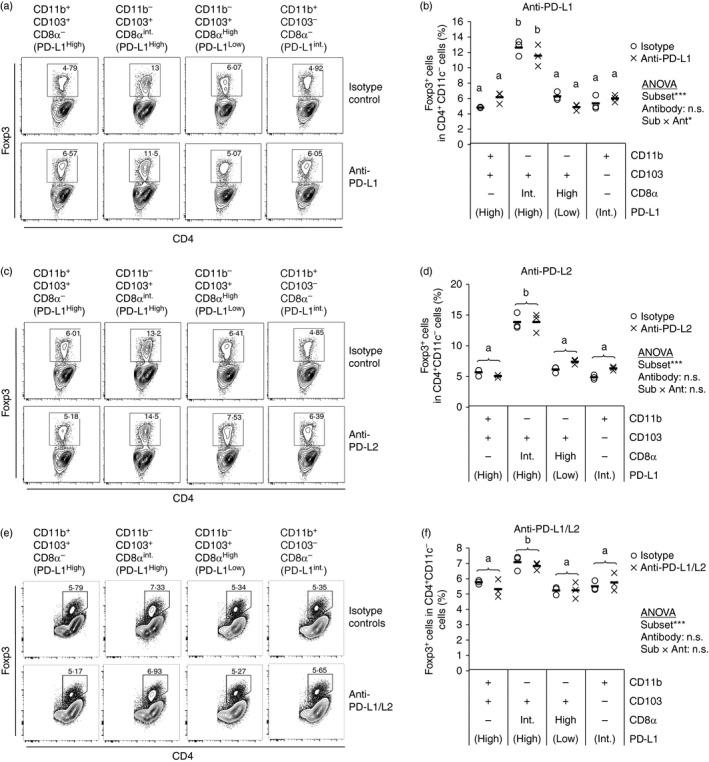
Neither programmed death ligand 1 (PD‐L1) nor PD‐L2 is critical for regulatory T (Treg) cell induction by CD11b− CD103+ PD‐L1High dendritic cells (DCs). DO11.10 CD4+ T cells (5 × 105 cells/ml) were co‐cultured with the indicated CD11c+ cell subsets (5 × 104 cells/ml) in the presence of ovalbumin peptide (OVAp; 10 nm) with anti‐PD‐L1 (a, b), anti‐PD‐L2 (c, d), or both anti‐PD‐L1 and anti‐PD‐L2 antibodies (e, f), and their isotype control antibodies (9 μg/ml). After 3·5 days, the cells in each well were collected and were analysed by flow cytometry. Two independent experiments were performed. (a, c, e) CD4 and Foxp3 expression on CD4+ CD11c− cells are shown. Data are from a representative well of each sample. (b, d, f) Proportion of Foxp3+ cells in CD4+ CD11c− cells [gates shown in (a), (c) and (e)] were analysed. The plot shows representative data from one experiment (n = 3). Symbols and horizontal bars indicate data from one well and mean of results from three wells, respectively. Statistical analysis was performed by two‐way analysis of variance and subsequent Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05). *P < 0·05, ***P < 0·001, n.s. (not significant): P ≥ 0·1.
Next, other factors involved in Treg cell induction by DCs were analysed. Treg cells are induced by TGF‐β, and this induction is synergistically enhanced by RA.19, 20, 21 TGF‐β has been reported to be activated by DCs through an integrin α v β 8‐dependent mechanism.26, 27, 28, 29 RA is produced by RALDH2 highly expressed in the intestinal CD103+ DCs.19, 22, 23 In addition, IDO is also involved in Treg cell induction.30 Therefore, expression of genes encoding TGF‐β, the TGF‐β‐activating integrins, RALDH2 and IDO was measured. Results are shown in Fig. 4. The Treg cell‐inducing CD11b− CD103+ PD‐L1High DCs highly expressed Aldh1a2 gene, which encodes RALDH2, and Itgb8 gene, which encodes integrin β 8. In contrast, there was little difference in the expression of Tgfb1 and Itgav genes, which encode TGF‐β and integrin α v, respectively. In addition, Ido1 gene, encoding IDO, was highly expressed by the CD11b+ CD103+ PD‐L1High subset, which implies that IDO activity is not critical for the difference in Treg cell induction among the subsets. These results suggested that the Treg cell induction by CD11b− CD103+ PD‐L1High subset is mediated by RA production through RALDH2 and/or TGF‐β activation through integrin α v β 8.
Figure 4.
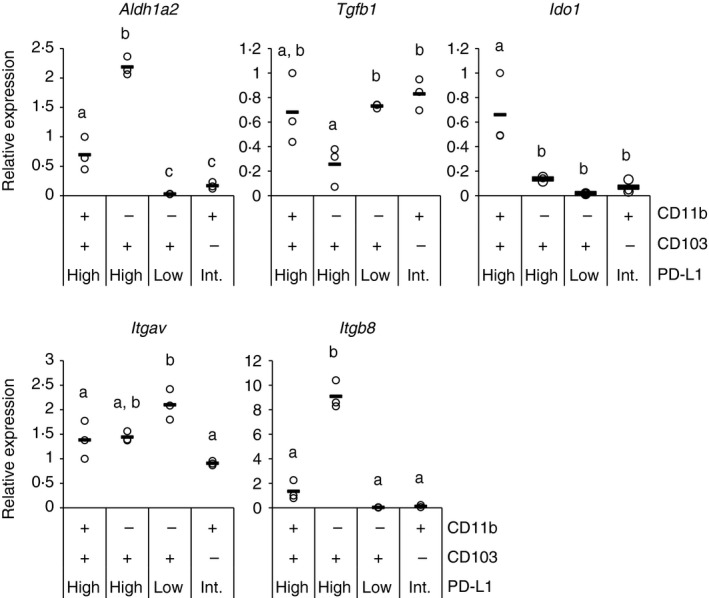
CD11b− CD103+ PD‐L1High dendritic cells (DCs) highly express RALDH2 and integrin β 8 genes. Relative gene expression of four indicated mesenteric lymph node (MLN) CD11c+ cell subsets was measured by quantitative PCR. cDNA from three independent experiments were analysed together, and relative values to expression in CD11b+ CD103+ PD‐L1High subset of one experiment are plotted. Circles and horizontal bars indicate data from one experiment and mean of data from the three experiments, respectively. Statistical analysis was performed by Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05).
To examine the involvement of RA and TGF‐β in the Treg cell induction by the DC subset, exogenous RA and TGF‐β were supplemented to the culture. Even in the presence of RA, the CD11b− CD103+ PD‐L1High DCs induced the highest proportion of Treg cells compared with the other subsets (Fig. 5a,b). This result suggests that RA is not critical for the Treg cell induction by the MLN DC subset. This was further confirmed using an antagonist of RA receptor, LE540. LE540 did not impair the Treg cell induction. In contrast to RA, exogenous TGF‐β abrogated the difference in Treg cell induction among the subsets (Fig. 5c,d). Therefore, TGF‐β is a critical factor for Treg cell induction by the CD11b− CD103+ PD‐L1High DCs.
Figure 5.
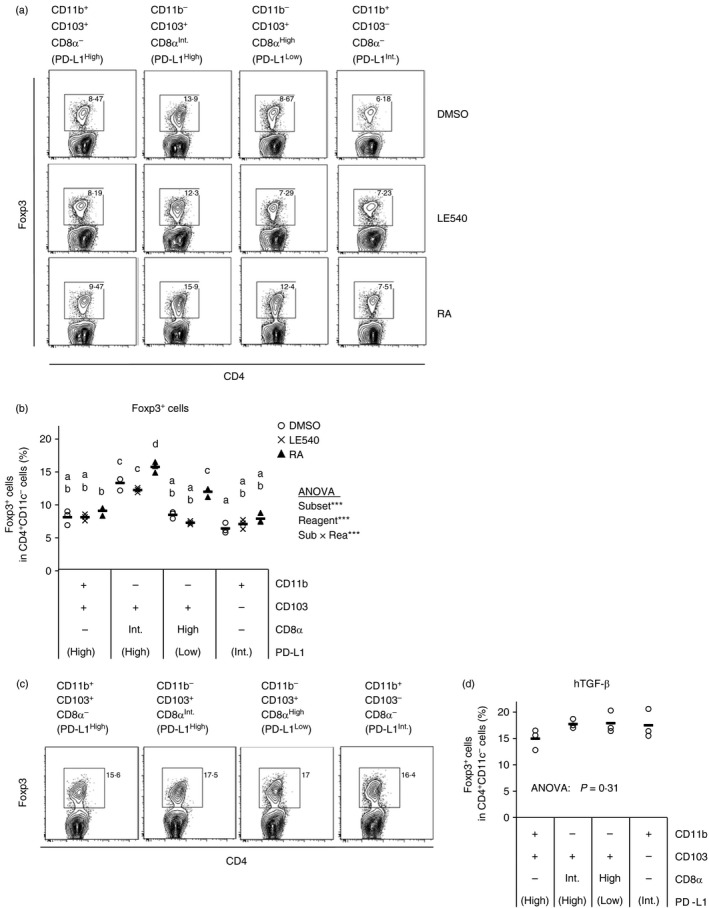
Transforming growth factor‐β (TGF‐β) is a determinant factor for regulatory T (Treg) cell induction by mesenteric lymph node (MLN) dendritic cell (DC) subsets. DO11.10 CD4+ T cells (5 × 105 cells/ml) were co‐cultured with the indicated CD11c+ cell subsets (5 × 104 cells/ml) in the presence of ovalbumin peptide (OVAp; 10 nm) with LE540 (1 μm), retinoic acid (RA) (1 μm), DMSO (a, b), and human TGF‐β (2 ng/ml), (c, d). After 3·5 days, the cells in each well were collected and were analysed by flow cytometry. Two independent experiments were performed. (a, c) CD4 and Foxp3 expression on CD4+ CD11c− cells are shown. Data are from a representative well of each sample. (b, d) Proportion of Foxp3+ cells in CD4+ CD11c− cells [gates shown in (a) and (c)] were analysed. The plot shows representative data from one experiment (n = 3). Symbols and horizontal bars indicate data from one well and mean of results from three wells, respectively. Statistical analysis was performed by two‐way analysis of variance and subsequent Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05). ***P < 0·001.
Taken together, the results suggest that MLN CD11b− CD103+ PD‐L1High DCs highly induce Treg cells by TGF‐β activation through integrin α v β 8.
Retinoic acid regulates the intestine‐homing receptors on Treg cells
Treg cells induced in MLNs should migrate into the intestine to suppress immune responses toward intestinal antigens and further to establish oral tolerance.14, 15 The migration to the small intestine requires CCR9 and integrin α 4 β 7 expression induced by RA.24 We compared expression of these homing‐receptors on Treg cells induced by the MLN CD11c+ cell subsets. As a result, the homing‐receptors on Treg cells induced by the subsets were barely different (Fig. 6). Both molecules were up‐regulated by exogenous RA and down‐regulated by LE540. Hence, RA plays a critical role in the induction of CCR9 and α 4 β 7 on Treg cells. In addition, these results certified that RA and LE540 indeed had activity to affect T‐cell responses, which confirms that RA is not critical for the Treg cell induction by CD11b− CD103+ PD‐L1High DCs in culture.
Figure 6.
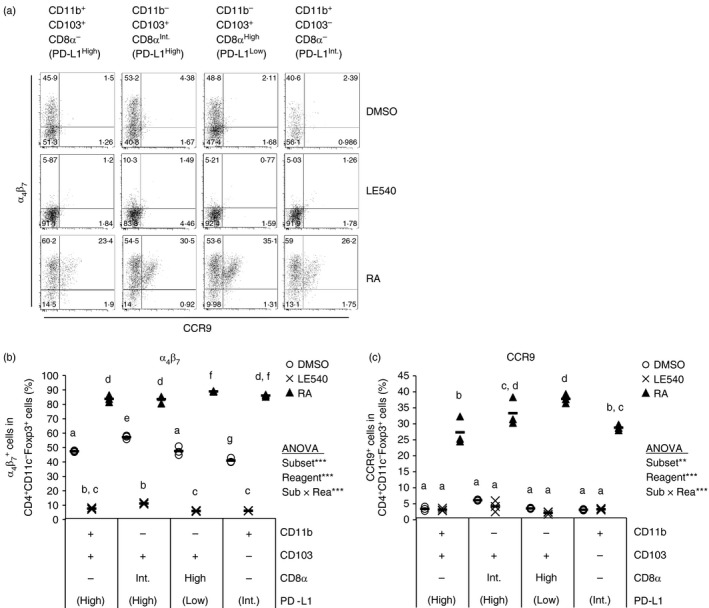
α 4 β 7 and CCR9 on regulatory T (Treg) cells could be induced by retinoic acid (RA). DO11.10 CD4+ T cells (5 × 105 cells/ml) were co‐cultured with indicated CD11c+ cell subsets (5 × 104 cells/ml) in the presence of ovalbumin peptide (OVAp; 10 nm) with LE540 (1 μm), RA (1 μm) and DMSO. After 3·5 days, the cells in each well were collected and were analysed by flow cytometry. Two independent experiments were performed. (a) α 4 β 7 and CCR9 expression on CD4+ CD11c− Foxp3+ cells are shown. Data are from a representative well of each sample. (b,c) Proportion of α 4 β 7 + cells (b) or CCR9+ cells (c) in CD4+ CD11c− Foxp3+ cells were analysed. The plot shows representative data from one experiment (n = 3). Symbols and horizontal bars indicate data from one well and mean of results from three wells, respectively. Statistical analysis was performed by two‐way analysis of variance and subsequent Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05). **P < 0·01, ***P < 0·001.
CD11b− CD103+ PD‐L1High DCs are migratory subsets bearing orally administered antigens
Lymph nodes (LNs) contain DCs of two different origins. One is a migratory DC, which migrates to the LNs from the periphery, and the other is resident DC, which is differentiated in the LNs from progenitors. Migration from the periphery is dependent on CCR7.35, 36, 37, 48 Thus, we examined CCR7 expression of the four subsets. The Treg cell‐inducing CD11b− CD103+ PD‐L1High DCs highly expressed Ccr7 gene and CCR7 on the cell surface (Fig. 7a,b left). This DC subset exhibited a phenotype of migratory DCs, MHC IIHigh (Fig. 7b right). These results suggest that CD11b− CD103+ PD‐L1High subset is a migratory DC subset. In addition, CD11b+ CD103+ PD‐L1High subset and some fraction of CD11b+ CD103− PD‐L1Int subset exhibited the migratory phenotypes whereas the CD11b− CD103+ PD‐L1Low subset did not.
Figure 7.
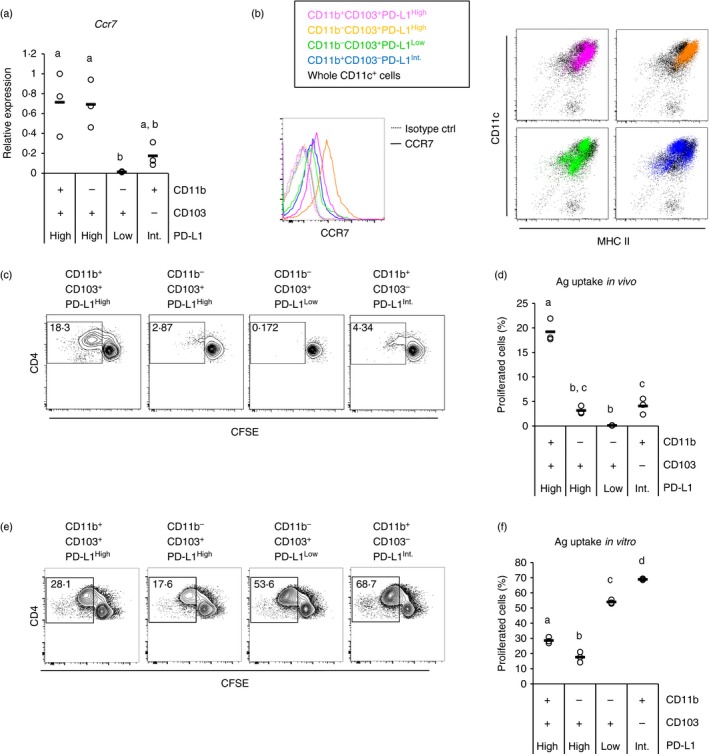
CD11b− CD103+ PD‐L1High dendritic cells (DCs) are a migratory DC subset. (a) Relative gene expression of four indicated mesenteric lymph node (MLN) CD11c+ cell subsets were measured by quantitative PCR. cDNA from three independent experiments were analysed together, and relative values to expression in CD11b+ CD103+ PD‐L1High subset of one experiment are plotted. Circles and horizontal bars indicate data from one experiment and mean of data from the three experiments, respectively. (b) Enriched MLN CD11c+ cells were analysed by flow cytometry, and CCR7 (left) histogram and MHC II‐CD11c plots (right) of the four indicated subsets are shown. Data are representative of three independent experiments. (c,e) Four MLN CD11c+ cell subsets from ovalbumin (OVA) ‐fed mice (c) or normal mice (e) were cultured with CFSE‐labelled CD4+ cells from DO11.10 mice (three wells/sample). In addition, OVA protein (100 ng/ml) was added in (e). After 3 days, the cells in the each well were collected, and CD4 and CFSE expression on CD4+ CD11c− cells were analysed by flow cytometry. Data are from a representative well of each sample. (d,f) Proportion of proliferated (CFSELow) cells in CD4+ CD11c− cells [gates shown in (c) and (e)] were analysed. The plot shows representative data from one experiment (n = 3). Circles and horizontal bars indicate data from one well and mean of results from three wells, respectively. Statistical analysis was performed by Tukey's honest significant difference test. Values not sharing a common letter are significantly different (P < 0·05).
We examined whether the migratory DC subsets in MLNs are able to obtain and present orally administered antigens. The four MLN CD11c+ cell subsets were purified from mice fed OVA in their drinking water and co‐cultured with CFSE‐labelled OVA‐specific T cells, and antigen presentation by the co‐cultured CD11c+ cell subsets was detected as a decrease in CFSE in the T cells, which indicates T‐cell proliferation. The results showed that proliferation of the OVA‐specific T cells was induced by the three MLN subsets that exhibited migratory phenotypes (Fig. 7c,d). Among the three subsets, CD11b+ CD103+ PD‐L1High and CD11b− CD103+ PD‐L1High subsets from mice fed control water induced little (< 0·5%) T‐cell proliferation (see Supplementary material, Fig. S1). Hence, at least, these two subsets may capture and carry orally administered antigens in vivo. In contrast, the CD11b+ CD103− PD‐L1Int subset induced relatively high antigen‐independent proliferation (see Supplementary material, Fig. S1). The proliferated cells reached the same level as those induced by the subset from OVA‐fed mice (Fig. 7c,d). Hence, this subset might not present orally administered antigens, at least in this assay. CD11b− CD103+ PD‐L1Low DCs barely induced T‐cell proliferation (Fig. 7c,d). On the other hand, the four subsets could capture and present OVA antigen supplemented in vitro (Fig. 7e,f). Hence, the inability of CD11b− CD103+ PD‐L1Low DCs to present the orally administered antigens is indeed due to its location in vivo, i.e. this subset is a resident DC subset.
The in vitro Treg induction assays revealed that PD‐L1 and PD‐L2 are not involved in Treg induction (Fig. 3), which is inconsistent with the previous study.38 This suggests that these molecules may be involved indirectly in Treg cell induction in MLN DCs. One possible process was antigen uptake. Hence, we examined whether PD‐L1 blocking can inhibit antigen uptake of the DC subsets in vitro. However, PD‐L1 neutralization did not inhibit the antigen uptake of MLN DC subsets, but slightly increased T‐cell proliferation (see Supplementary material, Fig. S2).
Collectively, CD11b− CD103+ PD‐L1High DCs capture antigens in the intestine, migrate to MLNs and highly induce Treg cells through TGF‐β activation. For this reason, this subset may play an important role in induction of oral tolerance.
Discussion
In the intestine, immune responses to non‐harmful antigens should be regulated. The present study showed that MLN CD11c+ cells contain four subsets expressing CD103 and/or PD‐L1. Among them, CD11b− CD103+ PD‐L1High DCs highly induced Treg cells. This Treg cell induction may be mediated by a high capacity to activate latent TGF‐β. The CD11b− CD103+ PD‐L1High subset highly expressed TGF‐β‐activating integrin β 8 gene, and furthermore, differences in Treg cell inducing efficiency among the subsets were abrogated by exogenous active TGF‐β. The CD11b− CD103+ PD‐L1High DCs exhibited features of migratory DCs, i.e. CCR7+ MHCIIHigh. Furthermore, this subset could present orally administered antigens. Collectively, the present study revealed that CD11b− CD103+ PD‐L1High DCs may obtain luminal antigens in the intestine, migrate into MLNs and highly induce Treg cells through TGF‐β activation.
Many studies have intensely focused on the intestinal DCs. Previous studies have reported that the intestinal CD11c+ cells consist of phenotypically and functionally distinct subsets. One remarkable marker that could identify characteristic DC subsets in the intestine is CD103. The present study found that MLN CD11b− CD103+ DCs consist of PD‐L1High and PD‐L1Low subsets. These two subsets had different origins. CD11b− CD103+ PD‐L1High DCs were a migratory subset whereas CD11b− CD103+ PD‐L1Low DCs were resident. These subsets were also identified by CD8α expression instead of PD‐L1, i.e. the CD8α Int subset was PD‐L1High whereas the CD8α High subset was PD‐L1Low in CD11b− CD103+ DCs. These results are consistent with a previous study that reported that migratory lymph‐borne CD11b− CD103+ DCs are CD8α Int whereas MLN‐resident CD11b− DCs are CD8α High.37 Migratory DCs in MLNs are derived from both the small and large intestines.49, 50 Both intestines are a source of MLN CD11b− CD103+ DCs whereas all CD11b+ CD103+ DCs in MLNs originate in the small intestine.50 In the large intestine, antigens in the distal colon are transported to caudal and iliac LNs, but not MLNs, and oral tolerance is induced at the sites.51 Hence, the MLN CD11b− CD103+ PD‐L1High DCs might induce Treg cells specific to antigens derived from both the small intestine and the proximal colon. In addition to CD8α, XCR1 expression also differed between PD‐L1High and PD‐L1Low subsets in CD11b− CD103+ DCs. In the intestine, XCL1, the ligand of XCR1, might be secreted from T cells.44 It is possible that XCR1 is down‐regulated in the PD‐L1High subset in order to migrate toward CCR7 ligands secreted in MLNs. XCR1‐expressing DCs in several tissues, including the intestine, function in antigen cross‐presentation to CD8+ T cells.41, 42, 43, 44, 45, 52 Migratory XCR1+ DCs in MLNs can present orally administered antigens to CD8+ T cells.42 On the other hand, CD8α‐expressing migratory DCs in MLNs, which also express XCR1, can present antigens in intestinal epithelial cells to CD8+ T cells.37 The present study revealed that these migratory XCR1+ and CD8α + DCs in MLNs highly express PD‐L1. This CD11b− CD103+ PD‐L1High XCR1+ CD8α Int DC subset might regulate CD8+ T cells specific to orally derived and intestinal epithelial cell‐derived antigens in the steady state.
The present study revealed that MLN CD11b− CD103+ PD‐L1High DC subset highly induces Foxp3+ Treg cells. This is probably due to efficient TGF‐β activation by integrin α v β 8. CD11b− CD103+ PD‐L1High DC subset highly expressed Itgb8 gene, and the difference in Treg cell induction was abrogated by exogenous TGF‐β. In contrast, Treg cell induction was not affected by RA supplementation and RA receptor inhibition, although RA has been reported to enhance TGF‐β‐mediated Treg cell induction.19, 20, 21 In the present study, exogenous RA could induce and LE540 could inhibit expression of CCR9 and integrin α 4 β 7 on T cells, which means that the activities of both reagents were sufficient. One possible explanation is that RA‐mediated Treg induction requires a proper dose of TGF‐β. Indeed, some previous studies have reported that RA can enhance Treg cell induction only in the presence of exogenous TGF‐β.20, 21, 26, 53
Retinoic acid plays an important role in mucosal immunity. The present study revealed that MLN CD11b− CD103+ PD‐L1High DCs highly express Aldh1a2 gene encoding RALDH2. Previously, this gene was reported to be expressed by both CD11b+ CD103+ and CD11b− CD103+ MLN DCs at the same level.54 CD11b− CD103+ DCs in the previous study contain both of PD‐L1High and PD‐L1Low subsets, and the PD‐L1Low subset barely expressed Aldh1a2 gene. Hence, the average level of Aldh1a2 gene expression in CD11b− CD103+ DCs may have appeared almost equal to that of the CD11b+ CD103+ subset. Consistent with the gene expression, the CD11b− CD103+ PD‐L1High DCs had the highest ALDH activity among the four subsets (Takano T. unpublished data). Therefore, this DC subset may be a large source of RA in MLNs. RA induces expression of gut‐homing receptors on Treg cells. The homing of induced Treg cells to the intestine is indispensable for oral tolerance.14, 15 Collectively, the present study suggests the possibility that MLN CD11b− CD103+ PD‐L1High DCs play a major role in such process inducing oral tolerance through Treg cells.
The present study revealed that PD‐L1 and PD‐L2 on the DC subset were not critical for Treg cell induction, although a previous study reported that MLN DCs from PD‐L1−/− or PD‐L2−/− mice could not induce Treg cells.38 PD‐L1 was not involved in direct antigen uptake of the DCs, either. It is possible that PD‐L1 and PD‐L2 are involved in processes before the antigen presentation to T cells, such as antigen transfer from other cells that first capture antigens in vivo, DC activation, and DC migration to MLNs. Consistently, it has been reported that migratory DCs highly express the gene encoding PD‐L1, which raises a possibility that this molecule is involved in migration.55 In addition, there are differences between the previous38 and the present studies in the culture conditions used, so it is also possible that PD‐L1 is required for Treg cell induction in a specific condition, e.g. a specific cytokine milieu or antigen concentration.
Oral tolerance, systemic immune hyporesponsiveness to orally administered antigens, is induced partly by oral antigen‐specific Treg cell induction in the intestinal immune system.2, 3 An intentional induction of oral tolerance could be a therapeutic strategy for allergy and autoimmune diseases. Therefore, the MLN CD11b− CD103+ PD‐L1High DC subset may be a potential target for such therapeutic strategy.
Disclosures
The authors declare no commercial and financial conflict of interest.
Supporting information
Figure S1 Potential of mesenteric lymph node dendritic cell subsets to induce T‐cell proliferation without antigens.
Figure S2 Programmed death ligand 1 is not involved directly in antigen uptake of mesenteric lymph node dendritic cell subsets.
Acknowledgements
AS, RK and TT performed the experiments, AS, RK, HNA and SH designed the study, and RK and SH wrote the paper. This work was supported by Grant‐in‐Aids for Scientific Research (B) 23380073 and 26292065 from the Japan Society for the Promotion of Science (JSPS) to SH.
References
- 1. Steele L, Mayer L, Cecilia Berin M. Mucosal immunology of tolerance and allergy in the gastrointestinal tract. Immunol Res 2012; 54:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weiner HL, Wu HY, Quintana F, Wu H. Oral tolerance. Immunol Rev 2011; 241:241–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol 2012; 5:232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faria AMC, Weiner HL. Oral tolerance: therapeutic implications for autoimmune diseases. Clin Dev Immunol 2006; 13:143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hori S. Lineage stability and phenotypic plasticity of Foxp3+ regulatory T cells. Immunol Rev 2014; 259:159–72. [DOI] [PubMed] [Google Scholar]
- 6. Morikawa H, Sakaguchi S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3‐centered view to an epigenome‐defined view of natural Treg cells. Immunol Rev 2014; 259:192–205. [DOI] [PubMed] [Google Scholar]
- 7. Sawant DV, Vignali DAA. Once a Treg, always a Treg? Immunol Rev 2014; 259:173–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T‐cell homeostasis: steady‐state maintenance and modulation during inflammation. Immunol Rev 2014; 259:40–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan X, Cheng G, Malek TR. The importance of regulatory T‐cell heterogeneity in maintaining self‐tolerance. Immunol Rev 2014; 259:103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S et al Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol 2013; 14:307–8. [DOI] [PubMed] [Google Scholar]
- 11. Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell‐dependent and ‐independent control of allergic inflammation. Immunity 2008; 29:114–26. [DOI] [PubMed] [Google Scholar]
- 12. Kim KS, Hong S‐W, Han D, Yi J, Jung J, Yang B‐G et al Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 2016; 351:858–63. [DOI] [PubMed] [Google Scholar]
- 13. Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G et al Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med 2006; 203:519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hadis U, Wahl B, Schulz O, Hardtke‐Wolenski M, Schippers A, Wagner N et al Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 2011; 34:237–46. [DOI] [PubMed] [Google Scholar]
- 15. Cassani B, Villablanca EJ, Quintana FJ, Love PE, Lacy‐Hulbert A, Blaner WS et al Gut‐tropic T Cells that express integrin α 4 β 7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 2011; 141:2109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masopust D, Schenkel JM. The integration of T cell migration, differentiation and function. Nat Rev Immunol 2013; 13:309–20. [DOI] [PubMed] [Google Scholar]
- 17. Salmi M, Jalkanen S. Lymphocyte homing to the gut: attraction, adhesion, and commitment. Immunol Rev 2005; 206:100–13. [DOI] [PubMed] [Google Scholar]
- 18. Scott CL, Aumeunier AM, Mowat AM. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol 2011; 32:412–19. [DOI] [PubMed] [Google Scholar]
- 19. Coombes JL, Siddiqui KRR, Arancibia‐Cárcamo CV, Hall J, Sun C‐M, Belkaid Y et al A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF‐β and retinoic acid‐dependent mechanism. J Exp Med 2007; 204:1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M et al Reciprocal Th17 and regulatory T cell differentiation mediated by retinoic acid. Science 2007; 317:256–60. [DOI] [PubMed] [Google Scholar]
- 21. Benson MJ, Pino‐Lagos K, Rosemblatt M, Noelle RJ. All‐trans retinoic acid mediates enhanced Treg cell growth, differentiation, and gut homing in the face of high levels of co‐stimulation. J Exp Med 2007; 204:1765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yokota A, Takeuchi H, Maeda N, Ohoka Y, Kato C, Song SY et al GM‐CSF and IL‐4 synergistically trigger dendritic cells to acquire retinoic acid‐producing capacity. Int Immunol 2009; 21:361–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guilliams M, Crozat K, Henri S, Tamoutounour S, Grenot P, Devilard E et al Skin‐draining lymph nodes contain dermis‐derived CD103‐ dendritic cells that constitutively produce retinoic acid and induce Foxp3+ regulatory T cells. Blood 2010; 115:1958–68. [DOI] [PubMed] [Google Scholar]
- 24. Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut‐homing specificity on T cells. Immunity 2004; 21:527–38. [DOI] [PubMed] [Google Scholar]
- 25. Jaensson E, Uronen‐Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL et al Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med 2008; 205:2139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Worthington JJ, Czajkowska BI, Melton AC, Travis MA. Intestinal dendritic cells specialize to activate transforming growth factor‐β and induce Foxp3+ regulatory T cells via integrin α v β 8 . Gastroenterology 2011; 141:1802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Païdassi H, Acharya M, Zhang A, Mukhopadhyay S, Kwon M, Chow C et al Preferential expression of integrin α v β 8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology 2011; 141:1813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melton AC, Bailey‐Bucktrout SL, Travis MA, Fife BT, Bluestone JA, Sheppard D. Expression of α v β 8 integrin on dendritic cells regulates Th17 cell development and experimental autoimmune encephalomyelitis in mice. J Clin Invest 2010; 120:4436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acharya M, Mukhopadhyay S, Paidassi H, Jamil T, Chow C, Kissler S et al α v Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. J Clin Invest 2010; 120:4445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, Puccetti P et al Gut CD103+ dendritic cells express indoleamine 2,3‐dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 2010; 59:595–604. [DOI] [PubMed] [Google Scholar]
- 31. Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E et al Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 2013; 38:581–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA. et al Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012; 483:345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity 2014; 40:248–61. [DOI] [PubMed] [Google Scholar]
- 34. Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA et al CX3CR1‐mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005; 307:254–8. [DOI] [PubMed] [Google Scholar]
- 35. Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW et al Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med 2009; 206:3101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cerovic V, Houston S, Scott C, Aumeunier A., Yrlid U, Mowat A. et al Intestinal CD103− dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol 2012; 6:104–13. [DOI] [PubMed] [Google Scholar]
- 37. Cerovic V, Houston SA, Westlund J, Utriainen L, Davison ES, Scott CL et al Lymph‐borne CD8α + dendritic cells are uniquely able to cross‐prime CD8+ T cells with antigen acquired from intestinal epithelial cells. Mucosal Immunol 2015; 8:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukaya T, Takagi H, Sato Y, Sato K, Eizumi K, Taya H et al Crucial roles of B7‐H1 and B7‐DC expressed on mesenteric lymph node dendritic cells in the generation of antigen‐specific CD4+Foxp3+ regulatory T cells in the establishment of oral tolerance. Blood 2010; 116:2266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo . Science 1990; 250:1720–3. [DOI] [PubMed] [Google Scholar]
- 40. Kotaki R, Wajima S, Shiokawa A, Hachimura S. Toll‐like receptor 2 suppresses Toll‐like receptor 9 responses in Peyer's patch dendritic cells. Immunobiology 2015; 220:734–43. [DOI] [PubMed] [Google Scholar]
- 41. Bachem A, Hartung E, Güttler S, Mora A, Zhou X, Hegemann A et al Expression of XCR1 characterizes the Batf3‐dependent lineage of dendritic cells capable of antigen cross‐presentation. Front Immunol 2012; 3:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Becker M, Güttler S, Bachem A, Hartung E, Mora A, Jäkel A et al Ontogenic, phenotypic, and functional characterization of XCR1+ dendritic cells leads to a consistent classification of intestinal dendritic cells based on the expression of XCR1 and SIRPα . Front Immunol 2014; 5:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S et al Selective expression of the chemokine receptor XCR1 on cross‐presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 2009; 31:823–33. [DOI] [PubMed] [Google Scholar]
- 44. Ohta T, Sugiyama M, Hemmi H, Yamazaki C, Okura S, Sasaki I et al Crucial roles of XCR1‐expressing dendritic cells and the XCR1‐XCL1 chemokine axis in intestinal immune homeostasis. Sci Rep 2016; 6:23505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I et al Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol 2013; 190:6071–82. [DOI] [PubMed] [Google Scholar]
- 46. Scott CL, Zangerle Murray TFP, Beckham KSH, Douce G, Mowat AMI. Signal regulatory protein α (SIRPα) regulates the homeostasis of CD103+CD11b+ DCs in the intestinal lamina propria. Eur J Immunol 2014; 44:3658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ et al CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1‐inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol 2012; 42:3150–66. [DOI] [PubMed] [Google Scholar]
- 48. Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K et al CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J Immunol 2006; 176:803–10. [DOI] [PubMed] [Google Scholar]
- 49. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014; 14:667–85. [DOI] [PubMed] [Google Scholar]
- 50. Houston SA, Cerovic V, Thomson C, Brewer J, Mowat AM, Milling S. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol 2016; 9:468–78. [DOI] [PubMed] [Google Scholar]
- 51. Veenbergen S, van Berkel LA, du Pré MF, He J, Karrich JJ, Costes LMM et al Colonic tolerance develops in the iliac lymph nodes and can be established independent of CD103+ dendritic cells. Mucosal Immunol 2016; 9:894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brewitz A, Eickhoff S, Dähling S, Quast T, Bedoui S, Kroczek RA et al CD8+ T cells orchestrate pDC‐XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 2017; 46:205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun C‐M, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR et al Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med 2007; 204:1775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Denning TL, Norris BA, Medina‐Contreras O, Manicassamy S, Geem D, Madan R et al Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol 2011; 187:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A et al Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 2012; 13:888–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Potential of mesenteric lymph node dendritic cell subsets to induce T‐cell proliferation without antigens.
Figure S2 Programmed death ligand 1 is not involved directly in antigen uptake of mesenteric lymph node dendritic cell subsets.


