Abstract
DNA interstrand crosslinks (ICLs) are covalent linkages between two strands of DNA and their presence interfere with essential metabolic processes such as transcription and replication. These lesions are extremely toxic and their repair is essential for genome stability and cell survival. In this review, we will discuss how the removal of ICLs requires interplay between multiple genome maintenance pathways and can occur in the absence of replication (replication-independent ICL repair) or during S phase (replication-coupled ICL repair), the latter being the predominant pathway used in mammalian cells. It is now well recognized that translesion DNA synthesis (TLS), especially through the activities of REV1 and DNA polymerase zeta (Polζ), is necessary for both ICL repair pathways operating throughout the cell cycle. Recent studies suggest that the convergence of two replication forks upon an ICL initiates a cascade of events including unhooking of the lesion through the actions of structure-specific endonucleases thereby creating a DNA double stranded break (DSB). TLS across the unhooked lesion is necessary for restoring the sister chromatid prior to homologous recombination (HR) repair. Biochemical and genetic studies implicate REV1 and Polζ as being essential for performing lesion bypass across the unhooked crosslink and this step appears to be important for subsequent events to repair the intermediate DSB. The potential role of Fanconi anemia pathway in the regulation of REV1 and Polζ-dependent TLS and the involvement of additional polymerases, including DNA polymerases kappa, nu, and theta, in the repair of ICLs is also discussed in this review.
Keywords: REV1, REV3, REV7, Interstrand cross link (ICL) repair, Fanconi anemia, Translesion DNA synthesis
Introduction
Interstrand crosslinks (ICLs) are formed when bifunctional electrophilic agents react with two bases on opposing strands of the DNA helix. This covalent linkage imposes an insurmountable block to essential DNA metabolic processes such as transcription and replication. The repair of these lesions is essential for cell survival. Because ICLs are so potent in blocking DNA replication, agents which form these lesions are frequently used to treat patients with cancer (e.g. mitomycin C, nitrogen mustards, and platinum-containing compounds like cisplatin), as well as automimmune diseases, the latter taking advantage of the immunosuppressive activity of these agents (e.g. cyclophosphamide). Therefore, understanding how cells resist these lesions is important to our ability to improve the therapeutic use of these drugs in the future of personalized medicine and targeted therapy, as well as understand variations in patient response and toxicity (Deans and West 2011).
The formation of an ICL by these agents is a relatively rare event when compared to the frequency of crosslinking two adjacent bases within the same strand of DNA (i.e. an intrastrand crosslink). The more prevalent intrastrand crosslinks are most commonly removed by the nucleotide excision repair (NER) pathway which involves recognition of the crosslink and excision of the affected strand (de Laat et al. 1999). The DNA helix is restored after the excised strand is replaced via repair synthesis using the complimentary strand as a template which is followed by DNA ligation. Alternatively, intrastrand crosslinks can be bypassed during replication by specialized DNA polymerases referred to as translesion DNA polymerases, making these lesions relatively less toxic to proliferating cells. ICLs cannot be repaired by this robust NER pathway alone or bypassed during replication due to the fact that both strands of the double helix are involved in a covalent linkage. Furthermore, excising the lesion present on both strands simultaneously will result in removal of the genetic information necessary for repair synthesis. Therefore, multiple DNA repair processes are engaged to remove ICLs in a manner that preserves genomic integrity.
ICL repair: from simple to complex
The first model of ICL repair was brought to light by the influential studies performed by Ronald S. Cole implicating the cooperation of both NER and homologous recombination (HR) during ICL repair in E. coli (Cole 1973; Cole et al. 1976). The NER endonuclease UvrABC recognizes an ICL formed in DNA (e.g. photoactivated psoralen) and makes incisions on each side of the ICL within the same strand of DNA, essentially “unhooking” the ICL such that the excised ICL forms a large monoadduct still attached to other strand. The excised region of the opposite strand is further processed into a large gap by DNA pol I exonuclease activity that then serves as a substrate for RecA-mediated homologous recombination with an undamaged chromosome (Sinden and Cole 1978a; Sinden and Cole 1978b; Cheng et al. 1988; Sladek et al. 1989; Cheng et al. 1991). If recombination is not possible (for example in a recA mutant), an alternative approach is to fill in the gap opposite the unhooked crosslink by translesion DNA synthesis (TLS) through the activity of a specialized polymerase (such as PolII, PolIV or PolV in E. coli) (Berardini et al. 1999; Tang et al. 2000; Goodman 2002; Friedberg et al. 2005). However, this alternative pathway, also referred to as recombination-independent ICL repair, can be associated with an increased risk for mutagenesis due to the potential for miscoding and the error-prone nature of lesion bypass polymerases. After recombination or lesion bypass, the unhooked crosslink or monoadduct is ultimately removed through a second round of NER repair thus restoring the DNA helix to its original state.
The results from these early studies in bacteria serve as the foundation for current models of ICL repair. The basic scheme of unhooking the ICL and resolution of the unhooked crosslink by recombination or TLS combined with NER to remove the unhooked ICL after gap repair appears to be conserved in yeast, Drosophila, Xenopus, and vertebrates (McVey 2010). However, the levels of regulation and the number of pathways recruited into the process dramatically increases in complexity, depending upon whether repair is occurring in the absence of replication (i.e. replication-independent ICL repair) or during S phase (i.e. replication-coupled ICL repair). Important differences between prokaryotes and eukaryotes are now well characterized. First, in higher eukaryotes (e.g. vertebrates), ICL repair appears to be completed primarily during the S phase of the cell cycle as opposed to in the absence of DNA replication, such as in G1 (Akkari et al. 2000; Niedernhofer et al. 2004; Rothfuss and Grompe 2004). Second, the only NER proteins that have been strongly associated with promoting resistance to interstrand crosslinking agents are XPF and ERCC1 that together comprise a structure-specific endonuclease, suggesting that XPF-ERCC1 has a specific role in removing ICLs from DNA that is independent of its function in NER (Jachymczyk et al. 1981; Hoy et al. 1985; Andersson et al. 1996; De Silva et al. 2000; Clingen et al. 2005; Zhang et al. 2007). Third, both TLS and HR cooperate during the resolution of unhooked ICLs in a manner which requires DNA replication, rather than a competition between the TLS and HR pathways to resolve gap remaining after unhooking the crosslink as suggested by the early work in prokaryotes. These added layers of complexity necessitate the involvement of a higher order complex of proteins to recruit and regulate the multiple enzymatic activities involved in ICL repair. Proteins in this complex are encoded by genes that when mutated cause the rare disease Fanconi anemia (FA) characterized by bone marrow failure, developmental deficiencies, and predisposition to cancer (Niedernhofer 2007; Kee and D'Andrea 2010; Deans and West 2011)
Over the past decade, enormous progress has been made in defining the various steps operating during replication-coupled ICL repair (Muniandy et al. 2010). The current model of replication-coupled ICL repair now includes the actions of multiple pathways including NER, HR, mismatch repair, TLS, and structure specific endonucleases that are regulated at multiple levels by the BRCA1 tumor suppressor, the ataxia telangiectasia and Rad3 related (ATR) protein kinase and the FA pathway (Cantor and Xie 2010; Hlavin et al. 2010; Ho and Scharer 2010; Kee and D'Andrea 2010; Shen and Li 2010; Vasquez 2010; Wood 2010; Sengerova et al. 2011). Coordination of these multiple pathways is vital for maintaining genome stability throughout the process, in addition to determining relative hypersensitivity or resistance to ICL generating chemotherapeutic drugs. It is now well recognized that TLS, especially through the activities of REV1 and DNA polymerase zeta (Polζ), is necessary for both ICL repair pathways operating throughout the cell cycle and is the primary subject of this review.
REV1 and Polζ: DNA polymerases that promote mutagenesis
In 1971, Jeffrey Lemontt performed a screen for mutants conferring a ‘reversionless’ phenotype in Saccharomyces cerevisiae and identified two genes, Rev1 and Rev3, that were actively involved in UV-induced mutagenesis (Lemontt 1971). A similar screening later led to the identification of the Rev7 gene (Lawrence et al. 1985). Rev7 was later shown to form a heterodimeric complex with Rev3 forming the accessory and catalytic subunits of what is now referred to as Polζ (Nelson et al. 1996b). Rev1-deleted yeast cells show similar phenotypes to those lacking Rev3 or Rev7 suggesting that Rev1 is necessary for Polζ-dependent mutagenesis. Thus, the genetic epistasis observed between Rev1, Rev3, and Rev7 in yeast contributed to the prevailing model that Rev1 and Polζ cooperate to perform lesion bypass and are considered the main players participating in an error prone pathway of post-replication repair (Chang and Cimprich 2009; Waters et al. 2009). It is important to note here that Rev1 and Polζ-dependent TLS not only occurs during replication, but also after DNA replication has completed, being responsible for filling in gaps left behind after stalled replication forks resume replication downstream of a replication-blocking lesion, hence the term ‘postreplication repair’ was created to describe this process (Lopes et al. 2006; Jansen et al. 2009a; Jansen et al. 2009b; Diamant et al. 2012). The fact that single-stranded DNA gaps created after replication fork stalling can persist in cells well into the G2 phase of the cell cycle suggests that mechanisms exist to restart replication downstream of a blocking lesion. How this ‘repriming’ process is regulated remains largely unclear.
REV3 belongs to the B-family of polymerases which includes the highly accurate replicative DNA polymerases including Polδ, Polɛ and Polα (Morrison et al. 1989; Lawrence 2004). Polζ possesses lower processivity and a higher fidelity than the Y-family of TLS polymerases, even though REV3 is devoid of the 3’→5’ proofreading exonuclease activity present in most B-family DNA polymerases (Morrison et al. 1989; Nelson et al. 1996b; Lawrence 2004). Previous studies indicate that Polζ is not efficient in replicating through most DNA lesions and can perform bypass only at certain lesions such as cis-syn TT dimers where it has been reported to perform both the insertion and extension steps opposite a thymine glycol lesion in an error-free manner (Nelson et al. 1996b; Johnson et al. 2003). Rather than inserting nucleotides opposite DNA adducts, Polζ appears to be particularly specialized to extend distorted base pairs, such as mismatches that might result from an inaccurate base insertion by a TLS polymerase or a base pair involving a bulky DNA lesion (Lawrence 2004; Prakash et al. 2005). Therefore, the primary role of Polζ in TLS is proposed to be performing an extension reaction from nucleotides inserted opposite the lesion by another TLS polymerase (Prakash and Prakash 2002; Prakash et al. 2005). This proficiency in extending through mismatches along with a relatively high error rate for base substitutions is associated with the mutagenic properties of Polζ (Lawrence 2004; Zhong et al. 2006). Although, Rev3 alone is capable of polymerization, association of Rev3 with Rev7, the accessory subunit of Polζ, has been shown to stabilize and significantly enhance the polymerase activity of Rev3 by 20–30 fold, suggesting that Rev7 functions as a processivity factor for Polζ (Nelson et al. 1996b).
Yeast Rev1 was the first member of the Y-family proteins characterized as a deoxycytidyl (dCMP) tansferase rather than a processive DNA polymerase, whose activity appears to be restricted to inserting dCMPs opposite guanines and abasic sites in template DNA (Nelson et al. 1996a). Additional insights made from the crystal structure of the polymerase domain of S. cerevisiae Rev1 bound to a primer template and an incoming dCTP revealed that Rev1 uses a novel catalysis mechanism whereby the incoming dCTP pairs with an arginine rather than a template base and the template guanine is evicted from the DNA helix (Nair et al. 2005). This limited activity does not seem to be important for most functions of Rev1 since catalytic inactive Rev1 can still promote Rev3-dependent mutagenesis in yeast (Nelson et al. 2000; Lawrence 2002). Instead, Rev1 appears to play a more important role in regulating Polζ-dependent TLS.
The primary regulator of TLS in both E. coli and yeast is the sliding clamp that functions to promote processivity of DNA polymerases during replication (Friedberg et al. 2005). In E. coli, the homodimer, polIII-associated sliding β clamp (encoded by the dnaN gene) plays a central role in mediating the switch from normal DNA replication to TLS (Fujii and Fuchs 2004). In eukaryotes, the homotrimeric sliding clamp, proliferating cell nuclear antigen (PCNA), plays a similar role in mediating the switch between replication and TLS with an added level of regulation whereby TLS is activated by monoubiquitination of PCNA (Hoege et al. 2002). In S. cerevisiae, genetic studies have shown that genes belonging to the Rad6 epistasis group are involved in the replication of damaged DNA, a process referred to as post-replication repair (Jentsch et al. 1987; Bailly et al. 1997; Broomfield et al. 1998). Of these, Rad6 and Rad18 are required for both error-free and error-prone replication bypass of UV-induced DNA lesions, while Rad5, Mms2, and Ubc13 play a role in regulating the replication past DNA adducts in an error-free manner using a TLS polymerase-independent mechanism (Xiao et al. 2000). Rad6 and the Mms2-Ubc13 complex function as E2 ubiquitin conjugating enzymes in association with the Rad18 and Rad5 E3 ubiquitin ligases respectively, and regulate the ubiquitination state of PCNA (Jentsch et al. 1987; Bailly et al. 1997; Hoege et al. 2002).
In response to replication fork stalling and the generation of long stretches of single-stranded DNA, Rad6 and Rad18 monoubiquitinate PCNA on lysine-164 (K164) triggering post-replication repair (Davies et al. 2008). One potential outcome is mutagenic lesion bypass by Rev1 and Polζ. Alternatively, the other TLS polymerase expressed in yeast, Polη (encoded by the Rad30 gene) can accurately insert nucleotides opposite thymine dimers thus preventing UV-induced mutatagenesis. The ubiquitin conjugated to PCNA on K164 can be further extended forming a K63-linked ubiquitin chain attached to PCNA in a reaction mediated by Mms2–Ubc13 and Rad5 (Hofmann and Pickart 1999; Hoege et al. 2002; Andersen et al. 2008). While the monoubiquitination of PCNA mediates the switch to TLS, evidence suggests that polyubiquitination of PCNA channels postreplication repair into an uncharacterized error-free damage avoidance pathway that involves template switching (Stelter and Ulrich 2003).
All eukaryotic Y-family polymerases including REV1 possess ubiquitin binding motifs (UBM) or ubiquitin-binding zinc finger (UBZ) domains which increase their affinity for ubiquitinated PCNA (Kannouche et al. 2004; Watanabe et al. 2004; Bienko et al. 2005; Plosky et al. 2006; Acharya et al. 2008; Sabbioneda et al. 2008; Bomar et al. 2010). Although the ubiquitination of PCNA is necessary for TLS in yeast, recent studies suggest that alternative pathways may regulate polymerase switching in vertebrates. Analysis of the replication of damaged DNA in chicken DT40 cells demonstrated a predominant role for PCNA ubiquitination in promoting the filling in of post-replication gaps (Edmunds et al. 2008). However, REV1-dependent TLS across a TT 6-4 photoproduct (a dominant UV lesion) in DT40 cells carrying a PCNA K164 mutation appears to be normal as measured by a plasmid system (Szüts et al. 2008). Recent work from the Livneh group utilized mouse embryonic fibroblasts in which specific TLS genes associated with ubiquitination of PCNA were manipulated. These studies showed that eliminating expression of REV3, Polη or REV1 in PCNAK164R/K164R mouse embryo fibroblasts further increased their sensitivity to UV-radiation indicating the existence of a TLS pathway that is independent of PCNA ubiquitination (Hendel et al. 2011). At least in DT40 cells, this non-canonical TLS pathway appears to be largely dependent on REV1 (Edmunds et al. 2008; Szüts et al. 2008). Recent studies identified the Fanconi anemia core complex as being an important regulator of REV1 localization to stalled replication forks resulting from UV or cisplatin treatment (Mirchandani et al. 2008; Hicks et al. 2010; Kim et al. 2012).
REV1 and Polζ: collaborators in completing TLS across many DNA lesions
As discussed above, REV1 contains two UBMs that are important for recruitment to blocked DNA replication forks or gaps remaining in DNA after the completion of replication (Bienko et al. 2005; Guo et al. 2006). REV1 also possesses a unique polymerase interacting domain that makes direct contact with REV7 and a variety of TLS DNA polymerases including Polη and REV3 (Murakumo et al. 2001; Guo et al. 2003; Masuda et al. 2003; Ohashi et al. 2004; Tissier et al. 2004; Acharya et al. 2006; D'Souza and Walker 2006). Today it is has become clear that for most DNA adducts, more than one TLS polymerase is needed to accomplish a single lesion bypass event in cells (Shachar et al. 2009; Hicks et al. 2010). Current models suggest a two step process where a Y family polymerase inserts the first nucleotide opposite a damaged base and a second polymerase performs the extension step, even if the resulting primer/template pair is highly distorted (Prakash and Prakash 2002). Polζ is thought to be the ‘universal extender’ due to its ability to extend a wide variety of distorted or mismatched primer templates (Johnson et al. 2000b; Haracska et al. 2001). REV1 is thought to primarily be a regulator of TLS by orchestrating polymerase switching events between the first TLS polymerase and the universal extender, Polζ (Guo et al. 2003; Acharya et al. 2006). This dependence of Polζ on REV1 is consistent with the fact that Polζ lacks a UBD to preferentially localize to regions where PCNA has been monoubiquitinated by the RAD18 ubiquitin ligase.
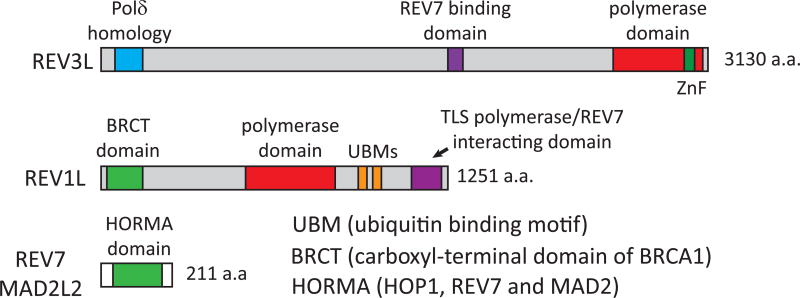
Until recently, very little was known about how mammalian REV1 interacts with Polζ and other Y-family TLS polymerases.. Earlier studies have characterized potential binding sites between REV1, REV3 and REV7 using yeast two hybrid assays and co-immunoprecipitation analyses, but it remained unclear whether these three protein form a heterotrimer in cells (Murakumo et al. 2000; Murakumo et al. 2001; Guo et al. 2003; Ohashi et al. 2004). We found that REV1 is capable of co-immunoprecipitating with REV3 in REV7-depleted cells suggesting that full length REV3 and REV1 have an additional mode of interaction yet to be identified (Sharma et al. 2012a). Based on the recent crystal structure between REV7 and the REV7 binding site in REV3 (amino acids 1847–1898)(Hara et al. 2010), Hara et al suggest that REV7 binds both REV3 and REV1 simultaneously where binding of REV7 to REV3 alters the conformation of REV7 in a way that reveals an additional REV1 binding site (Hara et al. 2010). This is an intriguing result since it has always been assumed that REV7 interactions with REV1 and REV3 were mutually exclusive. In fact, recent structural and biochemical studies now indicate that the C-terminal domain of REV1 can simultaneously interact with REV7 as well as the REV1-interacting region of other Y family polymerases (like Polη and Polκ) (Kikuchi et al. 2012b; Kikuchi et al. 2012a; Pozhidaeva et al. 2012; Wojtaszek et al. 2012a; Wojtaszek et al. 2012b). These studies provide clues about the possible role of the REV1 C-terminal domain in facilitating TLS polymerase switching by demonstrating that REV1 does in fact provide a scaffold for both inserter (e.g. Polη) and extender polymerases (Polζ) to bind during the replicative bypass of complicated lesions. (Kikuchi et al. 2012a; Kikuchi et al. 2012b) Future studies will be needed to fully disclose how REV1 performs its function as a TLS polymerase ‘switcher’ during complex lesion bypass events. Figure 1 illustrates the various domains within REV3, REV1, and REV7 (MAD2L2) that have been characterized to date.
Figure 1. Physical representation of the functional domains of the human REV3, REV1 and REV7 proteins.
REV3 has a region in the N-terminus that is homologous to Polδ, a REV7 binding domain, and a conserved Zinc finger motif in the catalytic domain. REV1 possesses a BRCT domain that interacts with PCNA or DNA, two ubiquitin binding motifs that mediate localization to stalled replication forks, and a C-terminal TLS polymerase and REV7-binding domain. REV7 is almost entirely composed of a HORMA domain that is thought to mediate interactions with chromatin.
REV1 and Polζ: essential for TLS during ICL repair
The specialized function of REV1 and Polζ in ICL repair came to light when the extreme sensitivities to agents capable of forming ICLs were noted in eukaryotic cells lacking either one of these two polymerases (Simpson and Sale 2003; Sonoda et al. 2003; Niedzwiedz et al. 2004; Wu et al. 2004; Zander and Bemark 2004; Nojima et al. 2005; Okada et al. 2005; Cheung et al. 2006; Lin et al. 2006; Sarkar et al. 2006; Wittschieben et al. 2006; Roos et al. 2009; Wang et al. 2009; Hicks et al. 2010; Sharma et al. 2012b). In comparison, the sensitivities of cells lacking REV1 or Polζ to agents that create primarily monoadducts in DNA are relatively benign suggesting REV1 and Polζ play a specialized role in ICL repair.
In yeast, the importance of ICL repair during the G1 phase or in stationary phase is well documented. ICL repair in G1 is initiated following the recognition and unhooking of the ICL by the NER pathway in an analogous fashion as first described in bacteria. The TLS pathway became implicated following the observations that ICLs are highly mutagenic and mutations in Rev3 (Pso1) or Rad6 (Pso8), the E2 ubiquitin conjugating enzyme associated with Rad18 during post-replication repair, are associated with hypersensitivities to psoralen interstrand crosslinks (Henriques and Moustacchi 1980; Cassier and Moustacchi 1981; Henriques and Moustacchi 1981; Barre et al. 1999; De Silva et al. 2000). The replication-independent model was then studied in greater detail revealing that yeast Rev3 mutants are specifically hypersensitive to ICL-generating agents in G1 (McHugh et al. 2000; Sarkar et al. 2006). Genetic and biochemical evidence suggest that following unhooking of an ICL by the NER machinery, normal replicative polymerases such as Polδ attempt to fill in the gap opposite the adduct but stall, initiating the Rad18-dependent PCNA ubiquitination pathway that directs Rev1 and Polζ to perform TLS. Consistent with findings by others, Polζ-dependent TLS requires the yeast Polδ subunit Pol32, and in the case for ICL repair, Pol32 is required for PCNA ubiquitination and Rev7 recruitment to chromatin in an ICL-dependent manner (Huang et al. 2000; Gibbs et al. 2005; Sarkar et al. 2006; Acharya et al. 2009). Whether a similar pathway operates in vertebrates to repair ICLs in G1 is unknown. However, reporter assays designed to measure recombination-independent ICL repair in vertebrates do strongly suggest that NER endonuclease XPF-ERCC1, Polζ and RAD18 cooperate to repair ICLs in a manner that does not require DNA replication, at least in an extrachromosomal context (Wang et al. 2001; Shen et al. 2006). In intact cells, the global genome NER protein XPC is rapidly recruited to psoralen ICLs in G1 cells and their removal was defective in XPC deficient cells (Muniandy et al. 2009). Orlando Schärer’s group have recently implicated the transcription-coupled NER protein CSB, as well as the general NER proteins XPA, XPF, XPG, in the repair of cisplatin ICLs independently of DNA replication (Enoiu et al. 2012). Similar to yeast, REV1 and Polζ are crucial for this repair pathway executed during the G1 phase of the cell cycle. The initial recognition of the ICL involves either the transcription-coupled or global genome NER pathway depending upon the lesion present (Figure 2).
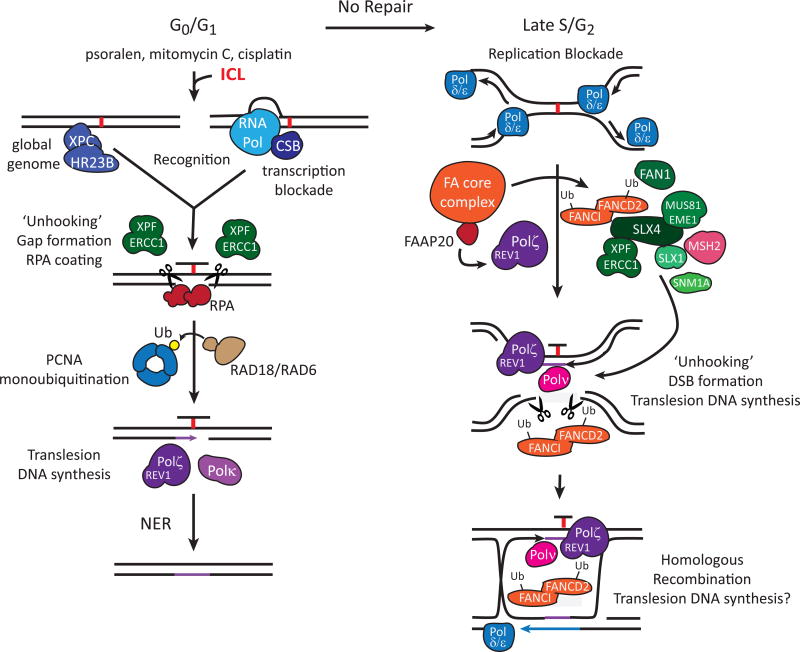
Figure 2. A model for human ICL repair.
Nonproliferating cells are capable of removing ICLs in a manner that is independent of DNA replication. Depending upon the type of ICL or whether the ICL has interfered with RNA transcription, the XPC/HR23B (global genome NER) or CSB (transcription-coupled NER) protein recognizes the lesion and stimulates ‘unhooking’ by the XPF-ERCC1 endonuclease. A single stranded DNA gap is formed that attracts Replication Protein A (RPA) which in turn promotes RAD18/RAD6-dependent monoubiquitination of PCNA. Monoubiquitinated PCNA recruits REV1/Polζ and/or Polκ to the site where they perform TLS opposite the unhooked lesion, the latter presumably removed by a second set of NER reactions to return the DNA duplex to its original state. If the lesion is not removed in G1 (left panel), convergence of DNA polymerases on an unrepaired ICL during DNA replication initiates the Fanconi anemaia (FA) pathway involving detection of the ICL by the FA core complex followed by ubiquitination of the FANCD2/FANCI heterodimer in the late S/G2 phase of the cell cycle (right panel). Ubiquitinated FANCD2/FANCDI promotes excision or ‘unhooking’ of the ICL by structure-dependent endonucleases and likely promotes TLS opposite the ‘unhooked’ ICL in preparation for homologous recombination (HR). MUS81-EME1, SNM1A, FAN1, XPF-ERCC1 and SLX1-SLX4 are structure specific endonucleases thought to be involved in unhooking the ICL, generating ICL-associated double strand breaks (DSBs) and nucleolytic processing of the ICL during repair. SLX4 has also been shown to be an endonuclease scaffold protein and mutations in the SLX4 gene cause Fanconi anemia. FAAP20 interacts with the FA core complex and directly binds to REV1. The mismatch repair protein MSH2 is implicated in ICL repair through an unknown mechanism. The FA pathway and REV1/Polζ can promote HR repair of a direct DSB. We speculate that REV1 and Polζ are not only involved in TLS across the ‘unhooked ICL’ but also perform DNA synthesis during HR repair when the template is incompatible for normal replicative DNA polymerases.
The link between DNA replication, homologous recombination, and ICL repair was established with the findings that ICL-generating agents cause DNA double stranded breaks (DSBs) in a manner dependent upon DNA replication and cells deficient in HR repair are extremely sensitive to these agents (Muniandy et al. 2010). Based on these observations, models explaining ICL repair in mammals evolved to include unhooking, which can occur in the G1 or S phases of the cell cycle, and collision of the DNA replication machinery on the processed ICL. The unhooked lesion causes replication fork stalling and release of one arm of the replication fork to generate a ‘one ended’ DSB. Repair of this replication-associated DSB is channeled into the HR repair pathway (McHugh et al. 2001; Hinz 2010; Deans and West 2011). Earlier studies monitoring the generation of DSBs in cells following treatment with mitomycin C using γ-H2AX as a surrogate marker for DSBs, as well as pulsed-field gel electrophoresis, first implicated the structure-specific MUS81-EME1 endonuclease as being required for DSB generation in S phase (Hanada et al. 2006). Multiple endonucleases, including XPF-ERCC1, SLX4, FAN1, and SNM1A are also thought to play important roles in the initial lesion processing events required for ICL repair (Fekairi et al. 2009; Muñoz et al. 2009; Svendsen et al. 2009; Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010; Yang et al. 2010; Kim et al. 2011; Stoepker et al. 2011; Wang et al. 2011; Yamamoto et al. 2011). Together, these studies strongly suggest that the generation of DSBs in cells treated with ICL generating agents is an active process and not caused by regression of a stalled fork resulting in collapse. After incision and DSB formation upon collision of a replication fork with an ICL, multiple factors are involved in the completion of repair, including REV1 and Polζ, the Fanconi anemia pathway, the mismatch repair MSH2 protein, XPF-ERCC1, and HR repair (Zhang et al. 2007). A common phenotype in cells deficient in any of these proteins or pathways is an inability to resolve ICL-induced DSBs in a timely manner (Rothfuss and Grompe 2004; Hicks et al. 2010; Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Wang et al. 2011).
Additional studies performed in mammalian cells support the model that REV1 and Polζ perform an essential step that allows ICL-associated DSBs to be channeled into the HR repair pathway. Human cancer cells depleted of REV1, REV3 or REV7 are significantly more sensitive to cisplatin or mitomycin C as compared to those depleted of RAD18 or Polη (Hicks et al. 2010). This hypersensitivity correlates with a defect in the resolution of replication-associated DSBs and the accumulation of chromosomal aberrations following drug treatment. These deficiencies in resolving ICL-associated DSBs and preventing clastogenic effects associated with interstrand crosslinking agents were very similar to cells lacking RAD51, which is essential for HR repair, or the many other factors associated with ICL repair. Although PCNA monoubiquitination by RAD18 is important for localization of both Polη and REV1 to facilitate bypass of cisplatin intrastrand croslinks, the absence of RAD18 failed to duplicate the deficiencies in ICL observed in cells lacking REV1 or Polζ suggesting an alternative mechanism exists that promotes TLS during ICL repair, at least in the cancer lines examined (Hicks et al. 2010). It is important to note here that agents that cause non-distorting interstrand crosslinks (e.g. mitomycin C) or create DSBs in DNA (ionizing radiation or topoisomerase poisons) are not potent inducers of PCNA monoubiquitination per se (Shiomi et al. 2006; Niimi et al. 2008; Brown et al. 2009; Hicks et al. 2010). However, RAD18 may play additional roles in regulating the Fanconi anemia pathway and ICL repair – by facilitating the activation of the Fanconi anemia FANCD2/FANCI heterodimer (Geng et al. 2010; Song et al. 2010; Palle and Vaziri 2011; Williams et al. 2011).
Xenopus laevis extracts have been an extraordinary useful system for studying the biochemistry of DNA replication and recently, ICL repair of a single cisplatin ICL placed in a plasmid capable of undergoing replication. The landmark work by Johannes Walter provided unequivocal evidence that Polζ (as inferred by immunodepletion of Rev7) plays a crucial role in performing TLS across an unhooked crosslink after convergence of two replication forks upon the ICL (Raschle et al. 2008). The model proposed from these studies involves incision of the ICL on both sides of the sister chromatid generating a two ended DSB which is unique from previous models suggesting the generation of a one ended DSB. In order for HR repair to proceed, the collaborative actions of Rev1 and Polζ are needed to insert a nucleotide opposite the unhooked ICL followed by extension to restore the other sister chromatid. Interestingly, recent biochemical studies have characterized Polζ as being inefficient at mediating TLS across artificial ‘unhooked’ crosslinks in vitro (Ho et al. 2011).
The FA pathway: regulator of REV1 and Polζ
At least 16 FA or FA-interacting genes have been identified and these ‘FA’ proteins have been shown to form several complexes to orchestrate ICL repair (Moldovan and D'Andrea 2009; Deans and West 2011; Kim et al. 2012). Eight of these proteins (FANCA, -B, -C, -E, -F, -G, -L and -M) along with five FA-associated proteins (FAAP100, FAAP24, HES1, MHF1, MHF2 and the newly identified FAAP20) form the nuclear core complex (Gurtan and D'Andrea 2006; Huang et al. 2010; Kee and D'Andrea 2010; Ali et al. 2012; Constantinou 2012; Kim et al. 2012; Leung et al. 2012). The core complex functions as an E3 ubiquitin ligase by monoubiquitinating the FANCD2-FANCI heterodimer in response to DNA damage (Garcia-Higuera et al. 2001; Dorsman et al. 2007; Sims et al. 2007; Smogorzewska et al. 2007). Monoubiquitination takes place constitutively in the S-phase and is dramatically increased upon DNA damage (Garcia-Higuera et al. 2001; Taniguchi et al. 2002). Upon damage, the monoubiquitinated FANCD2-I complex is then recruited to the chromatin where it interacts with downstream FA proteins [FANCD1/BRCA2, FANCJ/BACH1, FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4 along with the FA-associated nuclease 1 (FAN1)] that are all involved in promoting HR repair (Howlett et al. 2002; Litman et al. 2005; Reid et al. 2007; Xia et al. 2007; Shen et al. 2009; Liu et al. 2010; Smogorzewska et al. 2010; Vaz et al. 2010; Crossan et al. 2011; Stoepker et al. 2011). Monoubiquitinated FANCD2 also provides binding sites for SLX4 and FAN1, which are structure specific endonucleases that play roles in unhooking the ICL, HR repair, and potentially facilitating TLS (Liu et al. 2010; Cybulski and Howlett 2011; Sengerova et al. 2011; Yamamoto et al. 2011). Deficiencies in any of these proteins results in the characteristic deficiency in DSB resolution following exposure to DNA crosslinking agents, as well as reductions in gene conversion efficiencies, identical to what has have observed in REV1 or Polζ-depleted cells (Hicks et al. 2010; Sharma et al. 2012a).
Ketan Patel first demonstrated the epistatic relationship between the FANCC gene, which encodes one of the fanconi anemia core proteins, and REV1 or REV3 in chicken DT40 cells treated with cisplatin thus directly implicating REV1 and REV3 in ICL repair (Niedzwiedz et al. 2004). The authors found little difference in the sensitivities of FANCC −/− DT40 cells compared to FANCC−/− REV1 −/− cells or FANCC−/− REV3 −/− cells to loss in viability or the accumulation of chromosomal aberrations following cisplatin treatment thus establishing an epistatic relationship between these genes. Sonoda’s laboratory confirmed these observations (Nojima et al. 2005). The direct evidence regarding the involvement of the FA pathway in ICL repair was once again obtained using the Xenopus leavis egg extract system, wherein it was shown FANCD2-FANCI ubiquitination promotes the incision and TLS steps of ICL repair (Knipscheer et al. 2009). This work led to the proposal that like PCNA ubiquitination, FANCD2-I ubiquitination plays a role in recruiting TLS polymerases to ICLs via their UBM/UBZ domains. Also, a direct interaction between the putative PIP domain of FANCD2 and PCNA has been demonstrated (Howlett et al. 2009). However to date, no evidence exists that suggest a direct interaction between FANCD2 and REV1. Regardless, these studies brought forth the most current model for ICL repair that involves the cooperation between the FA pathway, translesion DNA synthesis by REV1 and Polζ, and homologous recombination.
The hallmark of FA patient cells, as well as cells deficient in the breast cancer and HR repair associated-proteins BRCA1 or BRCA2, or the TLS polymerases REV1 and Polζ, is hypersensitivity to the clastogenic effects of DNA interstrand crosslinking agents such as cisplatin and mitomycin C (Garcia-Higuera et al. 2001; Howlett et al. 2002; Venkitaraman 2004). The formation of aberrant radial chromosomes in patient cells treated with DNA crosslinking agents is a common diagnostic test for fanconi anemia and these specific aberrations have been identified in REV1-depleted mammalian cells (Mirchandani et al. 2008; Auerbach 2009). Given the fact that the formation of radial chromosomes has not been identified in cells deficient in other TLS polymerases treated with ICL-generating agents (the exception being DNA polymerase nu and kappa (Minko et al. 2008; Moldovan et al. 2010), underscores the importance of REV1 and Polζ in ICL repair.
Recent work from the D’Andrea group identified a novel FA core component – FAAP20 – which directly binds to FANCA, while the C-terminal UBZ4 domain of FAAP20 directly binds to REV1, thereby providing a functional link between the two pathways (Kim et al. 2012). Although these studies suggest that FAAP20 specifically promotes REV1-dependent TLS across replication stalling lesions like UV-induced thymine dimers, FAAP20 also plays an important role in facilitating ICL repair by facilitating FANCD2/FANCI monoubiquitination and activation of the ICL repair pathway (Ali et al. 2012; Leung et al. 2012). These studies support the idea that FAAP20 may directly promote REV1-dependent TLS during ICL repair. A model for mammalian ICL repair is presented in Figure 2.
Do REV1 and Polζ act alone during ICL repair?
DNA polymerase kappa
DNA polymerase kappa, Polκ, a homologue of the E. coli DinB (DNA Pol IV) gene found in higher eukaryotes, belongs to the Y-family of DNA polymerases (Ogi et al. 1999; Johnson et al. 2000a; Gerlach et al. 2001; Ohmori et al. 2001). Polκ is known to be involved in error-free bypass of bulky minor groove N2-deoxyguanine adducts and plays a critical role in limiting mutagenesis from these lesions (Ogi et al. 2002; Zhang et al. 2002; Avkin et al. 2004; Choi et al. 2006; Jarosz et al. 2006; Temviriyanukul et al. 2012). Polκ has also been shown to process N2-N2 guanine minor groove ICLs and the efficiency of this bypass increases with the shortening of non-template crosslinked strand (Minko et al. 2008). It has also been demonstrated in vitro that error-free bypass of a model acrolein-mediated N(2)-dG ICL involves the simultaneous action of two polymerases, wherein REV1 contributes to the bypass by inserting dC opposite the cross-linked dG while primer extension is performed by Polκ (Klug et al. 2012). Supporting a role in ICL repair, siRNA-mediated depletion of Polκ in GM639 cells led to increased hypersensitivity and increased chromosomal damage in the form of radial formation upon treatment with MMC (Minko et al. 2008), a characteristic phenotype of Fanconi anemia patient cells. On the other hand, Polκ-deficient chicken DT40 cells display similar sensitivity to cisplatin as the wild type cells (Nojima et al. 2005). Recently, a role for Polκ in performing replication-independent repair of a helix-distorting trimethylene ICL has been described in Xenopus egg extracts and efficient repair can be measured in the absence of Polζ (Williams et al. 2012). As previously mentioned, replication-independent repair of a cisplatin ICL in mammalian cells requires REV1 and Polζ, whereas Polκ deficient cells show a minor defect in the repair of this specific lesion (Enoiu et al. 2012). Additional studies are needed to understand whether these differences in results can be explained by the type of ICL lesion being repaired or the cellular context (or model system) being examined, or whether Polκ collaborates with REV1 and Polζ during ICL repair in resting cells.
DNA polymerase theta and nu
Genetic studies performed in Drosopila melanogaster identified the mus308 gene as playing an important role in the resistance to DNA crosslinking agents such as nitrogen mustards and cisplatin (Boyd et al. 1981; Boyd et al. 1990). Mus308 is composed of both an N-terminal helicase-like domain and a C-terminal A-family polymerase domain (Harris et al. 1996; Oshige et al. 1999; Pang et al. 2005). Polθ is the closest relative of mus308 in vertebrates and possesses both the C-terminal A-family DNA polymerase domain and the N-terminal helicase-like domain (Sharief et al. 1999; Seki et al. 2003). POLθ is a low fidelity DNA polymerase capable of efficient bypass of abasic sites and thymine glycols as well as extension of mismatched primer termini (Seki et al. 2004; Arana et al. 2008; Seki and Wood 2008; Hogg et al. 2011; Hogg et al. 2012). Experiments in avian DT40 cells have shown that the deletion of POLθ alone or in combination POLN or HEL308 does not convey hypersensitivity to DNA crosslinking agents (Yoshimura et al. 2006). However, POLθ-deficient cells display spontaneous, radiation and Mitomycin C- induced chromosomal abnormalities implicating a possible role of POLθ in ICL or HR repair (Shima et al. 2003; Shima et al. 2004; Goff et al. 2009). Studies in Caenorhabditis elegans have also identified a role for POLQ-1 (a POLθ homolog) in ICL repair (Muzzini et al. 2008). In addition to a role in ICL repair, POLθ also promotes base excision repair of oxidative damage (Yoshimura et al. 2006) and somatic hypermutation (Masuda et al. 2005; Seki et al. 2005; Masuda et al. 2007; Martomo et al. 2008; Kohzaki et al. 2010).
Polν (POLN) and HEL308 (HELQ) are smaller Drosophila mus308 gene homologs in vertebrates and separately make up the polymerase and the helicase-like domain, respectively, of mus308. HEL308 displays 3’ → 5’ helicase activity (Marini and Wood 2002) whereas POLν is a low fidelity A-family DNA polymerase that generates a high error rate when incorporating nucleotides opposite dG (Marini and Wood 2002) (Marini et al. 2003). Polν is capable of DNA-templated synthesis and can bypass thymine glycols, a product of oxidative stress, with a high fidelity in vitro, (Takata et al. 2006). A recent report showed that Polν can also perform efficient bypass of major groove ICLs with the linkage between N6-dAs in complementary DNA strands. On the other hand, Polν was blocked when the lesions were located in the minor groove via a N2-dG linkage, thus implying that the ability of Polν to bypass ICLs is structure dependent (Yamanaka et al. 2010). In vitro experiments also show that purified Polν conducts low efficiency non-mutagenic bypass of psoralen ICLs (Zietlow et al. 2009).
The most convincing evidence suggesting an important role for Polν in ICL repair were obtained analyzing the effects of Polν-depleted in human cells. HeLa cells deficient in Polν display increased sensitivity to MMC and cisplatin (Zietlow et al. 2009; Moldovan et al. 2010). Furthermore, Alan D’Andrea’s group identified HEL308 as a Polν-interacting protein and demonstrated that both of these proteins share an epistatic relationship with FANCD2 strongly suggesting that these proteins cooperate to promote ICL and HR repair (Moldovan et al. 2010). Similar to POLν, HEL308 also plays a role in ICL repair in C. elegans (Muzzini et al. 2008). However, like POLθ, DT40 cells lacking POLν do not display hypersensitivity to cisplatin or mitomycin C treatment (Yoshimura et al. 2006). These results indicate that important differences exist between DT40 and human cells in the roles TLS polymerases play in promoting genomic stability, underscoring the importance of studying these pathways in both model systems.
Translesion DNA synthesis: preparing the template for HR repair or participating in HR repair
The identification of BRCA2 and RAD51C mutations causing fanconi anemia-like syndromes provided a direct link between the FA pathway and regulation of homologous recombination repair (Howlett et al. 2002; Vaz et al. 2010). Other members of the FA pathway have also been implicated in regulating or participating in HR repair (Niedzwiedz et al. 2004; Nakanishi et al. 2005; Yamamoto et al. 2005; Smogorzewska et al. 2007; Zhang et al. 2007; Kratz et al. 2010; Pace et al. 2010). Deficiencies in the FA pathway are typically associated with partial defects in HR repair, the exception being BRCA2 deficiency since this protein is essential for RAD51 loading. We have also discovered that depletion of REV1, REV3, or REV7 is associated with a reduction in HR efficiency by approximately 50%, similar to FA cells, as opposed to 90–95% seen in cells deficient in the RAD51 protein (Sharma et al. 2012a). This same reduction in HR repair is observed in cells depleted of Polν or HEL308 and no additive reductions in gene conversion efficiencies are observed when FANCD2 co-depleted along with Polν, suggesting that Polν participates in the FA pathway regulating HR repair (Moldovan et al. 2010). Since these studies are measuring repair of a site specific DSB by HR, they imply that alternative DNA polymerases may be needed to perform a subset of these reactions and are not specifically confined to preparing the sister chromatid for HR repair after ICL unhooking. This idea has been recently confirmed in genetic studies using Drosophila melanogaster as a model system, demonstrating a specific requirement for Polζ during HR repair (Kane et al. 2012). Together, these observations suggest that REV1 and Polζ (possibly in collaboration with Polν) may be required to synthesize DNA during a subset of HR reactions that involve extension of distorted or mispaired primer templates that would otherwise cause stalling of normal DNA polymerases. At least in mammalian cells, the Fanconi anemia pathway may be important for regulating TLS during HR repair.
Conclusion
Both REV1 and Polζ have been implicated in promoting DSB repair and genomic stability (Sonoda et al. 2003; Okada et al. 2005; Wittschieben et al. 2006; Schenten et al. 2009; Sharma et al. 2012a). The results from these studies at least partially explain why REV3 is required for embryogenesis in mice, in addition to the apparently nonredundant function of REV3 as a TLS polymerase promoting replication across a multitude of DNA lesions, including those associated with oxydative stress (Bemark et al. 2000; Wittschieben et al. 2000; Wang et al. 2002; Lange et al. 2012). It is becoming increasingly clear that Polζ contributes to tumor suppression and is essential for cell proliferation of nontransformed mouse embryonic fibroblasts (Wittschieben et al. 2010; Lange et al. 2012). On the other hand, cultured human cancer cell lines appear to be far more sensitive to REV3 depletion compared to ‘normal’ cells, as measured by increased formation of DSBs and loss in clonogenic survival in the absence of treatment with genotoxic agents (Knobel et al. 2011). These are intriguing observations in that they suggest that inhibiting REV3 could be a viable approach to selectively killing tumor cells.
The importance of REV1 and Polζ in promoting resistance to DNA crosslinking agents has important clinical implications. The emergence of drug resistance to cisplatin and cyclophosphamide is linked to the activities of REV3 and REV1 in murine models of B-cell lymphoma and lung adenocarcinoma (Doles 2010; Xie et al. 2010). Rendering tumor cells REV1 or REV3-deficient using shRNA significantly sensitizes these tumors to treatment and limited the emergence of drug resistance, the latter thought to be attributed to mutagenic bypass of DNA crosslinks. Taken together, these data suggest that inhibition of REV1 or Polζ may have dual anti-cancer effects – sensitizing tumors to therapy and preventing the emergence of chemoresistance by limiting drug-induced mutagenesis. Before REV1 or Polζ can be considered as targets alone or for adjuvant therapy with crosslinking agents, the additional roles characterized for REV1 and Polζ in maintaining genomic stability will need to be better understood.
Acknowledgments
Grant Sponsor: National Cancer Institute; Grant Number CA133046
References
- Acharya N, Johnson R, Pages V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc Natl Acad Sci USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Johnson RE, Prakash S, Prakash L. Complex Formation with Rev1 Enhances the Proficiency of Saccharomyces cerevisiae DNA Polymerase ζ for Mismatch Extension and for Extension Opposite from DNA Lesions. Mol Cell Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkari YMN, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA Replication Is Required To Elicit Cellular Responses to Psoralen-Induced DNA Interstrand Cross-Links. Mol Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, Grassman E, Auerbach AD, Pang Q, Meetei AR. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119:3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- Andersson BS, Sadeghi T, Siciliano MJ, Legerski R, Murray D. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer Chemother Pharmacol. 1996;38:406–416. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA Repair Proteins Rad6 and Rad18 Form a Heterodimer That Has Ubiquitin Conjugating, DNA Binding, and ATP Hydrolytic Activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- Barre FX, Asseline U, Harel-Bellan A. Asymmetric recognition of psoralen interstrand crosslinks by the nucleotide excision repair and the error-prone repair pathways. J Mol Biol. 1999;286:1379–1387. doi: 10.1006/jmbi.1999.2550. [DOI] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase ζ ( Rev3 ) leads to embryonic lethality and impairs blastocyst development in vitro Curr Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-Binding Domains in Y-Family Polymerases Regulate Translesion Synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bomar MG, D'Souza S, Bienko M, Dikic I, Walker GC, Zhou P. Unconventional ubiquitin recognition by the ubiquitin-binding motif within the Y family DNA polymerases iota and Rev1. Mol Cell. 2010;37:408–417. doi: 10.1016/j.molcel.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JB, Golino MD, Shaw KE, Osgood CJ, Green MM. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics. 1981;97:607–623. doi: 10.1093/genetics/97.3-4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JB, Sakaguchi K, Harris PV. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc Natl Acad Sci USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Niimi A, Lehmann AR. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle. 2009;8:689–692. doi: 10.4161/cc.8.5.7707. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Xie J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environ Mol Mutagen. 2010;51:500–507. doi: 10.1002/em.20568. [DOI] [PubMed] [Google Scholar]
- Cassier C, Moustacchi E. Mutagenesis induced by mono- and bi-functional alkylating agents in yeast mutants sensitive to photo-addition of furocoumarins (pso) Mutat Res. 1981;84:37–47. doi: 10.1016/0027-5107(81)90048-8. [DOI] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Sancar A, Hearst JE. RecA-dependent incision of psoralen-crosslinked DNA by (A)BC exonuclease. Nucleic Acids Res. 1991;19:657–663. doi: 10.1093/nar/19.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Vanhouten B, Gamper HB, Sancar A, Hearst JE. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC exonuclease. J Biol Chem. 1988;263:15110–15117. [PubMed] [Google Scholar]
- Cheung HW, Chun ACS, Wang Q, Deng W, Hu L, Guan XY, Nicholls JM, Ling MT, Chuan Wong Y, Wah Tsao S, Jin DY, Wang X. Inactivation of Human MAD2B in Nasopharyngeal Carcinoma Cells Leads to Chemosensitization to DNA-Damaging Agents. Cancer Res. 2006;66:4357–4367. doi: 10.1158/0008-5472.CAN-05-3602. [DOI] [PubMed] [Google Scholar]
- Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- Clingen PH, De Silva IU, McHugh PJ, Ghadessy FJ, Tilby MJ, Thurston DE, Hartley JA. The XPF-ERCC1 endonuclease and homologous recombination contribute to the repair of minor groove DNA interstrand crosslinks in mammalian cells produced by the pyrrolo[2,1-c][1,4]benzodiazepine dimer SJG-136. Nucleic Acids Res. 2005;33:3283–3291. doi: 10.1093/nar/gki639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RS. Repair of DNA Containing Interstrand Crosslinks in Escherichia coli: Sequential Excision and Recombination. Proc Natl Acad Sci USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RS, Levitan D, Sinden RR. Removal of psoralen interstrand cross-links from DNA of Escherichia coli: Mechanism and genetic control. J Mol Biol. 1976;103:39–59. doi: 10.1016/0022-2836(76)90051-6. [DOI] [PubMed] [Google Scholar]
- Constantinou A. Rescue of replication failure by Fanconi anaemia proteins. Chromosoma. 2012;121:21–36. doi: 10.1007/s00412-011-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard PH, McIntyre RE, Gallagher F, Kettunen MI, Lewis DY, Brindle K, Arends MJ, Adams DJ, Patel KJ. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski KE, Howlett NG. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle. 2011;10:1757–1763. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza S, Walker GC. Novel Role for the C Terminus of Saccharomyces cerevisiae Rev1 in Mediating Protein-Protein Interactions. Mol Cell Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of Ubiquitin-Dependent DNA Damage Bypass Is Mediated by Replication Protein A. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NGJ, Hoeijmakers JHJ. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the Roles of Nucleotide Excision Repair and Recombination in the Repair of DNA Interstrand Cross-Links in Mammalian Cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant N, Hendel A, Vered I, Carell T, Reiβner T, de Wind N, Geacinov N, Livneh Z. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012;40:170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D, Mohan S, Schindler D, Arwert F, Pals G, Mathew CG, Waisfisz Q, de Winter JP, Joenje H. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE. PCNA Ubiquitination and REV1 Define Temporally Distinct Mechanisms for Controlling Translesion Synthesis in the Avian Cell Line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Enoiu M, Jiricny J, Schärer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res Jul 18. 2012 doi: 10.1093/nar/gks670. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR, Iii, Russell P, Fuchs RP, McGowan CH, Gaillard P-HL. Human SLX4 Is a Holliday Junction Resolvase Subunit that Binds Multiple DNA Repair/Recombination Endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi Anemia Proteins and BRCA1 in a Common Pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Geng LY, Huntoon CJ, Karnitz LM. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol. 2010;191:249–257. doi: 10.1083/jcb.201005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- Gibbs PEM, McDonald J, Woodgate R, Lawrence CW. The Relative Roles in Vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 Protein and Pol32 in the Bypass and Mutation Induction of an Abasic Site, T-T (6-4) Photoadduct and T-T cis-syn Cyclobutane Dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-Binding Motifs in REV1 Protein Are Required for Its Role in the Tolerance of DNA Damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtan AM, D'Andrea AD. Dedicated to the core: understanding the Fanconi anemia complex. DNA Repair. 2006;5:1119–1125. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strand breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M. Crystal Structure of Human REV7 in Complex with a Human REV3 Fragment and Structural Implication of the Interaction between DNA Polymerase ζ and REV1. J Biol Chem. 2010;285:12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, Myung K, Tateishi S, D'Andrea A, Jacobs H, Livneh Z. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques JAP, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980;95:273–288. doi: 10.1093/genetics/95.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques JAP, Moustacchi E. Interactions between mutations for sensitivity to psoralen photoaddition (pso) and to radiation (rad) in Saccharomyces cerevisiae. J Bacteriol. 1981;148:248–256. doi: 10.1128/jb.148.1.248-256.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential Roles for DNA Polymerases Eta, Zeta, and REV1 in Lesion Bypass of Intrastrand versus Interstrand DNA Cross-Links. Mol Cell Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- Hlavin EM, Smeaton MB, Miller PS. Initiation of DNA interstrand cross-link repair in mammalian cells. Environ Mol Mutagen. 2010;51:604–624. doi: 10.1002/em.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schärer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Schärer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Harney JA, Rego MA, Kolling FWt, Glover TW. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem. 2009;284:28935–28942. doi: 10.1074/jbc.M109.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA Cross-Link Removal in Chinese Hamster Cell Mutants Hypersensitive to Bifunctional Alkylating Agents. Cancer Res. 1985;45:1737–1743. [PubMed] [Google Scholar]
- Huang M-E, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D'Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachymczyk WJ, Borstel RC, Mowat MRA, Hastings PJ. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: The RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182:196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LHF, Haracska L, de Wind N. Separate Domains of Rev1 Mediate Two Modes of DNA Damage Bypass in Mammalian Cells. Mol Cell Biol. 2009a;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N. Mammalian polymerase ζ is essential for post-replication repair of UV-induced DNA lesions. DNA Repair. 2009b;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc Natl Acad Sci USA. 2000a;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000b;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta ( ζ ) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DP, Shusterman M, Rong Y, McVey M. Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila. PLoS Genet. 2012;8:e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Crystallization and X-ray diffraction analysis of the ternary complex of the C-terminal domain of human REV1 in complex with REV7 bound to a REV3 fragment involved in translesion DNA synthesis. Acta crystallographica Section F, Structural biology and crystallization communications. 2012a;68:962–964. doi: 10.1107/S1744309112032435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase zeta by interaction between REV1 and REV7. J Biol Chem Aug 2. 2012b doi: 10.1074/jbc.M112.396838. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D'Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases eta and iota. Chem Res Toxicol. 2012;25:755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel P, Kotov I, Felley-Bosco E, Stahel R, Marti T. Inhibition of REV3 Expression Induces Persistent DNA Damage and Growth Arrest in Cancer Cells. Neoplasia. 2011;13:961–970. doi: 10.1593/neo.11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki M, Nishihara K, Hirota K, Sonoda E, Yoshimura M, Ekino S, Butler JE, Watanabe M, Halazonetis TD, Takeda S. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Lange SS, Wittschieben JP, Wood RD. DNA polymerase zeta is required for proliferation of normal mammalian cells. Nucleic Acids Res. 2012;40:4473–4482. doi: 10.1093/nar/gks054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C, Das G, Christensen R. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci USA. 2012;109:4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Okuda T, Trang J, Howell SB. Human REV1 Modulates the Cytotoxicity and Mutagenicity of Cisplatin in Human Ovarian Carcinoma Cells. Mol Pharmacol. 2006;69:1748–1754. doi: 10.1124/mol.105.020446. [DOI] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 Acts with FANCI-FANCD2 to Promote DNA Interstrand Cross-Link Repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple Mechanisms Control Chromosome Integrity after Replication Fork Uncoupling and Restart at Irreparable UV Lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- MacKay C, Déclais A-C, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DMJ, Rouse J. Identification of KIAA1018/FAN1, a DNA Repair Nuclease Recruited to DNA Damage by Monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- Marini F, Wood RD. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2002;277:8716–8723. doi: 10.1074/jbc.M110271200. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair. 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, J OW. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and Enzymatic Properties of a Stable Complex of the Human REV1 and REV7 Proteins. J Biol Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:3425–3433. doi: 10.1128/mcb.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- McVey M. Strategies for DNA interstrand crosslink repair: Insights from worms, flies, frogs, and slime molds. Environ Mol Mutagen. 2010;51:646–658. doi: 10.1002/em.20551. [DOI] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, McCaffrey RM, D'Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair. 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G-L, D'Andrea AD. How the Fanconi Anemia Pathway Guards the Genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy PA, Thapa D, Thazhathveetil AK, Liu S-t, Seidman MM. Repair of Laser-localized DNA Interstrand Cross-links in G1 Phase Mammalian Cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais A-C, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DMJ, Rouse J. Coordination of Structure-Specific Nucleases by Human SLX4/BTBD12 Is Required for DNA Repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Roth T, Ishii H, Rasio D, Numata Si, Croce CM, Fishel R. A Human REV7 Homolog That Interacts with the Polymerase zeta Catalytic Subunit hREV3 and the Spindle Assembly Checkpoint Protein hMAD2. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair. 2008;7:941–950. doi: 10.1016/j.dnarep.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 Employs a Novel Mechanism of DNA Synthesis Using a Protein Template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996a;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine-Thymine Dimer Bypass by Yeast DNA Polymerase zeta. Science. 1996b;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ. The Fanconi Anemia Signalosome Anchor. Mol Cell. 2007;25:487–490. doi: 10.1016/j.molcel.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NGJ, Beverloo HB, Hoeijmakers JHJ, Kanaar R. The Structure-Specific Endonuclease Ercc1-Xpf Is Required To Resolve DNA Interstrand Cross-Link-Induced Double-Strand Breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple Repair Pathways Mediate Tolerance to Chemotherapeutic Cross-linking Agents in Vertebrate Cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Ogi T, Kato T, Jr, Kato T, Ohmori H. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells. 1999;4:607–618. doi: 10.1046/j.1365-2443.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple Roles of Vertebrate REV Genes in DNA Repair and Recombination. Mol Cell Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshige M, Aoyagi N, Harris PV, Burtis KC, Sakaguchi K. A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat Res. 1999;433:183–192. doi: 10.1016/s0921-8777(99)00005-1. [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 Corrupts DNA Repair in the Absence of the Fanconi Anemia Pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Palle K, Vaziri C. Rad18 E3 ubiquitin ligase activity mediates fanconi anemia pathway activation and cell survival following DNA topoisomerase 1 inhibition. Cell Cycle. 2011;10:1625–1638. doi: 10.4161/cc.10.10.15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, McConnell M, Fisher PA. The Drosophila mus 308 gene product, implicated in tolerance of DNA interstrand crosslinks, is a nuclear protein found in both ovaries and embryos. DNA Repair. 2005;4:971–982. doi: 10.1016/j.dnarep.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozhidaeva A, Pustovalova Y, D'Souza S, Bezsonova I, Walker GC, Korzhnev DM. NMR Structure and Dynamics of the C-Terminal Domain from Human Rev1 and Its Complex with Rev1 Interacting Region of DNA Polymerase η. Biochemistry. 2012;51:5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish SD, Lach FP, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew CG, Auerbach AD, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Roos WP, Tsaalbi-Shtylik A, Tsaryk R, Güvercin F, de Wind N, Kaina B. The Translesion Polymerase Rev3L in the Tolerance of Alkylating Anticancer Drugs. Mol Pharmacol. 2009;76:927–934. doi: 10.1124/mol.109.058131. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair Kinetics of Genomic Interstrand DNA Cross-Links: Evidence for DNA Double-Strand Break-Dependent Activation of the Fanconi Anemia/BRCA Pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda S, Gourdin AM, Green CM, Zotter A, Giglia-Mari G, Houtsmuller A, Vermeulen W, Lehmann AR. Effect of Proliferating Cell Nuclear Antigen Ubiquitination and Chromatin Structure on the Dynamic Properties of the Y-family DNA Polymerases. Mol Biol Cell. 2008;19:5193–5202. doi: 10.1091/mbc.E08-07-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, Rajewsky K. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair. 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengerova B, Wang AT, McHugh PJ. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011;10:3999–4008. doi: 10.4161/cc.10.23.18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reiszner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn J-Y, Glover TW, Canman CE. REV1 and polymerase ζ facilitate homologous recombination repair. Nucleic Acids Res. 2012a;40:682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Shah NA, Joiner AM, Roberts KH, Canman CE. DNA Polymerase ζ Is a Major Determinant of Resistance to Platinum-Based Chemotherapeutic Agents. Mol Pharmacol. 2012b;81:778–787. doi: 10.1124/mol.111.076828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, Wang W, Li L. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Jun S, O'Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 Play Major Roles in Recombination-independent Repair of DNA Interstrand Cross-links Mediated by Monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) J Biol Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- Shen X, Li L. Mutagenic Repair of DNA Interstrand Crosslinks. Environ Mol Mutagen. 2010;51:493–499. doi: 10.1002/em.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2006;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Cole RS. Repair of cross-linked DNA and survival of Escherichia coli treated with psoralen and light: effects of mutations influencing genetic recombination and DNA metabolism. J Bacteriol. 1978a;136:538–547. doi: 10.1128/jb.136.2.538-547.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Cole RS. Topography and kinetics of genetic recombination in Escherichia coli treated with psoralen and light. Proc Natl Acad Sci USA. 1978b;75:2373–2377. doi: 10.1073/pnas.75.5.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek FM, Munn MM, Rupp WD, Howardflanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5'-exonuclease of DNA polymerase I. J Biol Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated Translesion Synthesis of Bulky DNA Adducts Is Coupled to Activation of the Fanconi Anemia DNA Repair Pathway. J Biol Chem. 2010;285:31525–31536. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van GDC, Takata M, Takeda S. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW, Jaspers NG, Bettecken T, Joenje H, Schindler D, Rouse J, de Winter JP. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 Assembles A Holliday Junction Resolvase and Is Required for DNA Repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szüts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S–thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor J-S, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Temviriyanukul P, Meijers M, van Hees-Stuivenberg S, Boei JJ, Delbos F, Ohmori H, de Wind N, Jansen JG. Different sets of translesion synthesis DNA polymerases protect from genome instability induced by distinct food-derived genotoxins. Toxicol Sci. 2012;127:130–138. doi: 10.1093/toxsci/kfs074. [DOI] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck M-P, Lehmann AR, Fuchs RPP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase polη and REVl protein. DNA Repair. 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Vasquez KM. Targeting and processing of site-specific DNA interstrand crosslinks. Environ Mol Mutagen. 2010;51:527–539. doi: 10.1002/em.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Tracing the network connecting brca and fanconi anaemia proteins. Nat Rev Cancer. 2004;4:266–276. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- Wang AT, Sengerova B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, Gileadi O, Hartley JA, McHugh PJ. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25:1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang S-Y, Wang S, Lu J, Wu W, Weng L, Chen D, Zhang Y, Lu Z, Yang J, Chen Y, Zhang X, Chen X, Xi C, Lu D, Zhao S. REV3L confers chemoresistance to cisplatin in human gliomas: The potential of its RNAi for synergistic therapy. Neuro-Oncol. 2009;11:790–802. doi: 10.1215/15228517-2009-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kajiwara K, Kawamura K, Kimura M, Miyagishima H, Koseki H, Tagawa M. An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem Biophys Res Commun. 2002;293:1132–1137. doi: 10.1016/S0006-291X(02)00341-8. [DOI] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng HY, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic Translesion Polymerases and Their Roles and Regulation in DNA Damage Tolerance. Microbiol Mol Biol R. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Hannah L, Gottesman Max E, Gautier J. Replication-Independent Repair of DNA Interstrand Crosslinks. Mol Cell. 2012;47:140–147. doi: 10.1016/j.molcel.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood. 2011;117:5078–5087. doi: 10.1182/blood-2010-10-311761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MKK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. Loss of DNA Polymerase ζ Enhances Spontaneous Tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA Polymerase ζ Causes Chromosomal Instability in Mammalian Cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, D'Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa. J Biol Chem Aug 2. 2012a doi: 10.1074/jbc.M112.394841. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J, Liu J, D'Souza S, Wang S, Xue Y, Walker GC, Zhou P. Multifaceted recognition of vertebrate Rev1 by translesion polymerases ζ and κ. J Biol Chem. 2012b;287:26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. Mammalian Nucleotide Excision Repair Proteins and Interstrand Crosslink Repair. Environ Mol Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Lin X, Okuda T, Howell SB. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–8035. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, Buerstedde JM, Komatsu K, Thompson LH, Takata M. Fanconi Anemia Protein FANCD2 Promotes Immunoglobulin Gene Conversion and DNA Repair through a Mechanism Related to Homologous Recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci USA. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan G-L, D'Andrea AD. RAD18-dependent Recruitment of SNM1A to DNA Repair Complexes by a Ubiquitin-binding Zinc Finger. J Biol Chem. 2010;285:19085–19091. doi: 10.1074/jbc.M109.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander L, Bemark M. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair. 2004;3:743–752. doi: 10.1016/j.dnarep.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair. 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]