Abstract
Uptake of organic solutes and xenobiotics by mammalian cells is mediated by ATP-independent transporters, and four families of transporters have now been identified. To search for novel organic solute transporters, a liver cDNA library from an evolutionarily primitive marine vertebrate, the little skate Raja erinacea, was screened for taurocholate transport activity by using Xenopus laevis oocytes. In contrast to the organic anion transporters identified to date, a transport activity was identified in this library that required the coexpression of two distinct gene products, termed organic solute transporter α and β (Ostα, Ostβ). Ostα cDNA encodes for a protein of 352 aa and seven putative transmembrane (TM) domains. Ostβ contains 182 aa and has at least one and perhaps two TM domains. There is no significant sequence identity between Ostα and Ostβ, and only low identity with sequences in the databases; however, Ostα bears a resemblance to some G protein-coupled receptors, and Ostβ exhibits 22% amino acid identity with the C-terminal TM and intracellular domains of protocadherin-γ, a cell surface glycoprotein. Xenopus oocytes injected with the cRNA for both Ostα and Ostβ, but not each separately, were able to take up taurocholate, estrone sulfate, digoxin, and prostaglandin E2, but not p-aminohippurate or S-dinitrophenyl glutathione. Transport was sodium-independent, saturable, and inhibited by organic anions and steroids, including the major skate bile salt, scymnol sulfate. These results identify an organic anion transporter composed of a putative seven-helix TM protein and an ancillary membrane polypeptide.
Epithelial cells continuously extract large amounts of organic solutes, drugs, and other xenobiotics from circulating blood plasma. Some of the transporters responsible for this drug clearance recently have been characterized at the molecular level and include four families of ATP-independent transporters: the Na+-coupled bile acid transporters (NTCPs), the Na+-independent organic anion transporting polypeptides (OATPs), the organic anion transporters (OATs), and the organic cation transporters (OCTs) (1–5). The first member of each of these families was identified by expression cloning in Xenopus laevis oocytes, and additional members subsequently have been identified by homology screening. All of these transporters consist of single polypeptides, which, when expressed in heterologous systems, are able to mediate organic solute transport.
Because of the large number of endogenous and exogenous compounds that must be transported by the liver, kidney, intestine, and other tissues, it is likely that other transporters and transporter families also exist, but have not yet been described (5). In an attempt to identify novel organic anion transport proteins we used a comparative approach, screening a liver cDNA library from an evolutionarily ancient vertebrate species, the little skate Raja erinacea. This elasmobranch is thought to have evolved 200 million years ago, yet displays many physiological features of modern mammals. Of direct relevance to the present study, the livers of all vertebrates, including the skate, function as the primary site of clearance and metabolism of endogenous and exogenous lipophilic organic substances such as bile salts, steroids, eicosanoids, and natural toxins (6–11). We recently have identified the skate orthologue of the bile salt export pump (12), as well as putative members of the skate OATP, multidrug resistance protein, and multidrug resistance-associated protein families (unpublished observations).
To identify yet additional organic solute transporters (Osts) in skate liver we used the expression cloning strategy in Xenopus oocytes (13) and screened for bile salt ([3H]taurocholate) transport activity. As described in the present report, during the screening of various cRNA pools from a skate liver cDNA library it was noted that some of the cRNA-stimulated transport activity was not stable and was lost when positive pools of clones were divided into progressively smaller groups of clones. We hypothesized that this transport activity may be mediated by a polygenic transporter (e.g., a heteromultimeric protein complex), and therefore initiated studies to identify the two or more gene products that may be interacting to generate the active transport system. After multiple rounds of screening in Xenopus oocytes, this approach identified two genes that when coexpressed mediate organic solute transport. The cDNA and predicted amino acid sequences for Ostα and Ostβ most likely identify an additional family of membrane transporters.
Experimental Procedures
Materials and Animals.
[Glycine-2-3H]glutathione (50 Ci/mmol), [3H(G)]taurocholic acid (3.47 Ci/mmol), [3H(N)]estrone sulfate (53 Ci/mmol), [3H(G)]digoxin (19 Ci/mmol), [3H(N)]prostaglandin E2 (200 Ci/mmol), and p-[glycyl-2-3H]aminohippuric acid (4.08 Ci/mmol) were purchased from NEN. [3H]S-2,4-dinitrophenyl glutathione was synthesized enzymatically from [3H]glutathione and 1-chloro-2,4-dinitrobenzene as described (14). Scymnol sulfate was kindly provided by Gert Fricker, University of Heidelberg, Heidelberg, Germany. Other chemicals and reagents were obtained from either Sigma or J. T. Baker. Mature Xenopus were purchased from Nasco, Fort Atkinson, WI. Animals were maintained under a constant light cycle at a room temperature of 18°C.
Preparation of Fractionated Skate Liver Poly(A+) RNA.
Total RNA was isolated from skate liver by using guanidinum thiocyanate extraction followed by cesium chloride gradient centrifugation (15). Poly(A+) RNA was separated by running the total RNA fraction twice over an oligo(dT)-cellulose column. The poly(A+) RNA was size-fractionated by centrifugation through a linear sucrose gradient (6–26% wt/wt) by using a modification of the method of Hagenbuch et al. (16). The isolated poly(A+) RNA fractions were checked for taurocholate uptake activity by expression in Xenopus oocytes. The two fractions with the greatest uptake activity (1.2–2.3 kb and 0.6–1.5 kb) were combined, and this mRNA was used to synthesize a cDNA library.
Construction and Screening of cDNA Library.
An expression library was constructed by using the SuperScript Plasmid System kit (GIBCO/BRL). The synthesized cDNA was ligated into pSPORT1 vector and transformed into MAX Efficiency DH5α-competent cells (GIBCO/BRL). The cDNA library contained about 6 × 105 colonies, of which 3 × 105 were screened by a sib-selection procedure. So as not to lose a positive bacterial clone during amplification in liquid culture, pools of 1,000–10,000 colonies of the initial unamplified cDNA library were grown in liquid culture overnight. Plasmid DNA was isolated from each pool by using a Wizard DNA purification system (Promega). The DNA was linearized with NotI and used to synthesize cRNA via a T7 mMessage mMachine kit (Ambion, Austin, TX). Routinely, 50 nl of cRNA solution (1–50 ng) was injected per oocyte, and 3 days later transport activity was measured. Once a positive pool was identified it was further subdivided and screened until two positive clones (Ostα and Ostβ) were isolated.
Xenopus Oocyte Preparation, Microinjection, and Uptake Assays.
Isolation of Xenopus oocytes was performed as described by Goldin (17) and previously used in our laboratory (18–20). Stage V and VI defolliculated oocytes were selected and incubated at 18°C in modified Barth's solution [88 mM NaCl/1 mM KCl/2.4 mM NaHCO3/0.82 mM MgSO4/0.33 mM Ca(NO3)2/0.41 mM CaCl2/20 mM Hepes-Tris, pH 7.5], supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml). Oocyte medium was changed daily, and healthy oocytes, defined as those with a clean brown animal pole and a distinct equator line, were selected for experiments. Transport studies were performed as described (18, 19).
DNA Sequence and Hydropathy Analysis.
Double-stranded cDNA of detected clones were sequenced in both directions at the University of Rochester Medical Center Core Nucleic Acid Laboratory. The full-length sequences of both clones were obtained by using T7 and SP6 primers, as well as synthetic oligonucleotide primers. Sequence analysis was performed with the DNA and protein sequence analysis program lasergene from DNAstar (Madison, WI). Membrane topology and putative membrane-spanning domains were determined by hidden Markov model analysis (hmmtop; http://www.enzim.hu/hmmtop/index.html) and Kyte-Doolittle hydropathy analysis (21). The presence of possible signal peptides was evaluated with the SIGNALP V1.1 program (http://www.cbs.dtu.dk/services/).
Northern Blotting and Reverse Transcription–PCR (RT-PCR) Analysis.
Poly(A+) RNA isolated from various skate organs was separated on a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane (Hybond N), and hybridized with the full-length cDNAs (NotI/SalI fragment) of Ostα and Ostβ labeled by random priming (Stratagene). The blot was hybridized at 65°C overnight in ExpressHyb Hybridization Solution (CLONTECH) and washed twice at room temperature for 20 min in 2 × SSC, 0.05% SDS followed by two 20-min washes at 50°C in 0.1 × SSC, 0.1% SDS. RT-PCR analysis for Ostα and Ostβ was completed by using GIBCO's SuperScript One-Step RT-PCR with Platinum Taq kit. Primers for Ostα were designed to generate a 547-bp fragment corresponding to the coding region between base pairs 122 and 669 of Ostα's cDNA sequence. Similarly the primers for Ostβ were designed to produce a fragment of 399 bp from the coding region of its sequence between base pairs 116 and 515.
Statistical Analysis.
Kinetic data from experiments measuring uptake of radiolabeled substrate were fit to the Michaelis–Menten equation by nonlinear least-squares regression analysis using SIGMAPLOT 4.16. Vmax and Km values with standard errors were derived from these curves, and Ki values were calculated by using the equation: Ki = [I]/(Kmi/Kmo − 1). Comparison of data measuring initial rates of uptake of radiolabeled substrates in the presence and absence of inhibitors were performed by unpaired Student's t test and correlated to P < 0.05.
Results
Isolation and Structural Analysis of Ostα and Ostβ.
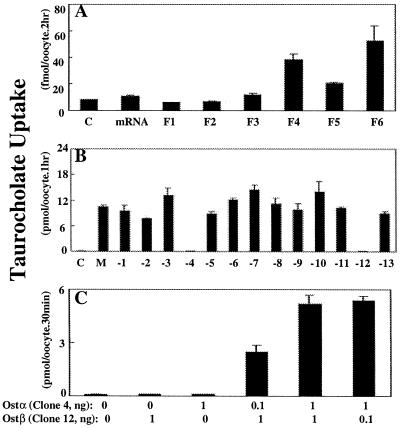
[3H]Taurocholate transport activity in oocytes injected with size-fractionated skate liver mRNA was highest in size fractions 4 (1.2–2.3 kb) and 6 (0.6–1.5 kb), and intermediate in fraction 5 (Fig. 1A). This bimodal distribution indicates that either two different-sized mRNA molecules are able to stimulate taurocholate transport or that two separate genes from these partially overlapping mRNA size fractions must be coexpressed to generate the transport signal. To evaluate these possibilities, mRNA fractions 4 and 6 were combined, a cDNA library was constructed, and the library was screened for [3H]taurocholate transport activity. After multiple rounds of screening, a pool containing 13 clones was identified that exhibited strong taurocholate transport activity (M in Fig. 1B). However, when cRNA from the individual clones of this pool were injected into oocytes they failed to stimulate transport (data not shown), indicating that two or more gene products may be required. To evaluate which of the 13 clones were required, a mixture of cRNA was prepared from either all 13 clones or from only 12 clones by sequentially deleting each clone from the mixture (Fig. 1B). Taurocholate uptake was observed under all conditions except when either clone 4 or clone 12 was deleted (Fig. 1B), indicating that both of these genes are required for transport. When the cRNA from clone 4 or clone 12 was injected individually, no transport was observed; however, when they were injected simultaneously in various ratios there was strong taurocholate transport activity (Fig. 1C). Clone 4 was denoted Ostα, and clone 12 was named Ostβ.
Figure 1.
Screening of skate liver mRNA for [3H]taurocholate-transporting activity in Xenopus oocytes. (A) Taurocholate uptake in oocytes injected with either water (control, C), total skate liver mRNA (mRNA, 50 ng), or size-fractionated mRNA (F1–F6, 50 ng). Total skate liver mRNA was separated on a linear 6–26% (wt/wt) sucrose gradient, and oocytes were injected with mRNA collected from fractions 1–6 (F1, 3–4 kb; F2, 2.5–3.5 kb; F3, 2–3 kb; F4, 1.2–2.3 kb; F5, 0.8–1.8 kb; F6, 0.6–1.5 kb). After 3 days in culture, uptake of 1 μM [3H]taurocholate was measured at 25°C for 2 h and is reported as fmol/oocyte per 2 h. Values are means ± SE (n = 8). (B) Taurocholate uptake in oocytes injected with either water, a mixture of cRNA from 13 separate clones (M, 5 ng), or cRNA mixtures of 12 clones of which one clone was sequentially removed (M-1 to M-13, 5 ng each). After 3 days in culture, uptake of 20 μM [3H]taurocholate was measured at 25°C for 1 h and is reported as pmol/oocyte per 1 h. Values are means ± SE (n = 8). (C) Effect of various cRNA ratios of clone 4 (Ostα) and clone 12 (Ostβ) on [3H]taurocholate uptake in oocytes. Uptake of 20 μM [3H]taurocholate was measured at 25°C for 30 min, at 3 days after injection of cRNA, and is reported as pmol/oocyte per 30 min. Values are means ± SE (n = 4).
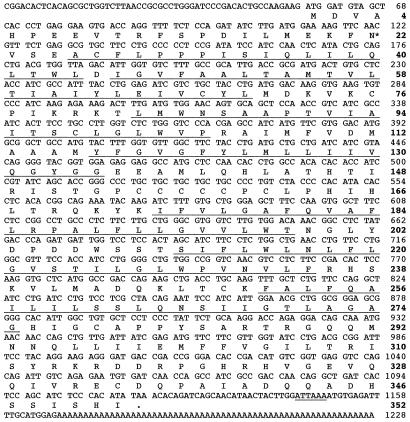
The DNA sequences and deduced amino acid sequences for Ostα and Ostβ are shown in Figs. 2 and 3, respectively. The total cDNA inserts of Ostα and Ostβ consist of 1,228 and 882 nt and predict polypeptides of 352 and 182 aa, with calculated molecular masses of ≈39 and 20 kDa. Ostα is predicted to have seven transmembrane (TM) domains, an intracellular carboxyl terminus, and a single N-glycosylation site (Asn-22) (Fig. 2). This polypeptide has a cluster of six cysteine residues in the hydrophilic loop between TM domains 4 and 5. In contrast, Ostβ is predicted to have two TM domains and two possible N-linked glycosylation sites (Asn-42 and Asn-48) (Fig. 3). However, analysis of Ostβ with the SIGNALP V1.1 program (www.cbs.dtu.dk/services/) indicated that its first 27 aa form a signal peptide. Because this region contains a putative TM domain, the mature Ostβ protein may have only one TM domain; however, this prediction has not yet been tested experimentally.
Figure 2.
Nucleotide and deduced amino acid sequence for Ostα. Predicted TM domains are underlined and numbered 1–7, and the single putative N-linked glycosylation site (Asn-22) is indicated by *. A 6-nt possible polyadenylation signal is also underlined, and the first in-frame termination codon is noted by a period. This sequence has been submitted to GenBank (accession no. AY027664).
Figure 3.
Nucleotide and deduced amino acid sequence for Ostβ. The two predicted TM domains are underlined and numbered. There are a total of four putative N-linked glycosylation sites (Asn-42, -48, -91, and -106); however, because two each are distributed on either side of one TM domain, only two can be extracellular. Asn-42 and -48 are predicted to be in an extracellular loop, and these are indicated by *. The 6-nt polyadenylation signal (AATAAA) also is underlined, and the first in-frame termination codon is noted by a period. This sequence has been submitted to GenBank (accession no. AY027665).
A search of the databases for both nucleotide and amino acid sequences (March 2001) failed to identify significant sequence identity for either Ostα or Ostβ, indicating that they are novel sequences. However, we identified proteins of unknown function in the human, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana databases that exhibit about 15–20% predicted amino acid identity with Ostα and up to 30% identity with each other. Many of these sequences also predict seven-helix proteins, suggesting that they may have a common membrane topology and perhaps a common evolutionary origin. The Ostβ sequence was also unique, although it does exhibit 22% amino acid identity with the C-terminal 200 aa (i.e., the single TM domain and intracellular C terminus) of protocadherin-γ (AF152506), a cell surface glycoprotein.
Northern Blot Analyses.
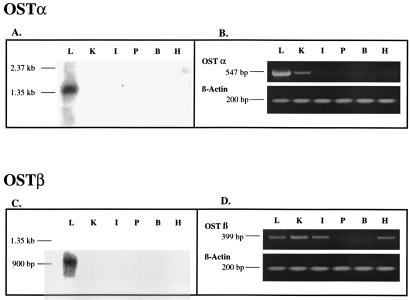
Northern blot analysis of Ostα showed a single major band at 1.4 kb in skate liver, but no significant expression was detected in any of the other skate tissues examined (Fig. 4A). RT-PCR identified a signal for Ostα in liver, as expected, and a weak signal in skate kidney, but all other tissues examined were negative (Fig. 4B). Ostβ also was detected only in skate liver by Northern blot, at a size of ≈0.9 kb (Fig. 4C), whereas RT-PCR analysis indicated its presence in several skate tissues including liver, kidney, intestine, and heart, but not pancreas or brain (Fig. 4D). The reason for this discrepancy is not clear, but it is possible that Ostβ or a closely related sequence may be expressed in other tissues at levels that cannot be detected by Northern blot using our full-length cDNA probe under high-stringency conditions.
Figure 4.
Northern blot and RT-PCR analysis of skate tissue RNA. (A and C) Five micrograms of mRNA from skate liver (L), kidney (K), intestine (I), pancreas (P), brain (B), and heart (H) were separated by 1.2% denaturing agarose gel electrophoresis, transferred to a nylon membrane, and UV-crosslinked. The blot was hybridized with 32P-labeled full-length DNA fragments of Ostα (A) and Ostβ (C) under high stringency conditions. (B and D) RT-PCR was conducted on the same tissues using 0.5 μg total RNA as a template. The predicted 547-bp PCR product for Ostα was observed in liver and to a lesser extent in kidney. The predicted PCR fragment for Ostβ was detected in liver, kidney, intestine, and heart.
Functional Characterization of Ostα/Ostβ-Mediated Transport.
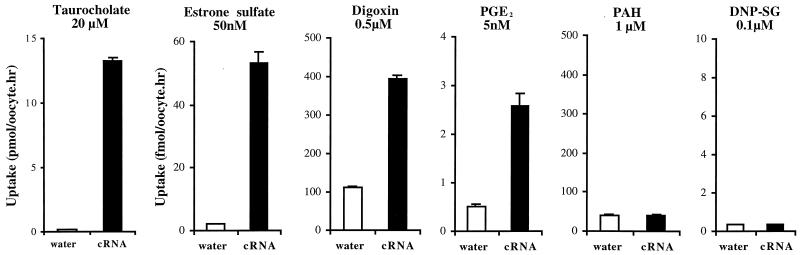
Oocytes injected with Ostα and Ostβ cRNA (1 ng each) were able to transport taurocholate, estrone sulfate, digoxin, and prostaglandin E2, but not p-aminohippurate or S-dinitrophenyl glutathione (Fig. 5), indicating that this transport system is multispecific and may participate in cellular uptake of conjugated steroids and eicosanoids. Transport was sodium-independent and saturable (data not shown), although the apparent Michaelis constants (Km) for taurocholate (785 ± 43 μM), estrone sulfate (85 ± 16 μM), and digoxin (148 ± 30 μM) were approximately an order of magnitude higher than those reported for the OATP transporters (1). The skate major bile salt scymnol sulfate was a competitive inhibitor of estrone sulfate transport (data not shown), with a Ki of 145 μM, indicating that scymnol sulfate may be an endogenous substrate for Ostα/Ostβ. However, because these Km and Ki values are relatively high, it is unlikely that these compounds are the preferred substrates for this transporter.
Figure 5.
Substrate selectivity of Ostα/Ostβ. Oocytes were injected with either water, or 1 ng Ostα cRNA plus 1 ng of Ostβ cRNA, in 50 nl of water per oocyte. After 3 days in culture, the uptake of radiolabeled compounds ([3H]taurocholate, 20 μM; [3H]estrone sulfate, 50 nM; [3H]digoxin, 0.5 μM; [3H]prostaglandin E2 (PGE2), 5 nM; [3H]p-aminohippuric acid (PAH), 1 μM; or [3H]S-2,4-dinitrophenyl glutathione (DNP-SG), 0.1 μM) was measured at 25°C for 1 h. Uptake values are reported as pmol/oocyte per h for taurocholate and as fmol/oocyte per h for the other substrates. Values are means ± SE (n = 3–4).
Ostα/Ostβ-mediated uptake of [3H]taurocholate and [3H]estrone sulfate was inhibited by a variety of bile salts, steroids, and other organic anions (Table 1). Although inhibition of transport does not necessarily indicate that these compounds are transported substrates, it does provide insight into possible substrates. In general, these inhibitors produced similar effects on [3H]taurocholate and [3H]estrone sulfate uptake; however, taurodeoxycholate, taurocholate, and bilirubin ditaurate all produced moderate inhibition of [3H]taurocholate uptake, but had no effect on [3H]estrone sulfate uptake (Table 1). Uptake of both substrates was strongly inhibited by sulfated steroids, including the major skate bile salt scymnol sulfate. A comparison of the inhibitory potency of bile salts indicates that taurine-conjugated bile salts are in general more effective than the corresponding glycine-conjugated compounds, and unconjugated bile salts are the least effective inhibitors (Table 1). The addition of a sulfate group further enhances inhibitory potency, such that bile salts containing both taurine and sulfate modifications are strong cis-inhibitors of taurocholate and estrone sulfate transport. Spironolactone was also an inhibitor of transport, as were sulfobromophthalein, probenecid, and indomethecin. The kinetics of inhibition were examined for taurolithocholate, taurolithocholate sulfate, spironolactone, and sulfobromophthalein by using [3H]estrone sulfate as the substrate. The first three of these compounds were found to be competitive inhibitors, with Ki values of 0.07 ± 0.03, 0.04 ± 0.01, and 0.20 ± 0.02 mM, respectively, whereas sulfobromophthalein was a noncompetitive inhibitor, with a Ki of 0.12 ± 0.02 mM.
Table 1.
Effect of bile salts, steroids, and anionic drugs on Ostα/Ostβ-mediated [3H]taurocholate and [3H]estrone sulfate uptake
| [3H]Taurocholate uptake (% control) | [3H]Estrone sulfate uptake (% control) | |
|---|---|---|
| Bile salts | ||
| Control | 100 ± 4 | 100 ± 4 |
| Lithocholic acid, 200 μM | 95 ± 6 | 94 ± 4 |
| Lithocholic acid sulfate, 200 μM | 40 ± 1* | 27 ± 1* |
| Glycolithocholic acid, 200 μM | 124 ± 8 | 107 ± 3 |
| Glycolithocholic acid sulfate, 200 μM | 72 ± 4* | 49 ± 2* |
| Taurolithocholic acid, 200 μM | 30 ± 2* | 36 ± 5* |
| Taurolithocholic acid sulfate, 200 μM | 24 ± 3* | 22 ± 4* |
| Scymnol sulfate, 200 μM | 54 ± 5* | 70 ± 5* |
| Glycodeoxycholic acid, 200 μM | 82 ± 6 | 110 ± 4 |
| Taurodeoxycholic acid, 200 μM | 39 ± 5* | 103 ± 5 |
| Taurocholate, 500 μM | 70 ± 6* | 95 ± 6 |
| Steroids | ||
| Control | 100 ± 4 | 100 ± 4 |
| Estrone sulfate, 200 μM | 60 ± 1* | 62 ± 5* |
| Spironolactone, 200 μM | 24 ± 1* | 36 ± 2* |
| Digoxin, 500 μM | 74 ± 5* | 56 ± 7* |
| Ouabain, 1 mM | 96 ± 8 | 100 ± 8 |
| 17β-Estradiol, 200 μM | 80 ± 3* | 100 ± 4 |
| Testosterone, 200 μM | 78 ± 7* | 84 ± 3* |
| Dexamethasone, 200 μM | 103 ± 6 | 97 ± 8 |
| Other anions | ||
| Control | 100 ± 4 | 100 ± 4 |
| Sulfobromophthalein, 100 μM | 44 ± 5* | 69 ± 2* |
| Bilirubin ditaurate, 500 μM | 43 ± 2* | 83 ± 9 |
| Probenecid, 1 mM | 53 ± 8* | 61 ± 8* |
| p-Aminohippurate, 500 μM | 71 ± 7* | 111 ± 3 |
| S-dinitrophenyl glutathione, 500 μM | 80 ± 7 | 99 ± 6 |
| Indomethacin, 200 μM | 47 ± 6* | 50 ± 3* |
Uptake of 20 μM [3H]taurocholate and 50 nM [3H]estrone sulfate was measured for 30 min and 1 hr, respectively, in the absence (control) and presence of the indicated compounds. Data are expressed as a percent of the control values ± SE (n = 3–4).
, Significantly different from control, P < 0.05.
Discussion
The present study describes the isolation and functional characterization of a multispecific organic solute and steroid transporter that requires the coexpression of two gene products, Ostα and Ostβ. Because the cDNA and predicted amino acid sequences of Ostα and Ostβ appear to be novel, they are likely to be members of an additional family of transporters.
Ostα and Ostβ were identified by using functional expression cloning in Xenopus oocytes, the technique that has revolutionized the molecular identification of plasma membrane transporters (13, 22–24). This powerful approach has been used to identify the initial members of essentially all known secondary-active transporter families. However, one of the limitations of expression cloning is that it generally fails to identify transporters that require two or more gene products to generate the active transport unit (22–24). Thus, expression cloning favors the discovery of transport proteins that function either as monomers, homomultimers, or those that can partner with native Xenopus oocyte proteins to form the active transport unit. For example, the identification of the heterodimeric amino acid transporters of the LAT family was facilitated by the presence of endogenous oocyte proteins that complement either member of the LAT heterodimer (25–28). Likewise, functional expression of two of the lactate transporters (MCT1 and MCT4) also requires an ancillary oocyte protein (29). Kirk and coworkers (29) demonstrated that a membrane glycoprotein (CD147) can fulfill this role for MCT1 and MCT2, but the mechanism by which this glycoprotein stimulates functional expression is unknown.
In contrast to the LAT or MCT transporters whose subunits appear to be complemented by endogenous oocyte proteins, neither Ostα nor Ostβ are functionally complemented in oocytes. That is, neither Ostα nor Ostβ generate a transport signal when expressed individually in oocytes (Figs. 1 and 2). Thus, identification of the active Ost transporter required us to simultaneously identify both genes. The present report provides an example of the use of expression cloning to simultaneously identify two components of a polygenic transporter.
Although the mechanism by which Ostα and Ostβ interact to generate transport activity is unknown, insight into this question is provided by comparing the predicted amino acid sequences and membrane topologies of Ostα and Ostβ with those of other multimeric proteins. Of primary importance is the prediction that both Ostα and Ostβ are integral membrane proteins. Thus, Ostα and Ostβ may interact directly at the level of the cell membrane to generate an active transporter complex. Ostα is predicted to have seven TM domains and its overall membrane architecture is reminiscent of the G protein-coupled receptors, with an extracellular amino terminus, seven TM helices, and an intracellular carboxyl terminus (Fig. 2). Ostβ is a smaller protein, but it has at least one and perhaps two TM domains (Fig. 3). The uncertainty in the number of Ostβ TM domains stems from the fact that its first 27 aa are predicted to encode a signal peptide. Because this region contains a putative TM domain (i.e., amino acids 2–20; Fig. 3), the mature Ostβ protein would have only a single TM domain. However, this prediction has not yet been tested experimentally.
Transport systems that function as heterodimers or heteromultimers include Na+/K+ ATPase, sensory rhodospins, and many ion channels and G protein-coupled receptors. Although Ostα and Ostβ do not share significant sequence identity with any of these transport systems, there are some structural features that are noteworthy. In the heterodimeric Na+/K+ ATPase, for example, the α-subunit contains 10 TM-spanning domains and functions as the catalytic portion of the pump, whereas the single TM β-subunit is required for pump assembly and translocation to the plasma membrane (30, 31). Thus, one possible function of the β-subunit of Ost is to chaperone or translocate the α-subunit to the cell surface. If correct, this hypothesis might explain the present observation that 0.1 ng of Ostβ cRNA was as effective as a 1 ng in stimulating transport activity induced by 1 ng of Ostα cRNA (Fig. 1). Although there are several possible explanations for this finding, it is consistent with the conclusion that either a limited amount of Ostβ is sufficient to deliver or activate Ostα at the cell surface, or that the stoichiometry of a putative Ostα/Ostβ protein complex is not 1:1. A putative chaperone function for Ostβ also might explain the more widespread organ distribution of Ostβ mRNA when compared with Ostα (Fig. 4). Ostβ was identified by RT-PCR in intestine and heart, whereas no Ostα was detected in these tissues. This broader tissue distribution indicates that Ostβ may have additional roles, including the possibility that it acts as a chaperone for other proteins with different organ specificity than Ostα. Additional studies are needed to evaluate these possibilities.
An additional and perhaps more relevant example of heterodimeric proteins is that of the sensory rhodopsins, which have a similar membrane topology as that predicted for Ostα/Ostβ, namely a seven-helix TM protein and a two-helix accessory protein. Sensory rhodopsins function as phototaxis receptors in archea and are composed of a seven-TM ion channel/receptor and a two-TM transducer protein that controls an intracellular phosphorylation cascade (32). Interestingly, the seven-helix motif of the rhodopsin molecule is conserved among all members of the large G protein-coupled receptor superfamily. However, despite this common evolutionary origin and conserved membrane architecture, the three families of G protein-coupled receptors exhibit remarkably low sequence identity (33, 34). The lack of sequence identity is explained in large part by the long evolutionary time scales involved, 2–3 billion years, which has allowed for considerable diversity in both sequence and function (33–36).
The structural similarities between Ostα/Ostβ and the sensory rhodopsins therefore suggest that Ostα may have evolved from an ancestral rhodopsin-like molecule, but has acquired the ability to transport steroids and eicosanoids, compounds that also function as ligands for some G protein-coupled receptors. Thus, Ostα may have evolved from a ligand-activated receptor into a ligand transporter. Although Ostα orthologues were not found in the databases, several proteins of unknown function were identified that exhibit 15–20% predicted amino acid identity with Ostα and up to 30% identity with each other. Most of these sequences also predict seven-helix proteins, suggesting a common membrane topology and possibly a common evolutionary origin, despite the relatively low amino acid identity.
Ostβ also appears unique in sequence, although it does exhibit 22% amino acid identity with the C-terminal 200 aa (i.e., the single TM helix and the cytosolic domain) of protocadherin-γ, a cell suface glycoprotein that belongs to the large cadherin superfamily (37, 38). Cadherins are involved in cell recognition, signaling, morphogenesis, and angiogenesis, but a role in organic solute transport has not yet been proposed. The protocadherin family itself is large and includes proteins from both vertebrates and lower multicellular organisms, suggesting that they are the ancestral cadherins (38).
An additional clue into the possible mechanism by which Ostα and Ostβ interact to generate the active transport complex is provided by the observation that both Ostα and Ostβ contain an Arg-X-Arg (RXR) sequence in their C terminus, putative cytosolic domains (Figs. 2 and 3). RXR sequences in hetero-oligomeric proteins recently have been demonstrated to function as retention or retrieval signals that must be masked before the corresponding protein complexes can be transported from the endoplasmic reticulum (39–41). This trafficking signal prevents cell surface expression of partially assembled or misfolded complexes (41). The RXR sequence is required for the assembly and membrane delivery of the hetero-oligomeric ATP-sensitive K+ channel (40) and for the heterodimerization and cell surface delivery of the γ-aminobutyric acid type B receptor (41), a seven-TM helix receptor. RXR sequences are also present in cytosolic domains of the cystic fibrosis transmembrane conductance regulator, where they regulate delivery of this ion channel to the plasma membrane (42) and may be involved in its interaction with other membrane proteins (43–45), but the precise mechanisms have not yet been identified.
Although the physiological substrates for Ostα/Ostβ are unknown, our functional characterization indicates that it can transport sterols and their metabolites. This observation is physiologically relevant because skates appear to excrete sterols mainly as sulfate conjugates (46, 47). The major bile salt in skate bile is scymnol sulfate, a sulfated bile alcohol that is rapidly removed from hepatic portal blood and transported into bile (47, 48). The overall substrate specificity of Ostα/Ostβ is most similar to that of the OATP family of Osts (1, 4), and in particular to that of human OATP-C (also known as LST1 and OATP-2). Ostα/Ostβ is able to transport taurocholate, estrone sulfate, digoxin, and prostaglandin E2, but not p-aminohippurate or S-dinitrophenyl glutathione (Fig. 5). Human OATP-C can transport all of these Ostα/Ostβ substrates, except for digoxin (1, 4). Additional studies are needed to evaluate the substrate specificity of Ostα/Ostβ, and in particular, to test whether other ligands for G protein-coupled receptors may be selective substrates for this transporter.
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK48823, ES06484, DK25636, and DK34989 and National Institute of Environmental Health Sciences Center Grants ES03828 and ES01247.
Abbreviations
- OATP
organic anion transporting polypeptide
- Ost
organic solute transporter
- RT-PCR
reverse transcription–PCR
- TM
transmembrane
Footnotes
References
- 1.Kullak-Ublick G A, Stieger B, Hagenbuch B, Meier P J. Semin Liver Dis. 2000;20:273–292. doi: 10.1055/s-2000-9426. [DOI] [PubMed] [Google Scholar]
- 2.Borst P, Evers R, Kool M, Wijnholds J. J Natl Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 3.Keppler D, Konig J. Semin Liver Dis. 2000;20:265–272. doi: 10.1055/s-2000-9391. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H, Sugiyama Y. Semin Liver Dis. 2000;20:251–263. doi: 10.1055/s-2000-8408. [DOI] [PubMed] [Google Scholar]
- 5.Saier M H. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer J L, Schwarz J, Smith N. Am J Physiol. 1976;230:974–981. doi: 10.1152/ajplegacy.1976.230.4.974. [DOI] [PubMed] [Google Scholar]
- 7.Smith D J, Grossbard M, Gordon E R, Boyer J L. Am J Physiol. 1987;252:G479–G484. doi: 10.1152/ajpgi.1987.252.4.G479. [DOI] [PubMed] [Google Scholar]
- 8.Boyer J L, Hagenbuch B, Ananthanarayanan M, Suchy F, Stieger B, Meier P J. Proc Natl Acad Sci USA. 1993;90:435–438. doi: 10.1073/pnas.90.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricker G, Dubost V, Finsterwald K, Boyer J L. Biochem J. 1994;299:665–670. doi: 10.1042/bj2990665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballatori N, Rebbeor J F, Connolly G C, Seward D J, Lenth B E, Henson J H, Sundaram P, Boyer J L. Am J Physiol. 2000;278:G57–G63. doi: 10.1152/ajpgi.2000.278.1.G57. [DOI] [PubMed] [Google Scholar]
- 11.Rebbeor J F, Connolly G C, Henson J H, Boyer J L, Ballatori N. Am J Physiol. 2000;279:G417–G425. doi: 10.1152/ajpgi.2000.279.2.G417. [DOI] [PubMed] [Google Scholar]
- 12.Cai, S.-Y., Wang, L., Ballatori, N. & Boyer, J. L. (2001) Am. J. Physiol., in press. [DOI] [PubMed]
- 13.Hediger M A, Coady M J, Ikeda T S, Wright E M. Nature (London) 1987;330:379–381. doi: 10.1038/330379a0. [DOI] [PubMed] [Google Scholar]
- 14.Ballatori N, Truong A T. J Biol Chem. 1995;270:3594–3601. doi: 10.1074/jbc.270.8.3594. [DOI] [PubMed] [Google Scholar]
- 15.Snutch T P, Mandel G. Methods Enzymol. 1992;207:297–309. doi: 10.1016/0076-6879(92)07019-k. [DOI] [PubMed] [Google Scholar]
- 16.Hagenbuch B, Lubbert H, Stieger B, Meier P J. J Biol Chem. 1990;265:5357–5360. [PubMed] [Google Scholar]
- 17.Goldin A L. Methods Enzymol. 1992;207:266–279. doi: 10.1016/0076-6879(92)07017-i. [DOI] [PubMed] [Google Scholar]
- 18.Ballatori N, Wang W, Li L, Truong A T. Am J Physiol. 1996;270:R1156–R1162. doi: 10.1152/ajpregu.1996.270.5.R1156. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Lee T K, Meier P J, Ballatori N. J Biol Chem. 1998;273:16184–16191. doi: 10.1074/jbc.273.26.16184. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Meier P J, Ballatori N. Mol Pharmacol. 2000;58:335–340. doi: 10.1124/mol.58.2.335. [DOI] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Gurdon J B, Wickens M P. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- 23.Sigel E. J Membr Biol. 1990;117:201–221. doi: 10.1007/BF01868451. [DOI] [PubMed] [Google Scholar]
- 24.Soreq H, Seidman S. Methods Enzymol. 1992;207:225–265. doi: 10.1016/0076-6879(92)07016-h. [DOI] [PubMed] [Google Scholar]
- 25.Palacin M, Estevez R, Bertran J, Zorzano A. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 26.Verrey F, Jack D L, Paulsen I T, Saier M H, Pfeiffer R. J Membr Biol. 1999;172:181–192. doi: 10.1007/s002329900595. [DOI] [PubMed] [Google Scholar]
- 27.Wells R G, Lee W S, Kanai Y, Leiden J M, Hediger M A. J Biol Chem. 1992;267:15285–15288. [PubMed] [Google Scholar]
- 28.Bertran J, Magagnin S, Werner A, Markovich D, Biber J, Testar X, Zorzano A, Kuhn L C, Palacin M, Murer H. Proc Natl Acad Sci USA. 1992;89:5606–5610. doi: 10.1073/pnas.89.12.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirk P, Wilson M C, Heddle C, Brown M H, Barclay A N, Halestrap A P. EMBO J. 2000;19:3896–3904. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fambrough D M, Lemas M V, Hamrick M, Emerick M, Renaud K J, Inman E M, Hwang B, Takeyasu K. Am J Physiol. 1994;266:C579–C589. doi: 10.1152/ajpcell.1994.266.3.C579. [DOI] [PubMed] [Google Scholar]
- 31.Blanco G, Mercer R W. Am J Physiol. 1998;275:F633–F650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 32.Spudich J L. Science. 2000;288:1358–1359. doi: 10.1126/science.288.5470.1358. [DOI] [PubMed] [Google Scholar]
- 33.Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, Mukohata Y. J Mol Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- 34.Gershengorn M C, Osman R. Endocrinology. 2001;142:2–10. doi: 10.1210/endo.142.1.7919. [DOI] [PubMed] [Google Scholar]
- 35.Milligan G. Science. 2000;288:65–67. doi: 10.1126/science.288.5463.65. [DOI] [PubMed] [Google Scholar]
- 36.Rocheville M, Lange D C, Kumar U, Patel S C, Patel R C, Patel Y C. Science. 2000;288:154–157. doi: 10.1126/science.288.5463.154. [DOI] [PubMed] [Google Scholar]
- 37.Wu Q, Maniatis T. Proc Natl Acad Sci USA. 2000;97:3124–3129. doi: 10.1073/pnas.060027397. . (First Published March 14, 2000, 10.1073/pnas.060027397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angst B D, Marcozzi C, Magee A I. J Cell Sci. 2000;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 39.Ellgaard L, Molinari M, Helenius A. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 40.Zerangue N, Schwappach B, Jan Y N, Jan L Y. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 41.Margeta-Mitrovic M, Jan Y N, Jan L Y. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 42.Chang X B, Cui L, Hou Y X, Jensen T J, Aleksandrov A A, Mengos A, Riordan J R. Mol Cell. 1999;4:137–142. doi: 10.1016/s1097-2765(00)80196-3. [DOI] [PubMed] [Google Scholar]
- 43.Naren A P, Quick M W, Collawn J F, Nelson D J, Kirk K L. Proc Natl Acad Sci USA. 1998;95:10972–10977. doi: 10.1073/pnas.95.18.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters K W, Qi J, Watkins S C, Frizzell R A. Am J Physiol. 1999;277:C174–C180. doi: 10.1152/ajpcell.1999.277.1.C174. [DOI] [PubMed] [Google Scholar]
- 45.Kunzelmann K, Schreiber R. J Membr Biol. 1999;168:1–8. doi: 10.1007/s002329900492. [DOI] [PubMed] [Google Scholar]
- 46.Tammar A R. Chem Zool. 1974;8:595–661. [Google Scholar]
- 47.Karlaganis G, Bradley S E, Boyer J L, Batta A K, Salen G, Egestad B, Sjovall J. J Lipid Res. 1989;30:317–322. [PubMed] [Google Scholar]
- 48.Fricker G, Wossner R, Drewe J, Fricker R, Boyer J L. Am J Physiol. 1997;273:G1023–G1030. doi: 10.1152/ajpgi.1997.273.5.G1023. [DOI] [PubMed] [Google Scholar]