Abstract
Expression profiles of primary breast tumors were investigated in relation to disseminated tumor cells (DTCs) in bone marrow (BM) in order to increase our understanding of the dissemination process. Tumors were classified into five pre‐defined molecular subtypes, and presence of DTC identified (at median 85 months follow‐up) a subgroup of luminal A patients with particular poor outcome (p=0.008). This was not apparent for other tumor subtypes. Gene expression profiles associated with DTC and with systemic relapse for luminal A patients were identified. This study suggests that DTC in BM differentially distinguishes clinical outcome in patients with luminal A type tumors and that DTC‐associated gene expression analysis may identify genes of potential importance in tumor dissemination.
Keywords: DNA microarrays, Breast cancer, Micrometastases in bone marrow, Molecular subtypes, Clinical outcome
1. Introduction
Genome‐wide expression profiling has demonstrated great power in deciphering molecular portraits of human breast tumors and has identified gene signatures that can be correlated to many different aspects of breast cancer such as tumor progression, prediction of outcome and sensitivity to therapy (Ayers et al., 2004, 2005, 2003, 2005, 2003, 2001, 2002, 2004). Distinct subtypes that are associated with significant differences in overall and disease‐free survival have been identified and validated in different patient cohorts (Bertucci et al., 2005; Sorlie et al., 2003; Sotiriou et al., 2003; Yu et al., 2004). Molecular profiling by microarray technologies will improve our understanding of primary tumor biology and the mechanisms behind tumorigenesis, and also provide improved tools for patient stratification to establish more tailored therapeutic strategies.
Presence of disseminated tumor cells (DTCs) in bone marrow (BM) as an independent prognostic factor for breast cancer patients receiving adjuvant systemic treatment as well as for untreated patients, is now well documented (Braun et al., 2000; Wiedswang et al., 2003; Diel et al., 1996), and supported by a large pooled analysis of more than 4700 patients (Braun et al., 2005). The occurrence of DTC in distant organs, such as BM, has traditionally been considered as rare and relatively late events during primary tumor progression. This has been challenged by recent expression profiling studies that recognized early primary tumors with a “metastatic phenotype” (Bernards and Weinberg, 2002; Ramaswamy et al., 2003; van't Veer et al., 2002; Weigelt et al., 2003), indicating that tumor dissemination may be an early event in tumorigenesis in some patients. Distinct biological properties of the primary tumors may affect the ability for disseminated tumor cells to settle in certain distant organs and subsequently be responsible for metastasis formation. Thus, combined analyses of primary tumors' gene expression patterns, other molecular parameters such as mutation in the TP53 gene and DTC status will increase our understanding of subsequent metastasis formation.
Previously, we reported the independent prognostic value of DTC in BM in early breast cancer in a large cohort of 817 patients (Wiedswang et al., 2003). From this cohort, gene expression patterns of 123 primary tumors prospectively collected during surgery have been examined by using cDNA microarrays. TP53 mutation status and HER2 amplification were also determined. The prognostic importance of DTC in BM was shown to be different among the various molecular subtypes, separating groups with significantly different outcome only in patients expressing the luminal A type tumors. Differentially expressed genes and functional gene sets that provide potentially useful markers for a more in‐depth study of the dissemination process were identified.
2. Results
2.1. Primary tumor characteristics vs DTC and relapse
In this sub‐cohort of 123 patients with early stage breast cancer, DTCs were detected in 24% of the cases (27/114; nine with missing data) (Table 1). To identify gene expression profiles associated with DTC in BM, we applied two‐class SAM, but were not able to find any genes that were significantly differentially expressed between tumors from patients with DTC in BM compared to those with no detectable DTC.
Table 1.
Molecular and clinicopathological data of the tumor material
| Characteristics | Number of patients | % (Excluding missing)a | |
|---|---|---|---|
| All | 123 | 100 | |
| DTC in BM | Yes | 27 | 23.7 |
| No | 87 | 76.3 | |
| Missing | 9 | ||
| Gene profile | Luminal A | 50 | 40.7 |
| Luminal B | 16 | 13.0 | |
| ERBB2+ | 21 | 17.1 | |
| Basal‐like | 17 | 13.8 | |
| Normal‐like | 15 | 12.2 | |
| Unclassified | 4 | 3.3 | |
| TP53 | Mutant | 38 | 32.5 |
| Wild type | 79 | 67.5 | |
| Missing | 6 | ||
| Her2 – FISH | Positive | 20 | 18.2 |
| Negative | 90 | 81.8 | |
| Missing | 13 | ||
| ER/PgR‐status | Positive | 80 | 66.1 |
| Negative | 41 | 33.9 | |
| Missing | 2 | ||
| Histology | Ductal | 97 | 78.9 |
| Lobular | 23 | 18.7 | |
| Other infiltrating | 2 | 1.6 | |
| DCIS | 1 | 0.8 | |
| Grade | I | 16 | 13.2 |
| II | 58 | 47.9 | |
| III | 47 | 38.8 | |
| Missing | 2 | ||
| Tumor status | pT1 | 55 | 44.7 |
| pT2 | 51 | 41.5 | |
| pT3‐4 | 13 | 10.6 | |
| Ptis | 1 | 0.8 | |
| pTXb | 3 | 2.4 | |
| Nodal status | Negative | 50 | 40.7 |
| pN1 | 35 | 28.5 | |
| pN2 | 21 | 17.1 | |
| pN3 | 10 | 8.1 | |
| pNXc | 7 | 5.7 | |
| Menopause | Pre | 40 | 32.5 |
| Post | 77 | 62.6 | |
| Unknown | 6 | 4.9 | |
| Adjuvant treatment | Yes | 74 | 61.7 |
| No | 46 | 38.3 | |
| Missing | 3 | ||
| Tamoxifen | 57 | 46.7 | |
| No Tamoxifen | 63 | 51.6 | |
| Missing | 3 | ||
| Systemic relapse | Yes | 39 | 32.5 |
| No | 81 | 67.5 | |
| Missing | 3 | ||
| Death | From breast cancer | 34 | 27.6 |
| Other cause | 21 | 17.1 | |
| No | 68 | 55.3 | |
Missing includes both non‐evaluable cases, not performed and information not available.
pTX defines those cases where no determination of tumor size was possible.
pNX defines those cases where no axillary dissection was performed but no clinical sign of nodal metastasis existed.
As breast tumors can be separated into different molecular subtypes with distinct biological characteristics accompanied with differences in survival, the association between DTC and molecular characteristics of the primary tumors within the previously defined gene expression‐based subtypes was explored. Tumors were assigned to a subtype using correlation to the expression centroids as previously described (Sorlie et al., 2003). As shown in the hierarchical clustering diagram in Figure 1A, all five subtypes of breast tumors were identified: 41% of the tumors were classified as luminal A, 13% were of the luminal B type, 17% were ERBB2+, 14% were basal‐like and 12% were characterized as normal breast tissue‐like (Table 1). Four tumors were labeled as unclassified (correlation <0.1). The most striking difference was between luminal A and basal‐like samples that were strongly anti‐correlated. However, for other samples, particularly a few normal breast‐like and ERBB2+ samples, the correlation of each tumor sample to its final designated subtype was not as convincing (Figure 1B).
Figure 1.
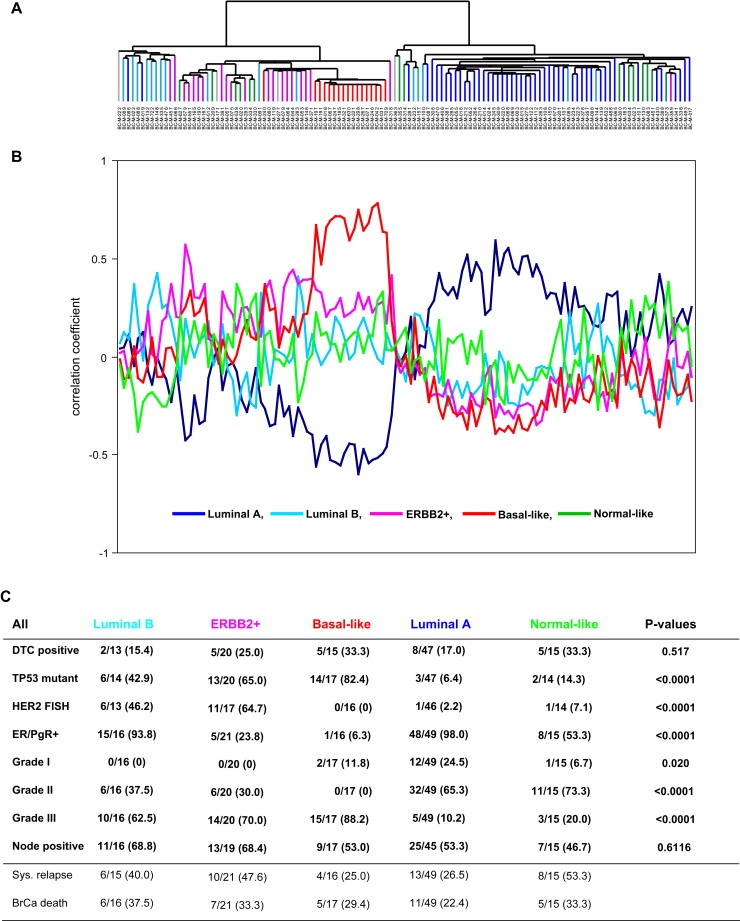
Subclassification of breast tumor samples using the “intrinsic” gene list. A. Hierarchical clustering dendrogram indicating the relationship between the tumor samples based on gene expression patterns from 494 intrinsic genes. Branches are color‐coded according to tumor subtype as indicated in 1B. Grey are unclassified B. Correlation of each sample to each of the five expression centroids as previously identified (Sorlie et al., 2003). C. Clinicopathological data by molecular tumor subtype. Cases with missing parameters within each analysis are excluded. The four unclassified tumors are also excluded.
Genomic DNA from 117 of these tumors was available for TP53 mutation screening. Mutations were found in 32.5% of the cases (Table 1), which is slightly higher than reported for breast cancer in general (IARC TP53 mutation database; Olivier et al., 2002). Of the 38 mutations detected, 27 were missense, three were nonsense, seven were deletions/insertions and one was a putative splice mutation downstream of exon 9 (Supplemental Table 1).
In the further analyses, the four patients with unclassified tumors and the one patient with ductal carcinoma in situ (DCIS) were excluded (n=118). DTC in BM was detected in all subgroups as shown in Figure 1C. As expected, there was a high frequency of ER/PgR‐expression in the luminal A and luminal B subtypes (98% and 94%, respectively) as compared to the ERBB2+ and basal‐like subtypes (Figure 1C). Also, a high frequency of TP53 mutations and histological grade 3 was observed among the luminal B, ERBB2+ and basal‐like subtypes (43% and 63%, 65% and 70%, 82% and 88%, respectively) compared to luminal A. Although the number of systemic relapses and breast cancer deaths were somewhat lower for patients with luminal A type tumors than for those with other tumor types, a considerable number of luminal A patients also experienced systemic relapse (27%) during the follow‐up time of median 85 months.
2.2. Survival analyses according to DTC and primary tumor molecular characteristics
Survival analyses stratified by tumor subtype and DTC status at 85 months median follow‐up (range 0.8–127, interquartile range 60) are shown in Figure 2A–J. The survival for the luminal A patients appeared more favorable than those for the other subtypes (Figure 2B), although significantly different only from the ERBB2+ subtype for distant disease‐free survival (P=0.04, log rank test). As shown in Table 2, similar systemic relapse‐ and breast cancer‐death patterns were observed when stratified for systemic treatment, but the small number of patients in each of the subgroups restricts further interpretation. Although clearly evident in the entire patient cohort (Wiedswang et al., 2003), DTC status did not significantly discriminate survival in this smaller sub‐cohort (Figure 2C and D). However, the presence of DTC was associated with significantly reduced distant disease‐free survival (DDFS) and breast cancer specific survival (BCSS) among the luminal A patients (Figure 2E and F; Table 2). Subdivision by treatment status showed unfavorable outcome for tamoxifen‐treated luminal A patients (n=20) with DTC‐positive status (p=0.002, log rank), whereas among the untreated patients (n=19), the low frequency of DTC‐positive cases (n=1) makes survival analysis impossible. The association between DTC and clinical outcome within the other subtypes is not clear because of limited sample size (Table 2). However, for patients with ERBB2+ and basal‐like tumors the systemic relapse‐ and breast cancer‐death frequencies appeared higher for the DTC‐negative than for DTC‐positive patients. Furthermore and in line with this, we found that among the patients with TP53 mutated tumors (71% of which were ERBB2+ or basal‐like), there was a trend towards association between the presence of DTC and improved DDFS, as compared to the absence of DTC (P=0.096, log rank). In contrast, DTC‐positive patients with TP53 wild type primary tumors experienced, at borderline significance, reduced DDFS as compared to the DTC‐negative patients with wild type tumors (P=0.046, log rank) (see Supplementary Figure 2).
Figure 2.
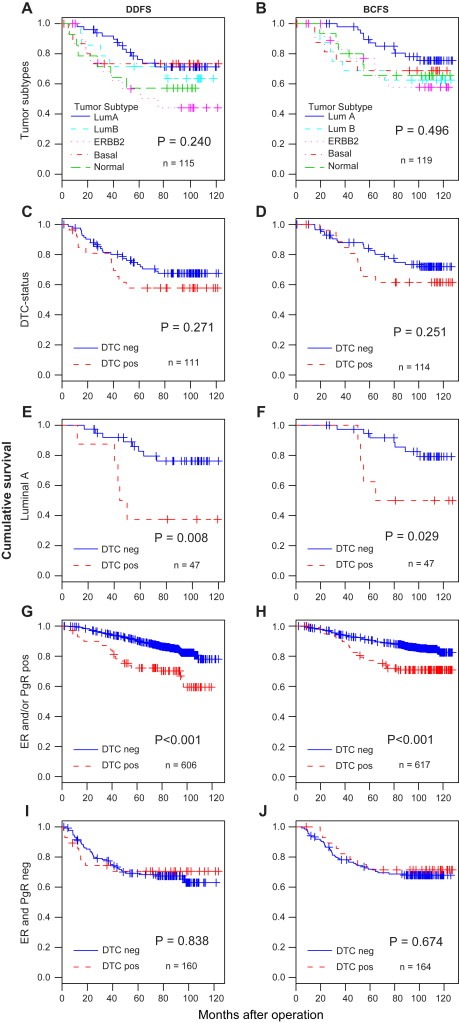
Overall and disease‐free survival analyses by DTC and tumor subtype. Analyses of distant disease free survival (DDFS) (A, C, E, G, I) and breast cancer specific survival (BCSS) (B, D, F, H, J) in patients according to (A, B) molecular tumor subtype; (C, D) DTC‐status for all patients subgroups; (E, F) DTC‐status for luminal A patients only; (G, H) DTC status in the entire 811‐cohort based on ER and/or PgR‐positive status; (I, J) DTC‐status in the 811‐cohort based on ER and PgR‐negative status.
Table 2.
The systemic relapse‐ and breast cancer‐death rates according to tumor subtypes, DTC status and adjuvant systemic treatment
| Subtype | Systemic relapse | Breast cancer death | ||
|---|---|---|---|---|
| Adjuvant− | Luminal A | 4/19 (21%) | 3/19 (16%) | |
| Luminal B | 0/3 (0%) | 0/3 (0%) | ||
| ERBB2+ | 2/6 (33%) | 2/6 (33%) | ||
| Basal‐like | 2/8 (25%) | 3/9 (33%) | ||
| Normal‐like | 3/8 (38%) | 2/8 (25%) | ||
| Adjuvant+ | Luminal A | 9/30 (30%) | 8/30 (27%) | |
| Luminal B | 6/12 (50%) | 6/13 (46%) | ||
| ERBB2+ | 8/15 (53%) | 5/15 (33%) | ||
| Basal‐like | 2/8 (25%) | 2/8 (25%) | ||
| Normal‐like | 3/5 (60%) | 3/5 (60%) | ||
| All patients | Luminal A | DTC+ | 5/8 (63%) | 4/8 (50%) |
| DTC− | 8/39 (21%) | 7/39 (18%) | ||
| Luminal B | DTC+ | 1/2 (50%) | 1/2 (50%) | |
| DTC− | 4/11 (36%) | 4/11 (36%) | ||
| ERBB2+ | DTC+ | 1/5 (20%) | 1/5 (20%) | |
| DTC− | 8/15 (53%) | 6/15 (40%) | ||
| Basal‐like | DTC+ | 1/5 (20%) | 1/5 (20%) | |
| DTC− | 3/9 (33%) | 3/10 (30%) | ||
| Normal‐like | DTC+ | 3/5 (60%) | 2/5 (40%) | |
| DTC− | 3/9 (33%) | 2/10 (20%) | ||
The observed difference in clinical significance of DTC between patients with tumors with different molecular characteristics was further explored by an updated analysis of the entire study population with known DTC status (the 817‐patient cohort). Survival analyses were performed separately for the ER‐ and/or PgR‐positive and the ER/PgR‐negative patient groups (assessed by IHC), as surrogate markers for luminal and the basal‐like/ERBB2+ subtypes, respectively. As illustrated in Figure 2G–J, a marked difference in both DDFS and BCSS was detected for the ER‐ and/or PgR‐positive patients stratified for DTC (P<0.001), whereas DTC status did not discriminate outcome in ER‐ and PgR‐negative patients. Subdivision according to ER/PgR status within the 123 patient cohort, revealed the same survival pattern as for the entire study population. No difference in outcome between DTC‐positive and DTC‐negative was detected in the ER‐ and PgR‐negative patients, whereas a marked survival difference was observed for the ER‐ and/or PgR‐positive patients according to DTC status (P=0.007, log rank).
2.3. Gene profiles associated with DTC and systemic relapse
The strong association between DTC and clinical outcome for the luminal A molecular subtype encouraged further analysis of genes associated with DTC within this group of patients. Two‐class SAM identified 20 genes (including two replicate probes) highly expressed in tumors associated with positive DTC status (20% FDR) (Table 3). Of these, several are involved in regulation of transcription, transporter activity and ATP binding. When expression data from these genes were used in a hierarchical clustering analysis, the luminal A samples formed two main branches and all eight tumors associated with DTC clustered together in one branch (Figure 3). Of these eight patients, five experienced systemic relapse of the disease. Similarly, SAM was also applied to identify genes correlated with distant relapse in the luminal A patients irrespective of DTC status. Here, we identified 64 genes (76 UniGene clusters) with an FDR of 5% that showed significantly different expression patterns between tumors from patients who experienced systemic relapse and those who did not (see Supplemental Figure 1 for the complete cluster diagram and Supplemental Table 2 for the list of genes). Only two of the 20 genes (LONP2 and MCM6) associated with DTC status for the luminal A patients were also among the genes associated with systemic relapse.
Table 3.
Gene information on the 20 genes correlating with DTC status from SAM analysis
| Symbol | Name | UG cluster | Clone ID | GeneID (LocusLink) | Cytoband | GO: molecular function/biological process |
|---|---|---|---|---|---|---|
| DIP2B | DIP2 disco‐interacting protein 2 homolog B (Drosophila) | Hs.505516 | IMAGE:489031 | 57609 | 12q13.12 | Ligase activity/metabolism |
| MCM6 | Minichromosome maintenance deficient 6 homolog (S. cerevisiae) | Hs.444118 | IMAGE:1587847 | 4175 | 2q21 | ATP binding/DNA replication |
| PLEK2 | Pleckstrin 2 | Hs.170473 | IMAGE:453710 | 26499 | 14q23.3 | |
| FOXI1 | Forkhead box I1 | Hs.87236 | IMAGE:1901720 | 2299 | 5q34 | Transcription factor activity/development |
| SYT13 | Synaptotagmin XIII | Hs.436643 | IMAGE:23443 | 57586 | 11p12‐p11 | Transporter activity/transport |
| PRDM6 | PR domain containing 6 | Hs.135118 | IMAGE:259607 | 93166 | 5q23.2 | DNA binding/transcription regulation |
| LONP2a | Lon peptidase 2, peroxisomal | Hs.651202 | IMAGE:181787 | 83752 | 16q12.1 | ATP binding/proteolysis |
| ZNF185 | Zinc finger protein 185 (LIM domain) | Hs.16622 | IMAGE:855079 | 7739 | Xq28 | Zinc ion binding |
| ENTPD4 | Ectonucleoside triphosphate diphosphohydrolase 4 | Hs.444389 | IMAGE:2272922 | 9583 | 8p21.3 | Hydrolase activity/nucleic acid metabolism |
| PPP2R2Ca | Protein phosphatase 2, regulatory subunit B (PR 52), gamma isoform | Hs.479069 | IMAGE:399593 | 5522 | 4p16.1 | Hydrolase activity/signal transduction |
| LANCL2 | LanC lantibiotic synthetase component C‐like 2 (bacterial) | Hs.595384 | IMAGE:429647 | 55915 | 7q31.1‐q31.33 | ATP binding/regulation of transcription |
| SFTPD | Surfactant, pulmonary‐associated protein D | Hs.253495 | IMAGE:1601355 | 6441 | 10q22.2‐q23.1 | Bacterial binding/alveolus development |
| TSGA10 | Testis specific, 10 | Hs.120267 | IMAGE:745499 | 80705 | 2q11.2 | Porin activity/spermatogenesis |
| B3GALT2 | UDP‐Gal:betaGlcNAc beta‐1,3‐galactosyltransferase, polypeptide 2 | Hs.518834 | IMAGE:1556976 | 8707 | 1q31 | Beta‐1,3‐GalTase activity/protein amino acid glycosylation |
| SEZ6L | Seizure related 6 homolog (mouse)‐like | Hs.194766 | IMAGE:2244016 | 23544 | 22q12.1 | |
| KCND2 | Potassium voltage‐gated channel, Shal‐related subfamily, member 2 | Hs.21703 | IMAGE:770427 | 3751 | 7q31 | Voltage‐gated potassium channel activity/cation transport |
| VSTM2 | V‐set and transmembrane domain containing 2 | Hs.335933 | IMAGE:1914847 | 222008 | 7p11.2 | |
| FANCL | Fanconi anemia, complementation group L | Hs.631890 | IMAGE:713058 | 55120 | 2p16.1 | Ligase activity/DNA repair |
| FDPS | Farnesyl diphosphate synthase | Hs.335918 | IMAGE:1655450 | 2224 | 1q22 | transferase activity/cholesterol biosynthesis |
| VRK2 | Vaccinia related kinase 2 | Hs.651156 | IMAGE:824117 | 7444 | 2p16‐p15 | Protein serine‐threonine kinase activity/amino acid phosphorylation |
The 20 genes were represented by 22 clones; LONP2 and PPP2R2C were represented twice each; and GO, gene ontology.
Figure 3.
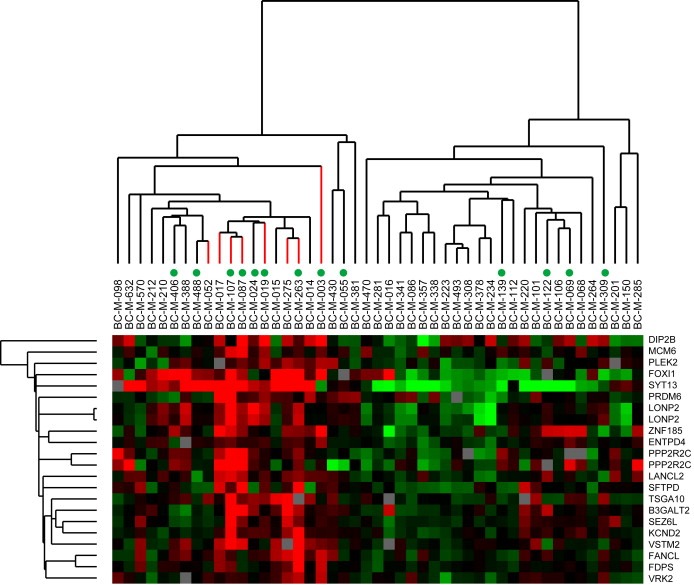
Hierarchical clustering diagram of genes associated with DTC. A. Cluster diagram showing the relationship of 20 the genes (22 clones) correlating with DTC status in 47 luminal A tumors, as identified by SAM. Samples associated with DTC in BM are color‐coded with red branches; green dots indicate tumors from patients who later experienced systemic.
To further explore the primary tumor characteristics of the luminal A type in association with DTC, we used Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) to identify potentially interesting regulatory modules or co‐expressed gene sets that may be important for cancer dissemination. A gene set representing the CREB pathway was identified (FDR=0.138) whose genes were enriched in tumors associated with positive DTC status. A similar analysis for distant relapse in luminal A tumors failed to identify any significant gene sets with an FDR<0.25.
Although the presence of DTC is a strong risk factor for future metastases, a significant portion of the patients do not experience relapse. Different biological properties of the disease might explain this difference. Hence, we searched for differentially expressed genes in tumors from patients with DTC who later experiences systemic relapse (n=5) compared to those who were free of the disease within the follow‐up time (n=3). Despite the small sample size, SAM identified one gene, CXXC4 which is a negative regulator of the Wnt receptor pathway, with q‐value 0% and that were considerably higher expressed in the tumors from the DTC‐positive patients with systemic relapse.
3. Discussion
One of the challenges when studying breast tumor dissemination is to understand the molecular and functional characteristics of a cancer disease that results in cells that reside in bone marrow and later develop into overt metastases. Associations between DTC and certain primary tumor characteristics such as size, differentiation and vascular invasion have previously been noted (Naume et al., 2001; Gerber et al., 2001; Mansi et al., 1999). This study focus on the association between gene expression patterns in primary breast tumors and presence of DTC in BM.
We observed that the presence of DTC in BM was associated with poor outcome in the luminal A subgroup while this was not found for the ER‐negative/TP53‐mutated group of tumors, which for a large part represents the basal‐like and ERBB2+ subtypes. One possible explanation for this difference is that non‐luminal A patients with ER‐negative/TP53‐mutated tumors and presence of DTC may respond better to systemic adjuvant treatment compared to luminal A/TP53 wild type DTC‐positive patients. It is known that ER‐negative patients achieve complete remissions more frequently than the ER‐positive patients in the neoadjuvant setting (Colleoni et al., 2004). On the other hand, ER‐negative patients have in general an increased risk of early relapse as compared to the ER‐positive patients (Colleoni et al., 2004; Thorpe et al., 1987), and TP53 mutation is associated with reduced effect of some chemotherapeutic agents (Aas et al., 1996; Andersson et al., 2005; Geisler et al., 2001). Furthermore, studies have indicated that DTC may be refractory to chemotherapy, although this was shown in very high‐risk patients (Braun et al., 2000). Another explanation might be that the ER‐negative/TP53‐mutated patients more frequently have clinically relevant tumor cell dissemination to other distant organs irrespective of the BM findings. Indeed, a higher frequency of visceral metastasis was observed among the ER‐negative/DTC‐negative patients than the ER‐negative/DTC‐positive patients (21% vs 14%) in the whole patient cohort. This is in contrast to the frequency of visceral metastasis among ER‐positive patients according to DTC status (6% in DTC‐negative vs 14% in DTC‐positive) (Naume et al., unpublished observations). Although we used pan‐cytokeratin monoclonal antibodies (AE1/AE3) to detect DTC, it cannot be excluded that tumorigenic cells in BM are differentially stained by the immunocytochemical technique across tumor subtypes due to different marker expression.
Another interesting hypothesis for the different significance of DTC among the subgroups could be a different frequency – or different relevance – of cancer stem cells or progenitor cells (Al Hajj et al., 2003). If cancer stem cells are present in the circulation or in BM in the ER‐negative/TP53‐mutated patients at extremely low frequencies, whereas differentiated, non‐tumorigenic cells shedded from the primary tumor are much more frequently present, the clinical implication of DTC in BM would be reduced. In contrast, if the luminal A subtype originates from a more differentiated cell type, it is possible that DTC in BM for this particular subtype more frequently represents more differentiated tumorigenic progenitor cells that have acquired ER expression. This corresponds well with bone as the most frequent metastatic site in ER+ breast cancers.
The disparate information of DTC in luminal A vs the ER‐negative/TP53‐mutated tumors initiated a further exploration of the gene expression patterns in the luminal A tumors. Among the 20 genes identified by SAM to be correlated with DTC status, there are genes involved in transport (SYT13 and KCND2), ATP‐binding (MCM6, LONP2 and LANCL2) and in regulation of transcription (FOXI1 and PRDM6) (source: Diehn et al., 2003). These were all highly expressed in DTC‐positive tumors, which is compatible with a more progressive phenotype.
Only two genes, LONP2 (LONPL) and MCM6, were common to both gene profiles associated with DTC and with systemic relapse. LONP2 (Lon peptidase 2) is a peroxisomal ATP‐dependent protease, possibly involved in the biogenesis of peroxisomes (Kikuchi et al., 2004). The relevance for tumor dissemination is unclear, but peroxisome proliferation is regulated by PPARs which belong to the steroid hormone receptor superfamily and signaling cross‐talk has been observed between PPARs and the estrogen receptor (Keller et al., 1995). Microsome maintainance protein 6 (MCM6) is involved in initiation of DNA replication. In support of our results, this protein has been shown to be overexpressed in several malignant diseases and associated with reduced survival and metastasis formation (Schrader et al., 2005; Winnepenninckx et al., 2006).
GSEA identified only one gene set, the CREB pathway, as enriched in luminal A tumors associated with DTC in BM. CREB binds the cyclic AMP response element (CRE) and activates transcription in response to a variety of extracellular signals. The CREB signaling pathway is involved in the induction of cyclin D1 protein levels (D'Amico et al., 2000), which is associated with estrogen receptor positive disease and reduced survival (Al‐Kuraya et al., 2004). Interestingly, it has been shown that CREB can increase the synthesis and deposition of non‐collagenous bone‐matrix proteins such as osteocalcin (OC) and bone sialoprotein (BSP) in prostate cancer (Huang et al., 2006), which may help sustain growth and survival of cancer cells residing in the bone marrow (Karadag et al., 2004).
Certain molecular characteristics of the individual disease might explain the ability for disseminated tumor cells to become tumorigenic. The limited number of DTC‐positive patients with luminal A type tumors in this data set (n=8), restricts our possibilities for a detailed exploration. However, CXXC4 (IDAX) was found to be highly expressed in the tumors from patients with DTC and a subsequent systemic relapse, as compared to the DTC‐positive patients without a subsequent relapse. This gene encodes a negative regulator of the Wnt‐signaling pathway by its binding to Dishevelled (DVL), which consequently reduces β‐catenin activation (Hino et al., 2001). CXXC4 is therefore an interesting candidate gene for further study of the metastatic process within the luminal A subtype.
A combined analysis of gene expression and DTC in BM in primary tumors using cDNA arrays was reported by Woelfle et al. (2003). A total of 12 DTC‐negative cases and seven DTC‐positive were selected for analysis in a retrospective study of ER‐positive stage pT2 tumors. The results showed differential expression of HIF‐1α, genes in the RAS pathway and certain cytokeratins. In contrast to our results, the signature for DTC was mainly characterized by transcriptional repression. This discrepancy between the genes identified in our study as compared to the results from Woelfle et al. may also be due to different experimental procedures and microarray platform used, differences in patient selection and statistical methods employed and the inherent nature of co‐variables in microarray studies (Ein‐Dor et al., 2005; Pusztai, 2006; Tan et al., 2003). The prospective design in our study and the use of whole genome cDNA arrays should yield the most optimal information. However, both studies are exploratory and the identified genes need to be evaluated in larger patient cohorts. Nevertheless, our results underline the importance of assessing the molecular heterogeneity of tumors when searching for associations between markers for progression and clinical outcome. Our results form a foundation for a planned further exploration of the identified genes, gene signatures and pathways in the entire cohort of 817 patients using tissue microarrays.
4. Experimental procedures
4.1. Patients, sampling of biomaterial and analysis of paraffin embedded primary tumors
Patients treated for localized breast cancer were included in this project (from 1995 to 1998) and have previously been described (Wiedswang et al., 2003). After informed, written consent, bone marrow (BM) aspiration was performed in general anesthesia, just prior to primary surgery for suspected breast cancer. If possible, parts of the tumor specimens were fresh frozen at −80°C immediately after surgery. The routine selection of patients to adjuvant treatment was based upon the prevailing National Guidelines, where postmenopausal hormone receptor (HR) positive patients received tamoxifen only, postmenopausal HR negative patients received CMF and premenopausal patients received CMF followed by tamoxifen if HR positive. Five patients received high dose chemotherapy and another five, preoperative chemotherapy due to large tumor size. After completed primary therapy, the patients were followed at 6–12 months intervals. A total of 920 patients were included in the study, clinical correlation to DTC status was originally reported from 817 (Wiedswang et al., 2003) and includes now an updated follow‐up of 811 patients (median follow‐up 85 months). Fresh frozen tissue samples were available from 123 individuals. Of these, 114 had known “disseminated tumor cell” (DTC) status. The remaining did not have a conclusive DTC result, but were included in the microarray analysis. One patient was diagnosed with ductal carcinoma in situ. The study was approved by the Regional Ethical Committee. The primary tumor and axillary lymph nodes collected during surgery were processed on a routine diagnostic basis, as previously described. Automatic immunostaining was performed using mouse monoclonal antibodies against ER and PgR (clones 6F11 and 1A6, respectively, Novocastra, Newcastle upon Tyne, UK). Immunopositivity was recorded if ≥10% of the tumor cell nuclei were immunostained. Amplification of the HER2 gene was assessed by FISH (fluorescence in situ hybridization) on tissue microarray sections using the PathVysion HER‐2 DNA Probe kit (Vysis Inc., Downers Grove, IL 60515, USA). Pretreatment was performed according to an in‐house protocol modified after Chin et al. (2003), and the remaining of the procedure, including scoring, was performed as described in the Kit package insert, defining a HER2/centromer 17 ratio of ≥2.0 as HER2 positivity. A complete listing of all experimental samples can be found in Supplemental Table 1.
4.2. Preparation of the bone marrow and immunocytochemical staining
A total of 40 ml BM was aspirated from anterior and posterior iliac crests bilaterally, 10 ml per site, and processed as described previously (Wiedswang et al., 2003). After separation by density centrifugation, mononuclear cells (MNCs) were collected and cytospins prepared (5×105 MNC/slide). The immunocytochemical staining was performed as previously described (Wiedswang et al., 2003). Briefly, four slides (2×106 BM MNC) were incubated with the anti‐cytokeratin monoclonal antibodies AE1 and AE3 (Sanbio, Uden, The Netherlands), and the same number of slides was incubated with an isotype‐specific irrelevant control monoclonal antibody. The visualization‐step included the alkaline phosphatase/anti‐alkaline phosphatase reaction, and the slides were counterstained with hematoxylin to visualize nuclear morphology.
4.3. Detection of DTC
The cytospins were manually screened by light microscopy using the 10× lens. All immunostained cells were closely evaluated by one of the pathologists (E.B.). Cells were scored as tumor cell (TC) only when the morphology was compatible with such, i.e. clearly enlarged nucleus as compared to surrounding hematopoietic cells and/or the presence of TC clusters. If lacking hematopoietic characteristics, cells with typically strong and/or irregular cytoplasmatic staining partially covering the nucleus, were also recorded as TC, according to published guidelines (Borgen et al., 1999). In difficult cases, a second pathologist (J.M.N.) was consulted and consensus obtained. The presence of positive cells classified as TC both in the AE1/AE3‐stained slides and in the corresponding negative control slides resulted in exclusion of nine patients from diagnostic conclusion.
4.4. TP53 mutation analysis
Genomic DNA was extracted by standard phenol/chloroform protocols on an automatic DNA isolation system (AB340, Applied Biosystems). The coding region covering exons 2 through 11 of the TP53 gene was screened for mutations by Temporal Temperature Gel Electrophoresis (TTGE) as described in detail elsewhere (Sorlie et al., 2004). Fragments with aberrant migration patterns were sequenced using the AB Prism 373 and 377 Automatic Sequencing systems (Applied Biosystems) to identify the nature of the mutations.
4.5. DNA microarray analysis
Total RNA was isolated from primary tumor tissues using the TRIzol reagent (Invitrogen). The integrity and quality of RNA samples were evaluated on the 2100 Bioanalyzer (Agilent) and the concentration measured using a NanoDrop spectrophotometer (NanoDrop Technologies). Amplification and labeling of RNA from tumor with Cy5 and from the Universal Human Reference (Stratagene) with Cy3 were performed as previously described (Zhao et al., 2002). Hybridization of labeled cRNA to arrays containing 42,000 features representing 24271 unique cluster IDs (UniGene Build Number 173) produced at the Stanford Functional Genomics Facility (http://www.microarray.org/sfgf/jsp/home.jsp) was performed at 65°C overnight as previously described. The hybridized arrays were scanned on an Agilent DNA microarray scanner and images analyzed by GenePix Pro v 4.1. All procedures are available at http://genome‐www.stanford.edu/breast_cancer/ and all raw data can be obtained from the Stanford Microarray Database (SMD) (http://genome‐www5.stanford.edu//). The data have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE3985. A subset of 20 tumors has been extensively analyzed using three different DNA microarray platforms (Sorlie et al., 2006).
4.6. Data processing
For the SAM and GSEA analyses, normalized (intensity dependent loess with print tip stratification) log2‐transformed gene expression ratios were retrieved from SMD (http://genome‐www5.stanford.edu/), filtered for spot intensity over background at least 1.5 in both channel 1 and channel 2, and finally, filtered for those genes that fulfilled the spot filter criteria in at least 85% of the experiments. For subtype classification, expression data for the intrinsic gene set (Sorlie et al., 2003) were extracted and both genes (559 clones representing 494 unique genes, weighted by UniGene clusters ID) and arrays were median‐centered and clustered using average‐linkage hierarchical clustering (Eisen et al., 1998). Based on the previously defined gene expression centroids, correlations with the five different breast tumor subtypes were calculated using the Correl function in Excel. These correlations were calculated based on the 549 clone IDs that were common between the two data sets. Each breast tumors sample was assigned to a tumor subclass based on the highest correlation value (<0.1 remained unclassified).
4.7. Statistical analyses
A database was established after permission from the Regional Committee of Ethics and the National Data Inspectorate. The endpoints for the survival analyses were breast cancer specific survival (BCSS) and distant disease‐free survival (DDFS) measured from the date of surgery to breast cancer‐related death or systemic relapse or otherwise censored at the time of the last follow‐up visit or at non‐cancer‐related death. Metastases to the skeleton, liver, lungs or brain were recorded as systemic relapses. Kaplan–Meier survival curves for time to BCSS and DDFS were constructed and P‐values were computed by the log rank test. Cox proportional hazard regression analysis was used for univariate estimation of prognostic impact of the relevant variables. For these analyses, the R software v. 2.4.0 was used. The Pearson chi‐square test was used to compare categorical variables. Furthermore, two‐class unpaired Statistical Analysis of Microarrays v. 3.0 (SAM) (Tusher et al., 2001) t‐statistics was performed with 400 permutations using the K‐nearest neighbor imputer (N=10) to identify differentially expressed genes between tumor groups. Finally, Gene Set Enrichment Analysis v. 2 (GSEA) (Subramanian et al., 2005) was applied to identify significantly differential expression of pre‐defined gene sets using the C2 collection of gene sets from the MSigDB.
Supporting information
Appendix Supplementary data
Supplemental Table 1 Complete listing of tumor samples with experimental, molecular and clinical information.
Supplemental Table 2 Gene information on the 76 clones identified by SAM as significantly associated with systemic relapse in luminal A tumors.
Supplemental Figure 1 Hierarchical cluster diagram of the 64 genes genes (represented by 76 clones) identified by SAM as significantly associated with systemic relapse in luminal A tumors. Green dots indicate tumors from patients who experienced systemic relapse.
Supplemental Figure 2 Kaplan–Meier survival curves stratified by TP53 mutation and DTC status. DDFS, distant disease‐free survival; BCSS, breast cancer specific survival.
Acknowledgments
We are grateful to Professor David Botstein for his contribution to this project. Support from the Stanford Microarray Database and the Stanford Functional Genomics Facility is highly appreciated. This work was supported by grants from the Norwegian Cancer Society (D99061), the Norwegian Research Council (155218/300) and the SalusAnsvar Award to ALBD. The project has received research funding from the Communities Sixth Frame Programme, project: DISMAL, contract no.: LSHC‐CT‐2005‐018911. The publication reflects only the authors' views and the Community is not liable for any use that may be made of the information contained therein.
Appendix A. Supplementary data 1.
1.1.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molonc.2007.03.004.
Naume Bjørn, Zhao Xi, Synnestvedt Marit, Borgen Elin, Russnes Hege Giercksky, Lingjærde Ole Christian, Strømberg Maria, Wiedswang Gro, Kvalheim Gunnar, Kåresen Rolf, Nesland Jahn M., Børresen-Dale Anne-Lise, Sørlie Therese, (2007), Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer, Molecular Oncology, 1, doi:10.1016/j.molonc.2007.03.004.
References
- Aas, T. , Borresen, A.L. , Geisler, S. , Smith-Sorensen, B. , Johnsen, H. , Varhaug, J.E. , Akslen, L.A. , Lonning, P.E. , 1996. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat. Med.. 2, 811–814. [DOI] [PubMed] [Google Scholar]
- Al Hajj, M. , Wicha, M.S. , Benito-Hernandez, A. , Morrison, S.J. , Clarke, M.F. , 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A.. 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kuraya, K. , Schraml, P. , Torhorst, J. , Tapia, C. , Zaharieva, B. , Novotny, H. , Spichtin, H. , Maurer, R. , Mirlacher, M. , Kochli, O. , 2004. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res.. 64, 8534–8540. [DOI] [PubMed] [Google Scholar]
- Andersson, J. , Larsson, L. , Klaar, S. , Holmberg, L. , Nilsson, J. , Inganas, M. , Carlsson, G. , Ohd, J. , Rudenstam, C.M. , Gustavsson, B. , 2005. Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann. Oncol.. 16, 743–748. [DOI] [PubMed] [Google Scholar]
- Ayers, M. , Symmans, W.F. , Stec, J. , Damokosh, A.I. , Clark, E. , Hess, K. , Lecocke, M. , Metivier, J. , Booser, D. , Ibrahim, N. , 2004. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J. Clin. Oncol.. 22, 2284–2293. [DOI] [PubMed] [Google Scholar]
- Bernards, R. , Weinberg, R.A. , 2002. A progression puzzle. Nature. 418, 823 [DOI] [PubMed] [Google Scholar]
- Bertucci, F. , Finetti, P. , Rougemont, J. , Charafe-Jauffret, E. , Cervera, N. , Tarpin, C. , Nguyen, C. , Xerri, L. , Houlgatte, R. , Jacquemier, J. , 2005. Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res.. 65, 2170–2178. [DOI] [PubMed] [Google Scholar]
- Borgen, E. , Naume, B. , Nesland, J.M. , Kvalheim, G. , Beiske, K. , Fodstad, Ø. , Diel, I. , Solomayer, E.F. , Theocharous, P. , Coombes, R.C. , 1999. Standardization of the immunocytochemical detection of cancer cells in BM and blood: I. establishment of objective criteria for the evaluation of immunostained cells. Cytotherapy. 1, 377–388. [DOI] [PubMed] [Google Scholar]
- Braun, S. , Pantel, K. , Muller, P. , Janni, W. , Hepp, F. , Kentenich, C.R. , Gastroph, S. , Wischnik, A. , Dimpfl, T. , Kindermann, G. , 2000. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N. Engl. J. Med.. 342, 525–533. [DOI] [PubMed] [Google Scholar]
- Braun, S. , Vogl, F.D. , Naume, B. , Janni, W. , Osborne, M.P. , Coombes, R.C. , Schlimok, G. , Diel, I.J. , Gerber, B. , Gebauer, G. , 2005. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med.. 353, 793–802. [DOI] [PubMed] [Google Scholar]
- Chang, H.Y. , Nuyten, D.S. , Sneddon, J.B. , Hastie, T. , Tibshirani, R. , Sorlie, T. , Dai, H. , He, Y.D. , van't Veer, L.J. , Bartelink, H. , 2005. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. U.S.A.. 102, 3738–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.C. , Wooten, E.C. , Tsimelzon, A. , Hilsenbeck, S.G. , Gutierrez, M.C. , Elledge, R. , Mohsin, S. , Osborne, C.K. , Chamness, G.C. , Allred, D.C. , 2003. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 362, 362–369. [DOI] [PubMed] [Google Scholar]
- Chin, S.F. , Daigo, Y. , Huang, H.E. , Iyer, N.G. , Callagy, G. , Kranjac, T. , Gonzalez, M. , Sangan, T. , Earl, H. , Caldas, C. , 2003. A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. Mol. Pathol.. 56, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni, M. , Viale, G. , Zahrieh, D. , Pruneri, G. , Gentilini, O. , Veronesi, P. , Gelber, R.D. , Curigliano, G. , Torrisi, R. , Luini, A. , 2004. Chemotherapy is more effective in patients with breast cancer not expressing steroid hormone receptors: a study of preoperative treatment. Clin. Cancer Res.. 10, 6622–6628. [DOI] [PubMed] [Google Scholar]
- D'Amico, M. , Hulit, J. , Amanatullah, D.F. , Zafonte, B.T. , Albanese, C. , Bouzahzah, B. , Fu, M. , Augenlicht, L.H. , Donehower, L.A. , Takemaru, K. , 2000. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J. Biol. Chem.. 275, 32649–32657. [DOI] [PubMed] [Google Scholar]
- Dai, H. , van't Veer, L. , Lamb, J. , He, Y.D. , Mao, M. , Fine, B.M. , Bernards, R. , van de Vijver, M. , Deutsch, P. , Sachs, A. , 2005. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res.. 65, 4059–4066. [DOI] [PubMed] [Google Scholar]
- Diehn, M. , Sherlock, G. , Binkley, G. , Jin, H. , Matese, J.C. , Hernandez-Boussard, T. , Rees, C.A. , Cherry, J.M. , Botstein, D. , Brown, P.O. , 2003. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res.. 31, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diel, I.J. , Kaufmann, M. , Costa, S.D. , Holle, R. , von, M.G. , Solomayer, E.F. , Kaul, S. , Bastert, G. , 1996. Micrometastatic breast cancer cells in bone marrow at primary surgery: prognostic value in comparison with nodal status. J. Natl. Cancer Inst.. 88, 1652–1658. [DOI] [PubMed] [Google Scholar]
- Ein-Dor, L. , Kela, I. , Getz, G. , Givol, D. , Domany, E. , 2005. Outcome signature genes in breast cancer: is there a unique set?. Bioinformatics. 21, 171–178. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B. , Spellman, P.T. , Brown, P.O. , Botstein, D. , 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A.. 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, S. , Lonning, P.E. , Aas, T. , Johnsen, H. , Fluge, O. , Haugen, D.F. , Lillehaug, J.R. , Akslen, L.A. , Borresen-Dale, A.L. , 2001. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res.. 61, 2505–2512. [PubMed] [Google Scholar]
- Gerber, B. , Krause, A. , Muller, H. , Richter, D. , Reimer, T. , Makovitzky, J. , Herrnring, C. , Jeschke, U. , Kundt, G. , Friese, K. , 2001. Simultaneous immunohistochemical detection of tumor cells in lymph nodes and bone marrow aspirates in breast cancer and its correlation with other prognostic factors. J. Clin. Oncol.. 19, 960–971. [DOI] [PubMed] [Google Scholar]
- Hino, S. , Kishida, S. , Michiue, T. , Fukui, A. , Sakamoto, I. , Takada, S. , Asashima, M. , Kikuchi, A. , 2001. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol. Cell Biol.. 21, 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.C. , Wu, D. , Xie, Z. , Zhau, H.E. , Nomura, T. , Zayzafoon, M. , Pohl, J. , Hsieh, C.L. , Weitzmann, M.N. , Farach-Carson, M.C. , 2006. beta2-Microglobulin is a signaling and growth-promoting factor for human prostate cancer bone metastasis. Cancer Res.. 66, 9108–9116. [DOI] [PubMed] [Google Scholar]
- Karadag, A. , Ogbureke, K.U. , Fedarko, N.S. , Fisher, L.W. , 2004. Bone sialoprotein, matrix metalloproteinase 2, and alpha(v)beta3 integrin in osteotropic cancer cell invasion. J. Natl. Cancer Inst.. 96, 956–965. [DOI] [PubMed] [Google Scholar]
- Keller, H. , Givel, F. , Perroud, M. , Wahli, W. , 1995. Signaling cross-talk between peroxisome proliferator-activated receptor/retinoid X receptor and estrogen receptor through estrogen response elements. Mol. Endocrinol.. 9, 794–804. [DOI] [PubMed] [Google Scholar]
- Kikuchi, M. , Hatano, N. , Yokota, S. , Shimozawa, N. , Imanaka, T. , Taniguchi, H. , 2004. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem.. 279, 421–428. [DOI] [PubMed] [Google Scholar]
- Ma, X.J. , Salunga, R. , Tuggle, J.T. , Gaudet, J. , Enright, E. , McQuary, P. , Payette, T. , Pistone, M. , Stecker, K. , Zhang, B.M. , 2003. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. U.S.A.. 100, 5974–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansi, J.L. , Gogas, H. , Bliss, J.M. , Gazet, J.C. , Berger, U. , Coombes, R.C. , 1999. Outcome of primary-breast-cancer patients with micrometastases: a long-term follow-up study. Lancet. 354, 197–202. [DOI] [PubMed] [Google Scholar]
- Naume, B. , Borgen, E. , Kvalheim, G. , Karesen, R. , Qvist, H. , Sauer, T. , Kumar, T. , Nesland, J.M. , 2001. Detection of isolated tumor cells in bone marrow in early-stage breast carcinoma patients: comparison with preoperative clinical parameters and primary tumor characteristics. Clin. Cancer. Res.. 7, 4122–4129. [PubMed] [Google Scholar]
- Olivier, M. , Eeles, R. , Hollstein, M. , Khan, M.A. , Harris, C.C. , Hainaut, P. , 2002. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat.. 19, 607–614. [DOI] [PubMed] [Google Scholar]
- Pusztai, L. , 2006. Chips to bedside: incorporation of microarray data into clinical practice. Clin. Cancer Res.. 12, 7209–7214. [DOI] [PubMed] [Google Scholar]
- Ramaswamy, S. , Ross, K.N. , Lander, E.S. , Golub, T.R. , 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet.. 33, 49–54. [DOI] [PubMed] [Google Scholar]
- Schrader, C. , Janssen, D. , Klapper, W. , Siebmann, J.U. , Meusers, P. , Brittinger, G. , Kneba, M. , Tiemann, M. , Parwaresch, R. , 2005. Minichromosome maintenance protein 6, a proliferation marker superior to Ki-67 and independent predictor of survival in patients with mantle cell lymphoma. Br. J. Cancer. 93, 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T. , Johnsen, H. , Vu, P. , Lind, G.E. , Lothe, R. , Borresen-Dale, A.L. , 2004. Mutation screening of the TP53 gene by temporal temperature gradient gel electrophoresis. Methods Mol. Biol.. 291, 207–216. [DOI] [PubMed] [Google Scholar]
- Sorlie, T. , Perou, C.M. , Tibshirani, R. , Aas, T. , Geisler, S. , Johnsen, H. , Hastie, T. , Eisen, M.B. , Van de, R.M. , Jeffrey, S.S. , 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A.. 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T. , Tibshirani, R. , Parker, J. , Hastie, T. , Marron, J.S. , Nobel, A. , Deng, S. , Johnsen, H. , Pesich, R. , Geisler, S. , 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U.S.A.. 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T. , Wang, Y. , Xiao, C. , Johnsen, H. , Naume, B. , Samaha, R.R. , Borresen-Dale, A.L. , 2006. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 7, 127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Neo, S.Y. , McShane, L.M. , Korn, E.L. , Long, P.M. , Jazaeri, A. , Martiat, P. , Fox, S.B. , Harris, A.L. , Liu, E.T. , 2003. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. U.S.A.. 100, 10393–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V.K. , Mukherjee, S. , Ebert, B.L. , Gillette, M.A. , Paulovich, A. , Pomeroy, S.L. , Golub, T.R. , Lander, E.S. , 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A.. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, P.K. , Downey, T.J. , Spitznagel, E.L. , Xu, P. , Fu, D. , Dimitrov, D.S. , Lempicki, R.A. , Raaka, B.M. , Cam, M.C. , 2003. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res.. 31, 5676–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, S.M. , Rose, C. , Rasmussen, B.B. , Mouridsen, H.T. , Bayer, T. , Keiding, N. , 1987. Prognostic value of steroid hormone receptors: multivariate analysis of systemically untreated patients with node negative primary breast cancer. Cancer Res.. 47, 6126–6133. [PubMed] [Google Scholar]
- Tusher, V.G. , Tibshirani, R. , Chu, G. , 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A.. 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer, L.J. , Dai, H. , van de Vijver, M.J. , He, Y.D. , Hart, A.A. , Mao, M. , Peterse, H.L. , van der, K.K. , Marton, M.J. , Witteveen, A.T. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415, 530–536. [DOI] [PubMed] [Google Scholar]
- Wang, Z.C. , Lin, M. , Wei, L.J. , Li, C. , Miron, A. , Lodeiro, G. , Harris, L. , Ramaswamy, S. , Tanenbaum, D.M. , Meyerson, M. , 2004. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res.. 64, 64–71. [DOI] [PubMed] [Google Scholar]
- Weigelt, B. , Glas, A.M. , Wessels, L.F. , Witteveen, A.T. , Peterse, J.L. , van't Veer, L.J. , 2003. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc. Natl. Acad. Sci. U.S.A.. 100, 15901–15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedswang, G. , Borgen, E. , Karesen, R. , Kvalheim, G. , Nesland, J.M. , Qvist, H. , Schlichting, E. , Sauer, T. , Janbu, J. , Harbitz, T. , 2003. Detection of isolated tumor cells in bone marrow is an independent prognostic factor in breast cancer. J. Clin. Oncol.. 21, 3469–3478. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx, V. , Lazar, V. , Michiels, S. , Dessen, P. , Stas, M. , Alonso, S.R. , Avril, M.F. , Ortiz Romero, P.L. , Robert, T. , Balacescu, O. , 2006. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl. Cancer Inst.. 98, 472–482. [DOI] [PubMed] [Google Scholar]
- Woelfle, U. , Cloos, J. , Sauter, G. , Riethdorf, L. , Janicke, F. , van Diest, P. , Brakenhoff, R. , Pantel, K. , 2003. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res.. 63, 5679–5684. [PubMed] [Google Scholar]
- Yu, K. , Lee, C.H. , Tan, P.H. , Tan, P. , 2004. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin. Cancer Res.. 10, 5508–5517. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Hastie, T. , Whitfield, M.L. , Borresen-Dale, A.L. , Jeffrey, S.S. , 2002. Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics. 3, 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Supplementary data
Supplemental Table 1 Complete listing of tumor samples with experimental, molecular and clinical information.
Supplemental Table 2 Gene information on the 76 clones identified by SAM as significantly associated with systemic relapse in luminal A tumors.
Supplemental Figure 1 Hierarchical cluster diagram of the 64 genes genes (represented by 76 clones) identified by SAM as significantly associated with systemic relapse in luminal A tumors. Green dots indicate tumors from patients who experienced systemic relapse.
Supplemental Figure 2 Kaplan–Meier survival curves stratified by TP53 mutation and DTC status. DDFS, distant disease‐free survival; BCSS, breast cancer specific survival.


